Abstract
High-throughput sequencing of cDNA libraries constructed from cellular RNA complements (RNA-Seq) naturally provides a digital quantitative measurement for every expressed RNA molecule. Nature, impact and mutual interference of biases in different experimental setups are, however, still poorly understood—mostly due to the lack of data from intermediate protocol steps. We analysed multiple RNA-Seq experiments, involving different sample preparation protocols and sequencing platforms: we broke them down into their common—and currently indispensable—technical components (reverse transcription, fragmentation, adapter ligation, PCR amplification, gel segregation and sequencing), investigating how such different steps influence abundance and distribution of the sequenced reads. For each of those steps, we developed universally applicable models, which can be parameterised by empirical attributes of any experimental protocol. Our models are implemented in a computer simulation pipeline called the Flux Simulator, and we show that read distributions generated by different combinations of these models reproduce well corresponding evidence obtained from the corresponding experimental setups. We further demonstrate that our in silico RNA-Seq provides insights about hidden precursors that determine the final configuration of reads along gene bodies; enhancing or compensatory effects that explain apparently controversial observations can be observed. Moreover, our simulations identify hitherto unreported sources of systematic bias from RNA hydrolysis, a fragmentation technique currently employed by most RNA-Seq protocols.
INTRODUCTION
Read abundances from RNA-Seq experiments reflect the quantities of different RNA molecules in the interrogated transcriptome (1). It is commonly accepted that gene expression profiles exhibit a similar shape across evolutionary distant organisms and functionally diverse cell types. Observations based on expressed sequence tags (2) show that most transcripts are rare, some are moderately abundant and only a small portion is very abundant. Such unbalanced distribution can be modelled using Zipf’s Law (3) which exhibits a characteristic linear behaviour in log–log (4). Furusawa and Kaneko (2) link the reason behind this observation to general thermodynamic diffusion constants that determine power law distributions in a large spectrum of biomolecules, whereas Ogasawara et al. (5) propose an evolutionary model.
However, experimental protocols are increasingly reported to generate deviations from the expected read distributions (6–8). Since the first ultra sequencing experiments on cellular transcriptomes (9,10), sample preparations for so-called RNA-Seq have evolved in multiple respects and generated a considerable repertoire of protocols, which however all stem from a common set of elementary components. First and foremost, because all current sequencing technologies can only handle DNA substrates, reverse transcription (RT) of RNA into cDNA has to be accomplished. In the first protocols to be proposed for library preparation, RT constituted the initial step, involving either poly-dT (for poly-A+ transcriptomes) or random primers (usually hexamers) to initiate first-strand synthesis. Poly-dT oligomers primarily bind in the region of the poly-A tail, which—especially for long transcripts—can result in template loss during RT of the entire molecule and thereby cause a loss of 5′-end information (9). Randomly primed first-strand synthesis of full-length RNA molecules, in contrast, can lead to a relative over-representation of the 5′-end information (11). To diminish RT-related biases, RNA-Seq concepts have changed towards protocols that postpone RT after fragmentation, which seems to prevent a strong bias of read abundances towards the 3′-end (1).
Second, fragmentation of transcripts is necessary because current sequencing platforms produce relatively short tags from the ends of much longer DNA molecules. Therefore, any attempt to sequence non-fragmented RNA populations would result in reads that exclusively reproduce the ends of transcripts. First RNA-Seq protocols relied on fragmentation by restriction enzymes (e.g. NlaIII or DpnII) to cleave reversely transcribed cDNA (9,12). Due to the sequence specificity of each restriction enzyme, however, the number of fragments produced by enzymatic digestion is not directly comparable between transcripts of different sequence compositions; for instance, ∼4% of known Drosophila melanogaster genes do not exhibit a NlaIII-recognition site (13), and even degradation by the endonuclease DNAseI—so far considered as unspecific—has recently been reported to exhibit strong sequence-selective characteristics (7). Therefore, efforts were soon directed towards the development of sequence-independent ‘random’ fragmentation protocols (13), at first by employing nebulisation, the physical shearing of cDNA molecules in a liquid medium (14). Although being cost-efficient and effective, nebulisation has been criticised for its inability to fragment DNA chains shorter than ∼700–800 nt and for producing suboptimal fragment size distributions when subsequent size selection steps are present (15). Consequently, current RNA-Seq protocols implement fragmentation by the controlled hydrolysis of RNA—usually catalysed by heat and acetate-complexed Mg2+ or Zn2+ ions (11)—which is generally considered to produce uniformly distributed fragments (1).
After RT, during the ‘final library preparation’, adapter sequences are ligated to both sides of the double-stranded DNA molecules, which mediate the binding of fragments to beads and harbour primer-binding sites for amplification. Randomly primed RT (7) and/or the adapter ligation process (16,17) promote sequence-selective biases which manifest as motifs at the fragment ends (7,8); promising RNA-ligation based protocols avoiding both steps have been demonstrated (17,18). Before sequencing, fragments of the primary library are often amplified by a polymerase chain reaction (PCR), because the most cost-efficient sequencing platforms to date do not accept single molecule substrates. Amplification efficiency is known to depend on the GC content of the respective molecule (17), although controversial reports on the correlation between GC content and RNA-Seq coverage have been published (6,17,19). Leading high-throughput technology providers therefore suggest a size selection step in order to keep amplification biases under control by making fragment lengths homogeneous: e.g. 300–1000 nt long fragments are recommended for the Roche’s pyrosequencer (20), and 200 nt ± 25 nt are usually suggested for Illumina sequencing experiments. Size selection in general is implemented by gel electrophoresis, which suffers from artefacts like molecule aggregates (21).
Finally, the ‘sequencing’ step obtains one arbitrary end (single reads) or both ends (paired-end reads) of the cDNA fragments in the library. Read sequences undergo modifications according to the technical limitations of the corresponding platform, e.g. insertions/deletions (indels) typically occur in reads produced by Roche pyrosequencing (22), whereas Illumina sequencing platforms mainly exhibit read sequences with an increased rate of nucleotide substitutions—and a correspondingly decreased quality—towards the read end (6). Additionally, the interplay between sequencing chemistry, sequencer machine calibration and the base calling algorithm employed during the downstream analysis of raw data determine subtle preferences in the so-called ‘crosstalk’, i.e. the misrecognition of chromophore-marked nucleotides (23).
