Background: The contribution of ribosomal proteins to ribosome assembly and function is often not well understood.
Results: L40 assembles within the cytoplasm into pre-60 S subunits and is required for Nmd3 and Rlp24 recycling.
Conclusion: L40 contributes to formation of 60 S subunits competent for subunit joining and translation elongation.
Significance: Our analysis of L40 function reveals an additional step during cytoplasmic pre-60 S maturation events.
Keywords: Nucleolus, Ribosomal RNA (rRNA), Ribosome Assembly, Ubiquitin, Yeast, Pre-rRNA Processing
Abstract
Most ribosomal proteins play important roles in ribosome biogenesis and function. Here, we have examined the contribution of the essential ribosomal protein L40 in these processes in the yeast Saccharomyces cerevisiae. Deletion of either the RPL40A or RPL40B gene and in vivo depletion of L40 impair 60 S ribosomal subunit biogenesis. Polysome profile analyses reveal the accumulation of half-mers and a moderate reduction in free 60 S ribosomal subunits. Pulse-chase, Northern blotting, and primer extension analyses in the L40-depleted strain clearly indicate that L40 is not strictly required for the precursor rRNA (pre-rRNA) processing reactions but contributes to optimal 27 SB pre-rRNA maturation. Moreover, depletion of L40 hinders the nucleo-cytoplasmic export of pre-60 S ribosomal particles. Importantly, all these defects most likely appear as the direct consequence of impaired Nmd3 and Rlp24 release from cytoplasmic pre-60 S ribosomal subunits and their inefficient recycling back into the nucle(ol)us. In agreement, we show that hemagglutinin epitope-tagged L40A assembles in the cytoplasm into almost mature pre-60 S ribosomal particles. Finally, we have identified that the hemagglutinin epitope-tagged L40A confers resistance to sordarin, a translation inhibitor that impairs the function of eukaryotic elongation factor 2, whereas the rpl40a and rpl40b null mutants are hypersensitive to this antibiotic. We conclude that L40 is assembled at a very late stage into pre-60 S ribosomal subunits and that its incorporation into 60 S ribosomal subunits is a prerequisite for subunit joining and may ensure proper functioning of the translocation process.
Introduction
Ribosomes are the huge ribonucleoprotein particles that catalyze protein synthesis. In all organisms, ribosomes are composed of two ribosomal subunits (r-subunits),3 the large one about twice the size of the small one (1). Over the last decade, the atomic structures of the small and large r-subunits have been determined from bacteria, archaea, and more recently from eukaryotes (2–14). These results have confirmed the long thought suggestion that the eukaryotic cytoplasmic ribosomes display considerable structural and functional similarities to their prokaryotic counterparts but are significantly larger and more complex (15, 16).
In eukaryotes, the synthesis of cytoplasmic ribosomes is a compartmentalized process that takes place largely in the nucleolus (reviewed in Refs. 17–19), although late steps occur in the nucleoplasm and even in the cytoplasm (reviewed in Ref. 20). Ribosome biogenesis is evolutionarily conserved throughout eukaryotes (21, 22), but it has been best studied in the yeast Saccharomyces cerevisiae (23). In the yeast nucleolus, the mature 18 S, 5.8 S, and 25 S rRNAs are co-transcribed as a single large precursor rRNA (pre-rRNA) by RNA polymerase I, whereas the pre-5 S rRNA is independently transcribed by RNA polymerase III (24). The yeast pre-rRNA processing pathway is currently almost fully known and requires a series of sequential endo- and exonucleolytic reactions (supplemental Fig. S1). Pre-rRNA processing occurs concomitantly to most rRNA modification reactions, folding of pre-rRNAs and assembly of most r-proteins to form pre-ribosomal particles (supplemental Fig. S2). Late pre-ribosomal particles are exported to the cytoplasm where they undergo final maturation (supplemental Fig. S2). It has been shown that more than 250 nonribosomal protein factors and 75 small nucleolar RNAs are involved in ribosome synthesis. All these factors, whose precise functions remain largely unknown, likely allow the ribosome maturation process to proceed with the required speed, accuracy, and directionality (25–27).
Ribosomal proteins are critically required for translation, most likely due to their role in directing the folding and maintaining the structure of the ribosome (28–32). Moreover, r-proteins are the binding sites for translation factors, ribosome-associated chaperones, the signal recognition particle, and the translocon (33–37); they can be targets for translation regulatory events (38, 39) and antibiotics (40, 41); some r-proteins have even extra-ribosomal roles (42–46). The consequences on ribosome biogenesis due to loss-of-function of most of the essential yeast r-protein genes has been reported (47, 48). One of the current challenges of the ribosome biogenesis field is defining the course of the assembly of r-proteins at a satisfactorily high resolution. A rough picture of this pathway has been obtained by pioneer work in yeast and HeLa cells (49, 50). These studies indicate that most r-proteins are incorporated into nuclear pre-ribosomal particles either early or late in the process. Few r-proteins were found to assemble in the cytoplasm. These results are consistent with the compositional analyses of early and late pre-ribosomal complexes purified by the tandem affinity purification method (51–53) and the specific study of the incorporation of tagged 40 S r-proteins into 90 S and 43 S pre-ribosomal particles (54). An equivalent study of the incorporation of 60 S r-proteins into pre-60 S r-particles has not yet been reported. There is only information on the timing of assembly for a few 60 S r-proteins, such as L5 and L11 (55), L10 (56), L17 (57), L24 (58), L26 (59), L35 (60), and P0 (61).
We are interested in understanding the contribution of specific 60 S r-proteins to ribosome biogenesis and function (59–62). In this study, we have undertaken the functional analysis of L40 in yeast ribosome synthesis. Homologues of L40 are found in archaea (archaeal L40e) but not in bacteria (63, 64). Interestingly, eukaryotic L40 is generated by proteolytic cleavage of a hybrid protein (Ubi1 or Ubi2), in which ubiquitin is fused to the N terminus of L40 (65, 66). There is another eukaryotic r-protein, S27a (yeast S31), that is synthesized as an ubiquitin-fusion precursor protein (yeast Ubi3) (65, 66). The role of the ubiquitin moiety of yeast Ubi3 has been addressed (66, 67); however, the role of Ubi1 and Ubi2 remains unexplored. Here, we show that depletion of L40 leads to a mild 27 SB and 7 S pre-rRNA processing delay and a nuclear retention of pre-60 S r-particles. Although depletion of L40 barely affects the net production of 60 S r-subunits, polysome profile analyses reveal a strong accumulation of half-mers. Noteworthy, we show that L40 assembles predominantly into cytoplasmic pre-60 S r-particles. The incorporation of L40 is a prerequisite for the release, and hence the subsequent recycling back into the nucle(ol)us, of Nmd3 and Rlp24 from cytoplasmic pre-60 S particles. Finally, we have monitored how L40 may influence the ribosome function during translation. Interestingly, rpl40a or rpl40b null mutants are hypersensitive to the antifungal sordarin derivative GM193663, whereas an HA-tagged L40 construct confers resistance to this antibiotic. Sordarin is a fungus-specific protein synthesis inhibitor that impairs the function of eukaryotic elongation factor 2 (eEF2) (68–70). Altogether, these data indicate that L40 is required for the ribosomal subunit joining process; moreover, L40 might be involved in the recruitment of the translation elongation factor eEF2 and thereby contribute to the proper functioning of the ribosomal translocation process.
EXPERIMENTAL PROCEDURES
Strains and Microbiological Methods
The yeast strains used in this study are listed in supplemental Table S1. Strain JDY919 was generated by PCR-based gene disruption of the RPL40A gene in the W303 strain. To this end, we used plasmid pFA6a, which contains the kanMX4 module, as a template (71) and the oligonucleotide pair RPL40A-kanUP/RPL40A-kanDOWN (supplemental Table S2). After transformation with the PCR product and selection of G418-resistant colonies, disruption was confirmed by PCR analysis on genomic DNA of several candidates with oligonucleotides RPL40A UP/DOWN. Strain JDY920 is a rpl40a::kanMX4 meiotic segregant of JDY919. To obtain strain JDY922, which is a rpl40b::kanMX4 meiotic segregant of JDY921, we followed a similar approach. First, we obtained diploid JDY921 by PCR-based gene disruption of the RPL40B gene in the W303 strain. In this case, oligonucleotides RPL40B-kanUP/RPL40B-kanDOWN (supplemental Table S2) were used to amplify the kanMX4 module of plasmid pFA6a and oligonucleotides RPL40B-UP/RPL40B-DOWN (supplemental Table S2) to confirm the disruption by PCR. Strain JDY711 is a rpl35b::kanMX4 mutant that was obtained by introducing this mutation from RBY139 (60) into the W303 background by three subsequent back-crosses with W303-1B.
Growth and handling of yeast and media followed standard procedures (72). Yeast cells were grown at 30 °C either in rich medium (1% yeast extract, 2% peptone) or in minimal medium (0.15% yeast nitrogen base, 0.5% ammonium sulfate) supplemented with the appropriate amino acids and bases as nutritional requirements and containing either 2% galactose (YPGal, SGal) or 2% glucose (YPD, SD). To prepare plates, 2% agar was added to the media before sterilization. When required, antibiotics were also added at the indicated concentration. Tetrad dissections were performed using a Singer MS micromanipulator. Escherichia coli DH5α strain was used for common cloning and propagation of plasmids (73).
Plasmids
The plasmids used in this study are listed in supplemental Table S3. To generate YCplac33-RPL40A and YCplac111-RPL40A, a PCR was performed using yeast genomic DNA as template and the oligonucleotides RPL40A-UP and RPL40A-DOWN (supplemental Table S2), placed ±1 kb upstream and downstream from the start-stop codon of the UBI1 ORF, respectively. The ∼2.8-kb PCR product was cloned into pGEM®-T Easy (Promega), then restricted with EcoRI and PstI, and cloned into YCplac33 and YCplac111 (74), which were also digested with the same enzymes. To generate YCplac33-RPL40B and YCplac111-RPL40B, we followed a similar strategy using the oligonucleotide pair RPL40B-UP/RPL40B-DOWN (supplemental Table S2). In this case, BamHI was used to subclone an ∼2.8-kb fragment from pGEM®-T Easy into YCplac33 and YCplac111. The resulting plasmids, whose correctness was confirmed by DNA sequencing, complemented both the rpl40a and rpl40b null alleles to the wild-type extent (see supplemental Figs. S3 and S4).
To construct pAS24-RPL40A, an ∼1.8-kb PCR product containing the UBI1 coding region and its intron was obtained by PCR using yeast genomic DNA as template and the GAL-RPL40A-UP and GAL-RPL40A-DOWN primers (supplemental Table S2). This product was cloned into pGEM®-T Easy, then restricted with SalI and PstI, and cloned into pAS24 (75), which was also digested with the same enzymes. The resulting plasmid, whose correctness was confirmed by DNA sequencing, complemented the rpl40 null strain to the wild-type extent in galactose-containing media (see Fig. 2).
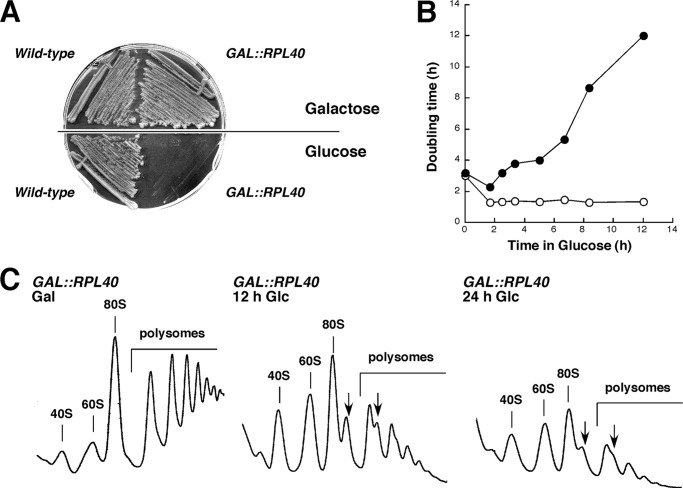
FIGURE 2.
Analysis of the GAL::RPL40 strain upon depletion of L40. A, growth comparison of the strains W303-1A (wild-type) and JDY925 [pAS24-RPL40A] (GAL::RPL40). The cells were streaked on YPGal (galactose) and YPD (glucose) plates and incubated at 30 °C for 3 days. B, growth curves of the wild-type (open circles) and GAL::RPL40 (closed circles) cells at 30 °C after shifting exponential cultures from liquid YPGal to liquid YPD medium for up to 12 h. Data are represented as the doubling time at the different times in YPD. C, polysome profile analysis of GAL::RPL40 cells grown in YPGal or shifted for 12 or 24 h to YPD. For details, see legend to Fig. 1B. The peaks of free 40 S and 60 S r-subunits, 80 S free couples/monosomes, and polysomes are indicated. Half-mers are labeled by arrows.
Plasmid YCplac33-UB-HA-RPL40A was generated by PCR using yeast genomic DNA as template and the oligonucleotide pairs HA-RPL40A-P1/HA-RPL40A-P2 and HA-RPL40A-P3/HA-RPL40A-P4. The corresponding PCR products P12 and P34 were cloned into pGEM®-T Easy. Fragment P12 was digested with EcoRI and KpnI and cloned into EcoRI/KpnI-restricted YCplac33 to generate YCplac33-P12. Then the fragment P34 was digested with KpnI and PstI and cloned into KpnI/PstI-restricted YCplac33-P12. The resulting plasmid, whose correctness was confirmed by DNA sequencing, partially complemented the rpl40 null (supplemental Fig. S5). YCplac111-UB-HA-RPL40A was constructed by using a similar approach.
