Abstract
Hypoxia drives malignant progression in part by promoting accumulation of the oncogenic transcription factor HIF-1α in tumor cells. Tumor aggressiveness also relates to elevation of the cancer stem cell-associated membrane protein CD24, which has been causally implicated in tumor formation and metastasis in experimental models. Here we link these two elements by showing that hypoxia induces CD24 expression through a functional hypoxia responsive element (HRE) in the CD24 promoter. HIF-1α overexpression induced CD24 mRNA and protein under normoxic conditions, with this effect traced to a recruitment of endogenous HIF-1α to the CD24 promoter. shRNA mediated-attenuation of HIF-1α or CD24 expression reduced cancer cell survival in vitro and in vivo at the levels of primary and metastatic tumor growth. CD24 overexpression in HIF-1α-depleted cancer cells rescued this decrease while HIF-1α overexpression in CD24-depleted cells did not. Analysis of clinical tumor specimens revealed a correlation between HIF-1α and CD24 levels and an association of their co-expression to decreased patient survival. Our results establish a mechanistic linkage between two critically important molecules in cancer, identifying CD24 as a critical HIF-1α transcriptional target and biological effector, strengthening the rationale to target CD24 for cancer therapy.
Keywords: CD24, HIF-1, hypoxia, metastasis
INTRODUCTION
Tumor hypoxia is a poor prognostic factor for patient outcome (1, 2). This is likely from hypoxic induction of angiogenic factors, chemokines, oncogenes and other drivers of tumor progression (3) that confer metastatic competence (4). CD24 is a mucin-like cell surface protein consisting of a short, heavily glycosylated core (5) linked to plasma membrane raft domains via glycosyl-phosphatidylinositol. In immune lineages, CD24 has been implicated in signal transduction and lymphocyte development (6, 7). Multiple studies have also demonstrated that CD24 is expressed in many human malignancies including breast, ovarian, colorectal, pancreatic, prostate, bladder cancer, and higher levels of expression are associated with increased aggressiveness and worse prognosis (5, 8). Although in breast cancer ambiguity exists as cells expressing CD44+CD24 (−/low) display more stem cell likeness (9), studies have shown that CD24+ cells are associated with poor prognosis in breast (9) and most other epithelial malignancies (5). Moreover, recent studies in pancreatic, colon and hepatocellular cancers have identified tumor initiating CD24+ populations that and are capable of self-renewal, differentiation and metastasis (10). These CD24 phenotypes have been delineated mechanistically by use of xenograft studies demonstrating how expression of CD24 in tumor cells enhances their metastatic ability (8, 11).
Despite its importance in cancer biology and clinical oncology (12, 13), little is known about the regulation of CD24. Having recently identified CD24 as an important transcriptional target (8) of the Ral GTPase pathway here we used bioinformatic analyses to initially associate a transcriptional signature of the Ral pathway with HIF-1α signaling. Based on these suggestive data, we then tested the hypothesis that CD24 might also be regulated by HIF signaling. Using bioinformatic, in vitro, in vivo, as well as analysis of human specimens in the present study, we demonstrate an important mechanistic link between two pivotal genes in tumor progression, HIF-1α and CD24, and identify the latter as an effector of HIF-1α-induced tumor growth and lung colonization.
MATERIALS AND METHODS
Statistics and Informatics
Group comparisons were made using one-way ANOVA and Newman-Keuls test unless otherwise indicated. Survival estimates in animal experiments were compared by Log Rank test of equality of survival distributions. For CD24 and HIF-1α staining, Spearman rank-based correlation was used to test correlation, while survival distributions were plotted and tested by the Log Rank test (Prism Version 5.0, GraphPad Software, La Jolla, CA) or Cox Proportional Hazards regression analysis (Matlab Version 2010b, The Mathworks, Natick, MA). Molecular Concepts Analysis (14) was performed using genes overexpressed at least 2-fold in a core transcriptional signature of the Ral GTPase pathway (n=32 unique genes, S.C.S. et al., manuscript in revision) (Figure S1) by uploading into Oncomine (15) as a custom concept. All Entrez gene IDs were used as the null set (n=42490). Pairwise associations of the Ral Signature with all molecular concepts in Oncomine (including Oncomine gene expression signatures, Gene Ontology, KEGG Pathways, Human Protein Reference Database, etc) were automatically computed. Overlap with molecular concepts in Oncomine was considered significant at p<1E-4, and selected concepts were exported into Cytoscape v2.8.2 for network visualization.
Cell culture and hypoxia
All cancer cell lines used were purchased from ATCC (Manassas, USA), cultured and profiled as described (16). UMUC-3 CD24 cells were generated by stably transfecting CD24 construct in pcDNA 3.1 vector. Short hairpin RNA (shRNA) expressing stable UMUC-3 cell lines were generated by transfecting CD24 shRNA or HIF-1α shRNA in PLKO1 vector (Sigma). For hypoxic treatment, cells were incubated with 1% O2 balanced with CO2 and Nitrogen in hypoxia chambers (Precision Scientific, Winchester, VA) as described (17).We identified a putative hypoxia responsive element (HRE) on CD24 promoter using a promoter scanning software Genomatix (18). To determine the HRE promoter activity, cells were lysed in passive lysis buffer (Promega) and assayed for firefly luciferase according to Promega Luciferase kit instructions. Normalization of nuclear material was achieved by a Cyquant cell proliferation assay performed using the same lysate following manufacturer’s instructions.
Cloning of the CD24 promoter, deletion mutation and reporter assays
Genomic DNA from several human cell lines described in results section, was isolated using the Puregene™ DNA isolation kit (Gentra Systems) according to the manufacturer’s instructions. The PCR reaction also included: 10X PfuUltra™ HF Reaction Buffer (Agilent), 100mM dNTP Mix (Agilent), 5% DMSO, 1M Betaine Solution (Sigma-Aldrich) and 100ng of DNA. The primers used were as follows: CD24 promoter 2344 F:5′-GAA-CCC-GGC-ACT-CCT-GAG-TCA-3′, R:5′-ACC-ATT-GCT-CTG-CCC-ATG-TCC-3′; CD24 promoter 1896 F:5′-GAT-CGC-TAG-CCA-CGC-CCG-GCC-AAA-GTA-TTT-C-3′, R:5′-GAT-CAA-GCT-TCA-GGA-TGC-TGG-GTG-CTT-GGA-G-3′. PCR products were isolated in a 1% agarose gel and purified using a Qiaquick® Gel Extraction Kit (Qiagen). Isolated products were digested at 37°C by the enzymes NheI and HindIII (New England Biolabs) and ligated into the pGL4.20 Luciferase Reporter Vector (Promega). CD24 promoters were sequenced at the University of Virginia DNA sequencing Core using the BigDye Terminator v3.1 (Applied Biosystems) chemistry on a 3730 DNA Analyzer (Applied Biosystems). Primers for sequencing were: RV primer 3: 5′-CTA-GCA-AAA-TAG-GCT-GTC-CCC-3′, pGL4.20 R: 5′-CTT-AAT-GTT-TTT-GGC-ATC-TTC-C-3′, CD24promSF1: 5′-CTC-CTC-TTT-GTG-CCG-GTT-CAT-T-3′, CD24prom SR1: 5′-CGG-TCC-TGG-AGC-AAG-TGC-A-3′. Sequences were submitted to GenBank with the following accession numbers:
To generate deletion fragments of the CD24 promoter, specific primers were designed to amplify increasingly larger segments of the promoter region, preserving the same NheI and HindIII cloning strategy for standardization. These were subcloned to yield reporter vectors encompassing 100bp, 223bp, 428bp, 680bp, and 942bp. The CD24 promoter deletion constructs isolated above were cloned into pGL4-Basic (Promega) and HRE binding site in the promoter was mutated with site-directed mutagenesis (Stratagene) kit instructions.
RNA isolation, real-time quantitative PCR
RNA was isolated and quantitative reverse transcription-PCR (RTPCR) done in the ABI Prism 7900HT Sequence Detection System from Applied Biosystems (Foster City, CA) as described (19). Primers used for CD24 were forward, 5V-CAATATTAAATCTGCTGGAGTTTCATG-3V and reverse, 5V-TCCATATTTCTCAAGCCACATTCA-3. Primers used for HIF-1α were 5′-TGCTTGGTGCTGATTTGTGA-3′ and reverse, 5′-GGTCAGATGATCAGAGTCCA-3′.