MATERIALS AND METHODS
Simulation of different fragmentation processes
Enzymatic digestion
In our implementation of in silico enzymatic digestion, position weight matrices are employed to capture the sequence selectivity of the corresponding enzyme (e.g. NlaIII or DpnII) and fragmentation points of cDNA molecules are determined by importance sampling.
Nebulisation
According to preliminary modelling attempts (24), potential breakpoints are distribtuted as a Gaussian function around the molecules’ midpoints and the breaking probability is drawn from an exponential of the ratio between the fragment size and the limiting size below which molecules are unlikely to be broken any further (λ = 700 nt for cDNA, Supplementary Methods).
Hydrolysis
Previously published models of hydrolysis are based on the assumption that fragment sizes produced by uniformly random fragmentation of molecules with the same length fall along a characteristic Weibull distribution, if the decay rate depends on molecule size (25). Here we propose a model for transcript populations of heterogeneous lengths, where we empirically derive a logarithmic dependence of the Weibull shape-parameter on the molecule’s length (see Supplementary Methods and Results section).
Simulation of reverse transcription
We model RT separately for first- and second-strand synthesis. The start point depends on the priming strategy (i.e. parameters Poly-dT or random) and optionally by a position weight matrix (PWM) describing the sequence bias. The primer extension length is parameterised by the minimum (RTmin) and maximum length (RTmax) of the expected cDNA molecules (Supplementary Methods).
Simulated size selection
As for the fragment sizes observed after gel electrophoresis, the Flux Simulator accepts parameterised normal distributions or empirical distributions. Fragments are subsampled according to such distributions, either by acceptance–rejection sampling or by the Metropolis–Hastings algorithm (26,27).
Simulated adapter ligation and PCR amplification
We simulate the reaction kinetics of the adapter ligation process—reflected by motifs of sequences that are preferred by the involved enzymes—as Bernoulli trials parameterised by a PWM representing the sequence bias. PCR-amplification is sensitive to the GC content of the amplified DNA stretches and in agreement with previous observations (17), we model PCR-efficiency as a quantity distributed normally about a GC-optimum (Supplementary Methods).
Simulated sequencing
During in silico sequencing, the fragments in the library are subsampled and the sequence of either an arbitrary end for single reads, or of both ends for paired mates, is obtained. The number of reads and their length may be specified; however, there cannot be more reads than the number of fragments in the library, nor can any read be longer than the fragment it comes from. The orientation of the reads is characteristic in sequencing-by-synthesis protocols (13,17) due to an intrinsic attribute of polymerases progressing strictly from 3′ to 5′ along the template (Supplementary Figure S1). For Illumina chemistry, we additionally implemented a quality-based error model (Supplementary Figure S2).
Simulated gene expression
In agreement with preliminary observations (2,5), our analysis of the reference data sets demonstrates that gene expression profiles estimated from RNA-Seq data exhibit an about linear shape in log–log space up to the first thousands of gene expression ranks (Supplementary Figure S3). By non-linear fitting to the experimental data, we deduced a modified Zipf’s Law, which we employ to assign randomised expression levels to genes and transcripts in our simulations (Supplementary Methods).
In the Flux Simulator, we also include the simulation of two biologically relevant modifications of annotated transcripts: transcripts with the same splicing structure, i.e. identical configuration of introns that are removed during the processing of nascent RNA, still may vary in their precise transcription start site and in the length of their poly-A tail (Supplementary Methods). These features can have a significant impact on the physical attributes of the corresponding molecules, playing an important role during library preparation.
Data source and basic processing
For our analysis, we employed publicly available read data (Supplementary Methods) from: Saccharomyces cerevisiae (9), Arabidopsis thaliana (28), Mus musculus (11), the same Homo sapiens sample sequenced with two different RNA-Seq protocols, i.e. flowcell RT-Seq (FRT) and standard hydrolysis (STD) protocol (17), and RNA control sequences spiked-in in high concentrations (29). In a first step, we mapped and split-mapped non-redundantly all the reads to the respective reference genome sequence using the GEM library (http://sourceforge.net/projects/gemlibrary); in the case of the cress data set, which is comparatively small, we also considered additional read mappings with long indels obtained with BLAT (30).
Subsequently, we focused on the distribution of reads that map to transcripts without alternatively processed forms. To define such transcripts, we considered a standard reference annotation of the transcriptome, i.e. the SGD annotation for yeast (31), the TAIR annotation for cress (32) and the murine as well as the human RefSeq annotation (33). This procedure provided us with mappings for 6 606 768 reads (47%) from yeast, 351 336 reads (65%) from cress and for 21 359 481 reads (68%) from mouse, and with 530 996 reads that map in proper pairs to the spike-in control sequences. Due to substantially different data set sizes (90 million versus 13 million reads), in the case of the human FRT- and the STD-Seq experiments, we extracted subsets of reads of suitable size before mapping to ensure comparability (Supplementary Table S1).