To generate YCplac33-RLP24-GFP, a PCR was performed using yeast genomic DNA as template and the oligonucleotides RLP24GFP-UP and RLP24GFP-DOWN (supplemental Table S2), placed 1 kb upstream from the start codon of the RLP24 ORF, and replacing the RLP24 stop codon with a BamHI restriction site, respectively. The ∼1.6-kb PCR product was cloned into pGEM®-T (Promega), then excised with EcoRI and BamHI, and cloned into YCplac33-yeGFP/TCYC1 (76), which was also digested with the same enzymes.
Construction of a GAL::RPL40 Allele and in Vivo Depletion of L40
Strain JDY920 harboring the YCplac33-RPL40A plasmid was crossed to JDY923, which is a MATα rpl40b::kanMX4 segregant of JDY921. The resulting diploid was sporulated, and tetrads were dissected. G418-resistant spore clones from at least three complete nonparental ditypes were transformed with pAS24-RPL40A, which allows expression of Ubi1 under the control of a GAL promoter. Transformants were streaked on SGal-Leu plates and subjected to plasmid shuffling on 5-fluoroorotic acid-containing SGal plates at 30 °C. One of these clones, JDY925 [pAS24-RPL40A], which we named the GAL::RPL40 strain, was used for further work.
For in vivo depletion of L40, the GAL::RPL40 strain was grown in liquid YPGal medium at 30 °C until mid-exponential phase (OD600 = 0.8) and then harvested, washed, and transferred to liquid YPD medium. Cell growth was monitored over a period of 24 h, during which the cultures were regularly diluted into fresh YPD medium. As control, the wild-type W303-1A strain was used. At different time points, samples were collected to perform protein and RNA extractions and polysome analysis.
Sucrose Gradient Centrifugation
Cell extracts for polysome and r-subunit analyses were prepared and analyzed as described previously (77) using an ISCO UA-6 system equipped to continuously monitor A254.
Protein Extractions and Western Blotting Analyses
Total yeast protein extracts were prepared and analyzed by Western blotting according to standard procedures (73). The following primary antibodies were used: mouse monoclonal anti-HA (HA.11 16B12, Covance); mouse monoclonal anti-Nop1 (MCA-28F2, EnCor Biotechnology); mouse monoclonal anti-GFP (Roche Applied Science); mouse monoclonal BH5 anti-stalk r-proteins (a gift from J. P. G. Ballesta) (78); mouse monoclonal anti-L3 (a gift from J. R. Warner) (42); mouse monoclonal 3AG10 anti-L12 (a gift from J. P. G. Ballesta) (79); rabbit polyclonal anti-L1 (a gift from F. Lacroute) (80); rabbit polyclonal anti-L5 (a gift from S. R. Valentini) (81); rabbit polyclonal anti-L10 (a gift from B. L. Trumpower) (82); rabbit polyclonal anti-L35 (a gift from M. Seedorf) (83), rabbit polyclonal anti-Mrt4 (a gift from J. P. G. Ballesta) (79); and rabbit polyclonal anti-Has1 (a gift from P. Linder) (84). Goat anti-rabbit or anti-mouse horseradish peroxidase-conjugated antibodies (Bio-Rad) were used as secondary antibodies. Immune complexes were visualized using an enhanced chemiluminescence detection kit (Pierce).
RNA Analyses
RNA extraction, Northern hybridization, and primer extension analyses were carried out according to standard procedures (85, 86). In all experiments, RNA was extracted from samples corresponding to 10 OD600 units of exponentially grown cells. Equal amounts of total RNA was loaded on gels or used for primer extension reactions (86). The sequences of oligonucleotides used for Northern hybridization and primer extension analyses have been described previously (87) and are listed in supplemental Table S2. Phosphorimager analysis was performed in a FLA-5100 imaging system (Fujifilm).
Pulse-chase labeling of pre-rRNA was performed exactly as described previously (88), using 100 mCi of [5,6-3H]uracil (PerkinElmer Life Sciences; 45–50 Ci/mmol) per 40 OD600 of yeast cells. Both the wild-type and GAL::RPL40 strains were first transformed with an empty YCplac33 plasmid to make them prototrophic for uracil. Then they were pre-grown in liquid SGal-Ura medium and shifted to liquid SD-Ura for 12 h. At this time point, the GAL::RPL40 strain was doubling every ∼10–12 h compared with ∼2 h for the wild-type strain. The cells were pulse-labeled for 2 min and then chased for 5, 15, 30, and 60 min with a large excess of nonradioactive uracil. Total RNA was extracted as above, and 20,000 cpm per RNA sample was loaded on gels and analyzed as described previously (88).
Immunoprecipitation of the HA-L40A Protein
For immunoprecipitation experiments, 150 ml of HA-tagged or untagged negative control cells were grown in YPD medium to an OD600 of 0.8, washed with cold water, harvested, and concentrated in 500 μl of ice-cold HA lysis buffer (50 mm Tris-HCl, pH 7.5; 5 mm (CH3COO)2Mg; 150 mm KCl; 0.2% Triton X-100) containing a protease inhibitor mixture (Complete, Roche Applied Science) and 1 mm PMSF. Cells were disrupted by vigorous shaking with glass beads in a Fastprep®-24 (MP Biomedicals) at 4 °C, and total cell extracts were obtained by centrifugation in a microcentrifuge at the maximum speed (∼16,100 × g) for 15 min at 4 °C. About 10 and 2% of the resulting supernatants were taken to analyze total RNA and protein, respectively. To the rest (about 450 μl), 2.5 μg of rat anti-HA high affinity antibody (Roche Applied Science) was added, and the mixture was incubated for 1 h at 4 °C with end-over-end tube rotation. Then 100 μl of protein A-Sepharose beads (GE Healthcare) were added, and the new mixture was incubated with end-over-end tube rotation for 4 h at 4 °C. After incubation, the beads were washed seven times with 1 ml of the HA lysis buffer at 4 °C and finally collected. Protein was extracted with Laemmli buffer from both total cell extract samples and 1/10th of the beads. The extracted proteins were analyzed by Western blotting using mouse monoclonal anti-HA antibody (HA.11 16B12, Covance). RNA was extracted from the total cell extract samples, and the rest of the beads as exactly described previously (89). The extracted RNA was analyzed by Northern blotting as described above.
Affinity Purification of GFP-tagged Proteins
GFP-tagged factors representative of 90 S and pre-60 S r-particles were purified following a one-step GFP-Trap®_A procedure (Chromotek), as described previously (90), with slight modifications. Briefly, GFP-tagged or untagged negative control cells harboring the YCplac33-HA-RPL40A plasmid were grown in 200 ml of YPD medium to an OD600 of 0.8. Cells were washed with cold water, harvested, and concentrated in 500 μl of ice-cold GFP lysis buffer (20 mm Tris-HCl, pH 7.5; 5 mm MgCl2; 150 mm CH3COOK; 1 mm DTT; 0.2% Triton X-100) containing a protease inhibitor mixture (Complete, Roche Applied Science). Cells were disrupted by vigorous shaking with glass beads in a Fastprep®-24 (MP Biomedicals) at 4 °C, and total extracts were clarified by centrifugation in a microcentrifuge at the maximum speed (∼16,100 × g) for 10 min at 4 °C. To each of the resulting supernatants, 25 μl of GFP-Trap®_A beads, equilibrated with the same buffer, were added, and the mixture was incubated for 2 h at 4 °C with end-over-end tube rotation. After incubation, the beads were washed six times with 1 ml of the GFP lysis buffer at 4 °C and resuspended in 100 μl of Laemmli buffer. Proteins were extracted by boiling for 10 min, separated in NuPAGE® SDS 4–12% gradient polyacrylamide gels (Invitrogen), and visualized by silver staining or Western blotting. To normalize the amount of purified complexes to be loaded for comparative studies, aliquots of the samples were first resolved in the same conditions and visualized by silver staining according to the manufacturer's instructions (Bio-Rad).
Fluorescence Microscopy
To test pre-ribosomal particle export, the appropriate strains were transformed with the pRS314-RFP-NOP1-RPL25-eGFP or the pRS314-RFP-NOP1-RPS3-eGFP plasmid (gifts from J. Bassler) (91), which harbors the reporters Nop1-mRFP and L25-eGFP or Nop1-mRFP and S3-eGFP, respectively (supplemental Table S3). Transformants were grown in SGal-Trp medium and shifted to SD-Trp for 9 h to deplete L40. Cells were washed, resuspended in PBS buffer (140 mm NaCl, 8 mm Na2HPO4, 1.5 mm KH2PO4, 2.75 mm KCl, pH 7.3), and examined with a Leica DMR fluorescence microscope equipped with a DC350F digital camera and its software. Images were processed with Adobe® Photoshop® CS2 (Adobe Systems Inc.).
To monitor whether L40 assembles in the nucleus, the JDY925 strain expressing an HA-tagged L40A from plasmid YCplac111-UBI-HA-RPL40A was transformed with plasmids pRS316-GAL-NMD3Δ100 (a gift from A. Jacobson) (92) and pRS314-RFP-NOP1-RPL25-eGFP (91). To monitor the localization of HA-tagged L40A, we performed immunofluorescence with cells grown in SD-Leu-Ura-Trp or shifted for 24 h to SGal-Leu-Ura-Trp toinduce the expression of the dominant-negative Nmd3Δ100 protein. Immunofluorescence was performed as described previously (93). Primary monoclonal mouse anti-HA antibodies (HA.11, Covance) at a dilution of 1:500 and secondary goat anti-mouse Cy3-coupled antibody (Jackson ImmunoResearch Laboratories) at 1:2000 were used to detect HA-L40A. 4′,6-Diamidino-2-phenylindole (DAPI, Fluka) was used to stain DNA in the nucleus. Fluorescently labeled cells were inspected and processed as described above. Cells from the same cultures were also inspected for the GFP and red fluorescent protein signal.
To study the subcellular location of Mex67, Mrt4, Nmd3, Rlp24, and Tif6 in the GAL::RPL40 strain, plasmids pRS316-MEX67-GFP (a gift from J. Bassler) (94), YCplac33-MRT4-eGFP (61), pAJ755 (a gift from A. W. Johnson) (95), YCplac33-RLP24-eGFP, and pTIF6-GFP (a gift from A.J. Warren) (96) were used (see supplemental Table S3). To determine the subcellular location of Arx1, strain JDY910, which harbors a genomically GFP-tagged Arx1 (see supplemental Table S1), was transformed with pAS24-RPL40A, and the resident YCplac33-UB-HA-RPL40A was counterselected on 5-fluoroorotic acid-containing plates. Cells were grown in SGal-Leu-Ura or subjected to in vivo depletion in SD-Leu-Ura for different times. When required, DAPI (1 μg/ml final concentration) was added to the cultures during the last 3 h of growth. Fluorescently labeled cells were inspected and processed as described above.
Antibiotic Sensitivity Assays
The sensitivity to different drugs impairing translation was tested as follows: the wild-type strain, the rpl40a or rpl40b null mutants, and the strain expressing the HA-tagged L40A protein were grown in YPD medium to an OD600 of 0.8 and diluted to an OD600 of 0.05. A series of 10-fold dilutions was done for each strain, and 5-μl drops were spotted on YPD plates containing the indicated antibiotics at the specified range of the following concentrations: anisomycin (from 5 to 15 μg/ml), cycloheximide (from 0.025 to 0.1 μg/ml), hygromycin B (from 5 to 20 μg/ml), paromomycin (from 0.1 to 2.5 mg/ml), and neomycin (from 0.5 to 5 mg/ml). Plates were incubated at 30 °C for 3–4 days.
The sordarin sensitivity assay was carried out in 96-well plates; the above strains were grown in YPD medium to an A600 of 0.8 and diluted to an A600 of 0.05 in 200 μl of YPD. Ten serial dilutions of the sordarin derivative GM193663 (Glaxo-Smith-Kline Research Center, Madrid; a gift from J. P. G. Ballesta) was done starting with the concentration of 4 μg/ml in the first well. The plates were incubated at 30 °C with shaking, and the A600 was measured regularly at intervals of 2 h during 24–48 h. Inhibition was measured as the percentage of growth in wells containing versus controls lacking sordarin.
Preparation of Ribosomes
Ribosomes were enriched as described previously (79). Briefly, 200 ml of cells expressing the HA-tagged or the untagged L40A r-protein were grown in YPD to an A600 of 0.8, washed, and concentrated in 500 μl of ice-cold buffer 1 (10 mm Tris-HCl, pH 7.4; 20 mm KCl; 12.5 mm MgCl2; 5 mm 2-mercaptoethanol) containing a protease inhibitor mixture (Complete EDTA-free, Roche Applied Science). Cells were disrupted by vigorous shaking with glass beads in a Fastprep®-24 homogenizer (MP Biomedicals) at 4 °C. An S30 fraction was obtained by centrifuging the extract at 16,100 × g for 20 min at 4 °C in an Eppendorf microcentrifuge. A crude ribosome pellet was prepared from the S30 fraction by centrifugation in a Beckman-Coulter OptimaTM Max using a TL110 rotor at 90,000 rpm for 60 min at 4 °C. The pellet was resuspended in buffer 2 (20 mm Tris-HCl, pH 7.4; 100 mm MgCl2; 5 mm 2-mercaptoethanol) and washed by centrifugation in a 20–40% discontinuous sucrose gradient in buffer 2 containing 500 mm ammonium acetate at 90,000 rpm for 120 min at 4 °C again in a TLA110 rotor. The pellet of washed ribosomes was dissolved in buffer 1 and stored at −80 °C. Ribosomal proteins were separated in SDS-NuPAGE 4–12% gradient polyacrylamide gels (Invitrogen) and visualized by Coomassie or Western blotting with selected antibodies of the above described.