Western blotting and Immunohistochemistry
Westerns were performed as previously described (20). Anti-CD24 mouse monoclonal antibody (SWA-11) was obtained as a gift from Dr. Peter Altevogt, mouse monoclonal anti-HIF-1α antibody (BD pharmingen, Cat# 610958) and HIF-2α (Novus Biologicals, Littleton, CO, Cat# NB100-132) was used at 1:2000 dilution, mouse monoclonal α-tubulin, (Abcam, UK) was used at 1:2000 dilution. Immunoblots were developed using Super Signal Femto (Pierce, Rockford, IL) and visualized using Alpha Innotech (San Leandro, CA) imaging. A tissue microarray containing well annotated clinical specimens with follow up (clinical and pathological data on specimens described in (21)) containing quadruplicate, representative cores from 151 zinc formalin-fixed, paraffin-embedded primary bladder cancers was evaluated. Slides were incubated with anti-CD24 (SWA-11) hybridoma supernatant with immunoperoxidase detection and DAB chromogen (22). Scoring of CD24 based on intensity was performed as described (8), ranging from negative to strong (0, 1+, 2+, and 3+). For staining with HIF-1α the monoclonal anti-HIF-1α antibody was used at 1:800 dilution for 30 minutes after antigen retrieval in a DAKO Pascal Pressure Chamber. Scoring for nuclear positivity was performed (0+: none, 1+: 1–10% nuclei, 2+: 10–50% nuclei, 3+: >50% nuclei) similar to reported (23). Nuclear staining was counted as positive at any intensity.
Cell viability assays
To monitor the viability of the cells, 4×103 cells grown in 96 well plates were stained with Calcein AM (stains live cells green) and Ethidium bromide homodimer (detects dead cells) using the “Live/Dead” assay system (Molecular probes, USA) following manufacturer’s instructions. Cells incubated with dyes for 15 minutes were exposed to fluorescence intensity measurements using excitation/ emission filter sets 495/520 nm (Calcein AM) and 530/642 nm (EthD). Mean fluorescence ±SEM (n=8 wells) of was estimated using BioTek Synergy 2 microplate reader (Winooski, VT). The “Viability Index” was calculated as the ratio of live fluorescence divided by dead fluorescence.
In Vivo Human Xenograft assays in immunocompromised mice
Six-week-old nude mice were injected subcutaneously with 106 cancer cells. Tumor dimensions were measured two times a week and tumor volumes calculated as described (24). Five- to six-week-old male (for PC3 cells) or female (for UMUC3 cells) athymic nude mice (Ncr nu/nu) were obtained from the National Cancer Institute (Frederick, MD). Animals were maintained according to the University of Virginia ICUC guidelines. To evaluate the ability of HIF-1 and CD24 expression-modified PC3 cells for metastasis, 2×105 cells in serum-free medium were inoculated in the left ventricle. Six-week-old nude mice were given tail vein injections with 2.5×106 UMUC3 cells as described. In addition, in separate experiments, 106 PC-3 cells were injected intra-prostatically as described (25). Bioluminescent in vivo imaging using IVIS 100 scanner (Xenogen Corp., Alameda, CA, USA) was used to monitor metastasis weekly as explained previously (25). D-Luciferin Firefly, potassium salt (30 mg/ml luciferin, Biosynth International, Naperville, IL, USA) was injected intraperitoneally at a dose of 150 mg/kg body weight approximately 10–15 min before imaging. Images were acquired in 5-min intervals using IVIS 100 scanner as described (24). The “survival” endpoint was defined according to ICUC criteria such as significant weight loss, pain unrelieved by analgesias, tumor erosion etc…). At euthanasia, the lungs of animals inoculated with UMUC3 cells were removed by dissection away from adjacent organs and examined as described (26). Genomic DNA was extracted from mouse lungs and 12p real time PCR performed as described (19).
Pimonidazole (Hypoxyprobe) immunohistochemistry
Six-week-old nude mice were injected subcutaneously with 106 UMUC-3 cells. After the tumors grew to a diameter of 1–1.5 cm, the animals were injected intravenously with pimonidazole hydrochloride (Hypoxyprobe) (60mg/kg). Ninety min after the injection, animals were sacrificed, tumors dissected into 2 or 3 portions and fixed with 10% neutral buffered formalin overnight, and embedded in paraffin. 4μm thick sections were processed and stained according to the manufacturer’s instructions as described (27). Consecutive sections from the same tumors were immunostained for CD24 as explained (8).
Chromatin Immunoprecipitation
UMUC-3 cells exposed to hypoxia for 24 hours were treated with 2% formaldehyde for 10 min at 25°C. Chromatin immunoprecipitation was performed using the ChIP-IT™ Express Kit Chromatin Immunoprecipitation Kit from Active Motif according to the manufacturer’s instructions. Briefly, genomic DNA was enzymatically sheared for 4 minutes at 37°C. Rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA), RNA Pol II (Active Motif), and HIF-1α (BD pharmingen Cat# 610958) and HIF-1β (Novus Biologicals, Littleton, CO, Cat# NB300-525) were incubated with the genomic DNA and magnetic beads for 4 hours at 4°C. After the recommended washings, the DNA-protein crosslinking was reversed and the protein was removed from the samples with proteinase K. Detection of CD24 promoter was performed using quantitative real-time PCR with the following primers, F: 5′-ACGGCTATTGTGGCTTTCCTGGTAT-3′; R: 5′-GCTTGGAGAACCGCTGGCTC-3′. Conditions for QRT-PCR included 40 cycles with an annealing temperature of 58° C. DNA amplification was measured using Sybr Green reaction mix supplemented with 1M Betaine and 5% DMSO on Applied Biosystems (GeneAmp PCR system 9700). DNA quantity was determined based on a standard curve and normalized based on the amount of input DNA from each sample.
Ethics
All research involving human participants was approved by the University of Virginia institutional review board. Informed consent was obtained and all clinical investigation conducted according to the principles expressed in the Declaration of Helsinki.
RESULTS
CD24 is induced by hypoxia in cancer cells and human tumor xenografts
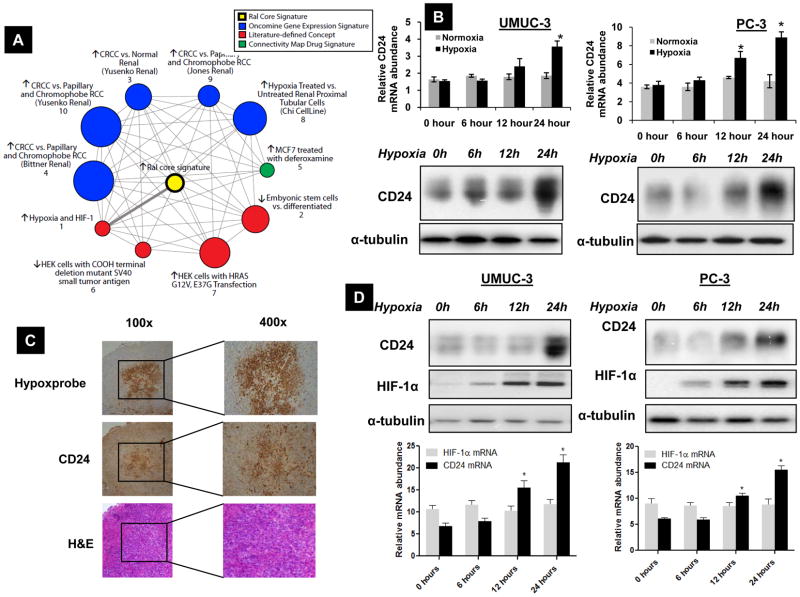
We recently identified CD24 as an important transcriptional target of signaling downstream from the Ral GTPase pathway, though little is known regarding the regulation of CD24. To generate testable hypotheses for regulators of CD24, we used Molecular Concepts Analysis (MCA) (14), implemented in Oncomine (15) to analyze a transcriptional signature of the Ral GTPase pathway (S. C. S. et al., manuscript in revision). Genes overexpressed from this signature, composed of a core set genes regulated two-fold by siRNA-mediated depletion of both RalA and RalB (28), were compared with other collections of biologically relevant gene sets (molecular concepts), for disproportionate overlap using MCA (14). Interestingly, several findings, shown in the molecular concepts map (Figure 1A) implicate hypoxia as a key theme among concepts significantly associated with the Ral signature, including known hypoxia targets, genes involved in clear cell renal cell carcinoma (a neoplasm driven by HIF signaling), and genes regulated by treatment of breast cancer cells with deferoxamine (a chemical hypoxia mimic). Figure S1 details the genes in the Ral transcriptional signature and their representation in individual molecular concepts.