RESULTS
Overview of the Flux Simulator RNA-Seq pipeline
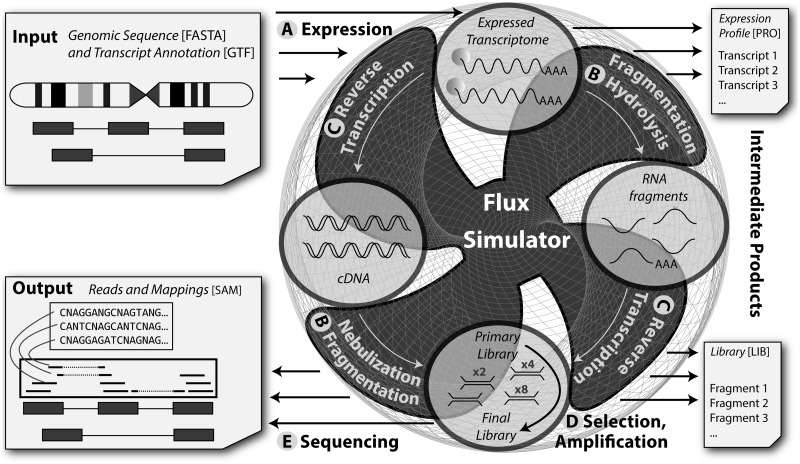
We implemented a computer pipeline for simulating RNA-Seq experiments—which we call the Flux Simulator—comprising explicit models for the processes that determine abundance and distribution of read tags according to the specified experimental protocol (see Figure 1 and Methods section). Starting from a genomic sequence for a certain species and a corresponding annotation of gene structures, the first step of this pipeline is, in fact, a transcriptome simulation (Figure 1A) where—if no pre-defined cell expression profile is available—annotated genes and transcripts are assigned randomised expression levels according to the general laws of gene expression (Supplementary Figure S3).
Figure 1.
Outline of the Flux Simulator pipeline. Provided the genomic sequence of an organism and a representative gene annotation as input, the initial step is a transcriptome simulation (A) to assign each transcript a randomised expression level according to general laws of gene expression. Subsequently, fragmentation (B) and RT (C) are carried out, either by first hydrolysing RNA and then transcribing the fragments into cDNA molecules (B and C, right) or by nebulisation respectively enzymatic digestion after reversely transcribing the entire RNA molecules (B and C, left). The simulated molecules of the primary library then get amplified by in silico PCR (D)—optionally after selecting a certain size range—and the final library then is subjected to simulated sequencing (E), including potential platform and sequencing chemistry specific error models. Finally, read sequences along with their genomic mappings are obtained.
Next, the in silico transcriptome undergoes RT/fragmentation (Figure 1B and C) according to the established experimental techniques: in one possible scenario, RNA molecules are first reversely transcribed into cDNA—adopting poly-dT or random primers—and afterwards fragmented by nebulisation or enzymatic digestion (Figure 1B and C, left); alternatively, fragmentation is carried out by RNA hydrolysis before fragments are transcribed into cDNA molecules by random priming (Figure 1B and C, right).
The Flux Simulator pipeline also provides optional steps to model the final library preparation, involving in silico ligation of adapter sequences, fragment size selection and PCR amplification (Figure 1D). Eventually high-throughput sequencing is simulated at the level of the single DNA molecule, offering the possibility to include platform-specific base calling errors (Figure 1E). The output comprises the read sequences and their genomic locations.
Physical properties of fragments produced by RNA-hydrolysis
By ‘uniform fragmentation’ we refer to the sequence-independent selection of breakpoints, as implemented for instance by DNA nebulisation or RNA hydrolysis, which is not to be confused with uniform breakpoint distributions along each transcript. In contrast to reports on the unequal representation of transcripts by nebulisation, fragmentation from RNA hydrolysis is considered to produce fragments of comparable lengths (11) without positional preferences (1). In this section, we study both hypotheses by simulating with our pipeline the experimental distributions observed for the so-called spike-in controls of known sequences (29). To this end, we extend a model proposed for uniform random fragmentation processes when the breaking probability depends on molecule size (25).
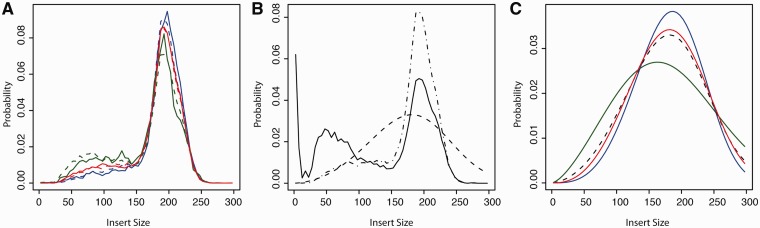
Paired-end reads generated from spike-in control sequences are particularly well suited to assess differences in fragment size distributions, as biases from incomplete transcript annotation can safely be excluded, and fragment sizes can be estimated straightforwardly by the distance between mapped read mates. Figure 2A demonstrates that fragments originating from three highly covered control sequences having substantially different lengths (i.e. 35 838 hydrolysis fragments from the 11 934 nt long Lambdaclone1-1, 472 364 fragments from the 1429 nt long OBF5, and 21 264 fragments from the 376 nt VATG sequence) also exhibit markedly different size distributions: when an arbitrary size of 150 nt is chosen as the threshold between short and long forms, we observe 36% fragments <150 nt for the short RNA control VATG (Figure 2A, green curve), whereas short fragments account for only 22% of the molecules in the case of the typical messenger-sized control OBF5 (Figure 2A, red curve), and their proportion drops to 15% for the long control sequence Lambdaclone1-1 (Figure 2A, blue curve).
Figure 2.
The size of an RNA molecule determines the hydrolysed fragments’ size distribution. (A) Theoretical and experimentally measured probability distributions of three control sequences with significantly distinct lengths: VATG (376 nt long, green lines), OBF5 (1429 nt, red lines) and Lambdaclone1-1 (11 934 nt, blue lines). The solid lines represent frequencies observed by the reads of an experiment, whereas the dashed lines are simulated results, obtained by the procedure depicted in B and C. Lambdaclone1-1—the longest control sequence—exhibits relatively less short fragments (blue curve), whereas VATG—the shortest control sequence—shows a comparatively lower fraction of long fragments (green curve). (B) The distribution used for simulated size filtering (solid line) is obtained by adjusting the insert size distribution observed across all sequences of the experiment (dashed-dotted line) with the combined distribution of insert sizes before filtering (dashed line), as estimated by simulated hydrolysis of the three spike-ins. During simulated size selection, weighted subsampling according to the filter distribution (solid line) is applied to the fragment size distributions (C) to derive the distributions predicted by the simulation after size filtering (A). (C) Computational prediction of fragment size distributions obtained from either control sequence after simulated hydrolysis, i.e. before size selection. According to the employed model, fragment sizes fall along Weibull distribution of same scale (= 200 nt) but different shapes (δ = 2.6, 3.2 and 4.1 for VATG, OBF5 and Lambdaclone1-1, respectively). Subjecting these simulated fragments to in silico size selection (B) can reproduce the differences observed in experimental results for the investigated controls (A).