RESULTS
Generation of Strains for the Phenotypic Analysis of the L40 Protein
Yeast L40 is an essential r-protein of 52 amino acids with a predicted mass of 6.0 kDa and a basic pI of 11.05. As most yeast r-protein genes (97), L40 is encoded by two independent paralogous genes, UBI1 (RPL40A and YIL148W) and UBI2 (RPL40B and YKR094C). Their coding regions result in identical hybrid proteins that are specifically cleaved to yield ubiquitin and the r-protein L40 (98).
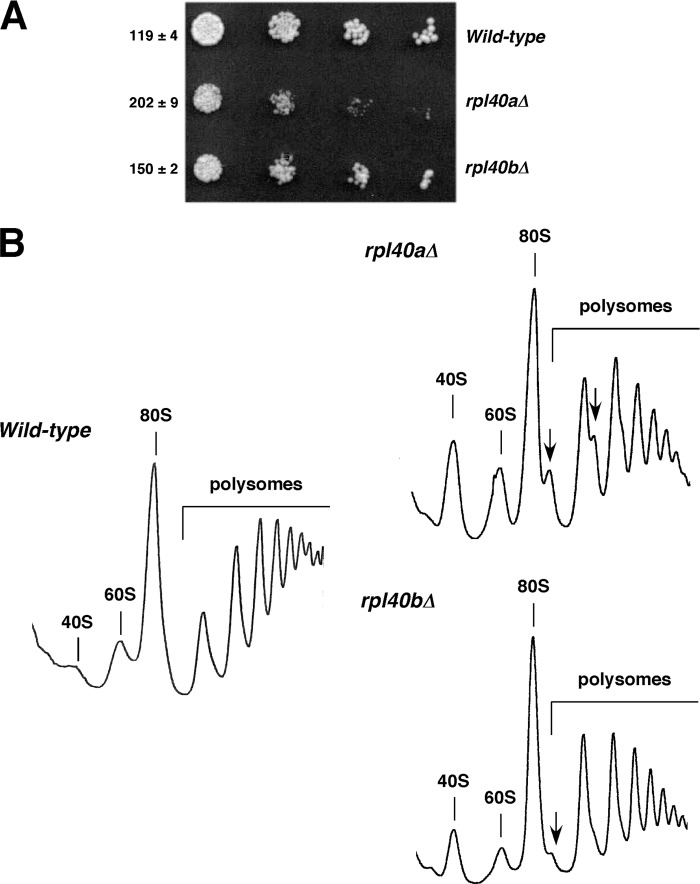
Early studies indicated that both UBI1 and UBI2 genes (hereafter RPL40A and RPL40B genes) are required for growth and normal accumulation of 60 S r-subunits (66). The impact of a 4 h depletion of L40 on 60 S r-subunit formation has been previously described in a systematic analysis of conditional mutants for expression of 26 distinct yeast essential 60 S r-proteins (47). To characterize in detail the contribution of L40 to ribosome biogenesis, we first studied the phenotypic consequences of the deletion of either the RPL40A or the RPL40B gene. As shown in Fig. 1A, the deletion of either RPL40A or RPL40B results in a mild growth defect, the one of RPL40A being slightly stronger. This result is in agreement with other reports (66, 99) and is consistent with genome-wide expression data that suggest that RPL40A might contribute to 58% and RPL40B to 42% of total mRNA levels (100). To address the contribution of each L40 protein to 60 S r-subunit production, we performed polysome profile analysis. As shown in Fig. 1B, both the rpl40aΔ and rpl40bΔ null strains showed a mild deficit in 60 S r-subunits with the rpl40aΔ strain being more affected than the rpl40bΔ strain. Quantification of total r-subunits by sucrose gradients indicated about a 5% reduction in the overall quantity of 60 S r-subunits relative to the wild-type strain in both rpl40aΔ and rpl40bΔ strains (data not shown). Recent studies have suggested that duplicated paralogous r-proteins could play specific nonredundant roles in ribosome biogenesis and assembly (101). However, few extra copies of RPL40A or RPL40B, which were provided from centromeric plasmids, fully restored a wild-type growth rate and polysome profiles to the rpl40bΔ and rpl40aΔ strains, respectively (supplemental Figs. S3 and S4). This suggests that there is no functional distinction between the L40A and L40B proteins at least under the laboratory conditions tested.
FIGURE 1.
Growth and polysome profile analysis of the rpl40a and rpl40b null mutants. A, growth phenotypes of rpl40 mutants. Strains W303-1A (Wild-type), JDY920 (rpl40aΔ), and JDY922 (rpl40bΔ), which carry a YCplac111 plasmid, were grown in minimal SD-Leu medium and diluted to an A600 of 0.05. Cells were spotted in 10-fold serial dilutions onto SD-Leu plates and incubated at 30 °C for 3 days. The doubling times in liquid SD-Leu at 30 °C are indicated in minutes. Values correspond to the mean ± S.D. of five independent experiments. B, polysome profiles of the above strains. These strains were grown in YPD medium at 30 °C to an A600 of around 0.8. Whole cell extracts were prepared, and 10 A260 values of each extract were resolved in 7–50% sucrose gradients. The A254 was continuously measured. Sedimentation is from left to right. The peaks of free 40 S and 60 S r-subunits, 80 S free couples/monosomes, and polysomes are indicated. Half-mers are labeled by arrows.
Depletion of L40 Leads to a Minor 60 S Ribosomal Subunit Shortage and a Subunit Joining Defect
To further examine the function of L40, we constructed a GAL::RPL40A rpl40bΔ strain (hereafter named the GAL::RPL40 strain; see “Experimental Procedures” and supplemental Table S1). Growth of this strain was not different from that of the isogenic wild-type strain on permissive YPGal medium but was progressively slowed after shifting to repressive YPD medium and ceased after ∼12 h (Fig. 2A). To address the contribution of L40 to 60 S r-subunit production, we first performed polysome profile analysis. As shown in Fig. 2B, and complementing previous data (47), the GAL::RPL40 strain displayed practically normal polysome profiles when grown in YPGal. When shifted for 12 h to YPD, it showed a clear decrease in polysomes and the appearance of half-mer polysomes. These features were even more pronounced upon a 24-h shift to YPD (Fig. 2B). Wild-type cells showed no alterations in the polysome profile when transferred to YPD (data not shown). Intriguingly, there was almost no decrease in the free 60 S/40 S ratio for the L40-depleted strain after the shift to YPD (Fig. 2B). This could be due to the accumulation of arrested pre-60 S r-particles to a simultaneous deficit of both r-subunits and/or, most likely, to a 60 S r-subunit joining defect in translation initiation upon depletion of L40 (see “Discussion”). Consistently, quantification of total r-subunits by run-off and low Mg2+ sucrose gradients showed that depletion of L40 leads to only 10% reduction of the 60 S to 40 S r-subunit ratio compared with the wild-type control upon 12 h of depletion (data not shown). We therefore conclude that depletion of L40 leads to a minor deficit in 60 S relative to 40 S r-subunits and a 60 S r-subunit joining defect.
Depletion of L40 Slightly Delays rRNA Processing at ITS2
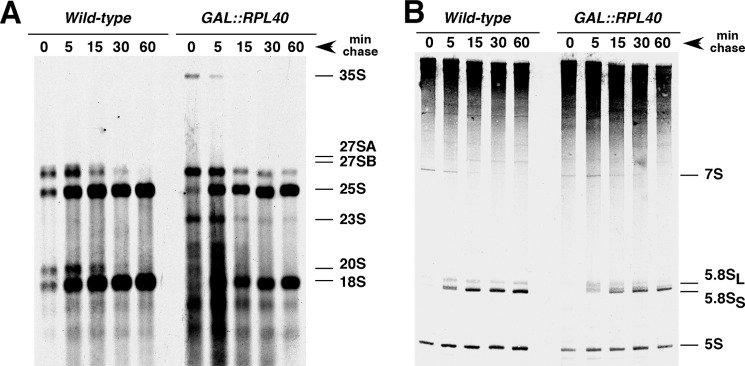
To determine whether depletion of L40 impairs ribosome biogenesis, we assessed the kinetics of rRNA accumulation by pulse-chase labeling experiments in wild-type and GAL::RPL40 cells shifted for 12 h to SD-Ura medium (see “Experimental Procedures”). Cells were pulse-labeled for 2 min with [5,6-3H]uracil and then chase for 5, 15, 30, and 60 min with a large excess of nonradioactive uracil. Compared with the wild-type cells, the GAL::RPL40 cells showed delayed processing of 35 S pre-rRNA as shown by the abundance and persistence of this precursor in early chase time points. This was combined with some loss of 27 SA2 and 20 S pre-rRNA and the appearance of the aberrant 23 S pre-rRNA species (Fig. 3A). More importantly, some 27 SB pre-rRNAs persisted until the last time point of the chase. However, a gradual decrease in 27 SB pre-rRNAs is observed over the chase, thus suggesting efficient turnover of these precursors. A minor accumulation of 7 S pre-rRNAs is also perceived (Fig. 3B). Interestingly, the production of mature rRNAs is minimally affected (Fig. 3). Thus, the minor 60 S r-subunit shortage detected in the L40-depleted cells is likely the consequence of the slight delay in 27 SB and 7 S pre-rRNA processing detected in these cells. We conclude that 60 S r-subunit biogenesis could continue at almost the wild-type rate in the absence of the L40 r-protein.
FIGURE 3.
Depletion of L40 slightly delays 27 SB pre-rRNA processing. A, strains W303-1A (wild-type) and JDY925 [pAS24-RPL40A] (GAL::RPL40) were transformed with YCplac33 and then grown at 30 °C in SGal-Ura and shifted for 12 h to SD-Ura. Cells were pulse-labeled with [5,6-3H]uracil for 2 min and then chased with an excess of unlabeled uracil for the times indicated. Total RNA was extracted, and 20,000 cpm were loaded and separated on a 1.2% agarose-6% formaldehyde gel (A) or a 7% polyacrylamide, 8 m urea (B), transferred to nylon membranes, and visualized by fluorography. The positions of the different pre-rRNAs and mature rRNA are indicated.
To assess whether L40 has a role in pre-rRNA processing, we analyzed the steady-state levels of pre- and mature rRNAs by Northern hybridization during a time course of L40 depletion. Consistent with the pulse-chase data, a certain decrease in the levels of mature rRNAs was observed after depletion of L40 especially at later time points (Fig. 4). The 35 S and the aberrant 23 S pre-rRNAs accumulated with ongoing depletion (Fig. 4A). These accumulations are consistent with delayed pre-rRNA processing at early cleavage sites. Probably, in part as a consequence of this delay, the levels of the 20 S pre-rRNA but more significantly those of the 27 SA2 pre-rRNA decreased (Fig. 4A). Finally, we detected almost no changes in levels of 27 SB pre-rRNAs (Fig. 4A) and a modest accumulation of 7 S pre-rRNAs (Fig. 4B).
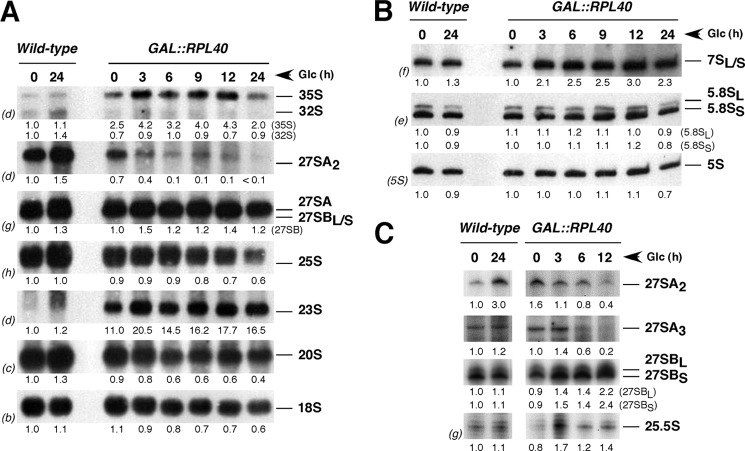
FIGURE 4.
Depletion of L40 does not significantly impair pre-rRNA processing. Strains W303-1A (wild-type) and JDY925 [pAS24-RPL40A] (GAL::RPL40) were grown in YPGal at 30 °C, shifted to YPD, and harvested at the indicated times. Total RNA was extracted from each sample. Equal amount of total RNA (5 μg) was subjected to Northern blot or primer extension analysis. A, Northern blot analysis of high molecular mass pre- and mature rRNAs separated on a 1.2% agarose, 6% formaldehyde gel. B, Northern blot analysis of low molecular mass pre- and mature rRNAs separated on a 7% polyacrylamide, 8 m urea gel. C, primer extension analysis of 27 S and 25.5 S pre-rRNAs. Probes used are indicated on the left of each panel in parentheses and described in supplemental Fig. S1A and supplemental Table S2. Signal intensities were measured by phosphorimager scanning; values (indicated below each lane) were normalized to those obtained for the wild-type control grown in YPGal and arbitrarily set at 1.0.
To increase the resolution of the pre-rRNA analysis, we also performed primer extension analyses upon depletion of L40 (Fig. 4C). The levels of 27 SA2 and 27 SA3 pre-rRNAs were clearly lower, as shown by the primer extension stop at site A2 and A3, respectively. In contrast, the levels of 27 SBL and 27 SBS modestly increased, as shown by the primer extension stops at B1L and B1S, respectively. Finally, primer extension analysis through site C2 displayed a clear increase in the level of the 25.5 S pre-rRNA upon depletion of L40.
Altogether, our results indicate that L40 is not essential for the maturation of rRNAs. Instead, L40 seems to be needed to optimize the pre-rRNA processing reactions occurring within the ITS2 region. Hence, the cleavage at site C2 and especially the further exonucleolytic trimming of 7 S and 25.5 S pre-rRNAs to mature 5.8 S and 25 S rRNAs, respectively, are slightly delayed upon L40 depletion.
