Abstract
2′-O-methylation is present within various cellular RNAs and is essential to RNA biogenesis and functionality. Several methods have been developed for the identification and localization of 2′-O-methylated sites in RNAs; however, the detection of RNA modifications, especially in low-abundance RNAs and small non-coding RNAs with a 2′-O-methylation at the 3′-end, remains a difficult task. Here, we introduce a new method to detect 2′-O-methylated sites in diverse RNA species, referred to as RTL-P [Reverse Transcription at Low deoxy-ribonucleoside triphosphate (dNTP) concentrations followed by polymerase chain reaction (PCR)] that demonstrates precise mapping and superior sensitivity compared with previous techniques. The main procedures of RTL-P include a site-specific primer extension by reverse transcriptase at a low dNTP concentration and a semi-quantitative PCR amplification step. No radiolabeled or fluorescent primers are required. By designing specific RT primers, we used RTL-P to detect both previously identified and novel 2′-O-methylated sites in human and yeast ribosomal RNAs (rRNAs), as well as mouse piwi-interacting RNAs (piRNAs). These results demonstrate the powerful application of RTL-P for the systematic analysis of fully or partially methylated residues in diverse RNA species, including low-abundance RNAs or small non-coding RNAs such as piRNAs and microRNAs (miRNAs).
INTRODUCTION
The presence of post-transcriptional RNA modifications, which are catalyzed by numerous specific enzymes or ribonucleoprotein (RNP) particles, is a characteristic feature of most cellular RNAs (1). In ribosomal RNAs (rRNAs) and small nuclear RNAs (snRNAs), 2′-O-methylation of the backbone ribose is the most common and conserved type of RNA modification (2). Human rRNAs contain approximately 105 2′-O-methylated sites (3), which are predominantly directed by box C/D small nucleolar RNAs (snoRNAs) through 1 or 2 10–21 nt antisense elements that base pair to the specific region of the rRNA (2). Most 2′-O-methylated sites are clustered around functionally important regions of rRNAs and may influence ribosome structure and function (4). Ribose 2′-O-methylation also occurs within the cap structure of mRNAs and provides a molecular signature for the distinction of self versus non-self mRNA by the RNA sensor Mda5 (5). Recently, several lines of evidence have shown that small RNAs, including piwi-interacting RNAs (piRNAs), endogenous small interfering RNAs (endo-siRNAs) and plant microRNAs (miRNAs), are 2′-O-methylated at their 3′-ends by methyltransferases (6). The 3′-terminal methylation in the small RNAs protects the molecules from uridylation (7) and degradation by exonucleases (8) and may also regulate specific RNA interference (RNAi) pathways. The detection of 2′-O-methylated nucleotides and the mechanistic study of this post-transcriptional modification are important for the understanding of RNA biogenesis and function as well as the mechanisms regulating gene expression.
Many biochemical approaches have been developed to detect and analyze nucleotide modifications including 2′-O-methylation. Liquid chromatography coupled with mass spectrometry (LC/MS) (9) and two-dimensional thin-layer chromatography (2D-TLC) approaches (10,11) can yield extensive information regarding the modifications in a given RNA; however, the procedures are complex, labor intensive and usually require a large amount of starting material or the purification of an individual RNA to a homogeneous state. An alternative to these techniques is primer extension by reverse transcriptase, which does not require RNA purification, as specificity for the RNA of interest is determined by annealing specific oligonucleotide DNA primers. Two types of methods based on reverse transcription (RT) have also been developed to study RNA modifications. The first exploits the tendency of 2′-O-methyl groups to impede reverse transcriptase at a low dNTP concentration, and the second utilizes the resistance of the phosphodiester bonds adjacent to the 2′-O-methyl groups to alkaline hydrolysis (3). These traditional RT-based methods are easily preformed but are laborious and time-consuming due to the required radioactive labeling, polyacrylamide gel electrophoresis and sequencing reaction steps. In some cases, a large amount of total RNA is also needed for primer extension due to the low sensitivity of the method. Recently, the detection sensitivity of RT-based methods has been improved by using fluorescent primers followed by capillary electrophoresis (12), or by using fluorescent dye in PCR (13). A site-specific method based on the resistance of 2′-O-methylated sites to cleavage by RNase H has also been developed (14,15). This assay is highly sensitive for the detection of methylated sites and particularly applicable for the analysis of low-abundance RNAs; however, this method also requires a radiolabeling step. Saikia et al. (16) also developed a ligation-based approach for the high-throughput analysis of RNA modifications using 32P-labeled oligonucleotides. Recently, a method using RNA-cleaving deoxyribozymes (DNAzymes) has been reported that is easily executed and requires no radioactive labeling to detect 2′-O-ribose methylations in rRNAs (17); however, this method is inapplicable for the analysis of modified residues in RNAs of low abundance or modifications near the ends of the RNA molecules.
