Background: FTase recognizes and modifies many proteins with C-terminal CA1A2X sequences.
Results: Mutating active site residues Trp-102β and Trp-106β significantly alters FTase peptide selectivity both in vitro and in vivo.
Conclusion: FTase substrate selectivity includes negative discrimination that can be relaxed/altered without losing activity.
Significance: Deciphering FTase peptide recognition allows creation of bioengineered prenylation pathways and provides a model for other multispecific enzymes.
Keywords: Enzyme Catalysis, Enzymes, Post Translational Modification, Protein Engineering, Protein Farnesylation, Enzyme Specificity, Multispecific Enzymes
Abstract
Post-translational modifications play essential roles in regulating protein structure and function. Protein farnesyltransferase (FTase) catalyzes the biologically relevant lipidation of up to several hundred cellular proteins. Site-directed mutagenesis of FTase coupled with peptide selectivity measurements demonstrates that molecular recognition is determined by a combination of multiple interactions. Targeted randomization of these interactions yields FTase variants with altered and, in some cases, bio-orthogonal selectivity. We demonstrate that FTase specificity can be “tuned” using a small number of active site contacts that play essential roles in discriminating against non-substrates in the wild-type enzyme. This tunable selectivity extends in vivo, with FTase variants enabling the creation of bioengineered parallel prenylation pathways with altered substrate selectivity within a cell. Engineered FTase variants provide a novel avenue for probing both the selectivity of prenylation pathway enzymes and the effects of prenylation pathway modifications on the cellular function of a protein.
Introduction
Posttranslational modifications are essential for the proper function of a large portion of the eukaryotic proteome. These modifications are estimated to increase the complexity of the proteome by 1–2 orders of magnitude beyond that provided by the open reading frames in the human genome (1), complicating the task of translating genomic information to the biological function of proteins. Enzymes that catalyze posttranslational modifications, ranging from phosphorylation to acylation, face a common task of selecting the correct amino acid(s) to modify from among a host of potential sites similar in both structural context and chemical reactivity. Understanding how these enzymes achieve this molecular recognition is essential to defining the full extent of posttranslational modification within the proteome and developing inhibitors targeting these enzymes for use as therapeutics.
Protein farnesyltransferase (FTase)3 is a model system for studying interactions involved in substrate recognition by posttranslational modification enzymes. FTase is a member of the prenyltransferase family of sulfur alkyltransferases (for review, see Refs. 2 and 3) that catalyzes the covalent attachment of a 15-carbon farnesyl group from farnesyl diphosphate (FPP) to a cysteine residue near the C terminus of a protein substrate. The attached lipid aids in localization of proteins to cellular membranes and enhances protein-protein interactions (4, 5). Prenylation is required for the proper function of many proteins, including members of the Ras and Rho superfamilies of small GTPases (2, 6). FTase is known to modify a large number of proteins within the cell (7–10); recent experimental and theoretical/computational studies using small peptide substrates suggest that several hundred proteins within the human proteome may be farnesylated (11–14). Based on peptide reactivity and structural studies of FTase-substrate complexes, a minimal substrate recognition motif for FTase is a peptide or protein containing a cysteine four amino acids from the C terminus (-CXXX). Analysis of known prenylated proteins has constrained this model further, suggesting that the best substrates for FTase contain a C-terminal “CA1A2X” sequence (9, 15–18). In this model, C refers to a cysteine residue three residues removed from the C terminus that is prenylated at the thiol group to form a thioether, A refers to any aliphatic amino acid, and X refers to a subset of amino acids that are proposed to determine specificity for FTase (methionine, serine, glutamine, alanine) or a related enzyme, protein geranylgeranyltransferase type I (leucine, phenylalanine). Expanding upon the CA1A2X box paradigm, bioinformatics analysis and biochemical studies of known substrates and related proteins indicate that sequences immediately upstream of the conserved cysteine residue modulate substrate selectivity (10, 19, 20).
Defining FTase substrate selectivity remains an area of intense interest, as many prenylated proteins play key roles in signaling pathways and cell function (2, 3, 6). Surveys of naturally prenylated proteins suggest that FTase favors a subset of moderately sized hydrophobic amino acids (valine, isoleucine, leucine, methionine, and threonine) at the A2 position (10), consistent with substrate preferences revealed by statistical analysis of reactivity with a peptide library (11). Functional studies of substrate selectivity at the A2 position of the CA1A2X sequence reveal that FTase recognizes both steric volume and polarity of this residue (21). Additionally, selectivity at A2 is also dependent on the identity of the amino acid at the adjacent X position, with the steric discrimination relaxed when the X residue is methionine or glutamine (21).
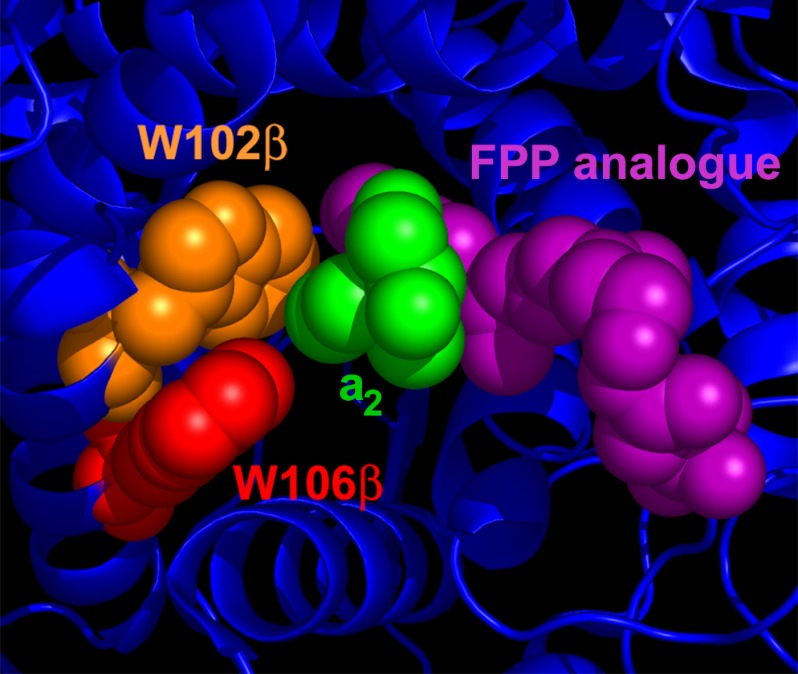
Crystallographic structures of FTase and geranylgeranyltransferase type I complexed with peptide substrates and isoprenoid mimetic inhibitors illuminate the active site environment that leads to the A2 selectivity (9). The binding site surrounding the A2 residue is composed of the side chains of two tryptophan (Trp-102β and Trp-106β) and one tyrosine (Tyr-361β) residues as well as the third isoprenoid unit of the bound FPP mimetic inhibitor (Fig. 1) (9), presenting a hydrophobic and closely packed environment consistent with the preference for moderately sized nonpolar amino acids at this site. Mutation of amino acids contacting the A2 residue can relax selectivity of FTase for the prenyl donor cosubstrate (22, 23). The structural studies predict that these same mutations should also affect peptide selectivity, but this possibility has not been explored. Changes in the structure of the isoprenoid tail of the prenyl donor cosubstrate can also alter FTase peptide selectivity (24), providing further functional evidence for a network of interactions within the FTase binding site that acts in concert to recognize peptide substrates.
FIGURE 1.
FTase active site structure. Structure of a peptide substrate bound to FTase illustrate the A2 residue binding pocket. The A2 residue of the peptide substrate KKKSKTKCVIM (green) is surrounded by residues Trp-102β (orange), Trp-106β (red), and Tyr-361β (not shown) within the active site of FTase. The A2 residue also contacts the isoprenoid tail of the FPP analog inhibitor FPT-II (purple). The figure was derived from PDB ID 1D8D and adapted from Reid et al. (9).
In this study we redesigned the peptide substrate selectivity of FTase by altering the active site contacts with the A2 residue of the CA1A2X sequence. Given the discrimination that FTase exhibits against large and charged amino acids at the A2 position, we first re-engineered the substrate specificity of FTase by substituting either a smaller (alanine, valine) or polar (histidine) amino acid at Trp-102β and/or Trp-106β. These substitutions significantly and specifically increase the reactivity of FTase with substrates containing tryptophan and aspartate at the A2 residue, respectively. Based on this success, we then created a library of mutants at Trp-102β and Trp-106β of FTase using saturation mutagenesis and screened for variants with altered specificity. Excitingly, we identified mutants that increase reactivity by up to 104-fold with target peptides containing either lysine or aspartate at A2, demonstrating catalytic efficiencies equivalent to that of WT FTase with natural substrates. Furthermore, these variants expressed in tissue culture cells catalyze in vivo prenylation of proteins containing non-natural CA1A2X sequences. The altered substrate specificity exhibited by members of the 102/106 library reveals that FTase selectivity is highly tunable through mutation of only two amino acids. Surprisingly, this enhancement in reactivity does not necessitate loss of reactivity with natural substrates, suggesting that the conserved Trp-102β and Trp-106β side chains decrease the substrate promiscuity of FTase mainly by discriminating against non-substrate sequences. These findings suggest that the complete conservation of Trp-102β and Trp-106β in FTase reflects a requirement for maintenance of substrate selectivity rather than catalytic activity. Additionally, the FTase variants developed in this work will serve as important tools for studying the activity, selectivity, and biological function of the in vivo prenylation pathway.