Depletion of L40 Impairs Export of Pre-60 S r-particles from the Nucleus to the Cytoplasm
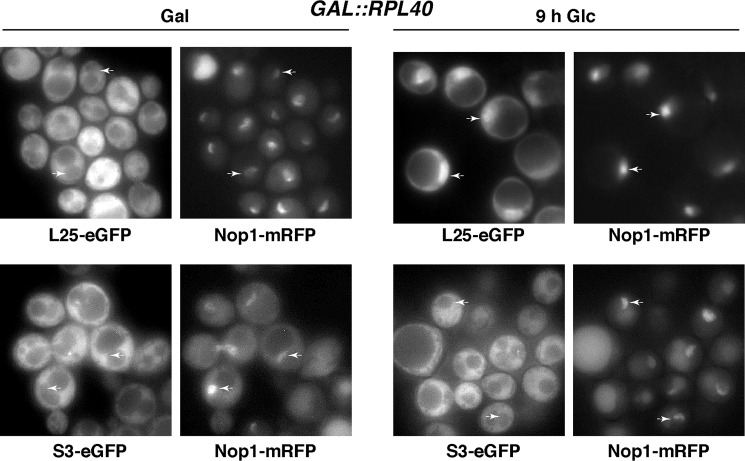
To determine whether L40 depletion impairs nuclear export of pre-60 S r-particles, we analyzed the location of the L25-eGFP 60 S reporter (91) in the GAL::RPL40 strain. Under permissive culture conditions (SGal-Ura) or after short shifts to nonpermissive conditions (3–6 h in SD-Ura), L25-eGFP was found predominantly in the cytoplasm in the GAL::RPL40 strain (Fig. 5 and data not shown). However, following a longer shift to glucose-containing medium (9–12 h in SD-Ura), GAL:RPL40 cells exhibited an accumulation of the L25-eGFP fluorescence signal all over the nucleus without evident restriction to the nucleolus (Fig. 5). Similar results were obtained when the L11B-eGFP 60 S reporter was used (data not shown) (102). These observations are specific for the large r-subunit because the location of the S2-eGFP and S3-eGFP 40 S r-subunit reporters was cytoplasmic in either SGal-Ura medium or upon a long shift to SD-Ura medium (Fig. 5 and data not shown). The wild-type strain was also examined; no nuclear fluorescence accumulation for any of the above r-reporters was observed either in SGal-Ura or in SD-Ura medium (data not shown). We conclude that pre-60 S r-particles are retained in the nucle(ol)us at later time points of L40 depletion.
FIGURE 5.
Depletion of L40 leads to nuclear retention of the 60 S r-subunit reporter L25-eGFP. GAL::RPL40 cells co-expressing Nop1-mRFP and either L25-eGFP or S3-eGFP from their cognate promoters were grown at 30 °C in SGal-Trp or shifted to SD-Trp for up to 9 h. The subcellular localizations of the GFP-tagged ribosomal proteins and the Nop1-mRFP nucleolar marker were analyzed by fluorescence microscopy. Arrows point to nucleolar fluorescence. Approximately 200 cells were examined for each reporter, and practically all cells gave the results shown in the pictures.
L40 Assembles in the Cytoplasm
Assembly of r-subunits occurs mainly in the nucle(ol)us, although few 60 S r-proteins appear to be loaded exclusively, such as L24 (49, 58), or preferentially, such as L10 or P0 (56, 61, 103, 104), onto cytoplasmic pre-60 S r-particles. Pulse-chase studies performed in the 1970s suggest that yeast L40 associates at a late stage of the 60 S r-subunit assembly process (49). The presence of a nuclear localization sequence (NLS) within L40 (cNLS mapper program (105)) suggests that L40 assembly may occur in the nucleus. To study precisely in which cellular compartment L40 assembles, we made use of an rpl40 null strain expressing HA-tagged L40A from a centromeric plasmid (see “Experimental Procedures” and supplemental Table S1). The resulting strain displayed almost wild-type growth and polysome profiles (supplemental Fig. S5, A and B). Moreover, the HA-L40A protein assembles efficiently into ribosomes (supplemental Fig. S5C).
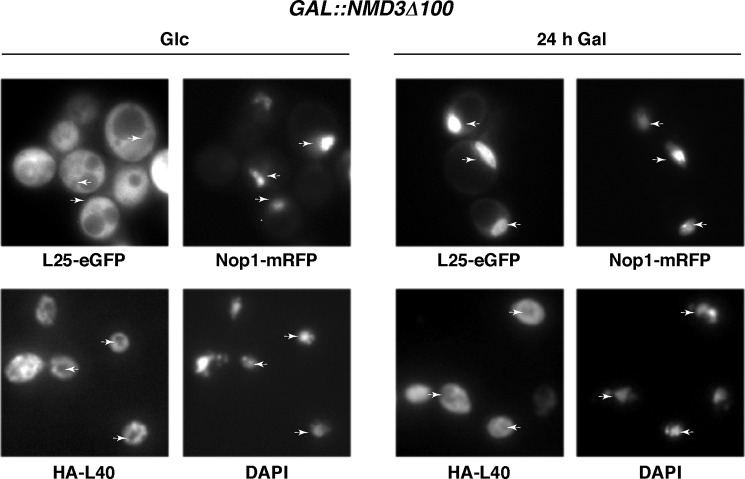
Because export of pre-60 S r-particles is largely dependent on the exportin Crm1/Xpo1, which is one of several pre-60 S export factors identified in yeast (106–111), and its pre-60 S adaptor protein Nmd3 (106, 112), we monitored the cytoplasmic or nuclear localization of the HA-L40A protein upon overexpression of a wild-type Nmd3 protein or the dominant-negative truncated Nmd3Δ100 protein. The mutant Nmd3Δ100 protein essentially lacks the C-terminally located nuclear export sequence signals that mediate nuclear export, but it retains the ability to bind to pre-60 S r-particles; thus, it specifically traps these particles in the nucleus (92). Under noninducible conditions or when the wild-type Nmd3 protein was overexpressed, HA-L40A was detected in the cytoplasm of all cells examined (Fig. 6 and data not shown). Strikingly, no change in the cytoplasmic distribution of HA-L40A was found upon overexpression of the Nmd3Δ100 protein (Fig. 6). As a control, we used the L25-eGFP reporter, which was simultaneously co-expressed with HA-L40A in the rpl40 null strain. L25-eGFP has been described to assemble in the nucle(ol)us (60, 106, 113). As expected, this reporter remained cytoplasmic in noninduced cells but was clearly found in the nucleus of cells expressing the Nmd3Δ100 protein (Fig. 6). Altogether, these data suggest that, independently of the predicted presence of NLS sequences, L40 assembles predominantly at the level of cytoplasmic pre-60 S r-particles.
FIGURE 6.
HA-tagged L40A protein assembles in the cytoplasm. Localization of L25-eGFP, Nop1-mRFP, and HA-L40A upon induction of the dominant-negative NMD3Δ100 allele and rpl40 null cells expressing L25-eGFP and Nop1-mRFP from plasmid pRS314-RFP-NOP1-RPL25-eGFP and HA-L40A from plasmid YCplac111-UB-HA-RPL40A were transformed with the pRS316-GAL-NMΔ3Δ100 plasmid. Transformants were grown in SD-Leu-Ura-Trp or shifted for 24 h to SGal-Leu-Ura-Trp to fully induce the Nmd3Δ100 protein. The GFP and red fluorescent protein signals were simultaneously inspected by fluorescence microscopy. Cells from the same cultures were also inspected by indirect immunofluorescence. HA-L40A was detected with monoclonal mouse anti-HA antibodies, followed by decoration with goat anti-mouse Cy3-coupled antibody. Chromatin DNA was stained with DAPI. Arrows point to either nucleolus or the nucleoplasm depending if Nop1-mRFP or DAPI was used as a marker, respectively. Approximately 200 cells were examined for each reporter, and practically all cells gave the results shown in the pictures.
L40 Associates with Very Late/Cytoplasmic Pre-60 S Ribosomal Particles
To investigate in more detail the timing of the assembly of L40 with pre-60 S r-particles, we first immunoprecipitated HA-L40-containing complexes from whole cell extracts and assayed co-purified pre-rRNA species and r-proteins by Northern hybridization and Western blotting, respectively. As shown in Fig. 7, there was a significant co-precipitation of L1 and L35 r-proteins (Fig. 7A) and mature 5.8 S and 5 S rRNAs (Fig. 7B) with the tagged HA-L40A protein, indicating that r-particles rather than soluble HA-L40 were immunoprecipitated. The precipitation was specific because neither r-proteins nor RNA were enriched from extracts of an untagged strain (Fig. 7, A and B). Interestingly, no 7 S pre-rRNAs were detected, suggesting that either the HA tag is not accessible for immunoprecipitation when HA-L40 is present in nuclear pre-60 S r-particles or the HA-L40 protein assembles into late pre-60 S intermediates, which have completed the pre-rRNA processing reactions. To distinguish between these two possibilities, we screened for the presence of HA-L40 following purification of GFP-tagged proteins specific for 90 S (Nop58/Nop5) (53, 114), early, intermediate, late, and cytoplasmic pre-60 S r-particles (Ssf1, Nsa1, Nop7/Yph1, Arx1, and Kre35/Lsg1) (52, 115–119) and mature 60 S r-subunits (P0) (see supplemental Fig. S2) (120). Purified particles were analyzed by SDS-PAGE, and HA-L40 was detected by Western blotting using anti-HA antibodies. Antibodies against selected pre-ribosomal factors (Nop1, Has1, and Mrt4) and other 60 S r-proteins (L1 and L3) were also used to define the pre-60 S r-particles at the different stages of their maturation. This analysis revealed that HA-L40 was only significantly enriched in P0-eGFP containing particles, which mainly represent mature 60 S r-subunits (Fig. 8). HA-L40 was practically absent from early (Ssf1-eGFP and Nop7-eGFP) and intermediate pre-60 S particles (Nsa1-eGFP) and very modestly represented in 90 S (Nop58-eGFP) and late (Arx1-eGFP) and cytoplasmic (Arx1-eGFP and Kre35-eGFP) pre-60 S particles (Fig. 8). In clear contrast, L1 and L3 were present, as reported previously (61, 90, 116), in all tested pre-ribosomal particles. Finally, and as expected, Nop1 is mainly associated with 90 S pre-ribosomal particles (114), Has1 with 90 S and early and intermediate pre-60 S r-particles (90), and Mrt4 with intermediate and late/cytoplasmic pre-60 S r-particles (61). Altogether, these results reveal that L40 associates with late/cytoplasmic pre-60 S r-particles but is only stably assembled into mature cytoplasmic 60 S r-subunits.
FIGURE 7.
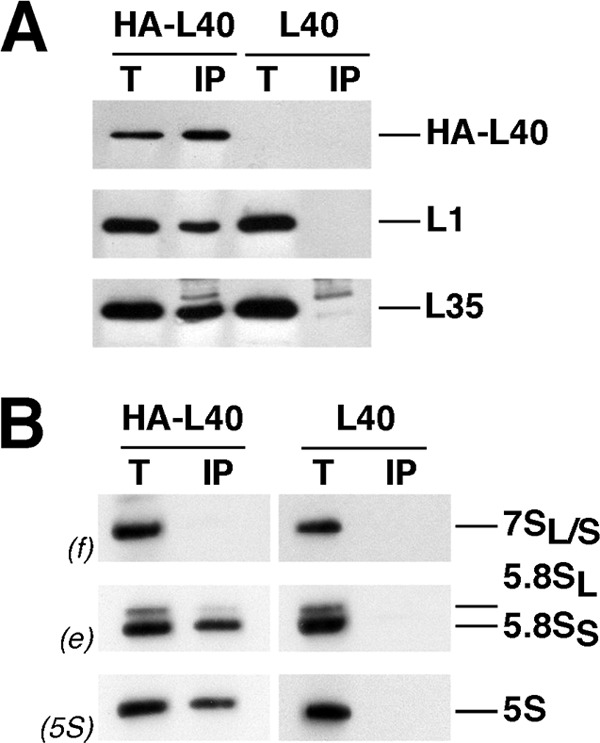
HA-L40A associates with almost mature pre-60 S ribosomal particles. HA-tagged L40A was affinity-purified with anti-HA antibodies as outlined under “Experimental Procedures” from total cell extracts of rpl40 null cells expressing HA-L40A (HA-L40). Wild-type cells (L40) were used as an untagged L40 control. A, proteins were extracted from the pellets obtained after immunoprecipitation (lanes IP) or from an amount of total extracts corresponding to 0.5% of that used for purification (lanes T) and subjected to SDS-PAGE and Western blotting using specific antibodies against the HA tag, L1, and L35 r-proteins. B, RNA was also extracted from the above IP and T samples and subjected to Northern analysis of 7 S pre-rRNA and mature 5.8 S and 5 S rRNAs. Probes, in parentheses, are described in supplemental Fig. S1A and supplemental Table S2.
FIGURE 8.