The analysed experiment employs a gel segregation step in which exclusively fragments with the overall size attributes shown in Figure 2B are selected. Therefore, one cannot computationally cast back to the intermediate size distribution of fragments after fragmentation and before size selection. However, a previously published model for uniformly random fragmentation processes in molecules having the same length predicts that the sizes of the produced fragments follow a Weibull distribution—which is specified by two characteristic parameters, the shape (δ) and the scale (η). According to Figure 2B we conducted an exhaustive search within the relevant parameter space followed by simulated size selection (Supplementary Materials and Methods) and we found that the differences observed for fragment sizes can be qualitatively reproduced employing a constant decay rate (η = 200 nt), with the further prescription that the shape parameters depend logarithmically on the molecule length (Supplementary Figure S4).
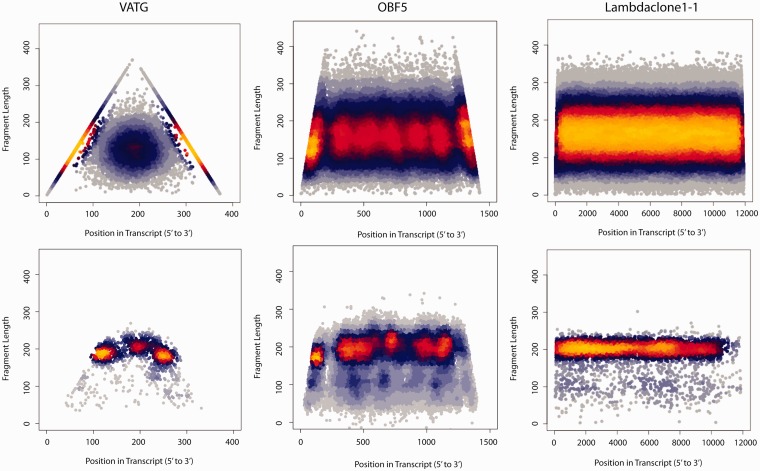
With our parameterised hydrolysis model (η = 200 nt, δ∼2.6 for VATG, δ∼3.2 for OBF5 and δ∼4.1 for Lambdaclone1-1) we then investigated the abundance distribution of fragments observed along transcript bodies. To avoid biases that have been demonstrated to impact on the ends of fragments in the considered experimental protocol (7,8), we focused during our analysis on the distribution of fragment midpoints along the RNA molecule they have been derived from. The top panels of Figure 3 show the density of such fragment centres produced by in silico hydrolysis along the three transcript bodies of Lambdaclone1-1, OBF5 and VATG (primary axis), segregated by the respective fragment size (secondary axis). The corresponding bottom panels depict the experimental outcome, which is sensitive to additional influences from other steps (e.g. size selection).
Figure 3.
RNA hydrolysis produces characteristic patterns of fragment accumulations. Distribution of fragment midpoints along RNA control sequences added to an RNA-Seq experiment that involves fragmentation by hydrolysis. The x-axis resolves the positions of fragment centres along the RNA control molecules VATG (right, 376 nt), OBF5 (centre, 1429 nt) and Lambdaclone1-1 (left, 11 934 nt long), whereas the y-axis segregates the obtained fragments according to their lengths. Data are shown as density scatter plots, with observed frequencies increasing from blue to red to yellow. The top panels show the distributions predicted by simulation employing the hydrolysis model implemented in the Flux Simulator, whereas the corresponding experimental outcome is shown in the bottom panels. Distinct points of fragment accumulations are notable in the short reference VATG and in the mRNA-sized control OBF5.
Albeit there are differences, the positional biases predicted by the hydrolysis simulation reproduce qualitatively the patterns of fragment concentrations observed in the experiment: the short VATG control exhibits three such distinct points (Figure 3, left), whereas the mRNA-sized OBF5 control shows seven fragment accumulations (Figure 3, centre)—and in both cases such points are located with remarkable symmetry about the centre of the reference molecule. Density fluctuations of Lambdaclone1-1 (Figure 3, right), the longest of the spike-in sequences considered, fall below the resolution limit of the diagram (Supplementary Figure S5).
Convolution of physical with chemical biases
After elucidating positional preferences caused by physical attributes of RNA molecules, we set off to establish computational models for capturing biases caused by a molecule’s sequence composition. Some sensitivity of RNA-Seq coverage to the GC content had already been noted earlier (6), especially in protocols involving PCR-amplification (17). In agreement with these previous studies we found that empirical PCR amplification efficiency can be appropriately modelled by a Gaussian distribution centred around a GC content of 50% (mean = 0.5, SD 0.1; Supplementary Figure S6).
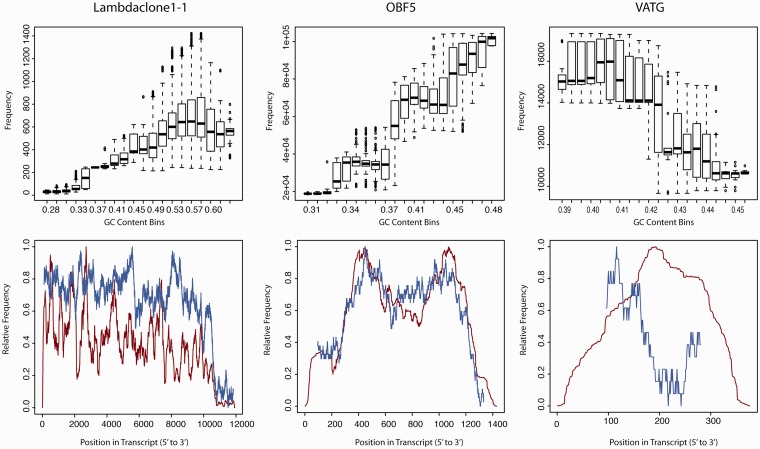
In the case studies of spike-in sequences described in the previous section, we assessed the correlation between the number of fragments covering a certain position and the GC content in a window of 192 nt (the mean fragment size) centred at that position (Figure 4, top panels): for the Lambdaclone1-1 and the OBF5 controls we found a high correlation (Pearson coefficient of 0.91 and 0.97, respectively) between binned GC fraction and the respective fragment coverage, whereas in the VATG control both attributes strongly anti-correlated (Pearson coefficient −0.88). These apparently contradictory observations cannot be satisfactorily explained just by a significantly larger range of GC content in the former two controls (ranging from ∼30% to >50% GC) as opposed to a quite tight spectrum (39–45% GC) in the latter case.