Figure 1.
A) Molecular concepts map (14, 15) of a core transcriptional signature of the Ral GTPase pathway (“Ral core signature” shown as (yellow ringed node), generated from genes expressed 2-fold higher in control as compared to Ral siRNA treated cells (n=32). Each node represents a molecular concept, or set of biologically related genes, while node size is proportional to the number of genes in the concept. The concept color indicates the concept type according to the legend. Each edge represents a significant enrichment (p<1E-4), with the thick edge representing the most significantly enriched concept. Several concepts implicate hypoxia (Nodes #1, 3, 4, & 5), while others are likely reflective of known functions of Ral, including acting downstream of Ras (Node #7) or small T antigen (Node #6). B). UMUC-3, PC-3 cells were exposed to hypoxia for various time periods as indicated. RNA extracted from these samples was analyzed by real-time quantitative PCR for CD24 mRNA expression. Paired lysates were analyzed for CD24 protein expression by western. *Significant difference compared to samples at 0h (p<0.01). C) Immunohistochemical (IHC) evaluation of CD24 expression in UMUC-3 tumor xenograft sections and corresponding Pimonidazole (Hypoxyprobe) staining and H&E (hematoxylin & eosin) staining in serial sections. Magnification indicated. D) UMUC-3 and PC-3 cells exposed to hypoxia for various time periods as indicated examined for HIF-1α and CD24 protein and mRNA expression. *Significant difference to the samples at 0h (p<0.01). * Significant difference to the samples at 0h (p<0.05). B, C and D) Blots are representative of three separate experiments. Error bars are SD of triplicate samples from one of three independent experiments.
More specifically implicating hypoxia in regulation of CD24, a previous study identified CD24 among a multitude of genes induced by hypoxia in human umbilical cord vein endothelial cells (29). To determine whether a similar regulatory circuit is operative in cancer cells from two common malignancies showing CD24 overexpression (8, 30), we exposed human PC-3 (prostate cancer) and UMUC-3 (bladder cancer) cell lines to hypoxia, and evaluated CD24 mRNA and protein levels. Exposure of cells to hypoxia increased CD24 mRNA by 12 hours, reaching a maximum at 24 hours (Figure 1B, S2A). Protein levels were seen to parallel this increase in mRNA in UMUC-3, PC-3 and two additional cell lines, KU-7 and LNCaP, derived from human bladder and prostate cancers, respectively (Figure 1B, S2B). Next, we evaluated the association of tumor hypoxia to CD24 expression in vivo. Immunohistochemistry revealed the pattern of CD24 expression as a function of Pimonidazole (Hypoxyprobe) staining in human bladder cancer xenografts. Supporting the in vitro findings above, CD24 protein expression was found to be elevated in hypoxic areas of the tumor (Figure 1C).
Isolation of the human CD24 gene promoter
Exposure of UMUC-3, PC-3 cells to hypoxia also led to an increase in HIF-1α protein but not mRNA and this preceded the increase in CD24 expression (Figure 1D, S2A). Since elemental iron is a critical factor for proline hydroxylation of HIF-1α, iron chelators such as Deferoxamine (31) are widely utilized in HIF-1 studies as surrogates of hypoxia (32). We hence treated UMUC-3 cells with 50μM DFO, which led to a 3-fold increase in CD24 protein expression after 12 hours (Figure S2C).
These findings suggested the presence of hypoxia responsive cis-acting elements (HRE) in the CD24 promoter. There have been two published CD24 promoter sequences, the first from a pooled sample of human DNA (33) while the second from a population of B lymphocytes (34). Unfortunately, these two promoters had considerable mismatched sequences between each other and the partial promoter from GenBank (Figure S3). Therefore, to determine the consensus CD24 promoter sequence in UMUC3 cells we cloned a 1139 base pair region that encompassed the first 79 base pairs of the CD24 5’UTR. Comparing the UMUC3 sequence to the previously reported sequences for the CD24 promoter several mismatches and deletions were observed (Figure S3). To ensure a consensus CD24 promoter sequence we cloned the same region in four additional cancer cell lines (LUL2, EJ, J82, and LNCaP) and two non-cancerous cell lines: telomerase (TERT) immortalized urothelial cell line and human embryonic kidney cells (293T). Here we report and then use of a consensus promoter whose sequence we is deposited in GenBank (Accession numbers are included in the Methods section).
CD24 gene is a transcriptional target of Hypoxia Inducible Factor 1α (HIF-1α) in hypoxia
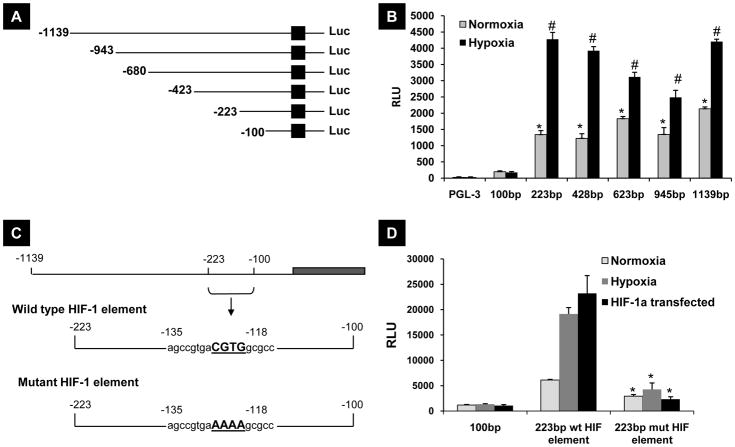
Using the cloned promoter, we generated and transiently transfected a series of CD24 5 ′-flanking sequence deletion mutants driving a luciferase reporter (Figure 2A) into UMUC-3 cells and incubated these under normoxia and hypoxia for 24 hours. Figure 2B indicates that the sequence between −100bp and −223bp is critical for CD24 transcriptional induction in hypoxia. To determine whether these sequences harbored candidate HIF binding sites, we used Genomatix software (35) to determine if any canonical HIF-1 binding sites existed from −118 to −135. We then used site-directed mutagenesis to obliterate the putative site that was found (CGTG to AAAA) in the luciferase reporter plasmid (Figure 2C) and transfected it into UMUC-3 cells. The mutation of the HIF-1 binding site completely abolished the responsiveness to hypoxia (Figure 2D), and also reduced basal expression, probably due to reduction of detectable levels of HIF-1 expression in normoxic cells. These results suggest that this putative HRE is necessary for the induction of CD24 gene in cancer cells.
Figure 2.