Here, we describe a new approach, RTL-P, for the detection of 2′-O-methylated sites in RNAs that uses reverse transcription at Low dNTP concentrations followed by PCR. By designing specific RT primers, RTL-P was successfully applied to identify both previously characterized and novel 2′-O-methylated sites in human rRNA, yeast rRNA and mouse piRNAs. In contrast to previously developed methods, RTL-P is highly sensitive and easy to perform. RTL-P is applicable for the high-throughput analysis of fully or partially methylated residues in diverse RNA species, including low-abundance RNAs or small non-coding RNAs. This new tool will help map the numerous post-transcriptional modifications in cellular RNAs.
MATERIALS AND METHODS
Sample preparation and RNA isolation
HEK293T (Human embryonic kidney 293T) cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco) and incubated at 37°C in a humidified chamber with 5% CO2. Total RNA was extracted from the HEK293T cells and adult C57BL/6 mouse testes using TRIzol (Invitrogen). The testes small RNA (<40 nt) was fractionated using the flashPAGE system (Ambion). Schizosaccharomyces pombe wild-type cells and the mutant haploid strain sp972 (△snR61), which was constructed by our laboratory, were grown in rich yeast extract, peptone, dextrose (YPD) medium at 30°C. Total RNA was isolated from S. pombe as previously described (18). The RNA samples were treated with DNase I (Takara) to remove residual DNA contamination.
The detection of ribose-methylated nucleotides by RTL-P
For the detection of 2′-O-methylated sites in the target rRNAs, RT-PCR was performed according to conventional methods with the following modifications. RT was performed in 25 μl reaction mixture containing 2–200 ng of total RNA, 1 μl (10 mM) specific RT primers and a low (0.5–4 μM) or high (40 μM–1 mM) concentration of dNTPs. The primer/RNA mixture was denatured at 70°C for 5 min and then chilled on ice. Following an initial annealing step at 42°C for 10 min, 200 U of M-MLV reverse transcriptase (Promega) and 0.5 U RNasin Ribonuclease Inhibitor (Promega) were added. The reaction was incubated at 42°C for 1 h and then heated at 75°C for 15 min to deactivate the reverse transcriptase. The resulting yeast extract, peptone, dextrose (YPD) product was directly added to subsequent amplification reactions.
For the detection of methylated sites at the 3′-ends of mouse miRNAs or piRNAs, the small RNA fraction (<40 nt) was isolated using the flashPAGE™ Fractionator (Ambion) and ligated to a 3′ RNA adapter using T4 RNA ligase (Takara). The ligation product was then reverse-transcribed into complementary DNA (cDNA) using a low (0.4 μM) or high (40 μM) dNTP concentration with or without anchored RT primers that were designed to anchor the modified nucleotide. The cDNA was subsequently amplified by PCR with specific primers under the same reaction conditions.
For the detection of partially methylated sites in RNA species, the RNA oligonucleotides with or without a 2′-O-methylated adenine nucleotide were synthesized (Guangzhou RiboBio Co., Ltd) and mixed at different ratios in three experimental runs. RT was performed in 25 μl reaction mixture containing 2 ng of total RNA oligonucleotides, 1 μl (10 mM) specific RT primers and a low (0.5 μM) concentration of dNTPs. The cDNA was amplified by PCR with specific primers for 13 cycles.