Figure 4.
Convolution of hydrolysis biases with sensitivity to GC content. The GC content of the three spike-in controls has been measured per position considering a surrounding window corresponding to the mean observed insert size (192 nt). To estimate coverage, the number of fragments that include a respective position is taken into account. Top panels: for each of 20 equally sized GC bins (x-axis), a box plot summarizes the distribution of fragment coverage (y-axis). Within their specific spectrum of GC content, Lambdaclone1-1 and OBF5 show a positive correlation of coverage with GC content, whereas for the VATG control, GC content and coverage anti-correlate. Bottom panels: the GC content (blue curve, normalised to [0;1] for each transcript) along the molecules (x-axis) is shown in comparison with the relative fragment coverage (red curve). Lambdaclone1-1 and OBF5 exhibit synchrone variations in GC content and coverage, whereas the relative GC content of VATG scores minimal around the molecule’s centre, where the mutual overlap of hydrolysis products provokes a peak in coverage (Figure 3).
The reasons behind this seemingly paradoxical dependence on GC content become clearer when considering GC-biases together with positional biases caused by fragmentation (Figure 4, bottom panels): in Lambdaclone1-1 and OBF5 fragments, coverage (red curve) declines where GC content (blue curve) drops; in VATG (Figure 4, right bottom panel), on the other hand, GC content shows a drop about the centre of the molecule where—consistent with our hydrolysis model (Figure 3)—the mutual overlap of fragment accumulations causes a coverage peak (Figure 4, left bottom panel). Similar observations hold for other control sequences from the same experiment. Interestingly, the transcript AGP, which has a length similar to that of VATG (325 nt versus 376 nt), exhibits—in contrast to VATG—a pronounced dependency of fragment coverage on GC: this is due to the fact that in the case of AGP, GC-distribution along the molecule and positional preferences of hydrolysis mutually amplify about the molecule’s centre (Supplementary Figure S7).
Sequence-selectivity at the ends of sequenced fragments
RNA-Seq is known to introduce biases not only in relation to the fraction of G and C nucleotides present in a sequence, but also for certain nucleotides towards the ends of a sequenced fragment, manifesting in motifs of bases preferred at specific positions (7). In agreement with earlier reports that predict fragment end positions by employing correspondingly observed motifs modelled as position weight matrices (PWMs), we found only moderate correlations between the observed fluctuations and the predictions based on PWMs (8). Supplementary Figure S8 depicts the effect of sequence-selectivity—which has been attributed to the enzyme–subtrate kinetics of randomly-primed reverse transcription process (7) and/or adapter ligation to cDNA molecules (16).
To alleviate such biases, a modified hydrolysis protocol is sometimes performed, where the ligation of adapter sequences to the RNA molecule comes before RT and the latter is carried out with primers specifically targeting anchor sequences in the adapters. Variants of such ‘RNA-ligation’ based methods differ in the way adapter sequences are attached to the respective 5′- and 3′-ends of RNA fragments, e.g. by the use of standard RNA ligase (17) or by poly-A polymerase and special circular ligase (18).
Both methods have been reported to improve the uniformity of read coverage along transcripts. Here we evaluate our computational models by analysing the difference between simulations with PWMs (derived from RNA-Seq data sets produced by the standard hydrolysis protocol) and the experimental results of the RNA-ligation method called FRT-Seq (as RT is performed on an Illumina flowcell), in the case of a human placental sample. In addition to the difference in substrate when ligating adapters, the FRT-protocol is PCR-free and employs no specific size selection step (17).
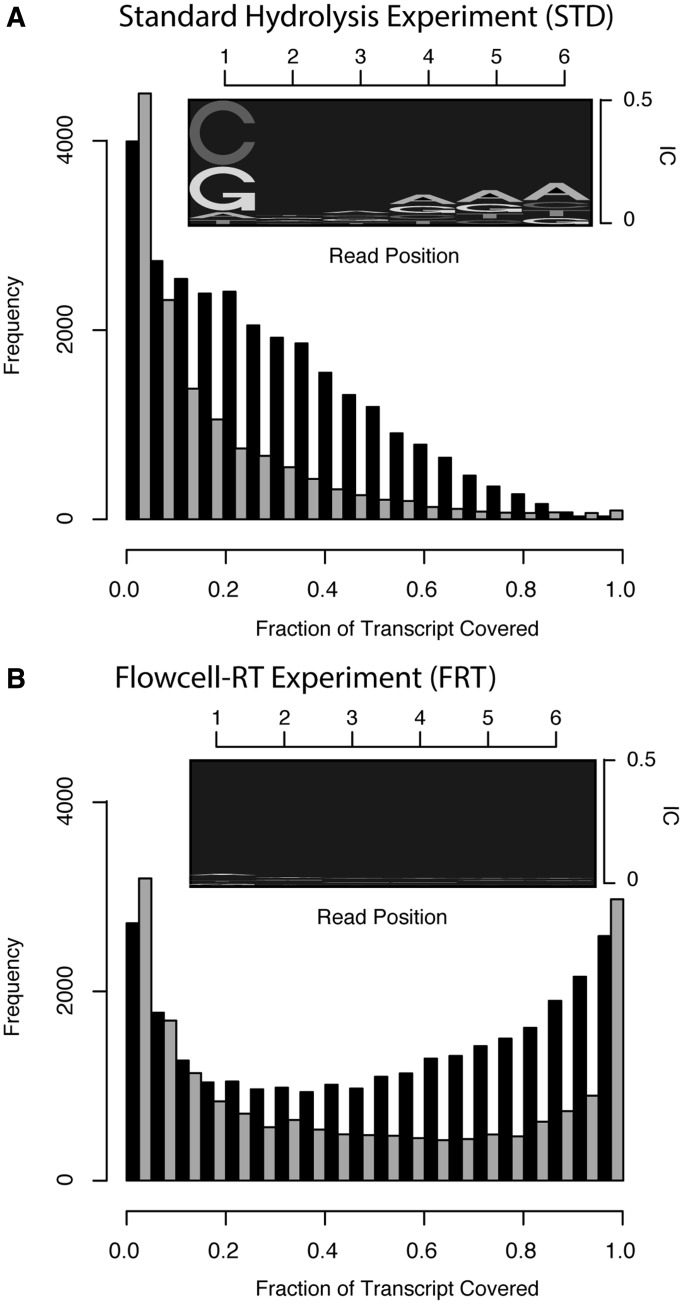