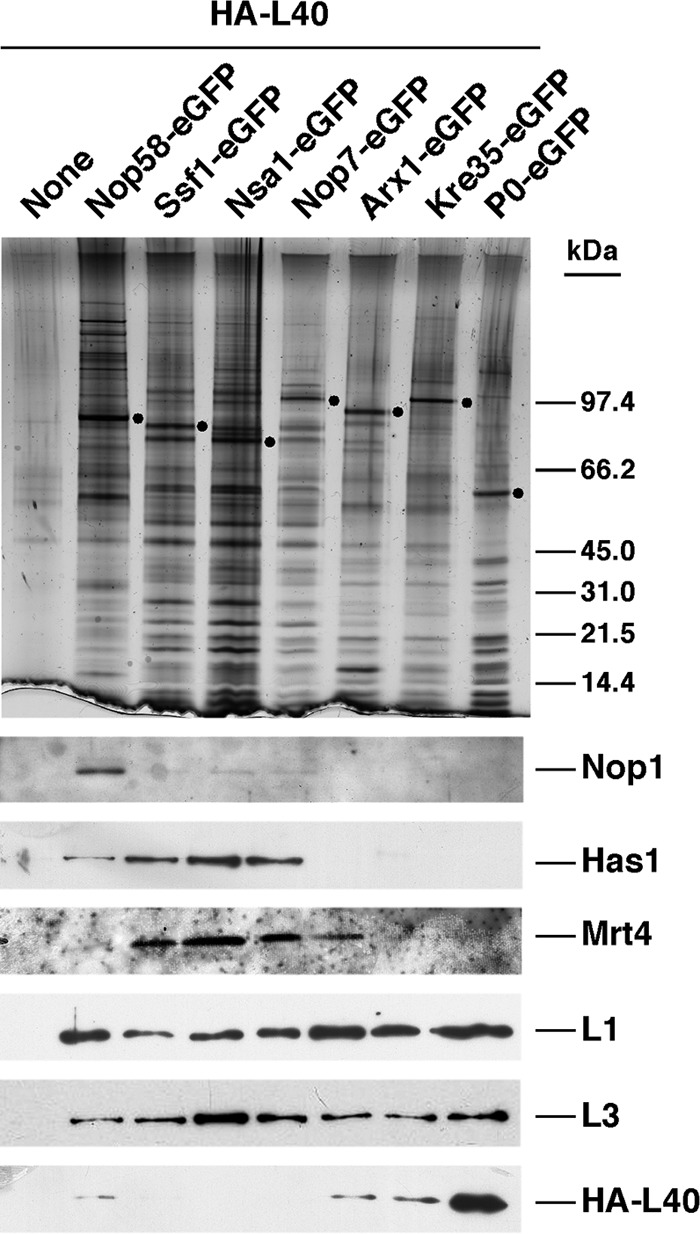
HA-L40A protein assembles onto late cytoplasmic pre-60 S ribosomal particles. The indicated GFP-tagged bait proteins were affinity-purified by using the GFP-Trap®_A procedure (see “Experimental Procedures”) from cells expressing HA-L40A from its cognate promotor. Cells not expressing a GFP-tagged bait protein were used as a negative control (None). Equivalent amounts of the corresponding purified complexes were separated on 4–12% gradient SDS-polyacrylamide gels and subjected to silver staining or Western blot analysis using specific antibodies against the HA epitope, the Nop1, Has1, and Mrt4 ribosome biogenesis factors and the L1 and L3 ribosomal proteins. Dots indicate the positions of bait proteins.
L40 Is Required for the Release of Nmd3 and Rlp24 from Cytoplasmic Pre-60 S Ribosomal Particles
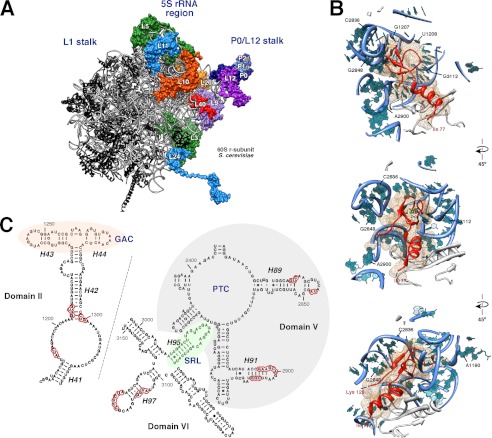
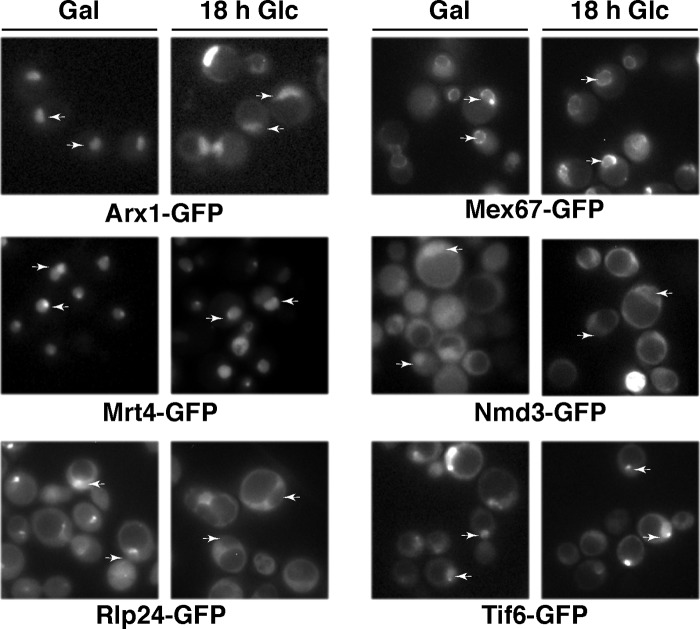
Recent reports on the complete structure of the eukaryotic large r-subunit have allowed the visualization of the L40 protein within mature ribosomes (7, 10). These structures reveal that L40 is positioned close to the binding site for the eEFs in the intersubunit joining face of the mature 60 S r-subunit (Fig. 9A) (7, 10). Hence, L40 binds to RNA residues in proximity to the GTPase-associated center in domain II of 25 S rRNA, the peptidyltransferase center in domain V, and the sarcin-ricin loop in domain VI of 25 S rRNA (Fig. 9, B and C). Several other r-proteins are clear neighbors of L40, among them are L9, L10, L12, and L20 and the ribosomal P-protein stalk (Fig. 9A). Interestingly, Sengupta et al. (121, 122) have mapped the binding site of Nmd3 in a region that might overlap with the binding sites of L40 and its closest neighbors. This site has been described to be close to the position of Tif6, which binds on the edge of the 60 S r-subunit joining face (7, 123). Thus, we found it particularly pertinent to examine whether assembly of L40 was a prerequisite for recycling of Nmd3 and/or Tif6. To assess this point, GFP-tagged versions of Nmd3, Tif6, and other nucleo-cytoplasmic shuttling factors, such as Arx1, Mex67, Mrt4, and Rlp24, were expressed in the GAL::RPL40 strain, and their localization was studied before or after L40 depletion. As reported previously (52, 58, 61, 94, 112, 124), all these proteins, albeit to different extents, display both nuclear and cytoplasmic localization under permissive conditions (Fig. 10 and supplemental Fig. S6). Importantly, although no mislocalization of Arx1-, Mex67-, Mrt4-, and Tif6-GFP could be observed, Nmd3- and Rlp24-GFP were retained in the cytoplasm upon 18 h of depletion of L40 (Fig. 10 and supplemental Fig. S6). Because Nmd3 is equally distributed between cytoplasm and nucleoplasm at steady state, it is difficult to observe a clear-cut cytoplasmic redistribution of this protein (95, 103, 125, 126); however, and in agreement with its cytoplasmic retention, we observed nuclear exclusion of Nmd3-GFP upon L40 depletion (Fig. 10 and supplemental Fig. S6). To distinguish between an impairment of the release of Nmd3 and Rlp24 from cytoplasmic pre-60 S r-particles or a defect in recycling free Nmd3 and Rlp24 to the nucle(ol)us, we subjected extracts from the GAL::RPL40 strain expressing GFP-tagged versions of each of these factors, before or after an 18-h shift to glucose-containing medium, to sucrose gradient fractionation. Then fractions were analyzed by Western blot using anti-GFP antibodies to detect GFP-tagged Nmd3 or Rlp24, respectively. As shown in supplemental Fig. S7, we could not detect significant changes in the pattern of sedimentation of Nmd3-GFP or Rlp24-GFP upon L40 depletion, which were still found in the 60 S region of the gradients. This result indicates that these proteins remain bound to high molecular mass complexes, most likely aberrant cytoplasmic pre-60 S r-particles, upon L40 depletion. Taken together, our data indicate that Nmd3 and Rlp24 require L40 to be efficiently displaced from cytoplasmic pre-60 S r-particles to recycle back to the nucle(ol)us.
FIGURE 9.
Localization of L40 in the three-dimensional structure of the yeast 60 S ribosomal subunit. A, position of the ribosomal proteins L3, L5, L9, L10, L11, L12, L20, L24, L40, P0, P1α, and P2β are highlighted. The remaining ribosomal proteins are colored in black and the rRNA in pale gray. The representation was generated with the UCSF Chimera program (147), using the atomic model for the crystal structure of the yeast 80 S ribosome described by Ben-Shem et al. (Protein Data Bank codes 3U5I and 3U5H (10)). B, close-up view of the position of L40 in the context of its binding site in the 60 S ribosomal subunit. Only rRNA residues situated at or closer than 12 Å are shown (bases and phosphate backbone). Base numbering follows the 25 S rRNA sequence deposited for the yeast 60 S r-subunit (Protein Data Bank codes 3U5D (10)), respectively. Selected residues are labeled in black. The N- and C-terminal ends of the L40 protein (Ile-77 and Lys-128, respectively) are also indicated. C, secondary structure of the GTPase-associated center (GAC), the peptidyltransferase center (PTC), and the sarcin-ricin loop region (SRL) from S. cerevisiae 25 S rRNA domain II, V, and VI, respectively. The structures, residue, and helix (H) numbers were taken from The Comparative RNA Web Site (148). Red circles indicate rRNA residues situated in close proximity of L40 (closer than 5 Å).
FIGURE 10.
Nmd3 and Rlp24 are not properly recycled back to the nucleus upon depletion of L40. The GAL::RPL40 strain, JDY925 [pAS24-RPL40A], was transformed with CEN URA3 plasmids that allow expression of GFP-tagged versions of either Arx1, Mex67, Mrt4, Nmd3, Rlp24, or Tif6. Cells were grown at 30 °C in SGal-Leu-Ura (Gal) and shifted for 18 h to SD-Leu-Ura (18 h Glc). Then the subcellular localization of these GFP-tagged proteins was examined by fluorescence microscopy. Arrows point to the nucle(ol)us. Approximately 200 cells were examined for each reporter. About 85% cells expressing GFP-tagged Nmd3 or Rlp24 showed full cytoplasmic fluorescence upon L40 depletion. Practically all cells gave the results shown in the pictures for remaining reporters.
Expression of HA-L40A Leads to Sordarin Resistance, whereas rpl40aΔ and rpl40bΔ Cells Show Hypersensitivity to This Compound
Both the GTPase-associated center and the sarcin-ricin loop have been proposed to interact directly with the G domain of eEFs (see Refs. 36, 127 and references therein). Moreover, the r-stalk (P-proteins and L12), which is in close proximity of L40 (Fig. 9A), has also been previously identified to interact with eEF2 (36). Both eEF2 and r-stalk proteins have been described to be targets of members of the sordarin family of antifungal compounds (69, 70, 128, 129). To evaluate whether L40 could mediate inhibition by or resistance to sordarin, the single rpl40aΔ and rpl40bΔ strains or the rpl40 null mutant expressing as the sole source of L40 a HA-tagged version of L40A were screened for their ability to grow in the presence of different concentrations of the sordarin derivative GM193663. As a control, we used both an isogenic wild-type and rpl35bΔ strain. As shown in Fig. 11, we found that the strain expressing the HA-tagged L40A r-protein is highly resistant to sordarin resulting in a more than 30-fold increase in the 50% inhibitory concentration (IC50) value compared with its isogenic wild-type counterpart. In contrast, the single rpl40aΔ and specially the rpl40bΔ mutant were clearly much more sensitive to this drug than the wild-type strain; the IC50 value obtained for the rpl40aΔ and rpl40bΔ mutants was at least 10-fold lower than that of the wild-type strain. Interestingly, this sensitivity is not simply related to a lower dosage of an r-protein because the IC50 value of an rpl35bΔ mutant is practically identical to that of the wild-type strain. Moreover, the effect of sordarin seems to be very specific because we could not have observed enhanced sensitivity or resistance to other translation antibiotics (anisomycin, cycloheximide, hygromycin B, paromomycin, and neomycin) for the above strains when compared with their isogenic wild-type counterpart. Finally, to test if the sordarin resistance of the strain expressing the HA-L40A protein is the indirect result of the destabilization of r-proteins such as L12, L1, or P0, whose loss-of-function mutations have been reported to induce sordarin resistance (68, 128), we prepared samples enriched in pre-ribosomal particles and mature ribosomes containing either wild-type L40 or HA-tagged L40A (see “Experimental Procedures”), and we washed them with a high salt solution (0.5 m NH4Cl). As shown in supplemental Fig. S8, no differences were detected in the purification rate of a subset of r-proteins, including L12, L1, and P0. Thus, these r-proteins are stably associated with ribosomes both in wild-type and the HA-L40A-expressing strain. Taken together, these results suggest that L40 specifically and functionally interacts with eEF2 during translation elongation.
FIGURE 11.