EXPERIMENTAL PROCEDURES
Miscellaneous Methods
All assays were performed at 25 °C. All curve fitting was performed with Graphpad Prism (Graphpad Software, San Diego, CA). FPP was purchased from Sigma. Dansylated peptides were synthesized by Sigma-Genosys (The Woodlands, TX) in the Pepscreen® format. Peptide purities were ≥75%, with the majority of peptides examined exhibiting >90% purity, as determined by HPLC (Alltech Nucleosil C-18 column) (21). Major contaminants consist of smaller peptide fragments, as indicated by mass spectrometry, that are not efficient substrates for FTase (25, 26). Peptides were solubilized in absolute ethanol containing 10% (v/v) DMSO and stored at −80 °C. Peptide concentrations were determined spectrophotometrically using Ellman's reagent (27).
Preparation of Wild-type FTase and Single Site FTase Variants
Wild-type FTase and FTase variants were expressed in BL21(DE3) Escherichia coli using a pET23aPFT vector and purified as described previously (21, 25, 28). Mutations at Trp-102β and Trp-106β were introduced into the pET23aPFT plasmid using QuikChange XL methodology (Stratagene) and confirmed by sequencing.
Steady-state Kinetics
The initial velocity for farnesylation catalyzed by FTase was determined from a time-dependent increase in fluorescence (λex 340 nm, λem 520 nm) upon farnesylation of the dansylated peptide (29). Assays were performed with 0.2–10 μm dansylated peptide, 20–100 nm FTase, and 10 μm FPP in reaction buffer (50 mm HEPPSO, pH 7.8, 5 mm tris(2-carboxyethyl)phosphine (TCEP), 5 mm MgCl2, and 10 μm ZnCl2) at 25 °C in a 96-well plate (Corning). Fluorescence was measured as a function of time in a POLARstar Galaxy plate reader (BMG Labtechnologies, Durham, NC) to define both the initial linear velocity as well as the reaction end point. The total fluorescence change observed upon reaction completion was divided by the initial concentration of the peptide substrate to yield a conversion from fluorescence units to product concentration; these values were averaged over several peptide concentrations to produce an amplitude conversion (AmpConv). The linear initial rate, in fluorescence intensity per second, was then converted to a velocity (μm product produced/s) using the equation V = (R/AmpConv), where V is velocity in μm/s, R is the velocity of the reaction in fluorescence units/s, and AmpConv refers to the ratio described above in fluorescence units/μm product. The steady-state kinetic parameters were calculated from a fit of the Michaelis-Menten equation to the peptide concentration dependence of the initial velocity at saturating FPP.
Construction of FTase 102/106 Variant Plasmid Library
Randomized codons (NNK, where N = equal mixture of A, T, G, and C and K = G or T) were introduced at positions 102β and 106β in FTase using a modification of the QuikChange methodology (Stratagene). Library diversity was verified by sequencing >20 randomly selected colonies without observation of a repeat of codons at both positions 102 and 106.
Screening of the FTase Library
Plasmid DNA encoding the FTase 102/106 library was transformed into BL21(DE3) E. coli using electroporation followed by selection on LB agar plates containing 100 μg/ml ampicillin. Single colonies (1504 total) were inoculated into single wells of a 96-deep well (2.2 ml volume) plate containing 900 μl of LB media with 1% glucose, 100 μg/ml ampicillin, and 60 μm isopropyl β-d-1-thiogalactopyranoside. Each 96-well plate was inoculated with a colony transformed with wild-type FTase (well A01, positive control), and well A02 was not inoculated (negative control), yielding a total of 16 plates for the FTase 102/106 library. Plates were sealed with gas-permeable seals (Abgene) and shaken for 20–24 h (380 RPM) at 28 °C. After growth, glycerol stocks were prepared and stored at −80 °C.
Cell lysates were prepared by the addition of 100 μl of lysis solution (Fastbreak bacterial lysis reagent (Promega) supplemented with 2 mg/ml lysozyme, 125 units/ml Benzonase (Sigma), and 100 μg/ml phenylmethylsulfonyl fluoride) to the cell cultures in 96-well plates followed by shaking (380 rpm) at 28 °C for 20 min. Lysates were stored on ice until assayed.
FTase activity in the cell lysates was measured under steady-state conditions similar to those reported above for purified FTase, measuring the time-dependent increase in fluorescence (λex 340 nm, λem 520 nm) upon farnesylation of the dansylated peptide (29). Initial screens were performed with 3 μm dansylated peptide, 2 μl cell lysate, and 10 μm FPP or GGPP in reaction buffer at 25 °C in a 96-well plate (Corning model 3650). Peptides were incubated in reaction buffer for 20 min before initiation of the assay reactions by the addition of the peptide solution to a solution containing the cell lysate. Fluorescence was measured at time points (1 min, 30 min, 1 h, 2 h, 3 h, 4 h, and 5 h) in a POLARstar Galaxy plate reader (BMG Labtechnologies, Durham, NC). For each peptide (dns-GCVLS, dns-GCVDS, and dns-GCVKS), active variants were assigned as those that exhibited both a doubling in fluorescence intensity and a plateau in fluorescence within ∼1.5 h indicating reaction completion.
In the secondary screen, reactions were performed under the same conditions described above and monitored continuously to measure the initial velocity. The initial velocity was determined from the time-dependent fluorescence change as previously described for assaying the activity of purified FTase. The steady-state kinetic parameters were determined from a fit of the Michaelis-Menten equation to the dependence of the initial velocity on the peptide concentration.
Construction of pCAF Vectors
A vector allowing co-expression of FTase and a fluorescent fusion protein was constructed using the pACT vector (Promega) that was modified by removal of a HindIII restriction site and introduction of a SacII restriction site. The pCAF vector contains two open reading frames for mammalian protein expression, ORF1 under the control of a CMV promoter and ORF2 under the control of a SV40 promoter. pCAF plasmids with the fluorescent fusion protein cloned into ORF2 and FTase cloned into ORF1 are referred to as pCAF2 vectors. A plasmid map for the pCAF2 parent vector is included in supplemental Figs. S3–S5. The DNA encoding the α- and β-subunits of FTase was subcloned from the pET23aPFT vector (28), and the TagRFP fusion protein was constructed by cloning TagRFP from the TagRFP-N vector (Evrogen) with the addition of a 3′-extension coding for the last 20 amino acids of H-Ras terminating in the -GCVLS sequence. FTase variants and mutations of the -CVLS sequence at the C terminus of the TagRFP fusion protein were prepared in pCAF2 using QuikChange XL methodology (Stratagene) and confirmed by sequencing. A table of vectors listing vector name and encoded genes in each open reading frame of the pCAF vectors is included in supplemental Table S1.
Cell Culture, Transfection, and Imaging
HEK293T cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum and 1% v/v pen-strep (Invitrogen). For transfections, cells were cultured in 12-well tissue culture plates (Corning). Mammalian expression vectors were transfected into HEK293T cells using the FuGENE 6 transfection reagent (Promega) according to the manufacturer's protocol. After 48 h of transfection, cells were washed with 1× PBS, fixed in 3.7% formaldehyde in 1× PBS, and imaged in 1× PBS on a Nikon TE2000 inverted microscope.
RESULTS
Mutagenesis of Trp-102β and Trp-106β to Increase Reactivity with dns-GCVWS and dns-GCVWS
Structural models of CA1A2X peptide substrates bound to the FTase active site revealed that the A2 residue is contacted by several amino acids including Trp-102β and Trp-106β (Fig. 1) (9). The juxtaposition of these two tryptophan residues creates a closely packed hydrophobic pocket for the A2 residue, consistent with the recognition of the A2 side chain of the substrate based on both the volume and hydrophobicity of the side chain (21). This binding pocket discriminates against peptide substrates containing large or charged residues at the A2 position. For example, the reactivity of FTase with the peptide substrate dns-GCVDS (kcat/Km = 41 ± 5 m−1 s−1) decreased by 4000-fold compared with dns-GCVLS (21).