A) Deletion mutants of the 5’-flanking region of CD24 gene. B) Deletion mutants were transiently transfected in UMUC-3 cells and the effect of deletions on hypoxia mediated promoter activity was assessed by measuring luciferase activity as explained in Methods. * Compared to PGL-3 vector transfected cells in grown in normoxia (p<0.01), # Compared to PGL-3 vector transfected cells exposed to hypoxia (p<0.01). C) Site directed mutation of HIF-1 binding site on CD24 promoter. The region CGTG between −118 and −135 on CD24 promoter was mutated to AAAA. D) Functional activity of these mutations were measured by transfecting wild type and mutated HIF-1 element followed by exposing them to normoxia, hypoxia or by co-transfecting HIF-1α transgene together. The results are representative of three independent experiments. *Compared to wild type CD24 promoter reporter transfected cells exposed to normoxia, hypoxia or after introduction of HIF-1 transgene (p<0.01)
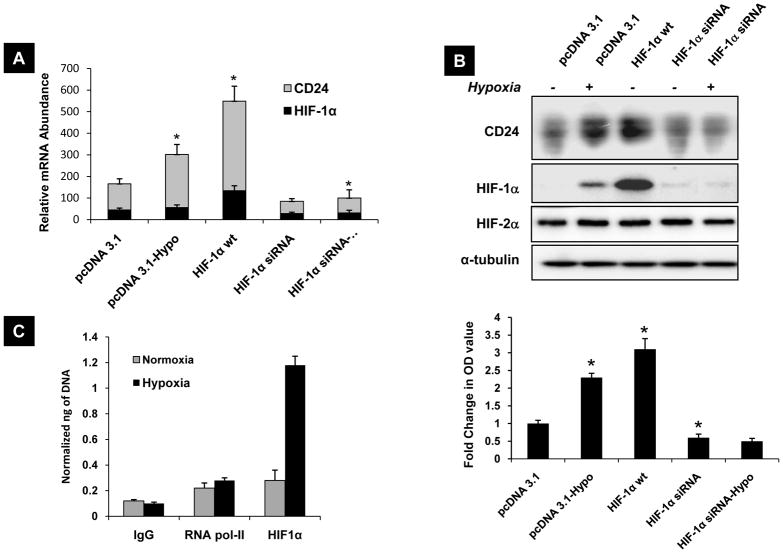
We next wanted to investigate the dependency of CD24 expression on HIF-1α levels. Transient overexpression of HIF-1α transgene in UMUC-3 cells showed a ~2 fold increase in CD24 mRNA expression and ~3 fold increase in CD24 protein expression in normoxia (Figure 3A,B). We evaluated four different shRNA oligos against HIF-1α, found similar effects and selected HIF-1α sh-3 for further studies (Figure S5D). Absence of appreciable change in HIF-2α levels after forced induction or silencing of HIF-1α indicates that CD24 expression is independent of HIF-2α status (Figure 3B). To determine the contribution of HIF-1α to hypoxic induction of CD24 expression, UMUC-3 cells transiently depleted of HIF-1α were exposed to hypoxia. HIF-1α depleted UMUC-3 cells failed to induce CD24 mRNA or protein expression after exposure to hypoxia (Figure 3A, B). To determine if HIF-1α regulates CD24 via a direct interaction with its promoter sequence, we performed chromatin immunoprecipitation. UMUC-3 cells were exposed to normoxia and hypoxia, and chromatin complexes immunoprecipitated with human HIF-1α or HIF-1β antibody, and PCR amplification was performed using specific primers to human CD24 promoter region encompassing identified HRE site. Results showed preferential binding of HIF-1α (Figure 3C), and HIF-1β (Figure S4) to CD24 promoter in hypoxia compared to normoxia.. This supports finding from previous studies that although HIF-1β (ARNT) is constitutively expressed, it heterodimerizes with HIF-1α and selectively bind to hypoxia responsive promoter elements of selected genes (36). These results also indicate direct transcriptional regulation of endogenous CD24 expression by HIF-1α.
Figure 3.
A) UMUC-3 cells transfected with pcDNA 3.1 vector, wild type HIF-1α and HIF-1α siRNA cells after exposure to 24 hours of hypoxia and mRNA expression of HIF-1α and CD24 assessed by quantitative realtime-PCR. * p<0.05, significantly different form pcDNA 3.1 transfected UMUC-3 cells. B) Corresponding proteins from A were analyzed for CD24 HIF-1α and HIF-2α expression by western blotting as explained in methods. The bar graph shows the fold change in expression of CD24 in all respective lanes normalized to α-tubulin OD values. * p<0.05, significantly different form pcDNA 3.1 transfected UMUC-3 cells. C) ChIP assay was performed on extracts from cells exposed to 12 hours of hypoxia and normoxia of UMUC-3 cells. HIF-1α immunoprecipitation was performed using specific anti-HIF-1α antibody. Primers spanning the HIF-1 binding region on CD24 promoter was used for quantitative real-time PCR amplification. The bars represent the normalized abundance of CD24 promoter region in these immunoprecipitated samples. IgG and Pol-II bars indicate the PCR amplification obtained when extracts were immunoprecipitated with a nonimmune mouse immunoglobulin and an antibody raised against the RNA polymerase II (Pol2 II). A–C) Blots are representative of three separate experiments. Error bars are SD of triplicate samples from one of three independent experiments.
CD24 is a critical determinant in HIF-1α driven tumor progression and metastasis
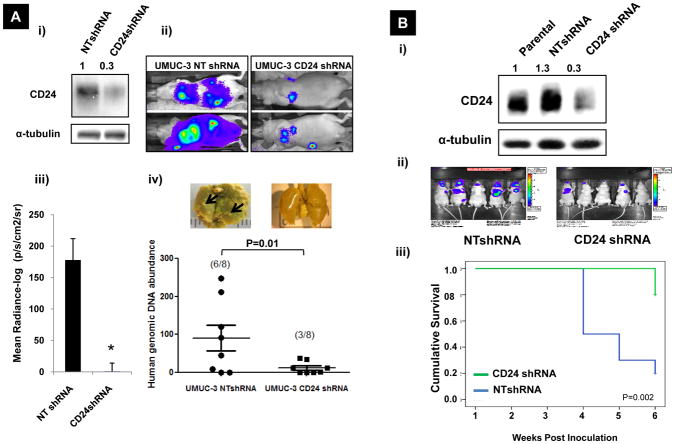
Both HIF-1α and CD24 overexpression are associated with poor prognosis in multiple human cancer types (37). We first evaluated the role of CD24 expression in experimental models of distant colonization of bladder and prostate cancer. The first using lung colonization of UMUC-3 cells after tail vein inoculation and the second using bone colonization after intraprostatic inoculation of PC3 cells. We silenced CD24 expression in UMUC-3 and PC-3 cells using lentiviral transduction of shRNA targeted to three different oligos against CD24 (CD24shRNA) or non-targeted scrambled control (NTshRNA). Expression of CD24 after transfecting these oligos were evaluated separately in UMUC-3 as well as PC-3 cells (Figure S5A) Effects of these shRNA’s on growth of UMUC-3 cells were assessed (Figure S5B). From these three different oligos we chose to use CD24-sh-3 for all further studies. Following injection of UMUC-3 cells we monitored the animals weekly using BLI (Bioluminescent Imaging) which was shown previously to track well with histology (52), and after 8 weeks sacrificed and evaluated their lungs by human-specific quantitative real-time PCR (26). At 8 weeks, mice injected with CD24-deficient UMUC-3 cells had lower levels of total photon radiance than control UMUC-3 NTshRNA cells and had significantly reduced numbers of residual tumor cells detectable by human specific quantitative real-time PCR (p=0.01, Figure 4A). Similarly, CD24 depleted PC-3 cells showed significantly lower photon radiance 3, 4 and 5 weeks after injection (Figure S5C) and higher cumulative survival compared to (p=0.002) mice inoculated with corresponding control PC-3 cells (Figure 4B). This result in prostate cells suggests a role of CD24 in lung colonization and supports our findings in bladder cancer cells (14) that CD24 expression affects lung retention of tumor cells.
Figure 4.
A) Effect of CD24 depletion on lung colonization of UMUC-3 cells: i) Western blots characterizing stable cell lines pre-inoculation; bands were quantified (numbers in figure) as described in the Methods ii) BLI images of representative animals; iii) Graph of mean radiance measured; iv) Human genomic DNA measured using 12p PCR in lung extracted from mice. Inset: Representative Bouin’s stained lungs metastasis development at 6 weeks. Arrows indicate surface metastasis sites. B) Effect of CD24 knock-down on metastasis of PC-3 cells: i) Western blots characterizing stable cell lines pre-inoculation; bands were quantified (numbers in figure) as described in the Methods ii) BLI images of representative animals; iii) Comparison of overall survival of mice injected with PC-3 CD24shRNA cells and non-target controls. (p=0.002 by Log Rank (Mantel-Cox) test of equality of survival distributions).