Figure 5 shows that the PWMs derived from read sequences differ substantially in the two cases. The information content, a logarithmic measure of the deviation from uniformly distributed nucleotide frequencies that correspond in the depicted sequence logos to the height of a stack of letters at every position, describes less severe biases in the FRT-Seq protocol (Figure 5A) than in the standard hydrolysis protocol (Figure 5B). Consequently, we observe a higher degree of transcript coverage in the FRT experiment (Figure 5A versus B, black bars). The trend can be reproduced in silico when providing the corresponding PWM and de-activating simulated PCR and size selection (Figure 5A and B, grey bars). Differences between the simulated and the experimental data set are mainly due to different mapping redundancies: on average ∼1.82 mappings per read are found for the experimental data set, whereas in the simulated data to every read exactly one mapping can be assigned.
Figure 5.
Correlation of sequence biases with transcript coverage in a standard hydrolysis experiment (A) and an RNA-ligation method (B). The panels above the bar plots depict sequence logos that capture biases as observed in the first six bases of sequenced reads, where the height of the letters are scaled according to the information content (IC), a logarithmic measurement that captures the divergence from equally distributed bases. The bar histogram in (A) and (B) show the corresponding transcript coverage by experimentally obtained reads (black bars), respectively, by correspondingly simulated reads (grey bars). The standard hydrolysis protocol exhibits stronger sequence selectivity that produces read stacks at specific positions and thereby reduces the overall transcript coverage.
Simulation of generic RNA-Seq experiments
We then employed the entire Flux Simulator pipeline to assess how well the models described so far—when combined—can mimic the overall distribution of reads along RNA molecules in populations of cellular transcripts. To allow the simulation of realistic transcript expression levels, we developed a transcriptome simulator: it is based on Zipf’s Law—which governs gene expression— and modified according to further empirical observations from RNA-Seq experiments (Supplementary Figure S3 and Supplementary Table S2).
Since sequencing-by-synthesis protocols produce reads whose first nucleotide identifies the fragment edge (i.e. the breakpoint) and whose mapping directionality further reveals the nature of the fragment edge (i.e. whether it constitutes a 5′- or 3′-end, Supplementary Figures S1 and S9), we separately focused on breakpoint distributions for reads mapping in sense and in antisense directions, thus preventing influences on sequence coverage by different read lengths. In our benchmark, we investigated four different experiments (i.e. the last four rows in Supplementary Table S1) that differ in species/tissue of the sequenced RNA, sample preparation and sequencing platform (9,11,28). For each data set, we provide a parameterised in silico model (Supplementary Table S3), and we compare the experimental observation with the simulation.
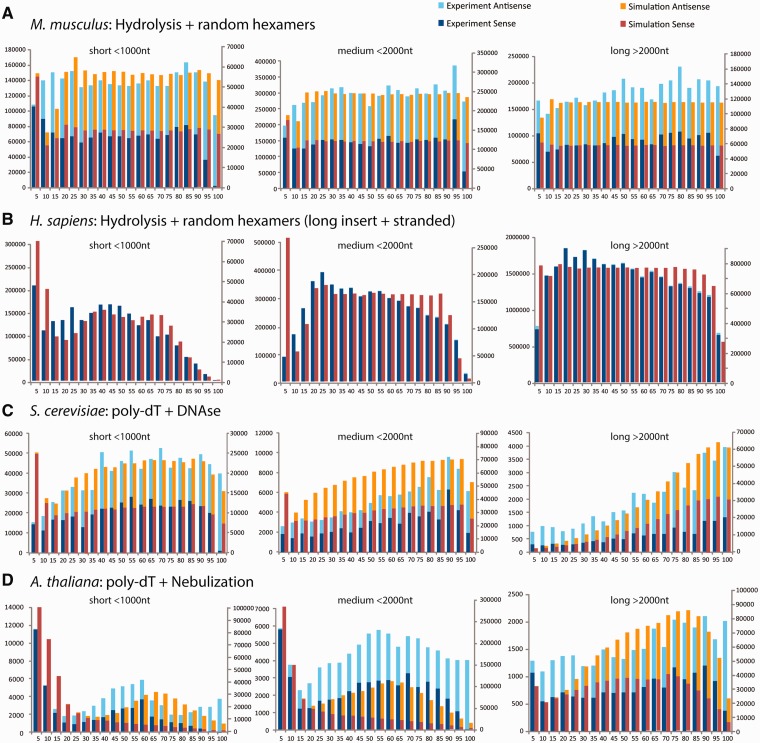
The results of our comparisons are summarized in Figure 6. As a general trend, they reproduce earlier observations (28) that reads from 5′-ends of fragments (sense mappings) generally increase close to the 5′-end of transcripts and decrease close to the 3′-end, whereas the 3′-ends of fragments (antisense mappings) exhibit an inverse effect (Figure 6). Our simulation reveals that the phenomenon is due to the fragmentation step, given that 5′/3′-ends of transcripts are also naturally included as 5′/3′-ends of some of the fragments produced from them; therefore, the fraction of transcription start sites preserved in the fragment population is higher for short transcripts that exhibit a comparatively lower number of breakpoints (Figure 6, left panels). A corresponding increase of antisense mappings is predicted by our simulations at the 3′-end of the transcripts, however, corresponding reads fall into the poly-A tail not included in Figure 6.