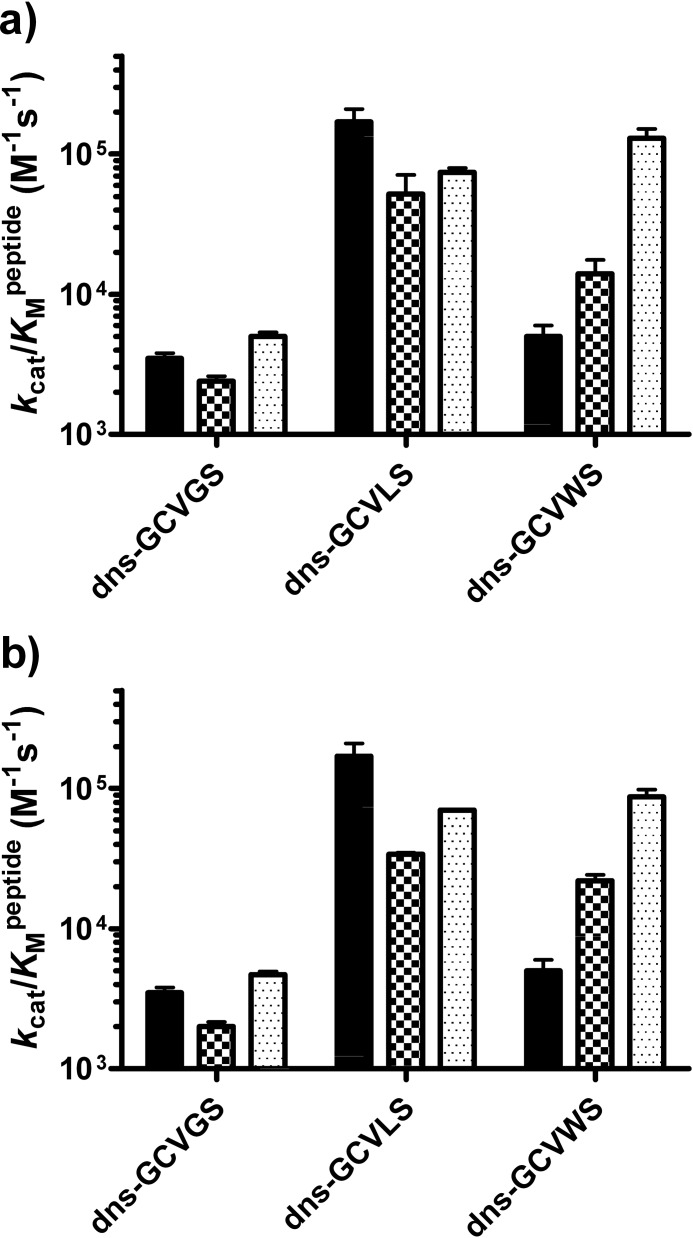
To enhance the reactivity of FTase with substrates containing large amino acids at the A2 position, Trp-102β and Trp-106β were individually mutated to alanine or valine to reduce steric bulk while maintaining a nonpolar environment. To analyze the substrate selectivity of these variants with three target peptides (dns-GCVGS, dns-GCVLS, and dns-GCVWS) the specificity constants, kcat/Km values, were measured, as this is the most relevant parameter for specificity in the presence of competing substrates (Fig. 2) (30). Mutation of either Trp-102β or Trp-106β to alanine or valine had minimal effects (<5-fold) on the value of kcat/Km for peptides bearing either glycine or leucine at the A2 position, demonstrating that these tryptophans are not essential for FTase activity. In contrast, reactivity with the dns-GCVWS peptide increased (up to 25-fold) as the steric bulk at Trp-102β or Trp-106β decreased from tryptophan (227.8 Å3) to valine (140 Å3) to alanine (88.6 Å3) (31). In fact, the kcat/Km value of the W102A or W106A variant for reaction with dns-GCVWS was comparable with that of dns-GCVLS and within 2-fold of the reactivity of WT FTase with dns-GCVLS. These data demonstrate that steric clash with the side chains of Trp-102β and Trp-106β discriminates against large amino acids at the A2 position of the substrate and plays an important role in FTase substrate recognition.
FIGURE 2.
Mutations at Trp-102β and Trp-106β that reduce steric bulk increase FTase reactivity with dns-GCVWS. a, shown is the kcat/Km value for prenylation of dns-GCVGS, dns-GCVLS, and dns-GCVWS peptides catalyzed by WT (solid bar), W102V (checkered bar), and W102A FTase (dotted bar). b, shown is the kcat/Km value for prenylation of dns-GCVGS, dns-GCVLS, and dns-GCVWS peptides catalyzed by WT (solid bar), W106V (checkered bar), and W106A FTase (dotted bar). The measured kcat/Km values are as follows: WT FTase, 3500 ± 300 m−1 s−1 (dns-GCVGS), 170,000 ± 40,000 m−1 s−1 (dns-GCVLS), and 5,000 ± 1,000 m−1 s−1 (dns-GCVWS); W102V FTase, 2,390 ± 210 m−1 s−1 (dns-GCVGS), 52,000 ± 19,000 m−1 s−1 (dns-GCVLS), and 14,000 ± 3,600 m−1 s−1 (dns-GCVWS); W102A FTase, 5,000 ± 350 m−1 s−1 (dns-GCVGS), 74,000 ± 5,400 m−1 s−1 (dns-GCVLS), and 130,000 ± 21,000 m−1 s−1 (dns-GCVWS); W106V FTase, 2,000 ± 160 m−1 s−1 (dns-GCVGS), 34,000 ± 770 m−1 s−1 (dns-GCVLS), and 22,000 ± 2,200 m−1 s−1 (dns-GCVWS); W106A FTase, 4,700 ± 250 m−1 s−1 (dns-GCVGS), 70,000 ± 500 m−1 s−1 (dns-GCVLS), and 87,000 ± 11,000 m−1 s−1 (dns-GCVWS).
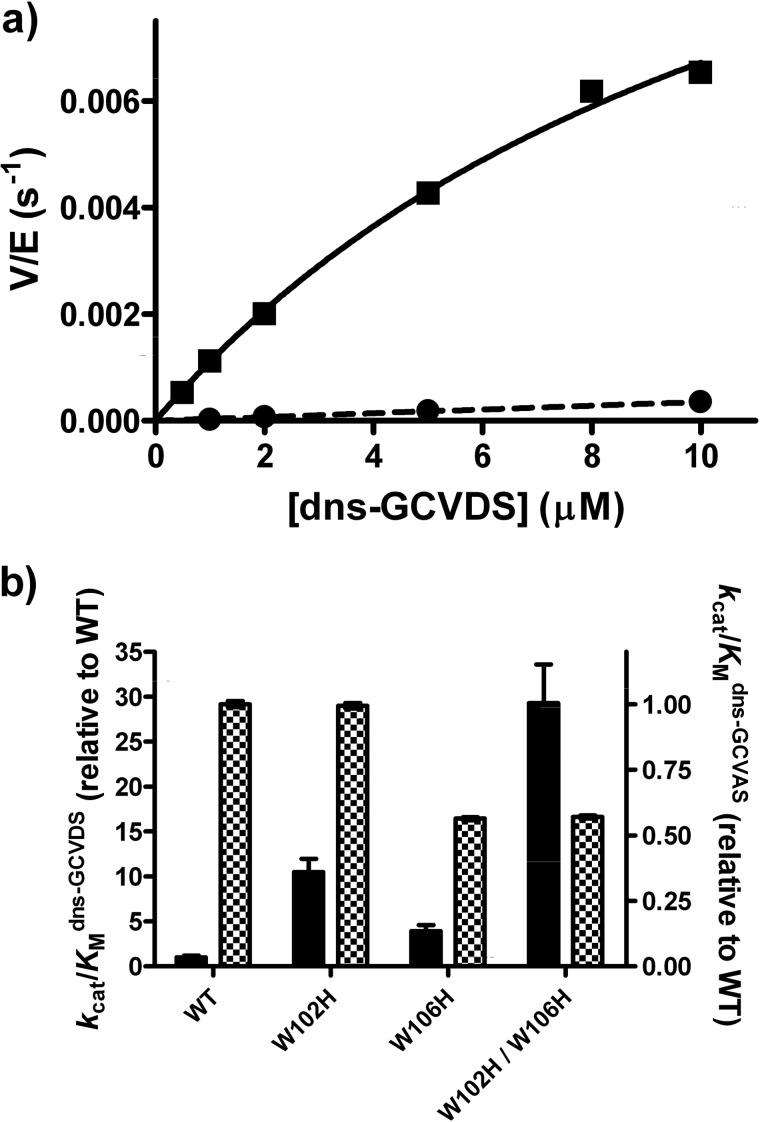
To investigate whether the recognition of substrates with polar side chains at the A2 site of the CA1A2X sequence could be enhanced by mutations at Trp-102β and Trp-106β, we substituted histidine for each tryptophan. Mutation of tryptophan to histidine introduces partial positive character while maintaining the planar geometry and aromatic character of the naturally occurring tryptophan. The reactivity of the W102H, W106H, and W102H/W106H FTase variants was comparable to that of WT FTase (<2-fold decrease) with dns-GCVAS (Fig. 3), a peptide substrate chosen to mimic dns-GCVDS without introduction of negative charge at the A2 position. In contrast, the W102βH and W106βH mutations increased the value of kcat/Km for farnesylation of dns-GCVDS by ∼10- and 4-fold, respectively, compared with WT FTase. The effects of these mutations are roughly additive. The value of kcat/Km for farnesylation of dns-GCVDS catalyzed by the double mutant (W102H/W106H) was ∼30-fold larger than that of WT FTase, leading to a significant alteration in substrate recognition by this mutant FTase.
FIGURE 3.