The PCR reaction mixtures contained 1.25 U ExTaq DNA polymerase (Takara), 0.5 μl of dNTPs (10 mM each), 0.5 μl of forward and reverse primers (10 mM), 20 μl 1 × PCR buffer and 2 μl of the cDNA template. PCR reactions were performed as follows: one cycle of 94°C for 4 min followed by cycles at 94°C for 20–30 s and at 60°C for 20–30 s. The number of PCR cycles performed was determined based on the template concentration. Generally, an optimum range of cycle number should be carefully checked by performing PCR assays with a serial of cycles. The PCR products were then equally loaded and separated on 1.5–2.5% agarose gels, stained with GelRed dye (Biotium) and visualized by UV trans-illumination. PCR signal intensities were analyzed using Bio-Rad Quantity One software or ImageMaster TotalLab software.
The detection of ribose-methylated nucleotides by primer extension
Total RNA from S. pombe was subjected to primer extension at varying concentrations of dNTPs to detect 2′-O-methylation as previously described (19,20). Briefly, ∼30 µg of total yeast RNA was mixed with the snR61-RT primer labeled with [γ-32P]ATP at the 5′-end in 20 μl of 1 × RT buffer (Promega). After a denaturation step at 65°C for 5 min, hybridization was performed at 42°C for 10 min. Primer extension with 200 U M-MLV reverse transcriptase (Promega) was performed on two aliquots of the RNA in parallel in the presence of either 4 μM or 0.15 mM dNTPs at 42°C for 1 h. The S. pombe 25S ribosomal DNA (rDNA) was cloned into the pMD-18T vector (Takara). The plasmid was then subjected to alkaline hydrolysis and sequenced with the corresponding primer used in the RT reaction. The cDNA products were separated on 8% polyacrylamide-8M urea gels with sequenced 25S rDNA in parallel and analyzed by a Storm 820 Phosphorimager (Molecular Dynamics).
Primers and oligonucleotides
The sequences of the primers and oligonucleotides used in this study are listed in Supplementary Table S1.
RESULTS
The principle of RTL-P for the detection of 2′-O-methylated RNA nucleotides
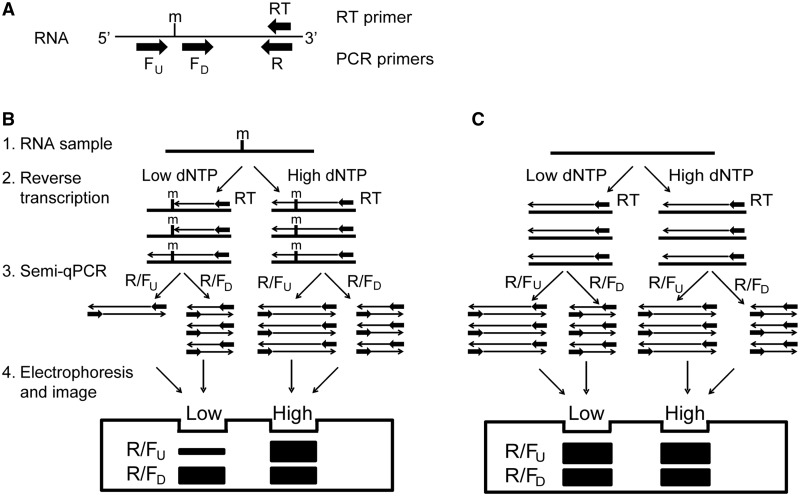
Reverse transcriptase pauses immediately preceding a 2′-O-methylated nucleotide and tends to terminate cDNA synthesis at a low dNTP concentration (19,21). Based on this principle, we developed a novel strategy to detect specific 2′-O-methylated sites in RNA by combining primer extension by reverse transcriptase at a low or high dNTP concentration with a subsequent PCR amplification (Figure 1).
Figure 1.