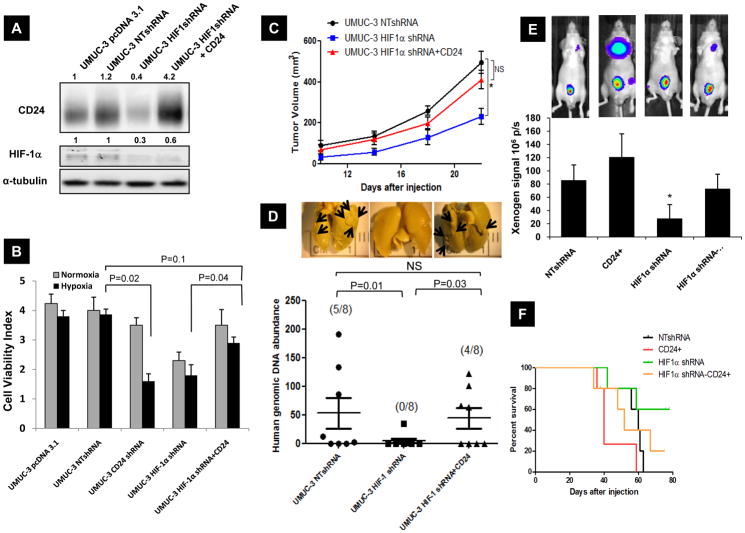
HIF-1α overexpression promotes metastasis while HIF-1α depletion reduces this phenotype via alteration of a cancer cell’s ability to survive hypoxic conditions (38). To study the contribution of CD24 in HIF-1α mediated viability during hypoxic stress in vitro and HIF-1α dependent metastasis in vivo, we generated HIF-1α short hairpin RNA (shRNA) expressing UMUC-3 cells We silenced HIF-1α expression in UMUC-3 cells using lentiviral transduction of shRNA targeted to three different oligos against HIF-1α or non-targeted scrambled control (NT). Expression of HIF-1α after transfecting these oligos were evaluated in UMUC-3 (Figure S5D). Effects of these shRNA’s on growth of UMUC-3 cells were assessed (Figure S5E). From these three different oligos we chose to use HIF-1α-sh-3 for all further studies. As expected, HIF-1α depleted UMUC-3 cells expressed significantly less CD24 compared to cells expressing scrambled control sequence (Figure 5A). CD24 depletion led to a reduction in cell viability in normoxia and an even more profound effect in hypoxic condition (p=0.0320). Importantly, this was comparable to that observed with HIF-1α depletion in these cells (Figure 5B). Furthermore, when CD24 expression was restored in HIF-1α depleted UMUC-3 cells using a CD24 transgene, this rescued the reduced survival observed after HIF-1α depletion in normoxia and hypoxia (p=0.04). When injected subcutaneously into mice, HIF-1α depleted UMUC-3 cells produced significantly decreased average tumor burden at a given time compared to UMUC-3 NTshRNA cells. Notably, cells with depleted HIF-1α but restored CD24 expression, displayed a comparable tumor burden at all tested times to UMUC-3 NTshRNA cells (p=0.13) (Figure 5C).
Figure 5.
A) Western blot of UMUC-3 cells transfected with pcDNA 3.1 vector, non-target control shRNA (NT-shRNA), HIF-1α shRNA, and CD24 overexpressed in HIF-1α shRNA transduced cells as explained in Methods. Bands were quantified as described in the Methods (numbers in figure). Data shown are representative blots of three separate experiments. B) Monolayer cell growth of 1000 cells / well in 96 well plate was estimated using “Live-Dead” assay (Molecular Probes) after 24 hours of exposed in normoxia and hypoxia as described. C) Subcutaneous tumor growth of engineered UMUC-3 cells in nude mice for a period of 25 days post injection. 10 mice were inoculated in each group. Tumor sizes were measured every fourth day and quantitated as in the Methods. * p=0.013, volume of tumors produced by UMUC-3-HIF-1α shRNA cells are significantly different form UMUC-3 NTshRNA cells. D) Quantitation of in vivo lung metastasis of HIF-1α and CD24 modified UMUC-3 cells by visual evaluation of surface lung metastases and total lung 12p quantitative PCR in mice injected via tail vein (n=8). Inset: Representative Bouin’s stained images of lung metastasis development at 8 weeks. Arrows indicate surface metastasis sites (visual quantitation of lung metastasis is shown in Supplementary Table 1). E) Quantitation of in vivo metastasis of HIF-1α and CD24 modified PC-3 cells injected orthotopically in prostate by Bioluminescence imaging (BLI) (n=8). Inset: Representative BLI images showing distant metastasis. Significant difference compared to BLI signal at 6 weeks post inoculation in NTshRNA group of animals (p<0.01). F) Kaplan Meyer curves indicating the survival as defined in materials and methods of nude mice injected with HIF-1α and CD24 modified PC-3 cells.
The engineered UMUC-3 cells described above were then inoculated via tail vein in vivo and lung metastasis assessed. Visual quantitation of lung metastases revealed that >50% of mice inoculated with emptyvector-transfected UMUC-3 cells had lesions at 8 weeks while none of the mice receiving UMUC-3 HIF-1α shRNA cells had detectable deposits (Supplementary Table 1). Notably, 50% of the mice injected with UMUC-3 HIF-1α shRNA cells stably overexpressing CD24 had visible lung metastasis. Evaluation of the lungs using quantitative real-time PCR with 12p human specific probe (26) showed a drastic reduction in human genomic DNA in HIF-1α depleted UMUC-3 cells compared to their non-target controls (SEM 47.8± 27 vs. 4320.2 ±1256 respectively, p=0.0081) while forced expression of CD24 in HIF-1α depleted cells rescued metastatic proficiency (p=0.031) (Figure 5D). Further, we sought to evaluate the role of HIF-1α and CD24 in spontaneous metastasis in an orthotopic prostate cancer model. Previously characterized luciferase expressing PC-3 cells with depleted HIF-1α and overexpressing CD24 were orthotopically implanted into prostate and assessed their metastatic ability by bioluminescence (BLI) imaging at various intervals. BLI signal and survival of animals (Figure 5E–F) showed that HIF-1α and CD24 expression are important for local and metastatic growth. Moreover, forced expression of CD24 partially rescued metastatic ability of HIF-1α depleted cells.
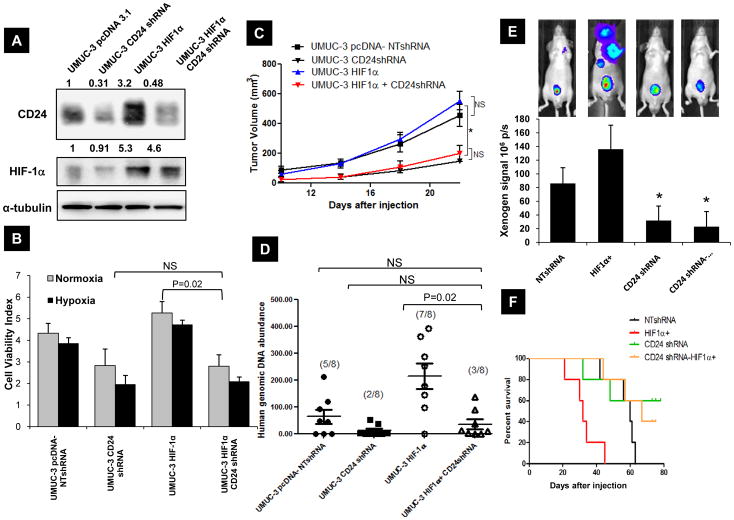
Next, to establish the requirement for CD24 in HIF-1α induced primary tumor growth and metastasis, we first generated stable HIF-1α overexpressing, and also CD24 depleted HIF-1α overexpressing UMUC-3 cells. HIF-1α overexpression in UMUC-3 cells led to a marginal increase in cell viability compared to NTshRNA cells while CD24 depletion in HIF-1α overexpressing cells reduced their survival (p=0.021) (Figure 6B). When injected subcutaneously into mice, HIF-1α overexpressing UMUC-3 cells marginally increased average tumor burden compared to UMUC-3 NTshRNA cells (Figure 6C). However, CD24 depletion even in the presence of forced HIF-1α expression produced smaller tumors (Figure 6C). These modified UMUC-3 cells were inoculated via tail vein in vivo and lung metastasis was assessed. As evaluated by visual assessment and quantitative real-time PCR with a 12p human specific probe, UMUC-3 cells stably overexpressing HIF-1α displayed more colonies in lung compared to UMUC-pcDNA NTshRNA cells, while CD24 depletion even in the presence of forced HIF-1α expression, reduced their ability to colonize to lungs (p=0.022) (Figure 6D). These results support the role of CD24 as an important downstream effector of HIF-1α mediated survival and metastasis. Further, PC-3 cells stably overexpressing HIF-1α, and also CD24 depleted HIF-1α overexpressing UMUC-3 cells were orthotopically implanted in prostate and assessed their metastatic ability by bioluminescence (BLI) imaging at various intervals. BLI signal and survival of animals (Figure 6E–F) showed that tumor bulk and animal survival following inoculation of PC-3 cells depends on elevated expression of HIF-1α, CD24 and that CD24 expression contributes to the ability of HIF-1α to promote metastasis. To our knowledge this is the first demonstration of the effect of CD24 and HIF-1α in a prostate orthotopic model that leads to spontaneous metastasis.