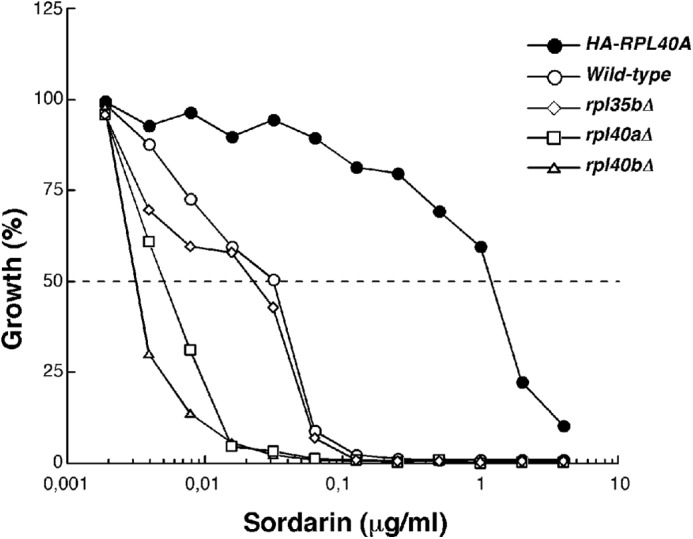
Cells expressing the HA-tagged L40A are resistant to sordarin. Growth inhibition assays were performed in liquid YPD medium with or without different amounts of the sordarin derivative GM193663 in a microtiter format. The relative growth after incubation for 18 h at 30 °C in the presence versus the absence of the antibiotic is the mean of four independent experiments. The strains evaluated were as follows: W303-1A (wild-type; open circles), JDY925 [YCplac33-UB-HA-RPL40A] (HA-RPL40A; closed circles), JDY919 (rpl40aΔ; open square), JDY922 (rpl40bΔ, open triangle), JDY711 (rpl35bΔ; open rhombus).
DISCUSSION
In this work, we have addressed the role of yeast L40 r-protein in 60 S r-subunit biogenesis and function. L40 was not modeled in the crystal structure of the 50 S r-subunit from Haloarcula marismortui (11), and it has only been recently localized in the structure of eukaryotic 60 S r-subunits from S. cerevisiae or Tetrahymena thermophila (see Fig. 9) (7, 10).
Two strain systems were used for the phenotypical analysis of L40 r-protein. First, we analyzed single rpl40a and rpl40b null mutants. Our results indicate that both genes contribute to growth and normal accumulation of 60 S r-subunits, with the role of RPL40A being apparently more important than that of RPL40B (Fig. 1). Although a functional specificity between many paralogous r-proteins has been described (101, 130, 131), we could not find any distinction between L40A and L40B at least for growth and polysome profile analyses under standard laboratory conditions (supplemental Figs. S3 and S4). Second, we established a conditional rpl40 null strain by disrupting the RPL40B gene and placing the ORF of RPL40A under the control of a GAL promoter, which allows genetic depletion of the L40 r-protein by transcriptional repression in cells growing with glucose as carbon source. Polysome profile analyses strongly indicate that 60 S r-subunits lacking L40 are defective in subunit joining (see below) and suggest that both r-subunits may have an increased susceptibility to degradation at longer time points of L40 depletion (Fig. 2); similar observations were made for the null rpl24 mutant and upon L1 depletion (47, 132, 133).
Consistent with the polysome profile results, pulse-chase labeling, Northern hybridization, and primer extension analyses in the L40-depleted strain indicate that L40 is practically dispensable for pre-rRNA processing. Thus, we could only observe a modest delay in 27 SB and 7 S pre-rRNA processing upon depletion of L40 (Fig. 3). Consequently, the relative amounts of these precursors slightly increase, although the steady-state levels of mature 25 S and 5.8 S rRNA only decrease at late time points of depletion (Fig. 4). In addition, the depletion of L40 leads to a mild delay in early cleavage events of pre-rRNA processing at the A0, A1, and A2 sites that have subsequent consequences for the levels of 20 S pre-rRNA and mature 18 S (Figs. 3 and 4); this delay has been extensively reported for many mutants in genes encoding 60 S r-subunit assembly factors and 60 S r-proteins and is likely due to inefficient recycling of trans-acting factors that cannot efficiently dissociate from aberrant pre-60 S r-particles (for further discussion see Refs. 23, 134). These data strongly suggest that L40 is not strictly required for the pre-rRNA processing reactions and that ribosomes lacking L40 are efficiently degraded. This is very unusual because all essential 60 S r-proteins studied so far, except perhaps L1 (47, 133) and L10 (82), play critical roles during ribosome maturation. Strikingly, pre-60 S particles are retained in the nucle(ol)us upon depletion of L40 (Fig. 5). Therefore, we must assume that, although pre-rRNA processing reactions are apparently taking place in a correct way, pre-60 S particles from L40-depleted cells are not fully competent for nucleo-cytoplasmic export. We have shown that an HA-tagged L40 is predominantly assembled in the cytoplasm (see below); thus, we are tempted to speculate that wild-type nuclear pre-60 S r-particles are devoid of L40, and most likely retention of these particles upon L40 depletion, as well as the abovementioned 23 S pre-rRNA accumulation, is occurring as a secondary effect due to inefficient recycling of a critical nucleo-cytoplasmic shuttling factor to the nucleus. A scenario similar to this one has already been demonstrated for the nuclear shuttling factors Nmd3, Mrt4, and Rlp24, which are recycled to the nucleus upon cytoplasmic assembly of L10 (56), P0 (61, 104, 135), and L24 (136), respectively (for further discussion see Refs. 20, 103, 126). Interestingly, our data demonstrate that L40 is an additional r-protein involved in the release of Nmd3 and Rlp24 from cytoplasmic pre-60 S r-particles (Fig. 10 and supplemental Figs. S6 and S7). How does the depletion of L40 block the release of these factors? One possibility is that L40 acts as an effector of the energy-consuming factors involved in the release of Nmd3 and Rlp24 from cytoplasmic pre-60 S r-particles. So far, the GTPase Kre35 (95) and the AAA-ATPase Drg1 (137) are the factors described to be required for the release of Nmd3 and Rlp24, respectively. Drg1, which is apparently one of the earliest acting factors during the cytoplasmic maturation of 60 S r-subunits, is required for the release of other factors in addition to Rlp24, among them Arx1, Mrt4, Nog1, and Tif6 (103, 126, 137); however, our results show that the recycling of Arx1 and Tif6 seems not to be affected by the depletion of L40 (Fig. 10). Kre35 is apparently one of the latest acting factors in this process (95, 103), and the release of Nmd3 by Kre35 also requires the r-protein L10. Thus, another possibility that could explain the lack of proper release of these factors upon L40 depletion is that assembly of L40 is coordinated to that of L10; indeed, although the structural analyses suggest that Nmd3 and L10 can bind simultaneously to pre-60 S r-particles (7, 10, 122), co-immunoprecipitation data are fully consistent with a model in which L10 could be preferentially loaded after Nmd3 release onto cytoplasmic pre-60 S particles in wild-type conditions (56). Interestingly, analyses of the 60 S r-subunit structure suggest that Nmd3 and L40 must sequentially associate with pre-60 S particles because their binding sites on 60 S r-subunits seem mutually exclusive (7, 10, 122). More puzzling is the role of L40 in the release of Rlp24 from cytoplasmic pre-60 S r-particles. It has been hypothesized that Rlp24 and L24 successively occupy the same rRNA-binding site during ribosome synthesis (58). If this hypothesis is correct, the binding site of Rlp24 would be placed about 10 nm away from that of L40, and therefore, a wave of conformational changes is required to explain our observation. Surprisingly, whether L10 is required to release Rlp24 from pre-60 S r-particles has so far not been addressed. Clearly, understanding the precise mechanism by which L40 participates in the recycling of Nmd3 and Rlp4 will require further work.
We have also addressed the course of assembly of L40. Previous data suggested that yeast L40 might associate with pre-60 S particles at a relatively late stage during the course of the 60 S r-subunit assembly (49). Here, we present several lines of evidence that yeast L40 may assemble in the cytoplasm, in complete agreement with the above described role in the release of shuttling factors (Figs. 6–8 and supplemental Fig. S2). However, we cannot fully exclude the possibility, as also suggested by the presence of predicted NLS sequences within L40, that some assembly could take place in the nucleus in wild-type conditions and that the HA tag could partially interfere with the import of the HA-L40 protein. Besides L40, few other r-proteins have been described to assemble exclusively (L24 and L10) or predominantly (P0) in the cytoplasm (56, 58, 61, 103, 104, 135, 136). Further experiments are required to discard the possibility that L40 may also assemble into pre-60 S particles within the nucleus.
Finally, we have addressed the function of L40 in translation. As stated by Wilson and Nierhaus (138), the assignment of a specific role, in terms of translational activity, to an individually given r-protein is extremely difficult. This is based on the highly cooperative nature of the interactions between rRNA and r-proteins and between the r-proteins themselves. Indeed, there are many reports describing that even small changes in the primary sequence of an r-protein have significant long distance effects on the rRNA structure (30, 59, 139–141). Our data suggest that L40 is required for the joining of 60 S and 40 S r-subunits and for the translocation process during translation. Both functions are fully consistent with the position of L40 on the mature 60 S r-subunit (7, 10). The r-subunit joining role of L40 is inferred from the analysis of the polysome profiles of rpl40aΔ and rpl40bΔ mutants but especially of those of L40-depleted cells. These polysome profiles showed a pronounced decrease in 80 S and polysomes and, more importantly, the accumulation half-mer polysomes without a net reduction in the levels of free 60 S r-subunits, as indicated by a ratio of free 60 S to 40 S r-subunits that is relatively similar to that of wild-type cells (Fig. 2). Interestingly, the L10 and L11 r-proteins, which are close neighbors of L40 (Fig. 9A) (7, 10), have been previously involved in r-subunit joining (82, 142). Moreover, a mutational change in the L33 r-protein, identified due to its Gcd− phenotype (derepression of Gcn4 translation), has been suggested to impede the r-subunit joining reaction (143). Consistent with this finding, our preliminary data suggest that both the rpl40aΔ strain and, to the lesser extent, the rpl40bΔ mutant confer a Gcd− phenotype that could be suppressed by overexpression of the translation initiation ternary complex components eIF2 and tRNAiMet.4 Because of the fact that Nmd3 is locked on cytoplasmic pre-60 S particles upon L40 depletion (see above), further experiments are needed to determine whether L40 has a direct role in the r-subunit joining reaction because the location of Nmd3 within the pre-60 S r-particles is apparently incompatible with the joining of 60 S and 40 S r-subunits (122). In agreement with this, it has been shown that Nmd3 does not bind to 80 S ribosomes or polysomes (144). In addition, we hypothesize that L40 participates in the translocation process during translation. This is deduced from the selective sordarin hypersensitivity of the rpl40aΔ and rpl40bΔ mutants and the increased resistance to this drug of cells expressing HA-tagged L40 as the sole source of L40 (Fig. 11). Sordarin derivatives inhibit fungal protein synthesis by stabilizing the eEF2-ribosome complex (69, 70), and many sordarin-resistant mutations, which allow eEF2 to function in translocation even when bound to a sordarin molecule, are linked to the genes that encode eEF2 and the r-stalk P0 and L12 proteins (68–70, 128, 129, 145). Thus, these observations support a role for the r-stalk proteins during translation elongation. Our observations that L40 is another target of sordarin strongly suggest that eEF2 and L40 may also interact functionally during the conformational switch that occurs during translation elongation. Whether L40 makes physical contact with eEF2 or whether the HA-L40 protein exerts sordarin resistance by affecting the functionality of r-stalk proteins or cytoplasmic assembly factors, e.g. the eEF2-related GTPase Efl1/Ria1 (124, 126, 146), remains to be elucidated.
In conclusion, we have shown that L40 assembles late into pre-60 S subunits and may participate in r-subunit joining and eEF2 function during the translation process. Future in-depth analyses will be required to understand the mechanistic details of the role of L40 during cytoplasmic 60 S r-subunit assembly as well as during the initiation and elongation steps of translation.
Supplementary Material
Acknowledgments
We are indebted to the colleagues mentioned in the text for their gift of material used in this study. We also thank M. Carballo, L. Navarro, and C. Reyes of the Biology Service (CITIUS) from the University of Seville for help with the phosphorimager analysis, F. Monje-Casas for valuable advice on immunofluorescence experiments, and J. P. G. Ballesta for valuable advice on sordarin assays.
This work was supported in part by Spanish Ministry of Science and Innovation and European Regional Development Grant BFU2010-15690, Andalusian Government Grant CVI-271, P08-CVI-03508 (to J. d. l. C.), and Swiss National Science Foundation Grant PP00P3_123341 (to D. K.).

This article contains supplemental Figs. S1–S8, Tables S1–S3, and additional references.
A. Fernández-Pevida and J. de la Cruz, unpublished results.
- r-subunit
- ribosomal subunit
- r-protein
- ribosomal protein
- pre-rRNA
- precursor rRNA
- eEF
- eukaryotic elongation factor
- NLS
- nuclear localization signal.