Histidine mutations at Trp-102β and Trp-106β enhance FTase reactivity with dns-GCVDS. a, dependence of activity on the peptide concentration catalyzed by WT (circle) and W102H/W106H (square) FTase is shown. Reactivity was measured with purified enzymes using the fluorescence-based assay as described under “Experimental Procedures.” b, shown is reactivity of W102H, W106H, and W102H/W106H FTases with dns-GCVDS (solid bar) and dns-GCVAS (checkered bar) relative to WT FTase. Relative reactivity was calculated as (kcat/Km)variant/(kcat/Km)WT with kcat/Km measured as described under “Experimental Procedures.” The measured kcat/Km values are 41 ± 5 and 15,800 ± 130 m−1 s−1 for dns-GCVDS and dns-GCVAS, respectively (WT FTase), 430 ± 30 and 15,700 ± 100 m−1 s−1 (W102H FTase), 160 ± 20 and 8,900 ± 50 m−1 s−1 (W106H FTase), and 1,200 ± 100 and 9,000 ± 50 m−1 s−1 (W102H/W106H FTase).
Randomization of Trp-102β and Trp-106β and Library Screening
Despite the enhanced reactivity of the W102H/W106H FTase variant, the catalytic efficiency for farnesylation of dns-GCVDS remains approximately 2 orders of magnitude below the reactivity of WT FTase with efficient substrates, such as the C-terminal sequence of H-Ras (-CVLS) (21). However, the significant increase in reactivity with dns-GCVDS demonstrates the importance of Trp-102β and Trp-106β in substrate recognition and suggests the potential for more substantial alterations in substrate recognition with other substitutions at these positions. Therefore, we constructed a library wherein these two amino acids were randomized through introduction of NNK codons coding for all 20 natural amino acids. From this library ∼1500 colonies were picked to provide coverage of at least 90% of the 400 possible double mutations at Trp-102β and Trp-106β (32). Individual colonies from the 102/106 variant library were grown in 96-well plates along with one well containing WT FTase (positive control) and one well containing non-inoculated media (negative control). Cell lysates were then assayed for prenylation activity using an adaption of the well established fluorescence-based prenylation assay (29). The farnesylation activity was measured using three peptides: dns-GCVLS, dns-GCVDS, and dns-GCVKS. dns-GCVLS serves as a control for the presence or absence of prenylation activity with a wild-type target protein sequence, whereas reactivity with dns-GCVDS or dns-GCVKS indicates variants capable of recognizing substrate sequences with a negative or positive charge at the A2 site, respectively. Each well was graded for observed prenylation activity, in comparison to WT FTase with dns-GCVLS, as described in the “Experimental Procedures” (supplemental Figs. S3–S5). The peptide concentration in these screening reactions (3 μm) constitutes kcat conditions for dns-GCVLS and kcat/Km conditions for dns-GCVDS and dns-GCVKS; although performing all screening reactions under kcat/Km conditions would be preferable, the peptide substrate concentrations were dictated by the fluorescence sensitivity of our cell lysate-based assay.
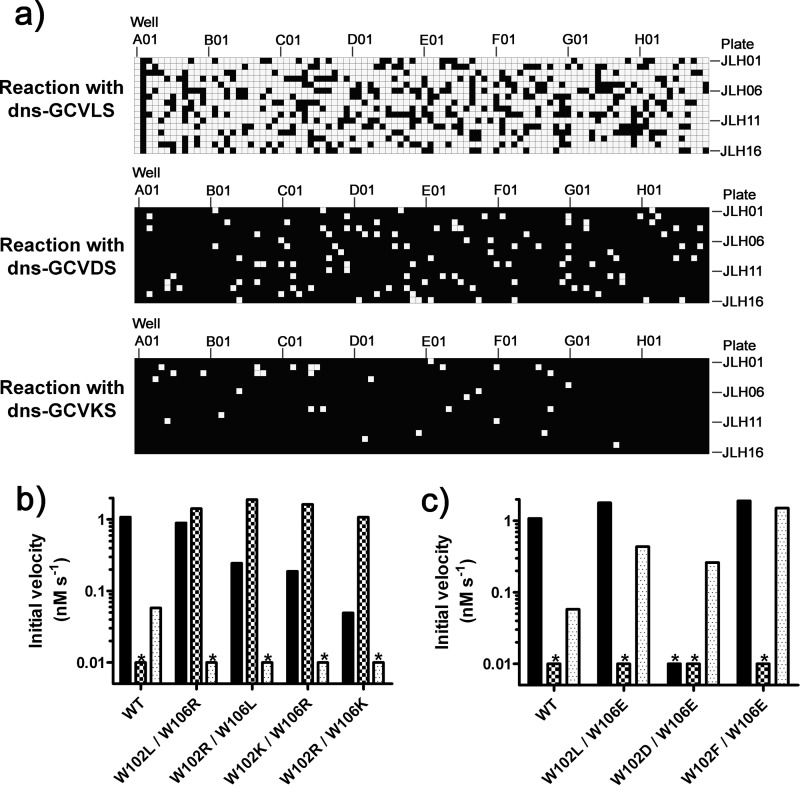
Unexpectedly, a large fraction (73%) of the variants isolated from the 102/106 library retain farnesylation activity with dns-GCVLS that is within 2-fold of the WT FTase activity (Fig. 4). This result indicates that the majority of mutations at Trp-102β and Trp-106β do not severely impact FTase folding, expression, substrate affinity, or catalytic activity. In contrast, a small fraction of the variants have gained high reactivity with the peptides containing a charged amino acid at A2; 111 variants (7% of the library) and 32 variants (2% of the library) catalyze farnesylation of dns-GCVDS and dns-GCVKS, respectively, with a rate that is within 2-fold of that observed for WT FTase with dns-GCVLS. After this initial screening, variants that demonstrated the highest activity with dns-GCVDS and dns-GCVKS were isolated and sequenced. A subset of these variants was then retransformed into bacteria, and the farnesylation activity in the lysates was re-measured; all of the hits were reproduced in this secondary screen (see supplemental Figs. S3–S5).
FIGURE 4.
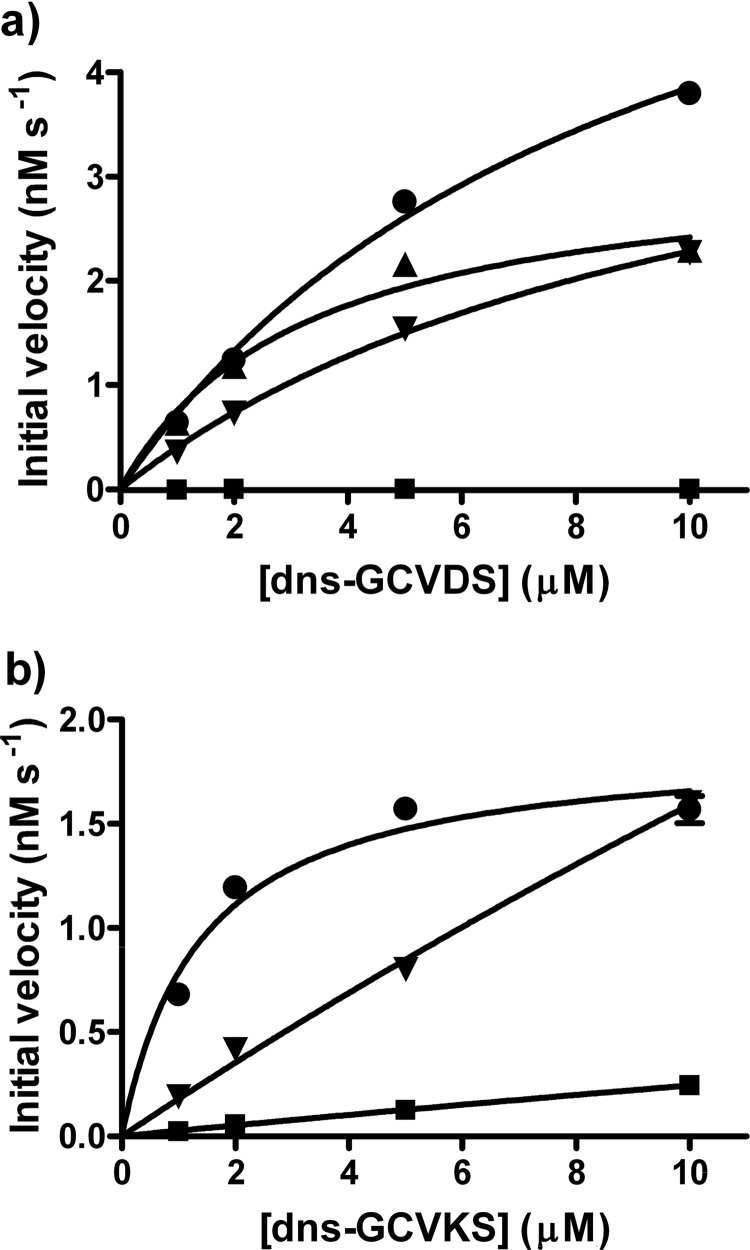
Activity assays of the 102/106 mutant library reveals both permissive and bioorthogonal variants. a, the reactivity of the 1504 variants in the 102/106 library is plotted as a heatmap, with wells A01-H12 across rows and plates JLH01-JLH16 across columns. White indicates a variant is active, satisfying the “hit” criteria outlined under “Experimental Procedures.” Each plate contains WT FTase in well A01 and media alone in well A02. For dns-GCVLS, 1095 of 1504 variants (73%) displayed activity; for dns-GCVDS, 111 of 1504 variants (7%) were active; for dns-GCVKS, 32 of 1504 (2%) were active. b and c, shown are initial rates for prenylation of dns-GCVLS (solid bar), dns-GCVDS (checkered bar), and dns-GVKS (dotted bar) measured with WT FTase, and active variants are identified in the initial screen. Bars marked with an asterisk (*) denote initial velocities below the detection threshold (<0.01 nm s−1) of the cell lysate-based assay. b, selectivity of variants that react with dns-GCVDS is shown. WT FTase displays activity with dns-GCVLS, no reaction with dns-GCVDS, and limited reactivity with dns-GCVKS. The W102L/W106L variant exhibits permissive activity with dns-GCVDS as defined under “Results”; the W102R/W102L, W102K/W106L, and W102R/W106K variants display bioorthogonal reactivity with dns-GCVDS. c, selectivity of variants with enhanced reactivity with dns-GCVKS. The W102L/W106E and W102F/W106E variants exhibit permissive reactivity with dns-GCVKS, whereas the Trp-102D/W106E displays bioorthogonal reactivity as defined under “Results.”