Figure 6.
A) Western blot of UMUC-3 cells transfected with pcDNA 3.1 vector, non-target control shRNA (NT-shRNA), HIF-1α overexpressing, and CD24 shRNA transduced in HIF-1α overexpressed cells as explained in Methods. Bands were quantified as described in the Methods (numbers in figure). Data shown are representative blots of three separate experiments. B) Monolayer cell growth of 1000 cells / well in 96 well plate was estimated using “Live-Dead” assay (Molecular Probes) after 24 hours of exposed in normoxia and hypoxia as described. C) Subcutaneous tumor growth of engineered UMUC-3 cells in nude mice for a period of 25 days post injection. 10 mice were inoculated in each group. Tumor sizes were measured every fourth day and quantitated as in the Methods. * p=0.024, volume of tumors produced by UMUC-3-HIF-1α CD24shRNA cells are significantly different form UMUC-3 NTshRNA or HIF-1α over expressing UMUC-3 cells. D) Quantitation of in vivo lung metastasis of HIF-1α and CD24 modified UMUC-3 cells by visual evaluation of surface lung metastases and total lung 12p quantitative PCR in mice injected via tail vein (n=8). E) Quantitation of in vivo metastasis of HIF-1α and CD24 modified PC-3 cells injected orthotopically in prostate by Bioluminescence imaging (BLI) (n=8). Inset: Representative BLI images showing distant metastasis. Significant difference compared to BLI signal at 6 weeks post inoculation in NTshRNA group of animals (p<0.01). F) Kaplan Meyer curves indicating the survival as defined in materials and methods of nude mice injected with HIF-1α and CD24 modified PC-3 cells.
Co-expression of CD24 and HIF-1α in human bladder cancers
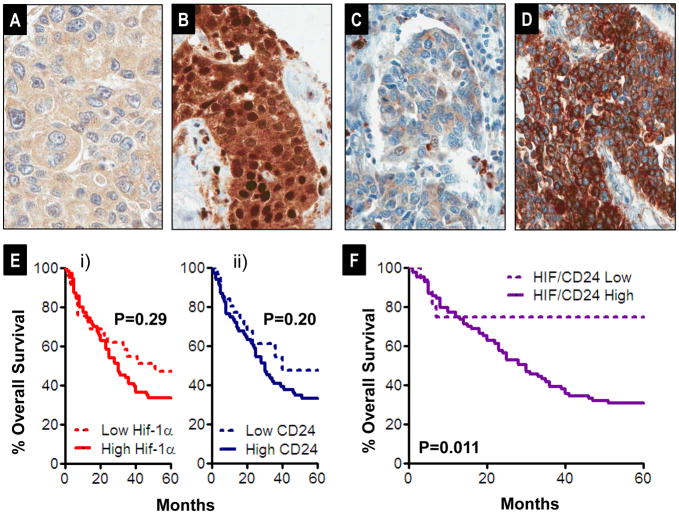
Finally, we sought to determine whether the pattern of expression of CD24 from human tumors might support the model that HIF-1α drives the metastatic phenotype in part via induction of CD24 expression, as indicated by the experiments described above. Hence we examined if there is any relationship between HIF-1α and CD24 protein expression by immunohistochemistry in human bladder cancer tissues, using a tissue microarray of archival tissues, using antibodies reported before (22, 39), and scored expression as detailed in the Materials and Methods. In total, 144 cases represented on the array showed interpretable staining for both antibodies; of these, 101 cases were urothelial carcinomas (the most common type of bladder cancer in Western countries and the histology from which bladder cell lines used experimentally herein were derived). As reported before (23), HIF-1α demonstrated highly variable nuclear positivity with frequent background moderate to strong cytoplasmic positivity (Figure 7A–B). Given its known function and prior reports in bladder cancer associating degree of nuclear positivity with key clinicopathologic variables, HIF-1α staining was scored as % nuclear positivity. CD24 showed variable staining among cases of urothelial carcinoma with cytoplasmic immunoreactivity (scored as 0+,1+,2+,3+ on overall intensity as reported before (11) as examples shown in Figure 7C–D).
Figure 7.
A–B) Representative 0+ (%0 nuclear positivity) and 3+ (>50% nuclear positivity) staining of HIF-1α on tissue microarray cores of human urothelial carcinoma, respectively. C–D) Representative 1+ (weak intensity) and 3+ (diffuse, intense) CD24 staining, respectively. E) Kaplan Meier analysis of overall survival post cystectomy of 101 urothelial carcinomas from the tissue microarray, stratified as a function of i) CD24 level (low CD24 (0+, 1+, N=31) and high CD24 (2+ and 3+, N=70)). Differences evaluated by Log Rank test. ii) Similar analysis to that of i) but comparing survival stratified by low HIF-1α (0+ and 1+ nuclear positivity, N=29) and high HIF-1α (2+ and 3+, N=72). F) Similar analysis to that in E but examining survival as a function of HIF-1α and CD24 combined (HIF+CD24) score, comparing low scores (0+, 1+, and 2+, N=16) to high scores (3+ to 6+, N).
Among urothelial cases we observed a significant, positive correlation between CD24 staining and HIF-1α nuclear positivity, rs=0.29, P=0.003. In contrast, in non-urothelial cases, CD24 and nuclear HIF-1α were not correlated significantly rs=0.036, P=0.84. Among the urothelial carcinomas, the degree of correlation between CD24 and HIF-1α was related to stage: among cases stage pTa, pT1, and pT2 (primary tumor that has not extended beyond the bladder), CD24 and HIF-1α were not significantly correlated (rs=0.001, P=1.0). In contrast, cases showing extravesical extension of the primary tumor (stages pT3 and pT4), showed the highest degree of correlation (rs=0.49, P=0<0.001). Importantly, the most prevalent pattern of expression of these proteins was of expression of both at greater than 2+ level (N=55), as opposed to many fewer cases showing expression of both proteins at low level (0+ or 1+, N=14).
Given our observations regarding roles for HIF-1α, and downstream, CD24, in our experimental metastasis model, we were interested in whether staining patterns for these proteins were associated with overall survival in the first five years post-cystectomy, where recurrence (metastasis) free survival contributes most strongly to overall survival (40). For these analyses, we tested association of HIF-1α and CD24 staining with overall survival at 60 months by the log rank test. For HIF-1α, we observed a non-significant trend toward decreased survival in cases showing 2+ or 3+ nuclear staining (HIF-1α High) as opposed to cases showing 0+ and 1+ (HIF-1α Low) (Figure 7E i). For CD24, we observed a non-significant trend toward decreased survival in cases showing 2+ or 3+ staining (CD24 High) as opposed to cases showing 0+ and 1+ (CD24 Low) (Figure 7Eii).
Given the observed pro-metastatic regulatory cascade of HIF-1α on CD24 expression observed above, we also tested the relationship between survival and a total, combined HIF/CD24 score (i.e., HIF-1α + CD24). Comparing cases showing staining consistent with induction of this pro-metastatic cascade (HIF/CD24 total >=3, High, N=85) to cases showing lack of induction of either or both of these proteins (HIF/CD24 total score <3, Low, N=16), we found a significant difference in survival (Figure 7F, P=0.01). Importantly, given the aforementioned association between increased correlation of HIF-1α and CD24 and high primary tumor stage, we tested the relationship between HIF/CD24 total score class (Low versus High) and survival at the 60 month endpoint, alone and with stage in uni- and multivariate Cox proportional hazards regressions models. Strikingly, not only was HIF/CD24 class significantly associated with survival (univariate Cox P=0.02), but it maintained independent, significant association with survival, adjusting for primary tumor stage (multivariate Cox P=0.01). The correlation of HIF-1α and CD24 expression in tumor tissues and association of their coexpression with poorer survival outcomes supports the clinical relevance of model system findings above that HIF-1α drives the metastatic phenotype in part via CD24 induction.
DISCUSSION
The results presented here are the first to show that CD24 is transcriptionally regulated by HIF-1α and that this former molecule is a critical downstream effector of HIF-1α effects on tumor growth and lung colonization. Furthermore the clinical relevance of this pathway was supported by data from human bladder tumors indicating that the clinical impact of HIF-1α expression is enhanced by CD24 co-expression. By linking CD24 with hypoxia, these findings can provide one explanation why CD24 expression is such a powerful prognostic factor in bladder and prostate cancer.