REFERENCES
- 1. Schmeing T. M., Ramakrishnan V. (2009) What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 2. Spahn C. M., Beckmann R., Eswar N., Penczek P. A., Sali A., Blobel G., Frank J. (2001) Structure of the 80 S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell 107, 373–386 [DOI] [PubMed] [Google Scholar]
- 3. Chandramouli P., Topf M., Ménétret J. F., Eswar N., Cannone J. J., Gutell R. R., Sali A., Akey C. W. (2008) Structure of the mammalian 80 S ribosome at 8.7 Å resolution. Structure 16, 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taylor D. J., Devkota B., Huang A. D., Topf M., Narayanan E., Sali A., Harvey S. C., Frank J. (2009) Comprehensive molecular structure of the eukaryotic ribosome. Structure 17, 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Armache J. P., Jarasch A., Anger A. M., Villa E., Becker T., Bhushan S., Jossinet F., Habeck M., Dindar G., Franckenberg S., Marquez V., Mielke T., Thomm M., Berninghausen O., Beatrix B., Söding J., Westhof E., Wilson D. N., Beckmann R. (2010) Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80 S eukaryotic ribosome. Proc. Natl. Acad. Sci. U.S.A. 107, 19754–19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Armache J. P., Jarasch A., Anger A. M., Villa E., Becker T., Bhushan S., Jossinet F., Habeck M., Dindar G., Franckenberg S., Marquez V., Mielke T., Thomm M., Berninghausen O., Beatrix B., Söding J., Westhof E., Wilson D. N., Beckmann R. (2010) Cryo-EM structure and rRNA model of a translating eukaryotic 80 S ribosome at 5.5 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 19748–19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klinge S., Voigts-Hoffmann F., Leibundgut M., Arpagaus S., Ban N. (2011) Crystal structure of the eukaryotic 60 S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948 [DOI] [PubMed] [Google Scholar]
- 8. Rabl J., Leibundgut M., Ataide S. F., Haag A., Ban N. (2011) Crystal structure of the eukaryotic 40 S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736 [DOI] [PubMed] [Google Scholar]
- 9. Ben-Shem A., Jenner L., Yusupova G., Yusupov M. (2010) Crystal structure of the eukaryotic ribosome. Science 330, 1203–1209 [DOI] [PubMed] [Google Scholar]
- 10. Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529 [DOI] [PubMed] [Google Scholar]
- 11. Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 [DOI] [PubMed] [Google Scholar]
- 12. Wimberly B. T., Brodersen D. E., Clemons W. M., Jr., Morgan-Warren R. J., Carter A. P., Vonrhein C., Hartsch T., Ramakrishnan V. (2000) Structure of the 30 S ribosomal subunit. Nature 407, 327–339 [DOI] [PubMed] [Google Scholar]
- 13. Schluenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., Yonath A. (2000) Structure of functional activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623 [DOI] [PubMed] [Google Scholar]
- 14. Selmer M., Dunham C. M., Murphy F. V., 4th, Weixlbaumer A., Petry S., Kelley A. C., Weir J. R., Ramakrishnan V. (2006) Structure of the 70 S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 15. Verschoor A., Warner J. R., Srivastava S., Grassucci R. A., Frank J. (1998) Three-dimensional structure of the yeast ribosome. Nucleic Acids Res. 26, 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wool I. G. (1979) The structure and function of eukaryotic ribosomes. Annu. Rev. Biochem. 48, 719–754 [DOI] [PubMed] [Google Scholar]
- 17. Henras A. K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65, 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. (2003) Ribosome assembly in eukaryotes. Gene 313, 17–42 [DOI] [PubMed] [Google Scholar]
- 19. Mélèse T., Xue Z. (1995) The nucleolus. An organelle formed by the act of building a ribosome. Curr. Opin. Cell Biol. 7, 319–324 [DOI] [PubMed] [Google Scholar]
- 20. Panse V. G., Johnson A. W. (2010) Maturation of eukaryotic ribosomes. Acquisition of functionality. Trends Biochem. Sci. 35, 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gerbi S. A., Borovjagin A. V. (2004) in Nucleolus (Olson M. O. J., ed) pp. 170–198, Landes Biosciences/Eurekah.com, Georgetown, TX [Google Scholar]
- 22. Eichler D. C., Craig N. (1994) Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49, 197–239 [DOI] [PubMed] [Google Scholar]
- 23. Venema J., Tollervey D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33, 261–311 [DOI] [PubMed] [Google Scholar]
- 24. Nomura M., Nogi Y., Oakes M. (2004) in Nucleolus (Olson M. O. J., ed) pp. 128–153, Landes Bioscience/Eurekah.com, Georgetown, TX [Google Scholar]
- 25. Strunk B. S., Karbstein K. (2009) Powering through ribosome assembly. RNA 15, 2083–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kressler D., Hurt E., Bassler J. (2010) Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 [DOI] [PubMed] [Google Scholar]
- 27. Dinman J. D. (2009) The eukaryotic ribosome. Current status and challenges. J. Biol. Chem. 284, 11761–11765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dresios J., Panopoulos P., Suzuki K., Synetos D. (2003) A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J. Biol. Chem. 278, 3314–3322 [DOI] [PubMed] [Google Scholar]
- 29. Dresios J., Derkatch I. L., Liebman S. W., Synetos D. (2000) Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39, 7236–7244 [DOI] [PubMed] [Google Scholar]
- 30. Petrov A., Meskauskas A., Dinman J. D. (2004) Ribosomal protein L3. Influence on ribosome structure and function. RNA Biol. 1, 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meskauskas A., Dinman J. D. (2007) Ribosomal protein L3. Gatekeeper to the A site. Mol. Cell 25, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meskauskas A., Dinman J. D. (2001) Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA 7, 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kramer G., Boehringer D., Ban N., Bukau B. (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 [DOI] [PubMed] [Google Scholar]
- 34. Valásek L., Mathew A. A., Shin B. S., Nielsen K. H., Szamecz B., Hinnebusch A. G. (2003) The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40 S ribosome in vivo. Genes Dev. 17, 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dalley J. A., Selkirk A., Pool M. R. (2008) Access to ribosomal protein Rpl25p by the signal recognition particle is required for efficient cotranslational translocation. Mol. Biol. Cell 19, 2876–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gomez-Lorenzo M. G., Spahn C. M., Agrawal R. K., Grassucci R. A., Penczek P., Chakraburtty K., Ballesta J. P., Lavandera J. L., Garcia-Bustos J. F., Frank J. (2000) Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80 S ribosome at 17.5 Å resolution. EMBO J. 19, 2710–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Briones E., Briones C., Remacha M., Ballesta J. P. (1998) The GTPase center protein L12 is required for correct ribosomal stalk assembly but not for Saccharomyces cerevisiae viability. J. Biol. Chem. 273, 31956–31961 [DOI] [PubMed] [Google Scholar]
- 38. Gingras A. C., Raught B., Sonenberg N. (2004) mTOR signaling to translation. Curr. Top Microbiol. Immunol. 279, 169–197 [DOI] [PubMed] [Google Scholar]
- 39. Ballesta J. P., Rodriguez-Gabriel M. A., Bou G., Briones E., Zambrano R., Remacha M. (1999) Phosphorylation of the yeast ribosomal stalk. Functional affects and enzymes involved in the process. FEMS Microbiol. Rev. 23, 537–550 [DOI] [PubMed] [Google Scholar]
- 40. McCoy L. S., Xie Y., Tor Y. (2011) Antibiotics that target protein synthesis. Wiley Interdiscip. Rev. RNA 2, 209–232 [DOI] [PubMed] [Google Scholar]
- 41. Yonath A. (2005) Antibiotics targeting ribosomes. Resistance, selectivity, synergism, and cellular regulation. Annu. Rev. Biochem. 74, 649–679 [DOI] [PubMed] [Google Scholar]
- 42. Vilardell J., Warner J. R. (1997) Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 17, 1959–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fewell S. W., Woolford J. L., Jr. (1999) Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18 S rRNA. Mol. Cell. Biol. 19, 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Warner J. R., McIntosh K. B. (2009) How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lindström M. S. (2009) Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem. Biophys. Res. Commun. 379, 167–170 [DOI] [PubMed] [Google Scholar]
- 46. Kapasi P., Chaudhuri S., Vyas K., Baus D., Komar A. A., Fox P. L., Merrick W. C., Mazumder B. (2007) L13a blocks 48 S assembly. Role of a general initiation factor in mRNA-specific translational control. Mol. Cell 25, 113–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pöll G., Braun T., Jakovljevic J., Neueder A., Jakob S., Woolford J. L., Jr., Tschochner H., Milkereit P. (2009) rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS ONE 4, e8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferreira-Cerca S., Pöll G., Gleizes P. E., Tschochner H., Milkereit P. (2005) Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18 S rRNA and ribosome function. Mol. Cell 20, 263–275 [DOI] [PubMed] [Google Scholar]
- 49. Kruiswijk T., Planta R. J., Krop J. M. (1978) The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta 517, 378–389 [DOI] [PubMed] [Google Scholar]
- 50. Lastick S. M. (1980) The assembly of ribosomes in HeLa cell nucleoli. Eur. J. Biochem. 113, 175–182 [DOI] [PubMed] [Google Scholar]
- 51. Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. (2003) The path from nucleolar 90 S to cytoplasmic 40 S pre-ribosomes. EMBO J. 22, 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nissan T. A., Bassler J., Petfalski E., Tollervey D., Hurt E. (2002) 60 S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21, 5539–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D., Gavin A. C., Hurt E. (2002) 90 S pre-ribosomes include the 35 S pre-rRNA, the U3 snoRNP, and 40 S subunit processing factors but predominantly lack 60 S synthesis factors. Mol. Cell 10, 105–115 [DOI] [PubMed] [Google Scholar]
- 54. Ferreira-Cerca S., Pöll G., Kühn H., Neueder A., Jakob S., Tschochner H., Milkereit P. (2007) Analysis of the in vivo assembly pathway of eukaryotic 40 S ribosomal proteins. Mol. Cell 28, 446–457 [DOI] [PubMed] [Google Scholar]
- 55. Zhang J., Harnpicharnchai P., Jakovljevic J., Tang L., Guo Y., Oeffinger M., Rout M. P., Hiley S. L., Hughes T., Woolford J. L., Jr. (2007) Assembly factors Rpf2 and Rrs1 recruit 5 S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21, 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. West M., Hedges J. B., Chen A., Johnson A. W. (2005) Defining the order in which Nmd3p and Rpl10p load onto nascent 60 S ribosomal subunits. Mol. Cell. Biol. 25, 3802–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sahasranaman A., Dembowski J., Strahler J., Andrews P., Maddock J., Woolford J. L., Jr. (2011) Assembly of Saccharomyces cerevisiae 60 S ribosomal subunits. Role of factors required for 27 S pre-rRNA processing. EMBO J. 30, 4020–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Saveanu C., Namane A., Gleizes P. E., Lebreton A., Rousselle J. C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. (2003) Sequential protein association with nascent 60 S ribosomal particles. Mol. Cell. Biol. 23, 4449–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Babiano R., Gamalinda M., Woolford J. L., Jr., de la Cruz J. (2012) Saccharomyces cerevisiae ribosomal protein L26 is not essential for ribosome assembly and function. Mol. Cell. Biol. 32, 3228–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Babiano R., de la Cruz J. (2010) Ribosomal protein L35 is required for 27 SB pre-rRNA processing in. Saccharomyces cerevisiae. Nucleic Acids Res. 38, 5177–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rodríguez-Mateos M., García-Gómez J. J., Francisco-Velilla R., Remacha M., de la Cruz J., Ballesta J. P. (2009) Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 7519–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rosado I. V., Kressler D., de la Cruz J. (2007) Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis. Nucleic Acids Res. 35, 4203–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu B., Lukin J., Yee A., Lemak A., Semesi A., Ramelot T. A., Kennedy M. A., Arrowsmith C. H. (2008) Solution structure of ribosomal protein L40E, a unique C4 zinc finger protein encoded by archaeon Sulfolobus solfataricus. Protein Sci. 17, 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakao A., Yoshihama M., Kenmochi N. (2004) RPG. The ribosomal protein gene database. Nucleic Acids Res. 32, D168–D170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chan Y. L., Suzuki K., Wool I. G. (1995) The carboxyl extensions of two rat ubiquitin fusion proteins are ribosomal proteins S27a and L40. Biochem. Biophys. Res. Commun. 215, 682–690 [DOI] [PubMed] [Google Scholar]
- 66. Finley D., Bartel B., Varshavsky A. (1989) The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338, 394–401 [DOI] [PubMed] [Google Scholar]
- 67. Lacombe T., García-Gómez J. J., de la Cruz J., Roser D., Hurt E., Linder P., Kressler D. (2009) Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40 S ribosomal subunits. Mol. Microbiol. 72, 69–84 [DOI] [PubMed] [Google Scholar]
- 68. Gómez-Lorenzo M. G., García-Bustos J. F. (1998) Ribosomal P-protein stalk function is targeted by sordarin antifungals. J. Biol. Chem. 273, 25041–25044 [DOI] [PubMed] [Google Scholar]
- 69. Justice M. C., Hsu M. J., Tse B., Ku T., Balkovec J., Schmatz D., Nielsen J. (1998) Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 273, 3148–3151 [DOI] [PubMed] [Google Scholar]
- 70. Capa L., Mendoza A., Lavandera J. L., Gómez de las Heras F., García-Bustos J. F. (1998) Translation elongation factor 2 is part of the target for a new family of antifungals. Antimicrob. Agents Chemother. 42, 2694–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wach A., Brachat A., Pöhlmann R., Philippsen P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793–1808 [DOI] [PubMed] [Google Scholar]
- 72. Kaiser C., Michaelis S., Mitchell A. (1994) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 73. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 74. Gietz R. D., Sugino A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534 [DOI] [PubMed] [Google Scholar]
- 75. Schmidt A., Bickle M., Beck T., Hall M. N. (1997) The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88, 531–542 [DOI] [PubMed] [Google Scholar]
- 76. Rosado I. V., de la Cruz J. (2004) Npa1p is an essential trans-acting factor required for an early step in the assembly of 60 S ribosomal subunits in Saccharomyces cerevisiae. RNA 10, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kressler D., de la Cruz J., Rojo M., Linder P. (1997) Fal1p is an essential DEAD-box protein involved in 40 S-ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 7283–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vilella M. D., Remacha M., Ortiz B. L., Mendez E., Ballesta J. P. (1991) Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44′ have different functional roles. Eur. J. Biochem. 196, 407–414 [DOI] [PubMed] [Google Scholar]