Sequencing of the successful variants isolated from the 102/106 library revealed two common themes (Table 1). First and perhaps not surprisingly, at least one of the two mutations observed at Trp-102β or Trp-106β leads to charge complementation with the charged A2 residue present in the target peptide: arginine or lysine is observed in variants active with dns-GCVDS and glutamate or aspartate in variants that react with dns-GCVKS. Second, the charged amino acid is accompanied by mutation of the second tryptophan to either a second charged group or a smaller amino acid, such as leucine or phenylalanine. Taken together, these two mutations simultaneously provide for charge-matching with the A2 side chain while increasing the size of the active site pocket compared with wild-type FTase. The role of the decreased steric bulk in enhancing the reactivity of the FTase variants could potentially arise from multiple factors, such as allowing for solvation of the A2 residue, accommodating alternate side chain conformers at A2, and/or lowering energetic barriers involved in conformational changes that occur during the FTase reaction cycle (33).
TABLE 1.
102/106 library variants identified in primary and secondary screens that catalyze farnesylation of dns-GCVDS and dns-GCVKS with enhanced efficiency
Charged residues are highlighted in bold.
| Reactive with dns-GCDVS |
Reactive with dns-GCVKS |
||
|---|---|---|---|
| 102 | 106 | 102 | 106 |
| Leu | Arg | Leu | Glu |
| Arg | Leu | Asp | Glu |
| Lys | Arg | Phe | Glu |
| Asn | Arg | Thr | Gln |
| Lys | Tyr | Ser | Asp |
| Arg | Val | Ile | Asp |
| Lys | Asn | ||
| Lys | His | ||
| Lys | Ile | ||
| Ile | Lys | ||
Reactivity of Active Variants
We selected the most active variants with each target peptide under the screening conditions to measure the steady-state kinetic parameters, Vmax and Vmax/Km (Table 2). SDS-PAGE analysis of cell lysates indicates that the concentrations of all of the variants were within 2-fold of the concentration of wild-type FTase as a percentage of total cellular protein (see supplemental Fig. S2). Therefore, significant increases in the observed velocities for the variant enzymes arise from an enhancement in catalytic efficiency (higher values of kcat and/or kcat/Km) rather than increased expression leading to higher enzyme concentrations in the cell lysates.
TABLE 2.
Steady-state kinetic parameters for farnesylation of dns-GCVDS and dns-GCVKS catalyzed by WT and variant FTases
FTase variant concentrations in cell lysate reactions are estimated to be ∼5 nm, based on the initial velocity observed for the cell lysate-based prenylation of dns-GCVLS by WT FTase (1.1 nm s−1) at 3 μm dns-GCVLS and the measured value of kcat/Km of 1.7 × 105 m−1s−1 for WT FTase with dns-GCVLS (21). The dash denotes “not tested.” ND, not determined.
| FTase | Reaction with dns-GCVDS |
Reaction with dns-GCVKS |
||||
|---|---|---|---|---|---|---|
| Vmax/Km | Vmax | Km | Vmax/Km | Vmax | Km | |
| 10−3 s−1 | nm s−1 | μm | 10−3 s−1 | nm s−1 | μm | |
| WT | <0.001a | ND | ≫ 10 | 0.024 ± 0.001 | > 0.2 | ≫10 |
| W102R W106L | 0.8 ± 0.1 | 7 ± 1 | 9 ± 2 | − | − | − |
| W102L W106R | 1.0 ± 0.2 | 3.2 ± 0.5 | 3 ± 1 | − | − | − |
| W102R W106K | 0.44 ± 0.03 | 4.8 ± 04 | 11 ± 2 | − | − | − |
| W102L W106E | − | − | − | 0.15 ± 0.01 | > >2 | > 10 |
| W106F W106E | − | − | − | 1.4 ± 0.4 | 1.9 ± 0.2 | 1.4 ± 0.5 |
a WT FTase activity with dns-GCVDS estimated based on a limit of detection for initial veloity of 0.01 nm s−1 and assumption that reaction of WT FTase with dns-GCVDS is under subsaturating (Vmax/ Km) conditions at 10 μm dns-GCVDS.
The steady-state kinetic parameters for reaction of the mutant FTases with dns-GCVDS are significantly improved compared with the values for WT FTase. Purified WT FTase catalyzed farnesylation of dns-GCVDS with values of kcat/Km ∼ 40 m−1s−1 and Km ≫ 10 μm (Fig. 3); in the cell lysate-based assay, the reactivity of WT FTase with dns-GCVDS was lower than the detection limit for reaction velocity of ∼0.01 nm s−1 (yielding an upper limit for WT FTase reactivity with dns-GCVDS of Vmax/Km < 0.001 × 10−3 s−1, assuming that 10 μm dns-GCVDS is subsaturating). In contrast, the substrate-dependent activity of three variants with the highest reactivity with dns-GCVDS (W102R/W102L, W102L/W102R, and W102R/W102K) each show curvature with calculated Km values ranging from 3 to 11 μm and values for Vmax/Km ranging from 0.4 × 10−3 to 1 × 10−3 s−1 (Fig. 5 and Table 2). Therefore, mutations at these two positions can increase the catalytic efficiency of FTase for farnesylation of dns-GCVDS by >100-fold.
FIGURE 5.
Evaluation of the reactivity of WT FTase and FTase variants with target peptides. Initial velocities were measured for WT FTase and selected FTase variants in the cell lysate-based fluorescence assay at four peptide concentrations, as described under “Experimental Procedures.” a, initial velocity for reaction with dns-GCVDS is shown; squares, WT FTase; circles, W102R/W106L variant; triangles, W102L/W106L variant; inverted triangles, W106L/W106K variant. B, initial velocity for reaction with dns-GCVKS is shown; squares, WT FTase; circles, W102F/W106E variant; inverted triangles, W106L/W106E variant.
The initial velocity for farnesylation of dns-GCVKS-catalyzed by WT FTase is linearly dependent on the peptide concentration, indicating that the value of Vmax/Km = 0.024 × 10−3 s−1 with Km ≫ 10 μm. One of the two variants tested, W102L/W106E, also exhibits a linear dependence on the concentration of dns-GCVKS, reflecting Km ≫ 10 μm. However, the value of Vmax/Km for this variant is 0.15 × 10−3 s−1, a 6-fold increase compared with WT FTase. In contrast, the W102F/W106E variant exhibits a Km value for farnesylation of dns-GCVKS of 1.4 μm (Table 2), suggesting a significant enhancement in peptide binding affinity, and a Vmax/Km value of 1.4 × 10−3 s−1, a 60-fold increase compared with WT FTase.
To prenylate non-natural sequences, such as CVDS or CVKS, the substrate selectivity of FTase must either expand or alter. Expansion leads to a “permissive” FTase variant, capable of prenylating both natural substrate sequences, such as CVLS, and non-natural target sequences. For example, the W102L/W106L variant catalyzed prenylation of dns-GCVLS with an initial velocity of 0.9 nm s−1 that was comparable to the initial velocity of 1.1 nm s−1 observed in parallel reactions with WT FTase using a comparable enzyme concentration. In contrast, W102L/W106L FTase catalyzed farnesylation of dns-GCVDS >100-fold faster than WT FTase (Fig. 4, Table 2). This mutation allows more permissive substrate recognition, illustrating that mutations do not necessarily lead to the loss of reactivity with naturally occurring substrates. Alternatively, in a “bioorthogonal” variant, FTase switches substrate specificity to the target peptide at the expense of reactivity with the natural substrate, as exemplified by the W102R/W106L variant. For this enzyme the initial velocity for farnesylation of the dns-GCVDS target peptide was increased from undetectable with WT FTase to 1.9 nm s−1, whereas the reactivity with the dns-GCVLS control peptide was decreased 4-fold (0.24 versus 1.1 nm s−1 for WT FTase), leading to an alteration in the ratio of reactivity with the two substrates that is >30,000-fold.4 Furthermore, the differential reactivity of these two variants indicates that the roles of Trp-102β and Trp-106β in substrate recognition are not redundant within the FTase active site.