Figure 6.
Comparison of simulated reads with experimental evidence in different sequencing protocols. For each experiment, transcripts from a reference annotation of the corresponding species have been classified into short (<1000 nt, left panels), intermediate (1000–2000 nt, centre panels), and long forms (>2000 nt, right panels). Red and orange bars show reads from the experiment that align in sense and antisense, respectively, to the directionality of transcription, the corresponding in silico results are shown as dark and light blue bars, respectively. (A) Read tag distributions from an RNA hydrolysis protocol in M. musculus sequenced on the Illumina GA2 platform. (B) A different hydrolysis experiment carried out with the recent HiSeq2000 technology (Illumina), producing longer reads that exclusively map in sense orientation, so called ‘dir RNA-Seq’). (C) A complementary Illumina experiment employing poly-dT primed RT and subsequent DNAse digestion of the (poly-A+) transcriptome of S. cerevisisae. (D) Results from an experiment in A. thaliana where poly-dT primed RT products are fragmented by nebulisation.
In Figure 6A, we assessed the distribution of reads for the hydrolysis protocol investigated in detail in a previous section. In agreement with earlier reports (1), the transcript-specific biases we pinpoint are not identifiable when sufficiently heterogeneous molecule groups are considered together (11). Only the small reduction of reads next to the 5′-bin in transcripts with <2000 nt reflects a cumulative effect of fragments that fall along sufficiently similar Weibull distribution (left and centre panel of Figure 6A).
In Figure 6B, we compare these results with a recent adaptation of the hydrolysis protocol that has been employed to produce the Illumina Body Map 2 (accession number ERP000546 in the European Nucleotide Archive). The experiment produced reads exclusively from the sense strand of RNAs obtained from a mixture of 16 tissues (libraries HCT20170 and HCT20173). Therefore, only spurious amounts of antisense mappings can be observed which, in agreement with previous reports about antisense transcription, can be found especially at the 5′-/3′-ends of long transcripts (Figure 6B, right panel).
In this protocol, the longer reads (100 nt)—and therefore also larger fragments—cause a more accentuated drop of read density towards the 5′-end. Moreover, the use of a so-called ‘ribofree’ technology allows extracting RNA species without relying on the presence of a poly-A tail. We therefore expect the downstream ends of 3′-most fragments—which would be represented by antisense mappings absent from this experiment—are at (or close to) the respective cleavage sites. Consequently, we observe the frequency of sense mappings to decrease at positions closer than the average fragment size to 3′-end of the transcribed sequence. The effect is marginally stronger in experimental data than when reproduced in silico, indicating that additional mechanisms might play a role here. However, our simulations are able to qualitatively reproduce that 3′-regions which suffer from such read under-representation are comparatively larger in short and medium-sized transcripts (Figure 6B, left and centre panel).
To simulate the experiment depicted in Figure 6C, we replaced the uniformly random fragmentation model by enzymatic digestion with DNAseI (9) and moved it after RT, which in this protocol has been realised by poly-dT priming on the original transcript templates. Our models correctly predict the under-representation of 5′-end information in poly-dT primed RT due to the simulated template loss of the reverse transcriptase during first-strand synthesis (1)—and an increasing impact of the bias from short to long transcripts (Figure 6B, left versus centre versus right panel).
In Figure 6D, we compared simulation results to with an experiment employing cDNA nebulisation in contrast to fragmentation by DNAseI (28). Our model of mechanical breakage is able to reproduce the known bias of read distribution towards the centres of the transcripts (28), especially in shorter transcripts that break few times (Figure 6D, left and centre panels); multiple recursive breaks along the body of long transcripts thin out these points of sharp breakpoint accumulation (Figure 6D, right panel).
DISCUSSION
We present the Flux Simulator, a framework for simulating RNA-Seq experiments in silico that breaks down heterogeneous sample preparation protocols into their atomic steps (Figure 1). For each step, we provide tunable computational models with a minimal set of free parameters, whose values can be estimated by corresponding quantities observed in real experiments. The Flux Simulator pipeline implements these steps as modules that can be flexibly joined: this structure allows simulation of arbitrary protocols. In the present article we focus on several protocols employed for the currently popular Illumina and Roche 454 sequencing platforms, but the modularity of our simulation platform allows analysis of arbitrary sequencing technologies, as those announced for the future by the manufacturers Ion Torrent (34) and Pacific Biosciences (35). Although our models are largely of approximate nature and describe in a simplified way the underlying complex chemistry, we show that our simulations reproduce fairly well the read distributions observed in practice (Figure 6).
If we accept that our bioinformatics models capture the main origins of the experimental biases, our simulation enables us to investigate intermediate stages of sequencing protocols—usually hidden layers of RNA-Seq (Figures 2–4). Specifically, we give computational and experimental evidence as to why insert size distributions obtained by hydrolysis differ substantially between transcripts of different lengths (Figure 2). In the light of the uniform random fragmentation model we developed, the dependence of the RNA molecules’ geometry on their length can be interpreted as shorter molecules being more linearised when hydrolysed, whereas longer RNA polymers—in spite of strongly denaturing conditions—still tend to form higher order structures. Therefore, size filtering alters the way transcripts are represented in the library as a function of the length of the original RNA molecule.
In addition, our models show why fragments obtained by sequence-independent fragmentation processes, as for instance cDNA nebulisation or RNA hydrolysis, are not uniformly distributed along the fragmented molecule, but occur more frequently at rather specific points: the ends of nebulised fragments accumulate at the midpoints of recursively split molecules (Figure 6D), whereas fragment density obtained by RNA hydrolysis propagates from a transcript’s ends towards its centre in patterns produced by characteristic Weibull distribution of the obtained insert sizes (Figure 3). Onto these patterns one has to superimpose sequence-specific biases (Figures 4 and 5). If heterogeneous transcripts are considered together, however, the recognition of these biases on large scale is complicated (Figure 6).