The RTL-P approach for detecting the presence of 2'-O-methylation in RNA. (A) A schematic of the primer-design strategy for RTL-P. The orientations of the primers are indicated by the arrows. (B and C) A schematic illustration of the RTL-P procedure used to detect the presence (B) or absence (C) of 2'-O-methylated sites in RNA. (1) RNA preparation by DNase I treatment to remove the DNA contaminants. (2) RT with a RT primer at a low or high dNTP concentration. At a low dNTP concentration, the RT reaction is impeded by the 2'-O-methyl groups, resulting in shorter RT products. At a high dNTP concentration, the RT reaction is not impeded and therefore does not produce shorter cDNA products. (3) PCR with different PCR primer pairs to amplify the RT products. The PCR reaction with the R/FU primer pair only amplifies the longer cDNA products, but amplification with the R/FD primer pair generates both the longer and shorter cDNA products. For RNAs with methylation sites, the quantity of the R/FU PCR product is less than the R/FD PCR product when the RT products generated at a low dNTP concentration are used as the amplification template. However, the quantity of the PCR product generated by the R/FU primers is equal to the R/FD PCR product when the RT products generated at a high dNTP concentration are used as the template. (4) Gel electrophoresis of the RT-PCR products. If the 2'-O-methylated site is present in the RNA analyzed, the R/FU band is weaker than the R/FD band in the low dNTP lane. In contrast, the R/FU band is nearly equal to or stronger than the R/FD band in the high dNTP lane. The intensity difference depends on the length of the PCR products generated from the R/FU primers versus the R/FD primers. m = 2'-O-methylation.
A specific set of designed RT-PCR primers for RTL-P is shown in Figure 1A. The primer used in the RT reaction was downstream of the predicted 2′-O-methylated site. Two forward primers for the subsequent PCR amplification were designed near each other and located either downstream (FD) or upstream (FU) of the predicted 2′-O-methylated site. The reverse PCR primer (R) was downstream of the FD.
The principle of the RTL-P method is depicted in Figure 1B and C. DNase-treated RNA samples were reverse-transcribed into cDNA with the RT primer at either a low or high dNTP concentration. The modified residues should induce a pause in the RT profile at a low dNTP concentration; therefore, the RT reaction with a low dNTP concentration should produce shorter cDNA products than the RT reaction with a high dNTP concentration. Each RT product was then analyzed by PCR with the designed PCR primer pairs. During the PCR, the R/FU primer pair should only amplify the long cDNA products, whereas the R/FD primer pair should amplify both the long and short cDNA products. When the RT products that had been generated using a low dNTP concentration were used as the DNA template for the PCR, the quantity of the PCR product produced using the R/FU primer pair should be less than the quantity produced by the R/FD primer pair. In contrast, when the RT products that had been generated using a high dNTP concentration were used as the DNA template, the quantity of the PCR products should be equal for both primer sets. Finally, equal volumes of the R/FU PCR product and the R/FD PCR product from the RT templates produced with a low or high dNTP concentration, respectively, are mixed. The PCR product yield and quality is then analyzed by gel electrophoresis. If the ribose 2′-OH of the residue in the target RNA is methylated, the PCR band intensity of the longer product produced by the R/FU primer pair should be weaker than the shorter product produced by the R/FD primer pair in the lane containing DNA amplified from the RT with a low dNTP concentration. In contrast, the intensity of the longer product will be slightly stronger than the shorter product in the lane containing DNA amplified from the RT with a high dNTP concentration due to the larger incorporation of dye within a longer template (Figure 1B). However, if the ribose 2′-OH is unmodified, the band intensity of the R/FU PCR product will be stronger than that of the R/FD product with both of the RT reactions produced with a low or high dNTP concentration (Figure 1C).