As these data demonstrate, mutating Trp-102 and Trp-106 can drastically alter the peptide substrate selectivity of FTase. These same mutations may also relax the selectivity of FTase for FPP compared with GGPP. For example, mutation of Trp-102 to threonine enhances the reactivity of FTase with larger prenyl donor cosubstrates, such as GGPP or biotin-geranyl pyrophosphate (22, 23). To determine if the FTase variants selected in this work also exhibit broadened co-substrate selectivity, the reactivity of five variants with significantly altered peptide selectivity (W102R/W106L, W102L/W106R, W102R/W106K, W102L/W106E, and W102F/W106E) were measured with GGPP as the cosubstrate. None of the variants exhibited observable prenylation of dns-GCVLS, dns-GCVDS, or dns-GCVKS with GGPP, suggesting that the mutations at Trp-102 and Trp-106 in these variants do not significantly alter FTase selectivity for FPP over GGPP.
Testing Variant FTase Activity under in Vivo Conditions
The variants developed herein catalyze farnesylation of non-natural peptide sequences efficiently in in vitro assays. However, the screening conditions used to identify these variants are significantly different from those that would be encountered within mammalian cells. For example, the FPP concentration (10 μm) is several orders of magnitude higher than the best estimates for the in vivo FPP concentration (34). Similarly, the concentrations of the protein substrates in vivo are likely significantly lower than the μm range, and in vivo there is a complex mixture of potential substrate and non-substrate proteins within the mammalian proteome.
To evaluate farnesylation of substrates within a biologically relevant context, we imaged the localization of a fluorescent fusion protein expressed in HEK293T cells (Fig. 6). The fluorescent fusion protein consists of a red fluorescent protein (TagRFP) with an appended C-terminal tail containing the upstream sequence of H-Ras (-KLNPPDESGPGCMSC-) terminating with one of four GCA1A2X sequences: -GCVLS, -GSVLS, -GCVDS, -GCVKS. Similar fusion proteins constructed with GFP have been used successfully in other studies to follow protein localization in the presence and absence of prenylation pathway modifications (35–37). The TagRFP-CAAX fusion proteins were co-expressed with either WT FTase or FTase variants selected for reactivity with dns-GCVDS (W102R/W106L) or dns-GCVKS (W102F/W102E) using mammalian dual expression (pCAF2) vectors (see “Experimental Procedures” and supplemental Fig. S1).
FIGURE 6.
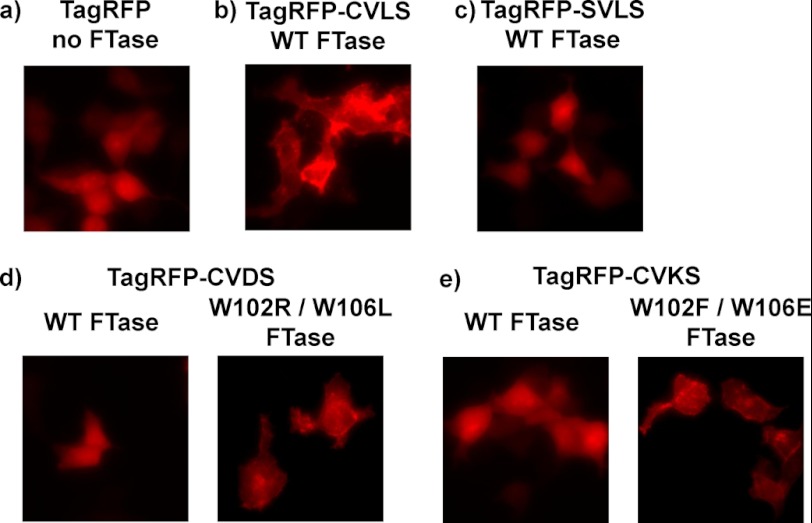
Fluorescent fusion protein localization confirms the in vivo activity of FTase variants. Mammalian expression vectors encoding TagRFP or TagRFP with a C-terminal extension terminating in a -CVA2S sequence in combination with a FTase variant were transfected into HEK293T cells and imaged as described under “Experimental Procedures.” a, shown is tag RFP alone, no FTase co-expressed. b, shown is TagRFP-CVLS co-expressed with WT FTase. c, shown is TagRFP-SVLS co-expressed with WT FTase. d, shown is TagRFP-CVDS co-expressed with either WT FTase or W102R W106L FTase. e, shown is TagRFP-CVKS co-expressed with either WT FTase or W102F/W106E FTase.
Expression of the unmodified TagRFP fluorescent protein yields diffuse fluorescence throughout the cell, consistent with the lack of cellular localization reported previously for this protein (Fig. 6) (38). The fusion protein terminating with -CVLS (TagRFP-CVLS) exhibits localization to cellular membranes, consistent with previous studies of prenylated protein localization using GFP fusion constructs (35–37). A fusion protein wherein the cysteine of the farnesylation target sequence is mutated to a serine to block farnesylation (TagRFP-SVLS) displays diffuse fluorescence, indicating that protein farnesylation is required for the localization observed with TagRFP-CVLS. When a fusion protein terminating in a sequence-containing aspartate at the A2 position (TagRFP-CVDS) is co-expressed with WT FTase, the fusion protein again displays diffuse fluorescence, consistent with an absence of farnesylation of this sequence (Fig. 6d). However, co-expression of TagRFP-CVDS with the W102R/W106L FTase variant results in a marked change in fluorescence localization, suggesting that introduction of the W102R/W106L variant leads to fusion protein farnesylation. Similarly, TagRFP-CVKS exhibits diffuse fluorescence when co-expressed with WT FTase, but localization to cellular membranes is observed when the W102F/W106E FTase variant is co-expressed (Fig. 6e). This functional complementation of the farnesylation defect caused by mutations at the A2 position confirms the ability of the FTase variants from the 102/106 library to function within the cellular environment. Furthermore, the ability of FTase variants to rescue prenylation of a protein unreactive with WT FTase constitutes the first step toward developing a synthetic prenylation pathway functioning in parallel to the natural pathway within the cell.
DISCUSSION
The preference of FTase for moderately sized nonpolar amino acids at the A2 position of the CA1A2X motif has been recognized from the earliest identification of prenylated proteins (3). The hydrophobic, tightly packed A2 binding pocket recognizes amino acids based on both polarity and size, leading to a preference for moderately sized nonpolar amino acids such as valine, isoleucine, and leucine (9, 21). The effects of altering the structure of this pocket on FTase peptide selectivity have remained a largely unanswered question. Given the biological role of FTase in which it must act upon a wide range of substrates while simultaneously avoiding aberrant modification of non-substrates, defining how peptide binding site mutations impact FTase substrate selectivity will aid in understanding the molecular recognition performed by this multisubstrate enzyme.
Mutagenesis of the conserved side chains, Trp-102β and Trp-106β, demonstrates that these side chains are important for the discrimination against large or polar amino acids at the A2 position of the canonical CA1A2X sequence. Furthermore, the substrate selectivity of FTase can be facilely tuned by two single site mutations within the FTase active site. This plasticity in substrate selectivity is remarkable in both the small number of mutations required to significantly alter specificity and the range of amino acids that can be tolerated at the A2 position of the substrate by FTase variants, including those bearing bulky and charged side chains. Furthermore, the ability of mutations at these two active site residues to radically alter FTase specificity without loss of activity suggests that other binding interactions (second shell or further) play a minor role in controlling FTase substrate selectivity at the A2 position.
Similar changes in substrate selectivity have been attempted in other protein-modifying enzymes such as trypsin, chymotrypsin, thrombin, and the bacterial endopeptidase OmpT (39–42). In comparison to FTase, altering the substrate selectivity in these proteases required a larger number of mutations, reflecting a more diffuse array of interactions involved in substrate recognition. For example, altering trypsin specificity to mirror chymotrypsin requires the substitution of two surface loops on trypsin with their chymotrypsin counterparts in addition to mutations in the substrate binding site that directly interact with the aromatic or positively charged amino acid defining the proteolytic cleavage site (39). The mutagenesis experiments with thrombin and OmpT focused on switching the enzyme specificity from the natural sequence to a new substrate. Consequently, the potential to broaden the specificity of these proteases to include new substrates while maintaining activity with natural substrates (the equivalent of the permissive FTase variants developed in this study) is unknown. In a study using phylogeny and gene synthesis to recreate an ancestral precursor of a group of serine proteases, Wouters et al. (43) found that a large number of mutations were required to generate a “primitive” serine protease with broadened specificity. Thus, the ease of altering FTase selectivity and the ability of this enzyme to expand its substrate manifold without losing overall activity is remarkable compared with other reported examples.
FTase is an example of a “multispecific” enzyme, as defined by Khersonsky and Tawfik (45) and others (44), that has evolved to act upon a wide range of potential substrates. The requirement to react with multiple substrates while maintaining selectivity against non-substrates presents a formidable molecular recognition challenge, one that requires both flexibility and fidelity. The first step toward understanding how FTase accomplishes this task is to delineate the interactions involved in recognizing protein substrates and to then define the contribution (positive or negative) of each interaction to binding and catalysis. Structural studies of FTase and geranylgeranyltransferase type I have provided a list of interactions proposed to participate in peptide substrate recognition (9), such as hydrogen-bonding interactions of the C-terminal acid group with conserved residues within the FTase and geranylgeranyltransferase type I active sites, the interaction of the cysteine side chain with the catalytic zinc ion, and the residues that form the binding sites contacting the A2 and X residues. Some of these interactions, including the cysteine-zinc coordination and the hydrogen bonding between the C-terminal carboxylate and conserved active site residues, provide energetic stabilization of substrate binding and can be considered “positive” in that they select for substrates (9, 46). In contrast, interactions with Trp-102β and Trp-106β lead to substrate selectivity mainly through unfavorable interactions with polar and large amino acids at the A2 position leading to “negative” discrimination against non-substrates. The observation that the large majority of the 102/106 variant library retains near-WT activity with dns-GCVLS indicates that the interactions with Trp-102β and Trp-106β are not essential for catalyzing prenylation of this natural sequence. Based on these findings, we propose that FTase combines positive (pro-substrate) and negative (anti-nonsubstrate) interactions to recognize a broad range of potential substrates to fulfill the functional requirement for multispecificity.