Friederichs et al. demonstrated that CD24 may contribute to metastatic dissemination as a ligand for the adhesion molecules P- and E-selectin (41), which are expressed on activated platelets and endothelial cells. Interestingly, hypoxia and re-oxygenation can induce P-selectin expression in endothelial cells (42) which may promote further cancer cell-endothelial interactions. Inhibition of CD24 by monoclonal antibodies or siRNAs has been reported to reduce primary tumor growth and metastasis of colon, pancreas and bladder cancers (16, 43). In the current study, CD24 depletion had a greater impact on cell viability in hypoxia-exposed than normoxic cells. This suggests CD24 expression serves to protect cells from hypoxia and allow them to survive in such adverse conditions in addition to promoting their local growth and metastatic colonization. Interestingly, the increase in CD24 mRNA in normoxia following HIF-1α transgene transfection was higher than that observed in the control transfected cells exposed to hypoxia. In contrast CD24 protein levels were similar despite higher HIF-1α in HIF-1α transgene transfected cells. This suggests CD24 protein levels are controlled by other factors or alternate limiting mediators of protein synthesis exist.
Immunohistochemistry and gene array based studies have shown CD24 expression is up-regulated in bladder and prostate cancers and its expression correlates with poor prognosis (8). Further this study showed, CD24 expression is critical for tumor growth and migration. Altevogt et al demonstrated that CD24 expression reduces SDF-1-mediated cell migration and signaling via CXCR4 (44) a mechanism by which CD24 may elicit tumor promoter effect. A recent study also suggested that CD24-dependent extracellular signal-regulated kinases and p38 mitogen-activated protein kinase activations are required for colorectal cancer cell proliferation in vitro and in vivo (45). CD24-bound oligosaccharides acts as ligand for P-selectin, a cell adhesion molecule present on endothelial cell wall and platelets shown to facilitate tumor passage through blood stream (46) Using a short-term lung colonization assay, we have shown that CD24 expressing cells preferentially bind to lungs compared to non-expressors (11). However, the mechanisms that drive CD24 expression in human cancer are unclear. In this study, automated comparison of the core Ral transcriptional signature (manuscript in revision) with all molecular concepts in Oncomine (including Oncomine gene expression signatures, Gene Ontology, KEGG Pathways, Human Protein Reference Database, etc) identified significant association with hypoxia related molecular concepts. Similarly, a previous study based on broad transcriptomic analysis of human umbilical cord vein endothelial cells (HUVEC) exposed in vitro to hypoxia, Scheurer et.al reported that CD24 is one of 65 genes whose mRNA increases with hypoxia (29). However, no further insights were provided as to whether this was a transcriptional effect or the mechanisms responsible. Here we observed hypoxia led to a significant up-regulation of CD24 mRNA and protein expression in a panel of bladder and prostate cancer cells. In addition, CD24 immunoreactivity was found in hypoxic areas of human xenografts indicating this association is also operative in vivo. Further studies performed in UMUC-3 bladder cancer cells uncovered a putative hypoxia responsive element in the 5’ flanking region of CD24 promoter. Using chromatin immunoprecipitation studies we demonstrated increase HIF-1 binding to the CD24 promoter after exposure to hypoxia. Together, this data suggests that at least for the length of promoter we had for our studies, the HRE site was a prime regulator of transcription.
As CD24, HIF-1α is elevated in advanced tumors and is associated with metastatic progression (2). HIF-1α mediated gene expression allows cells to respond to hypoxic insult by increasing oxygen delivery via secretion of angiogenic factors or confer a survival advantage to cells faced with decreased oxygen availability (47). In prostate (48) and bladder cancer (49) HIF-1α regulates multiple cytokines and their receptors. By regulating key target genes, HIF-1α promotes the tumor growth at primary and distant sites. Our data shows recovery of primary tumor growth and lung colonization (Figure 5C,D). Upon CD24 overexpression in HIF-1α depleted cells indicates that CD24 plays an important role in HIF-1 mediated tumor growth regardless of tumor site.
Finally, data from our clinical cohort support the relevance of our experimental findings to human tumors. We observed a significant correlation of HIF-1α and CD24, correlating in vitro findings of HIF-1α regulation of CD24 as initially suggested by molecular concepts mapping. Additionally, attempting to evaluate the output of this regulatory pathway by use of a total score adding scores for HIF-1α and CD24, we found that we could significantly stratify survival, independently of tumor stage. Importantly, the most prevalent pattern of expression of these proteins in bladder cancers was of coincident moderate to intense (2+ or 3+) staining of both, a finding of relevance to potential therapeutic strategies (2, 11), which appear rational and should be considered.
As many cancers are thought to develop and progress to metastasis from a small number of transformed, self-renewing “cancer stem cells” (50), these results implicate a role for the HIF-1α-CD24 axis in achieving that goal. In addition, the work presented here on human tumors shows the value of risk stratification based on HIF and CD24 protein expression and could serve to select patients for trials in the adjuvant or early metastatic setting with anti- HIF-1α or CD24 directed therapy.
Supplementary Material
Acknowledgments
Work was supported in part by National Institutes of Health grants CA075115 and CA104106 to DT. The authors wish to thank Drs. Chris Moskaluk and Pat Pramoonjago of the UVa Biorepository and Tissue Research Facility for their technical support.
Footnotes
AUTHOR CONTRIBUTIONS
S.T, M.A.H, S.C.S. S.A.T. and D.T. designed the experiments and analyzed the data. S.T, J.B.O, M.N, G.C, and H. F.F performed the experiments. S.T, S.C.S, S.A.T. and D.T wrote the manuscript.
Conflict of interests: None of the authors have any financial conflict of interest that might be construed to influence the results or interpretation of the manuscript.