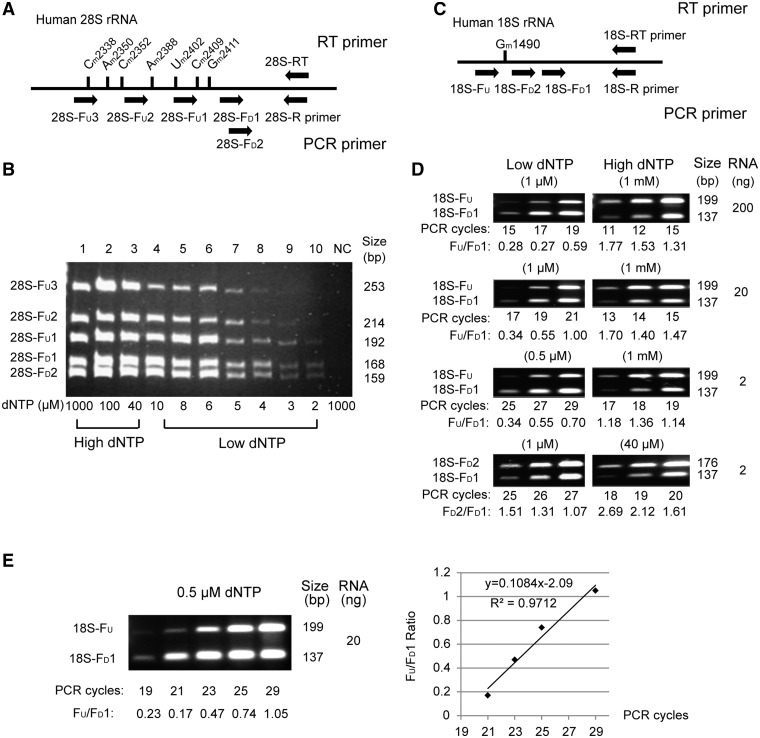
The detection of 2′-O-methylated nucleotides in rRNAs by RTL-P
To test the feasibility of our method, we first examined two cases, including a region of the human 28S rRNA with multiple methylation sites (Cm2338, Am2350, Cm2352, Am2388, Um2402, Cm2409 and Gm2411) and a single 2′-O-methylated site (Gm1490) in the human 18S rRNA from HEK293T cells. As shown in Figure 2A, we designed the 28S-RT primer downstream of the 2′-O-methylated nucleotides for the RT reaction as well as one reverse primer (28S-R primer) and five forward primers (28S-FD1, 28S-FD2, 28S-FU1, 28S-FU2 and 28S-FU3) for the semi-quantitative PCR. The 28S-FU1, 28S-FU2 and 28S-FU3 primers were located upstream of the various 2′-O-methylated sites, and the 28S-FD1 and 28S-FD2 primers were located downstream of the last 2′-O-methylated site, Gm2411. No 2′-O-methylated sites were located in the region between the binding sites of the 28S-FD1 and 28S-FD2 primers. The results of the semi-quantitative RT-PCR at varying dNTP concentrations (from 2 μM to 1 mM) are shown in Figure 2B. As expected, the tendency of the 2′-O-methyl groups to impede RT at a low dNTP concentration (<40 μM) was clearly detected. More 2′-O-methylated sites led to a reduced quantity of RT-PCR products at lower dNTP concentrations. No obvious differences between band intensities were observed between the RT-PCR products generated from the 28S-FD1 and 28S-FD2 primers as no 2′-O-methylated sites were located in the binding regions of these two primers. These results demonstrate that RTL-P can be applied for the detection of 2′-O-methylation in rRNA.
Figure 2.
The detection of 2′-O-methylation in human rRNAs by RTL-P. (A) A schematic diagram of the primer design for RTL-P to detect a region enriched with 2'-O-methylated nucleotides (Cm2338, Am2350, Cm2352, Am2388, Um2402, Cm2409, Gm2411) in the human 28 S rRNA. (B) The detection of several 2'-O-methylated sites in human 28 S rRNA by RTL-P at different dNTP concentrations. Twelve PCR cycles were used for the amplification. Approximately 200 ng of total RNA was used in each RT reaction. (C) A schematic of the primer-design for RTL-P to detect the Gm1490 methylation in the human 18 S rRNA. (D) The detection of Gm1490 by RTL-P under different template concentrations, dNTP concentrations and PCR cycles. (E) Correlation of signal intensity ratio of amplification products to the PCR cycle number for RTL-P assay at low dNTP concentrations. The ratios of PCR signal intensity are indicated beneath each lane. A linear range of signal ratios versus cycle number is shown in the right panel. All assays were independently performed at least three times with the representative results of a single experiment shown in the figure.