Protein prenylation is a widespread modification across eukaryotic biology, from animals to plants to yeast (2, 3, 47). The ability of mutations at Trp-102β and Trp-106β to alter FTase substrate selectivity suggests that sequence variation at these positions may provide for changes in FTase selectivity and divergent prenylated proteomes between different organisms. To explore this possibility, we performed a sequence alignment of available FTase β subunits to assess the conservation of Trp-102β and Trp-106β across eukaryotes (Fig. 7 and supplemental Fig. S6). ClustalW alignment of FTase β subunits from 59 organisms indicates high conservation of both Trp-102β (96%) and Trp-106β (100%), suggesting that these residues are functionally required for FTase activity. However, the robust expression and catalytic activity of the variants within the 102/106 library indicates that FTase can tolerate a wide range of mutations at Trp-102β and Trp-106β while maintaining activity with naturally occurring substrates. Furthermore, mutations at Trp-102β and Trp-106β expand the pool of FTase substrates by relaxing discrimination against large or polar amino acids at the A2 position. These effects on relative substrate reactivity rather than overall FTase activity suggest that the conservation of Trp-102β and Trp-106β reflects an evolutionary pressure to maintain substrate selectivity and to limit the extent of prenylation within the proteome.
FIGURE 7.

Trp-102β and Trp-106β exhibit high conservation across multiple eukaryotic kingdoms. ClustalW alignment of FTase β subunit sequences from 59 organisms spanning animals, plants, and fungi reveals that both Trp-102β (96%) and Trp-106β (100%) are highly conserved. In the alignment, Trp-102β and Trp-106β are highlighted in bold.
Protein prenylation modifications are essential for the proper function of many proteins involved in signaling pathways and other cellular processes. The prenylation pathway includes three steps: prenylation of the CAAX sequence followed by proteolysis of the -AAX sequence and then methylation of the C-terminal carboxylate. Although the enzymes in the prenylation pathway are well known, the specific impact of each modification on the localization and function of a specific target protein has been difficult to define. Numerous studies employing either inhibition and/or knockouts of prenylation pathway enzymes indicate that each modification step can be essential for substrate protein localization and function (35, 36, 48–50). However, these experiments eliminate modification of all of the prenylated proteins, leading to an undefined alteration of the membrane environment and cellular function. The effects of blocking prenylation of a single protein can be analyzed by substituting a serine for the cysteine in the CAAX sequence, but this does not allow individual analysis of each step in the prenylation pathway. Ideally, the effects of prenylation pathway modifications on a target protein should be analyzed in the background of an otherwise unperturbed cell. The FTase variants developed herein provide a gateway toward this approach by employing cognate non-natural CA1A2X sequence/FTase variant pairs to control the prenylation state of the target protein by transfection with a single vector. These FTase variants can also be used to “bypass” the natural FTase selectivity to probe the selectivity of downstream enzymes in the prenylation pathway (such as the CAAX protease Rce1) in a systemic fashion and to identify specific interactions involved in recognizing prenylated protein substrates. The “bioengineered” prenylation pathways will allow well defined studies of the effects of prenylation pathway modifications on protein structure, localization, and function.
Understanding the role of prenylation in controlling cellular function requires defining both the extent of the prenylated proteome and the effects of each prenylation pathway modification on protein localization and function. Extensive biochemical, structural, and computational studies have led to the identification of a large (and growing) pool of potential substrates for FTase and geranylgeranyltransferase type I, but development of improved models for predicting prenyltransferase substrates requires functional characterization of the specific enzyme-substrate interactions that engender substrate selectivity. Interactions between Trp-102β and Trp-106β in the FTase active site and the A2 residue of the CA1A2X sequence are the primary determinants for selectivity at this residue, with these interactions discriminating against large and polar amino acid side chains. Mutagenesis of Trp-102β and Trp-106β significantly expands the substrate selectivity of FTase, revealing that molecular recognition in this enzyme is exquisitely dependent on a small number of discrete active site interactions. These FTase variants provide an opportunity to investigate the extent, selectivity, and impact of individual prenylation pathway modifications on protein localization and function without global perturbation of the endogenous prenylation pathway. Such studies offer the potential to identify and functionally dissect the substrates responsible for the efficacy of prenylation pathway inhibitors on diseases such as cancer (6).
Supplementary Material
Acknowledgments
We thank members of the Fierke and Hougland groups for helpful comments and suggestions on this manuscript. We also thank Dr. Alejandra Yep for suggestions regarding development of the cell lysate-based screening assay.
This work was supported, in whole or in part, by National Institutes of Health Grant GM040602 (to C. A. F.) and National Institutes of Health Postdoctoral Fellowship GM078894 (to J. L. H.).

This article contains supplemental Table S1 and Figs. S1–S6.
Change in reactivity ratio is calculated as (Adns-GCVDS/Adns-GCVLS)variant/( Adns-GCVDS/Adns-GCVLS)WT, with A denoting farnesylation activity (e.g. initial velocity, kcat/KM−) of a given FTase with the target peptide. Initial velocities were used for library variants, and kcat/KM− values of 40 and 1.7 × 105 m−1s−1 were used for WT FTase activity with dns-GCVDS and dns-GCVLS, respectively.
- FTase
- protein farnesyltransferase
- FPP
- farnesyl diphosphate
- GGPP
- geranylgeranyl diphosphate
- HEPPSO
- 4-(2-hydroxyethyl)piperazine-1-(2-hydroxypropanesulfonic acid)
- dns
- dansyl
- ORF1
- opening reading frame 1 under control of the CMV promoter in the pCAF expression vector
- ORF2
- opening reading frame 2 under control of the SV40 promoter in the pCAF expression vector.
REFERENCES
- 1. Walsh C. T. (2005) Posttranslational Modification of Proteins: Expanding Nature's Inventory, Roberts and Co. Publishers, Englewood, CO [Google Scholar]
- 2. Zhang F. L., Casey P. J. (1996) Protein prenylation. Molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269 [DOI] [PubMed] [Google Scholar]
- 3. Benetka W., Koranda M., Eisenhaber F. (2006) Protein Prenylation. An (almost) comprehensive overview on discovery, history, enzymology, and significance in physiology and disease. Monatshefte für Chemie 137, 1241–1281 [Google Scholar]
- 4. Casey P. J. (1994) Lipid modifications of G proteins. Curr. Opin. Cell Biol. 6, 219–225 [DOI] [PubMed] [Google Scholar]
- 5. Marshall C. J. (1993) Protein prenylation. A mediator of protein-protein interactions. Science 259, 1865–1866 [DOI] [PubMed] [Google Scholar]
- 6. Berndt N., Hamilton A. D., Sebti S. M. (2011) Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer 11, 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Onono F. O., Morgan M. A., Spielmann H. P., Andres D. A., Subramanian T., Ganser A., Reuter C. W. (2010) A tagging-via-substrate approach to detect the farnesylated proteome using two-dimensional electrophoresis coupled with Western blotting. Mol. Cell. Proteomics 9, 742–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kho Y., Kim S. C., Jiang C., Barma D., Kwon S. W., Cheng J., Jaunbergs J., Weinbaum C., Tamanoi F., Falck J., Zhao Y. (2004) A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12479–12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reid T. S., Terry K. L., Casey P. J., Beese L. S. (2004) Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J. Mol. Biol. 343, 417–433 [DOI] [PubMed] [Google Scholar]