- 79. Rodríguez-Mateos M., Abia D., García-Gómez J. J., Morreale A., de la Cruz J., Santos C., Remacha M., Ballesta J. P. (2009) The amino-terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 37, 3514–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Petitjean A., Bonneaud N., Lacroute F. (1995) The duplicated Saccharomyces cerevisiae gene SSM1 encodes a eucaryotic homolog of the eubacterial and archaebacterial L1 ribosomal protein. Mol. Cell. Biol. 15, 5071–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zanelli C. F., Maragno A. L., Gregio A. P., Komili S., Pandolfi J. R., Mestriner C. A., Lustri W. R., Valentini S. R. (2006) eIF5A binds to translational machinery components and affects translation in yeast. Biochem. Biophys. Res. Commun. 348, 1358–1366 [DOI] [PubMed] [Google Scholar]
- 82. Eisinger D. P., Dick F. A., Trumpower B. L. (1997) Qsr1p, a 60 S ribosomal subunit protein, is required for joining of 40 S and 60 S subunits. Mol. Cell. Biol. 17, 5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Frey S., Pool M., Seedorf M. (2001) Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem. 276, 15905–15912 [DOI] [PubMed] [Google Scholar]
- 84. Emery B., de la Cruz J., Rocak S., Deloche O., Linder P. (2004) Has1p, a member of the DEAD-box family, is required for 40 S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 52, 141–158 [DOI] [PubMed] [Google Scholar]
- 85. Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (eds) (1994) Current Protocols in Molecular Biology, pp. 13.10.11–13.14.17, John Wiley & Sons, Inc., New York [Google Scholar]
- 86. Venema J., Planta R. J., Raué H. A. (1998) in Protein Synthesis: Methods and Protocols (Martin R., ed) pp. 257–270, Humana Press Inc., Totowa, NJ [Google Scholar]
- 87. Rosado I. V., Dez C., Lebaron S., Caizergues-Ferrer M., Henry Y., de la Cruz J. (2007) Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low molecular mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60 S ribosomal subunit biogenesis. Mol. Cell. Biol. 27, 1207–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kressler D., de la Cruz J., Rojo M., Linder P. (1998) Dbp6p is an essential putative ATP-dependent RNA helicase required for 60 S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 1855–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dez C., Froment C., Noaillac-Depeyre J., Monsarrat B., Caizergues-Ferrer M., Henry Y. (2004) Npa1p, a component of very early pre-60 S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyltransferase center modification. Mol. Cell. Biol. 24, 6324–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. García-Gómez J. J., Lebaron S., Froment C., Monsarrat B., Henry Y., de la Cruz J. (2011) Dynamics of the putative RNA helicase Spb4 during ribosome assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 31, 4156–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ulbrich C., Diepholz M., Bassler J., Kressler D., Pertschy B., Galani K., Böttcher B., Hurt E. (2009) Mechanochemical removal of ribosome biogenesis factors from nascent 60 S ribosomal subunits. Cell 138, 911–922 [DOI] [PubMed] [Google Scholar]
- 92. Belk J. P., He F., Jacobson A. (1999) Overexpression of truncated Nmd3p inhibits protein synthesis in yeast. RNA 5, 1055–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Valerio-Santiago M., Monje-Casas F. (2011) Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J. Cell Biol. 192, 599–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Segref A., Sharma K., Doye V., Hellwig A., Huber J., Lührmann R., Hurt E. (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16, 3256–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hedges J., West M., Johnson A. W. (2005) Release of the export adapter, Nmd3p, from the 60 S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24, 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Menne T. F., Goyenechea B., Sánchez-Puig N., Wong C. C., Tonkin L. M., Ancliff P. J., Brost R. L., Costanzo M., Boone C., Warren A. J. (2007) The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat. Genet. 39, 486–495 [DOI] [PubMed] [Google Scholar]
- 97. Mager W. H., Planta R. J., Ballesta J. G., Lee J. C., Mizuta K., Suzuki K., Warner J. R., Woolford J. (1997) A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 25, 4872–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ozkaynak E., Finley D., Solomon M. J., Varshavsky A. (1987) The yeast ubiquitin genes. A family of natural gene fusions. EMBO J. 6, 1429–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Steffen K. K., McCormick M. A., Pham K. M., MacKay V. L., Delaney J. R., Murakami C. J., Kaeberlein M., Kennedy B. K. (2012) Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191, 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., Green M. R., Golub T. R., Lander E. S., Young R. A. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 [DOI] [PubMed] [Google Scholar]
- 101. Komili S., Farny N. G., Roth F. P., Silver P. A. (2007) Functional specificity among ribosomal proteins regulates gene expression. Cell 131, 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Stage-Zimmermann T., Schmidt U., Silver P. A. (2000) Factors affecting nuclear export of the 60 S ribosomal subunit in vivo. Mol. Biol. Cell 11, 3777–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lo K. Y., Li Z., Bussiere C., Bresson S., Marcotte E. M., Johnson A. W. (2010) Defining the pathway of cytoplasmic maturation of the 60 S ribosomal subunit. Mol. Cell 39, 196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kemmler S., Occhipinti L., Veisu M., Panse V. G. (2009) Yvh1 is required for a late maturation step in the 60 S biogenesis pathway. J. Cell Biol. 186, 863–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kosugi S., Hasebe M., Tomita M., Yanagawa H. (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. U.S.A. 106, 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ho J. H., Kallstrom G., Johnson A. W. (2000) Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151, 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hackmann A., Gross T., Baierlein C., Krebber H. (2011) The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits. EMBO Rep. 12, 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yao Y., Demoinet E., Saveanu C., Lenormand P., Jacquier A., Fromont-Racine M. (2010) Ecm1 is a new pre-ribosomal factor involved in pre-60 S particle export. RNA 16, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yao W., Roser D., Köhler A., Bradatsch B., Bassler J., Hurt E. (2007) Nuclear export of ribosomal 60 S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell 26, 51–62 [DOI] [PubMed] [Google Scholar]
- 110. Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., Sekimoto T., Baumgärtel V., Boese G., Bassler J., Wild K., Peters R., Yoneda Y., Sinning I., Hurt E. (2007) Arx1 functions as an unorthodox nuclear export receptor for the 60 S preribosomal subunit. Mol. Cell 27, 767–779 [DOI] [PubMed] [Google Scholar]
- 111. Hung N. J., Lo K. Y., Patel S. S., Helmke K., Johnson A. W. (2008) Arx1 is a nuclear export receptor for the 60 S ribosomal subunit in yeast. Mol. Biol. Cell 19, 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. (2001) Nuclear export of 60 S ribosomal subunit depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21, 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Hurt E., Hannus S., Schmelzl B., Lau D., Tollervey D., Simos G. (1999) A novel in vivo assay reveals inhibition of ribosomal nuclear export in Ran-cycle and nucleoporin mutants. J. Cell Biol. 144, 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Dragon F., Gallagher J. E., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., Wehner K. A., Wormsley S., Settlage R. E., Shabanowitz J., Osheim Y., Beyer A. L., Hunt D. F., Baserga S. J. (2002) A large nucleolar U3 ribonucleoprotein required for 18 S ribosomal RNA biogenesis. Nature 417, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kallstrom G., Hedges J., Johnson A. (2003) The putative GTPases Nog1p and Lsg1p are required for 60 S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23, 4344–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Kressler D., Roser D., Pertschy B., Hurt E. (2008) The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60 S particles. J. Cell Biol. 181, 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., Rout M., Meagher D., Imai B., Guo Y., Brame C. J., Shabanowitz J., Hunt D. F., Woolford J. L., Jr. (2001) Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8, 505–515 [DOI] [PubMed] [Google Scholar]
- 118. Wehner K. A., Baserga S. J. (2002) The σ(70)-like motif. A eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9, 329–339 [DOI] [PubMed] [Google Scholar]
- 119. Fatica A., Cronshaw A. D., Dlakič M., Tollervey D. (2002) Ssf1p prevents premature processing of an early pre-60 S ribosomal particle. Mol. Cell 9, 341–351 [DOI] [PubMed] [Google Scholar]
- 120. Santos C., Ballesta J. P. (1994) Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 269, 15689–15696 [PubMed] [Google Scholar]
- 121. Oeffinger M. (2010) Joining the interface. A site for Nmd3 association on 60 S ribosome subunits. J. Cell Biol. 189, 1071–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sengupta J., Bussiere C., Pallesen J., West M., Johnson A. W., Frank J. (2010) Characterization of the nuclear export adaptor protein Nmd3 in association with the 60 S ribosomal subunit. J. Cell Biol. 189, 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Gartmann M., Blau M., Armache J. P., Mielke T., Topf M., Beckmann R. (2010) Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J. Biol. Chem. 285, 14848–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Senger B., Lafontaine D. L., Graindorge J. S., Gadal O., Camasses A., Sanni A., Garnier J. M., Breitenbach M., Hurt E., Fasiolo F. (2001) The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8, 1363–1373 [DOI] [PubMed] [Google Scholar]
- 125. Hedges J., Chen Y. I., West M., Bussiere C., Johnson A. W. (2006) Mapping the functional domains of yeast NMD3, the nuclear export adapter for the 60 S ribosomal subunit. J. Biol. Chem. 281, 36579–36587 [DOI] [PubMed] [Google Scholar]
- 126. Bussiere C., Hashem Y., Arora S., Frank J., Johnson A. W. (2012) Integrity of the P-site is probed during maturation of the 60 S ribosomal subunit. J. Cell Biol. 197, 747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Spahn C. M., Gomez-Lorenzo M. G., Grassucci R. A., Jørgensen R., Andersen G. R., Beckmann R., Penczek P. A., Ballesta J. P., Frank J. (2004) Domain movements of elongation factor eEF2 and the eukaryotic 80 S ribosome facilitate tRNA translocation. EMBO J. 23, 1008–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Botet J., Rodríguez-Mateos M., Ballesta J. P., Revuelta J. L., Remacha M. (2008) A chemical genomic screen in Saccharomyces cerevisiae reveals a role for diphthamidation of translation elongation factor 2 in inhibition of protein synthesis by sordarin. Antimicrob. Agents Chemother. 52, 1623–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Justice M. C., Ku T., Hsu M. J., Carniol K., Schmatz D., Nielsen J. (1999) Mutations in ribosomal protein L10e confer resistance to the fungal-specific eukaryotic elongation factor 2 inhibitor sordarin. J. Biol. Chem. 274, 4869–4875 [DOI] [PubMed] [Google Scholar]
- 130. Horiguchi G., Mollá-Morales A., Pérez-Pérez J. M., Kojima K., Robles P., Ponce M. R., Micol J. L., Tsukaya H. (2011) Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 65, 724–736 [DOI] [PubMed] [Google Scholar]
- 131. Xue S., Barna M. (2012) Specialized ribosomes. A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Baronas-Lowell D. M., Warner J. R. (1990) Ribosomal protein L30 is dispensable in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 5235–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. McIntosh K. B., Bhattacharya A., Willis I. M., Warner J. R. (2011) Eukaryotic cells producing ribosomes deficient in Rpl1 are hypersensitive to defects in the ubiquitin-proteasome system. PLoS ONE 6, e23579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gadal O., Strauss D., Braspenning J., Hoepfner D., Petfalski E., Philippsen P., Tollervey D., Hurt E. (2001) A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60 S ribosomal subunits. EMBO J. 20, 3695–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lo K. Y., Li Z., Wang F., Marcotte E. M., Johnson A. W. (2009) Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol. 186, 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lebreton A., Saveanu C., Decourty L., Rain J. C., Jacquier A., Fromont-Racine M. (2006) A functional network involved in the recycling of nucleocytoplasmic pre-60 S factors. J. Cell Biol. 173, 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Pertschy B., Saveanu C., Zisser G., Lebreton A., Tengg M., Jacquier A., Liebminger E., Nobis B., Kappel L., van der Klei I., Högenauer G., Fromont-Racine M., Bergler H. (2007) Cytoplasmic recycling of 60 S preribosomal factors depends on the AAA protein Drg1. Mol. Cell. Biol. 27, 6581–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wilson D. N., Nierhaus K. H. (2005) Ribosomal proteins in the spotlight. Crit. Rev. Biochem. Mol. Biol. 40, 243–267 [DOI] [PubMed] [Google Scholar]
- 139. Petrov A. N., Meskauskas A., Roshwalb S. C., Dinman J. D. (2008) Yeast ribosomal protein L10 helps coordinate tRNA movement through the large subunit. Nucleic Acids Res. 36, 6187–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Briones C., Ballesta J. P. (2000) Conformational changes induced in the Saccharomyces cerevisiae GTPase-associated rRNA by ribosomal stalk components and a translocator inhibitor. Nucleic Acids Res. 28, 4497–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Meskauskas A., Russ J. R., Dinman J. D. (2008) Structure/function analysis of yeast ribosomal protein L2. Nucleic Acids Res. 36, 1826–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Rhodin M. H., Rakauskaitė R., Dinman J. D. (2011) The central core region of yeast ribosomal protein L11 is important for subunit joining and translational fidelity. Mol. Genet. Genomics 285, 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Martín-Marcos P., Hinnebusch A. G., Tamame M. (2007) Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol. Cell. Biol. 27, 5968–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ho J. H., Kallstrom G., Johnson A. W. (2000) Nascent 60 S ribosomal subunits enter the free pool bound by Nmd3p. RNA 6, 1625–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Santos C., Rodríguez-Gabriel M. A., Remacha M., Ballesta J. P. (2004) Ribosomal P0 protein domain involved in selectivity of antifungal sordarin derivatives. Antimicrob. Agents Chemother. 48, 2930–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bécam A. M., Nasr F., Racki W. J., Zagulski M., Herbert C. J. (2001) Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60 S subunit of the ribosome in Saccharomyces cerevisiae. Mol. Genet. Genomics 266, 454–462 [DOI] [PubMed] [Google Scholar]
- 147. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 148. Cannone J. J., Subramanian S., Schnare M. N., Collett J. R., D'Souza L. M., Du Y., Feng B., Lin N., Madabusi L. V., Müller K. M., Pande N., Shang Z., Yu N., Gutell R. R. (2002) The comparative RNA web (CRW) site. An on-line database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.