Next, we tested the sensitivity of this method by attempting to detect a single known 2′-O-methylated site, Gm1490, in the human 18S rRNA. As shown in Figure 2C, Gm1490 is between the binding sites of the 18S-FU and 18S-FD1 or 18S-FD2 primers, and no 2′-O-methylated sites were located in the region between the 18S-FD1 and 18S-FD2 primer-binding sites. Figure 2D shows the results of RTL-P detection of the 2′-O-methylation at Gm1490 using 200 ng, 20 ng or 2 ng of total RNA in each RT reaction at a low (0.5–1 μM) or high (40 μM–1 mM) dNTP concentration and the comparison of the RT-PCR products using three different PCR cycles. In each assay, the band intensity of the 18S-FU RT-PCR product was obviously weaker (signal ratio <1) than the 18S-FD1 RT-PCR product at a low dNTP concentration and stronger (signal ratio >1) with a high dNTP concentration. Since no 2′-O-methylation sites were located between the binding sites of the 18S-FD1 and 18S-FD2 primers, the signal intensity of the 18S-FD2 RT-PCR product was always stronger than the 18S-FD1 product (signal ratio >1) regardless of the dNTP concentration used during the RT reaction. These results demonstrate that only 2 ng of total RNA for the RT reaction was sufficient to detect the presence of a single 2′-O-methylated site in a RNA molecule using the RTL-P method.
To clarify the linearity during amplification at low dNTP concentrations, we further compared the signal intensity ratios of PCR products (FU/FD1) amplified with different number of cycles (Figure 2E). The PCR assay showed a linear increase in signal ratio values (0.17–1.05) with the incremental PCR cycle numbers from 21 to 29. The ratios of PCR signal intensity correlated well with the PCR cycles (R2 = 0.9712).
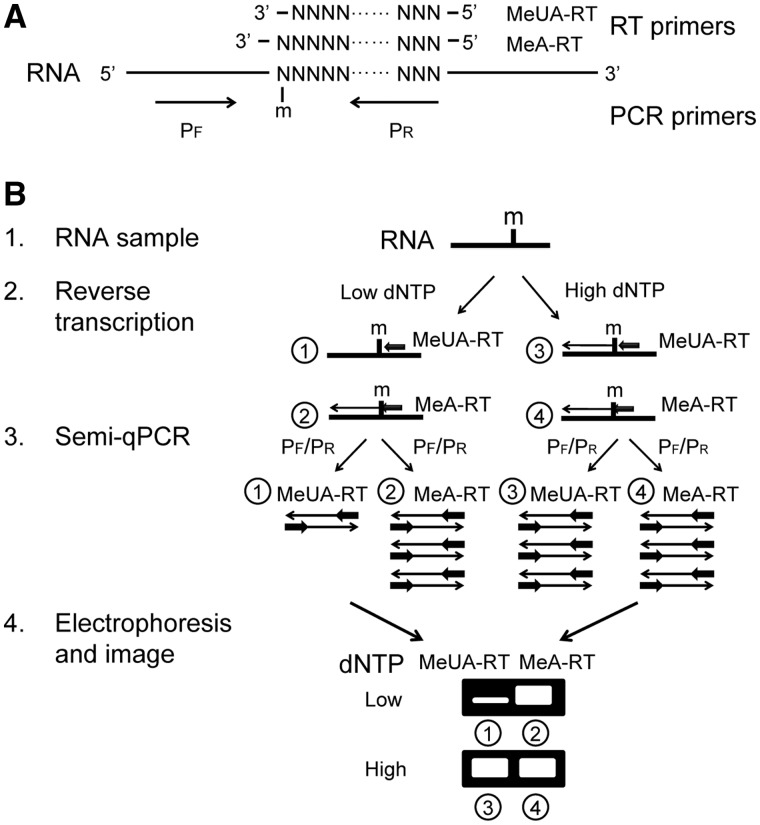
An alternative RTL-P strategy for determining the exact location of a 2′-O-methylated site within RNA
Although the method described above is sensitive and easily performed, it can only be used to detect the presence or absence of a 2′-O-methylated site within the region between the PCR primer-binding sites and cannot be used to determine which specific site within that region was 2′-O-methylated. To solve this problem, we developed a new RTL-P based strategy that used different RT primer sets to anchor the methylated site (Figure 3).
Figure 3.