As for the computational analysis of RNA-Seq experiments, we consider our simulation-based studies as a serious motivation to debunk the widespread belief that all biases should affect the interpretation of data negatively: in fact, well-understood biases of systematic nature are valuable as additional sources of information. Therefore, we are convinced that the critical evaluation of experiments mimicked in silico will have an increasing impact on design and evaluation of bioinformatics approaches to RNA-Seq.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–9, Supplementary Methods and Supplementary References [36–42].
AVAILABILITY
The Flux Simulator is implemented in platform-portable Java code (JDK compliance 1.6), source code and binaries are freely available via the webpage http://flux.sammeth.net.
FUNDING
European Science Foundation (to T.G.); Erasmus exchange grant of the European Community (to B.Z.); Post-doctoral fellowship of the Spanish Ministry of Science and Open Source license of Atlassian for their products Jira, Confluence and Fisheye (to M.S.); Spanish Ministry of Science (to R.G.) [BIO2006-03380 and CONSOLIDER CSD2007-00050]. Funding for open access charge: Bioinformatics and Genomics Program, Centre de Regulació Genòmica (CRG), 08003 Barcelona, Catalunya, Spain.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
M.S. initiated and developed the Flux Simulator, designed and performed the data analyses and wrote the manuscript. T.G. implemented the position-based error models and contributed to multiple analyses. B.Z. assessed biases of multiple data sets and developed scripts for the automatic classification of RNA-Seq experiments. P.R., E.R., V.L. and R.G. contributed with fruitful discussions. All authors approved the manuscript.
REFERENCES
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furusawa C, Kaneko K. Zipf's law in gene expression. Phys. Rev. Lett. 2003;90:088102. doi: 10.1103/PhysRevLett.90.088102. [DOI] [PubMed] [Google Scholar]
- 3.Zipf GK. Human Behavior and the Principle of Least Effort. Cambridge: Addison–Wesley; 1949. [Google Scholar]
- 4.Brakman S, Garretsen H, Van Marrewijk C, van den Berg M. The return of Zipf: towards a further understanding of the rank-size distribution. J. Regional Sci. 1999;39:739–767. [Google Scholar]
- 5.Ogasawara O, Kawamoto S, Okubo K. Zipf's law and human transcriptomes: an explanation with an evolutionary model. C. R. Biol. 2003;326:1097–1101. doi: 10.1016/j.crvi.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 6.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz S, Oren R, Ast G. Detection and removal of biases in the analysis of next-generation sequencing reads. PLoS One. 2011;6:e16685. doi: 10.1371/journal.pone.0016685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 11.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 12.Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, Gallusser FL, Izaurralde E, Rio DC, Dudoit S, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres TT, Metta M, Ottenwalder B, Schlotterer C. Gene expression profiling by massively parallel sequencing. Genome Res. 2008;18:172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surzycki S. Basic Techniques in Molecular Biology. Berlin: Springer; 2000. pp. 377–380. [Google Scholar]
- 15.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat. Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alon S, Vigneault F, Eminaga S, Christodoulou D, Seidman JG, Church GM, Eisenberg E. Bar-coding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011;21:1506–1511. doi: 10.1101/gr.121715.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mamanova L, Andrews RM, James KD, Sheridan EM, Ellis PD, Langford CF, Ost TW, Collins JE, Turner DJ. FRT-seq: amplification-free, strand-specific transcriptome sequencing. Nat. Methods. 2010;7:130–132. doi: 10.1038/nmeth.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lennon NJ, Lintner RE, Anderson S, Alvarez P, Barry A, Brockman W, Daza R, Erlich RL, Giannoukos G, Green L, et al. A scalable, fully automated process for construction of sequence-ready barcoded libraries for 454. Genome Biol. 2010;11:R15. doi: 10.1186/gb-2010-11-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory manual. NY: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1982. [Google Scholar]
- 22.Richter DC, Ott F, Auch AF, Schmid R, Huson DH. MetaSim: a sequencing simulator for genomics and metagenomics. PLoS One. 2008;3:e3373. doi: 10.1371/journal.pone.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SB, Hood LE. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 24.Iyengar SS, Quave SA. A computer model for hydrodynamic shearing of DNA. Comput. Prog. Biomed. 1979;9:160–168. doi: 10.1016/0010-468x(79)90029-1. [DOI] [PubMed] [Google Scholar]
- 25.Tenchov BG, Yanev TK, Tihova MG, Koynova RD. A probability concept about size distributions of sonicated lipid vesicles. Biochim. Biophys. Acta. 1985;816:122–130. doi: 10.1016/0005-2736(85)90400-6. [DOI] [PubMed] [Google Scholar]
- 26.Hastings WK. Monte Carlo sampling methods using Markov chains and their applications. Biometrika. 1970;57:97–109. [Google Scholar]
- 27.Metropolis N, Rosenbluth AW, Rosenbluth MN. Equations of state calculations by fast computing machines. J. Chem. Phys. 1953;21:1087–1092. [Google Scholar]
- 28.Weber AP, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 31.Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Feierbach B, Fisk DG, Hirschman JE, et al. Saccharomyces genome database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 35.Korlach J, Marks PJ, Cicero RL, Gray JJ, Murphy DL, Roitman DB, Pham TT, Otto GA, Foquet M, Turner SW. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc. Natl Acad. Sci. USA. 2008;105:1176–1181. doi: 10.1073/pnas.0710982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davidson EH. Gene Activity in Early Development. New York: Academic Press; 1976. [Google Scholar]
- 38.Martin KJ, Pardee AB. Identifying expressed genes. Proc. Natl Acad. Sci. USA. 2000;97:3789–3791. doi: 10.1073/pnas.97.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 40.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams JG. In: Genetic Engineering. Williamson R, editor. Vol. 1. Academic Press, New York; 1981. p. 2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.