References
- 1.Giaccia AJ, Simon MC, Johnson R. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev. 2004;18:2183–94. doi: 10.1101/gad.1243304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland RM. Tumor hypoxia and gene expression--implications for malignant progression and therapy. Acta Oncol. 1998;37:567–74. doi: 10.1080/028418698430278. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan R, Graham CH. Hypoxia-driven selection of the metastatic phenotype. Cancer Metastasis Rev. 2007;26:319–31. doi: 10.1007/s10555-007-9062-2. [DOI] [PubMed] [Google Scholar]
- 5.Kristiansen G, Sammar M, Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol. 2004;35:255–62. doi: 10.1023/b:hijo.0000032357.16261.c5. [DOI] [PubMed] [Google Scholar]
- 6.Kadmon G, Eckert M, Sammar M, Schachner M, Altevogt P. Nectadrin, the heat-stable antigen, is a cell adhesion molecule. J Cell Biol. 1992;118:1245–58. doi: 10.1083/jcb.118.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T, Kiyokawa N, Taguchi T, Sekino T, Katagiri YU, Fujimoto J. CD24 induces apoptosis in human B cells via the glycolipid-enriched membrane domains/rafts-mediated signaling system. J Immunol. 2001;166:5567–77. doi: 10.4049/jimmunol.166.9.5567. [DOI] [PubMed] [Google Scholar]
- 8.Smith SC, Oxford G, Wu Z, Nitz MD, Conaway M, Frierson HF, et al. The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res. 2006;66:1917–22. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 9.Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci U S A. 2008;105:14076–81. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee TK, Castilho A, Cheung VC, Tang KH, Ma S, Ng IO. CD24(+) Liver Tumor-Initiating Cells Drive Self-Renewal and Tumor Initiation through STAT3-Mediated NANOG Regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Overdevest JB, Thomas S, Kristiansen G, Hansel DE, Smith SC, Theodorescu D. CD24 offers a therapeutic target for control of bladder cancer metastasis based on a requirement for lung colonization. Cancer Res. doi: 10.1158/0008-5472.CAN-11-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaipparettu BA, Malik S, Konduri SD, Liu W, Rokavec M, van der Kuip H, et al. Estrogen-mediated downregulation of CD24 in breast cancer cells. Int J Cancer. 2008;123:66–72. doi: 10.1002/ijc.23480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taguchi T, Kiyokawa N, Mimori K, Suzuki T, Sekino T, Nakajima H, et al. Pre-B cell antigen receptor-mediated signal inhibits CD24-induced apoptosis in human pre-B cells. J Immunol. 2003;170:252–60. doi: 10.4049/jimmunol.170.1.252. [DOI] [PubMed] [Google Scholar]
- 14.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nature genetics. 2007;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 15.Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9:443–54. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–13. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Xie K, Mukaida N, Matsushima K, Fidler IJ. Hypoxia-induced elevation in interleukin-8 expression by human ovarian carcinoma cells. Cancer Res. 1999;59:5822–9. [PubMed] [Google Scholar]
- 18.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–42. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson BE, Frierson HF, Conaway MR, Seraj JM, Harding MA, Hampton GM, et al. Profiling the evolution of human metastatic bladder cancer. Cancer Res. 2004;64:7813–21. doi: 10.1158/0008-5472.CAN-04-0826. [DOI] [PubMed] [Google Scholar]
- 20.Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–20. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Moissoglu K, Wang H, Wang X, Frierson HF, Schwartz MA, et al. Src phosphorylation of RhoGDI2 regulates its metastasis suppressor function. Proc Natl Acad Sci U S A. 2009;106:5807–12. doi: 10.1073/pnas.0810094106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristiansen G, Machado E, Bretz N, Rupp C, Winzer KJ, Konig AK, et al. Molecular and clinical dissection of CD24 antibody specificity by a comprehensive comparative analysis. Lab Invest. 90:1102–16. doi: 10.1038/labinvest.2010.70. [DOI] [PubMed] [Google Scholar]
- 23.Tickoo SK, Milowsky MI, Dhar N, Dudas ME, Gallagher DJ, Al-Ahmadie H, et al. Hypoxia-inducible factor and mammalian target of rapamycin pathway markers in urothelial carcinoma of the bladder: possible therapeutic implications. BJU Int. 107:844–9. doi: 10.1111/j.1464-410X.2010.09517.x. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, McRoberts K, Berr SS, Frierson HF, Jr, Conaway M, Theodorescu D. Neuromedin U is regulated by the metastasis suppressor RhoGDI2 and is a novel promoter of tumor formation, lung metastasis and cancer cachexia. Oncogene. 2007;26:765–73. doi: 10.1038/sj.onc.1209835. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z, Owens C, Chandra N, Popovic K, Conaway M, Theodorescu D. RalBP1 is necessary for metastasis of human cancer cell lines. Neoplasia. 12:1003–12. doi: 10.1593/neo.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nitz MD, Harding MA, Theodorescu D. Invasion and metastasis models for studying RhoGDI2 in bladder cancer. Methods Enzymol. 2008;439:219–33. doi: 10.1016/S0076-6879(07)00417-X. [DOI] [PubMed] [Google Scholar]
- 27.Solomon B, Binns D, Roselt P, Weibe LI, McArthur GA, Cullinane C, et al. Modulation of intratumoral hypoxia by the epidermal growth factor receptor inhibitor gefitinib detected using small animal PET imaging. Mol Cancer Ther. 2005;4:1417–22. doi: 10.1158/1535-7163.MCT-05-0066. [DOI] [PubMed] [Google Scholar]
- 28.Oxford G, Smith SC, Hampton G, Theodorescu D. Expression profiling of Ral-depleted bladder cancer cells identifies RREB-1 as a novel transcriptional Ral effector. Oncogene. 2007;26:7143–52. doi: 10.1038/sj.onc.1210521. [DOI] [PubMed] [Google Scholar]
- 29.Scheurer SB, Rybak JN, Rosli C, Neri D, Elia G. Modulation of gene expression by hypoxia in human umbilical cord vein endothelial cells: A transcriptomic and proteomic study. Proteomics. 2004;4:1737–60. doi: 10.1002/pmic.200300689. [DOI] [PubMed] [Google Scholar]
- 30.Kristiansen G, Denkert C, Schluns K, Dahl E, Pilarsky C, Hauptmann S. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am J Pathol. 2002;161:1215–21. doi: 10.1016/S0002-9440(10)64398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradford TJ, Tomlins SA, Wang X, Chinnaiyan AM. Molecular markers of prostate cancer. Urologic oncology. 2006;24:538–51. doi: 10.1016/j.urolonc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 33.Shulewitz M, Soloviev I, Wu T, Koeppen H, Polakis P, Sakanaka C. Repressor roles for TCF-4 and Sfrp1 in Wnt signaling in breast cancer. Oncogene. 2006;25:4361–9. doi: 10.1038/sj.onc.1209470. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Liu BC, Lin GT, Lin CS, Lue TF, Willingham E, et al. Up-regulation of estrogen responsive genes in hypospadias: microarray analysis. J Urol. 2007;177:1939–46. doi: 10.1016/j.juro.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Werner T. Target gene identification from expression array data by promoter analysis. Biomol Eng. 2001;17:87–94. doi: 10.1016/s1389-0344(00)00071-x. [DOI] [PubMed] [Google Scholar]
- 36.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–4. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steeg PS, Theodorescu D. Metastasis: a therapeutic target for cancer. Nat Clin Pract Oncol. 2008;5:206–19. doi: 10.1038/ncponc1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudary N, Hill RP. Hypoxia and metastasis. Clin Cancer Res. 2007;13:1947–9. doi: 10.1158/1078-0432.CCR-06-2971. [DOI] [PubMed] [Google Scholar]
- 39.Filby CE, Hooper SB, Wallace MJ. Partial pulmonary embolization disrupts alveolarization in fetal sheep. Respir Res. 11:42. doi: 10.1186/1465-9921-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein JP, Lieskovsky G, Cote R, Groshen S, Feng AC, Boyd S, et al. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol. 2001;19:666–75. doi: 10.1200/JCO.2001.19.3.666. [DOI] [PubMed] [Google Scholar]
- 41.Friederichs J, Zeller Y, Hafezi-Moghadam A, Grone HJ, Ley K, Altevogt P. The CD24/P-selectin binding pathway initiates lung arrest of human A125 adenocarcinoma cells. Cancer Res. 2000;60:6714–22. [PubMed] [Google Scholar]
- 42.Closse C, Seigneur M, Renard M, Pruvost A, Dumain P, Belloc F, et al. Influence of hypoxia and hypoxia-reoxygenation on endothelial P-selectin expression. Haemostasis. 1996;26 (Suppl 4):177–81. doi: 10.1159/000217296. [DOI] [PubMed] [Google Scholar]
- 43.Sagiv E, Starr A, Rozovski U, Khosravi R, Altevogt P, Wang T, et al. Targeting CD24 for treatment of colorectal and pancreatic cancer by monoclonal antibodies or small interfering RNA. Cancer Res. 2008;68:2803–12. doi: 10.1158/0008-5472.CAN-07-6463. [DOI] [PubMed] [Google Scholar]
- 44.Schabath H, Runz S, Joumaa S, Altevogt P. CD24 affects CXCR4 function in pre-B lymphocytes and breast carcinoma cells. Journal of cell science. 2006;119:314–25. doi: 10.1242/jcs.02741. [DOI] [PubMed] [Google Scholar]
- 45.Wang W, Wang X, Peng L, Deng Q, Liang Y, Qing H, et al. CD24-dependent MAPK pathway activation is required for colorectal cancer cell proliferation. Cancer science. 2010;101:112–9. doi: 10.1111/j.1349-7006.2009.01370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, et al. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 2005;65:10783–93. doi: 10.1158/0008-5472.CAN-05-0619. [DOI] [PubMed] [Google Scholar]
- 47.Chan DA, Giaccia AJ. Hypoxia, gene expression, and metastasis. Cancer Metastasis Rev. 2007;26:333–9. doi: 10.1007/s10555-007-9063-1. [DOI] [PubMed] [Google Scholar]
- 48.Zhong H, Agani F, Baccala AA, Laughner E, Rioseco-Camacho N, Isaacs WB, et al. Increased expression of hypoxia inducible factor-1alpha in rat and human prostate cancer. Cancer Res. 1998;58:5280–4. [PubMed] [Google Scholar]
- 49.Theodoropoulos VE, Lazaris A, Sofras F, Gerzelis I, Tsoukala V, Ghikonti I, et al. Hypoxia-inducible factor 1 alpha expression correlates with angiogenesis and unfavorable prognosis in bladder cancer. Eur Urol. 2004;46:200–8. doi: 10.1016/j.eururo.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–72. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.