- 10. Maurer-Stroh S., Eisenhaber F. (2005) Refinement and prediction of protein prenylation motifs. Genome Biol. 6, R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hougland J. L., Hicks K. A., Hartman H. L., Kelly R. A., Watt T. J., Fierke C. A. (2010) Identification of novel peptide substrates for protein farnesyltransferase reveals two substrate classes with distinct sequence selectivities. J. Mol. Biol. 395, 176–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. London N., Lamphear C. L., Hougland J. L., Fierke C. A., Schueler-Furman O. (2011) Identification of a novel class of farnesylation targets by structure-based modeling of binding specificity. PLoS Comput. Biol. 7, e1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krzysiak A. J., Aditya A. V., Hougland J. L., Fierke C. A., Gibbs R. A. (2010) Synthesis and screening of a CaaL peptide library versus FTase reveals a surprising number of substrates. Bioorg. Med. Chem. Lett. 20, 767–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krzysiak A. J., Scott S. A., Hicks K. A., Fierke C. A., Gibbs R. A. (2007) Evaluation of protein farnesyltransferase substrate specificity using synthetic peptide libraries. Bioorg. Med. Chem. Lett. 17, 5548–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Omer C. A., Kral A. M., Diehl R. E., Prendergast G. C., Powers S., Allen C. M., Gibbs J. B., Kohl N. E. (1993) Characterization of recombinant human farnesyl-protein transferase. Cloning, expression, farnesyl diphosphate binding, and functional homology with yeast prenyl-protein transferases. Biochemistry 32, 5167–5176 [DOI] [PubMed] [Google Scholar]
- 16. Reiss Y., Seabra M. C., Armstrong S. A., Slaughter C. A., Goldstein J. L., Brown M. S. (1991) Nonidentical subunits of p21H-ras farnesyltransferase. Peptide binding and farnesyl pyrophosphate carrier functions. J. Biol. Chem. 266, 10672–10677 [PubMed] [Google Scholar]
- 17. Moores S. L., Schaber M. D., Mosser S. D., Rands E., O'Hara M. B., Garsky V. M., Marshall M. S., Pompliano D. L., Gibbs J. B. (1991) Sequence dependence of protein isoprenylation. J. Biol. Chem. 266, 14603–14610 [PubMed] [Google Scholar]
- 18. Fu H. W., Casey P. J. (1999) Enzymology and biology of CaaX protein prenylation. Recent Prog. Horm. Res. 54, 315–342 [PubMed] [Google Scholar]
- 19. Maurer-Stroh S., Koranda M., Benetka W., Schneider G., Sirota F. L., Eisenhaber F. (2007) Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput. Biol. 3, e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hicks K. A., Hartman H. L., Fierke C. A. (2005) Upstream polybasic region in peptides enhances dual specificity for prenylation by both farnesyltransferase and geranylgeranyltransferase type I. Biochemistry 44, 15325–15333 [DOI] [PubMed] [Google Scholar]
- 21. Hougland J. L., Lamphear C. L., Scott S. A., Gibbs R. A., Fierke C. A. (2009) Context-dependent substrate recognition by protein farnesyltransferase. Biochemistry 48, 1691–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Terry K. L., Casey P. J., Beese L. S. (2006) Conversion of protein farnesyltransferase to a geranylgeranyltransferase. Biochemistry 45, 9746–9755 [DOI] [PubMed] [Google Scholar]
- 23. Nguyen U. T., Guo Z., Delon C., Wu Y., Deraeve C., Fränzel B., Bon R. S., Blankenfeldt W., Goody R. S., Waldmann H., Wolters D., Alexandrov K. (2009) Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat. Chem. Biol. 5, 227–235 [DOI] [PubMed] [Google Scholar]
- 24. Reigard S. A., Zahn T. J., Haworth K. B., Hicks K. A., Fierke C. A., Gibbs R. A. (2005) Interplay of isoprenoid and peptide substrate specificity in protein farnesyltransferase. Biochemistry 44, 11214–11223 [DOI] [PubMed] [Google Scholar]
- 25. Hightower K. E., Casey P. J., Fierke C. A. (2001) Farnesylation of nonpeptidic thiol compounds by protein farnesyltransferase. Biochemistry 40, 1002–1010 [DOI] [PubMed] [Google Scholar]
- 26. Goldstein J. L., Brown M. S., Stradley S. J., Reiss Y., Gierasch L. M. (1991) Nonfarnesylated tetrapeptide inhibitors of protein farnesyltransferase. J. Biol. Chem. 266, 15575–15578 [PubMed] [Google Scholar]
- 27. Riddles P. W., Blakeley R. L., Zerner B. (1979) Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid). A reexamination. Anal. Biochem. 94, 75–81 [DOI] [PubMed] [Google Scholar]
- 28. Zimmerman K. K., Scholten J. D., Huang C. C., Fierke C. A., Hupe D. J. (1998) High level expression of rat farnesyl:protein transferase in Escherichia coli as a translationally coupled heterodimer. Protein Expr. Purif. 14, 395–402 [DOI] [PubMed] [Google Scholar]
- 29. Cassidy P. B., Dolence J. M., Poulter C. D. (1995) Continuous fluorescence assay for protein prenyltransferases. Methods Enzymol. 250, 30–43 [DOI] [PubMed] [Google Scholar]
- 30. Fersht A. (1999) Structure and Mechanism in Protein Science, pp. 110–111, W. H. Freeman and Co., New York [Google Scholar]
- 31. Zamyatnin A. A. (1972) Protein Volume in Solution. Prog. Biophys. Mol. Biol. 24, 107–123 [DOI] [PubMed] [Google Scholar]
- 32. Patrick W. M., Firth A. E., Blackburn J. M. (2003) User-friendly algorithms for estimating completeness and diversity in randomized protein-encoding libraries. Protein Eng. 16, 451–457 [DOI] [PubMed] [Google Scholar]
- 33. Pickett J. S., Bowers K. E., Hartman H. L., Fu H. W., Embry A. C., Casey P. J., Fierke C. A. (2003) Kinetic studies of protein farnesyltransferase mutants establish active substrate conformation. Biochemistry 42, 9741–9748 [DOI] [PubMed] [Google Scholar]
- 34. Tong H., Wiemer A. J., Neighbors J. D., Hohl R. J. (2008) Quantitative determination of farnesyl and geranylgeranyl diphosphate levels in mammalian tissue. Anal. Biochem. 378, 138–143 [DOI] [PubMed] [Google Scholar]
- 35. Roberts P. J., Mitin N., Keller P. J., Chenette E. J., Madigan J. P., Currin R. O., Cox A. D., Wilson O., Kirschmeier P., Der C. J. (2008) Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J. Biol. Chem. 283, 25150–25163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bergo M. O., Leung G. K., Ambroziak P., Otto J. C., Casey P. J., Young S. G. (2000) Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J. Biol. Chem. 275, 17605–17610 [DOI] [PubMed] [Google Scholar]
- 37. Chen Z., Otto J. C., Bergo M. O., Young S. G., Casey P. J. (2000) The C-terminal polylysine region and methylation of K-Ras are critical for the interaction between K-Ras and microtubules. J. Biol. Chem. 275, 41251–41257 [DOI] [PubMed] [Google Scholar]
- 38. Merzlyak E. M., Goedhart J., Shcherbo D., Bulina M. E., Shcheglov A. S., Fradkov A. F., Gaintzeva A., Lukyanov K. A., Lukyanov S., Gadella T. W., Chudakov D. M. (2007) Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods 4, 555–557 [DOI] [PubMed] [Google Scholar]
- 39. Kurth T., Ullmann D., Jakubke H. D., Hedstrom L. (1997) Converting trypsin to chymotrypsin. Structural determinants of S1′ specificity. Biochemistry 36, 10098–10104 [DOI] [PubMed] [Google Scholar]
- 40. Marino F., Pelc L. A., Vogt A., Gandhi P. S., Di Cera E. (2010) Engineering thrombin for selective specificity toward protein C and PAR1. J. Biol. Chem. 285, 19145–19152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Varadarajan N., Rodriguez S., Hwang B. Y., Georgiou G., Iverson B. L. (2008) Highly active and selective endopeptidases with programmed substrate specificities. Nat. Chem. Biol. 4, 290–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hedstrom L. (1996) Trypsin. A case study in the structural determinants of enzyme specificity. Biol. Chem. 377, 465–470 [PubMed] [Google Scholar]
- 43. Wouters M. A., Liu K., Riek P., Husain A. (2003) A despecialization step underlying evolution of a family of serine proteases. Mol. Cell 12, 343–354 [DOI] [PubMed] [Google Scholar]
- 44. Erijman A., Aizner Y., Shifman J. M. (2011) Multispecific recognition. Mechanism, evolution, and design. Biochemistry 50, 602–611 [DOI] [PubMed] [Google Scholar]
- 45. Khersonsky O., Tawfik D. S. (2010) Enzyme promiscuity. A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 [DOI] [PubMed] [Google Scholar]
- 46. Hightower K. E., Huang C. C., Casey P. J., Fierke C. A. (1998) H-Ras peptide and protein substrates bind protein farnesyltransferase as an ionized thiolate. Biochemistry 37, 15555–15562 [DOI] [PubMed] [Google Scholar]
- 47. Crowell D. N. (2000) Functional implications of protein isoprenylation in plants. Prog Lipid Res 39, 393–408 [DOI] [PubMed] [Google Scholar]
- 48. Hanker A. B., Mitin N., Wilder R. S., Henske E. P., Tamanoi F., Cox A. D., Der C. J. (2010) Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene 29, 380–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Allal C., Favre G., Couderc B., Salicio S., Sixou S., Hamilton A. D., Sebti S. M., Lajoie-Mazenc I., Pradines A. (2000) RhoA prenylation is required for promotion of cell growth and transformation and cytoskeleton organization but not for induction of serum response element transcription. J. Biol. Chem. 275, 31001–31008 [DOI] [PubMed] [Google Scholar]
- 50. Allal C., Pradines A., Hamilton A. D., Sebti S. M., Favre G. (2002) Farnesylated RhoB prevents cell cycle arrest and actin cytoskeleton disruption caused by the geranylgeranyltransferase I Inhibitor GGTI-298. Cell Cycle 1, 430–437 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.