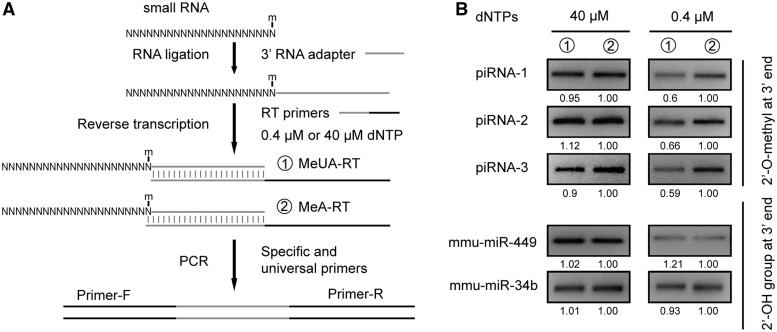
The RTL-P strategy for the detection of the exact location of a 2'-O-methylated site in RNA. (A) A schematic diagram of the primer design for the RT reaction and the PCR. m = 2'-O-methylation. (B) A schematic representation of the RTL-P procedure used to detect the exact location of the 2'-O-methylation. (1) RNA preparation by DNase I treatment to remove the DNA contaminants. (2) RT with the MeUA-RT or MeA-RT primers in different reaction tubes (①–④) at low or high dNTP concentrations, respectively. At a low dNTP concentration, the RT reaction with the UA-RT primer (①) is impeded by the 2'-O-methyl group and produces less cDNA products than that the MeA-RT primer (②). At a high dNTP concentration, the cDNA products generated with MeUA-RT primer (③) is equal to the MeA-RT primer (④). (3) PCR with the same PCR primer pair PF/PR to amplify the RT products. The quantity of PCR product generated from the MeUA-RT cDNA (①) is less than the quantity from the MeA-RT cDNA (②) when the residue of interest is 2'-O-methylated. In contrast, the quantity of PCR product generated from the MeUA-RT (③) and MeA-RT cDNAs (④) are equal when the residue analyzed is unmethylated. (4) Gel electrophoresis of the RT-PCR products. If the analyzed nucleotide is 2'-O-methylated, the band of the RT-PCR products generated from the MeUA-RT cDNA (①) is weaker than the band of the products generated from the MeA-RT cDNA (②) when a low concentration of dNTPs is used in the RT; however, the bands of the RT-PCR products generated from the MeUA-RT cDNA (③) and the MeA-RT cDNA (④) are equal when a high concentration of dNTPs is used. If the analyzed nucleotide is unmodified, no differences in band intensities of the RT-PCR products are observed regardless of the concentration of dNTPs used. N = any nucleotide base.
For this method, two RT primers, a methylated site unanchored primer (MeUA-RT) and a methylated site anchored primer (MeA-RT), are designed to the +1 and 0 nt downstream of the predicted methylated residue. The forward and reverse primers used for the PCR are located upstream and downstream of the methylated site (Figure 3A). The RNA samples are reverse-transcribed into cDNAs with the MeUA-RT or MeA-RT primers at a low or high dNTP concentration. At the low dNTP concentration, the extension of the MeUA-RT primer by reverse transcriptase pauses at the site with 2′-O-methylation, whereas the extension of the MeA-RT primer does not. This results in different concentrations of the RT products, which can be further amplified by PCR. In contrast, at a high dNTP concentration, the methylated residue usually does not affect the MeUA-RT primer extension; therefore, no significant concentration differences are seen between the RT or PCR products generated by the MeUA-RT and MeA-RT primer extension. If the ribose 2′-OH of the predicted residue was methylated, the band of the RT-PCR products generated using the different RT primers showed a dNTP concentration dependence as analyzed by gel electrophoresis. The band of the MeUA-RT product was much weaker than the MeA-RT product when a low dNTP concentration was used, but no significant differences in band intensities were observed between the MeUA-RT and MeA-RT products at a high dNTP concentration (Figure 3B). In contrast, if the ribose 2′-OH was unmodified, the band intensities of the MeUA-RT or MeA-RT products were independent of the dNTP concentration used and appeared equal on the gel. The exact location of the 2′-O-methylation site could be accurately determined by comparing the RT-PCR products generated by the MeUA-RT and MeA-RT primers at the low dNTP concentration.
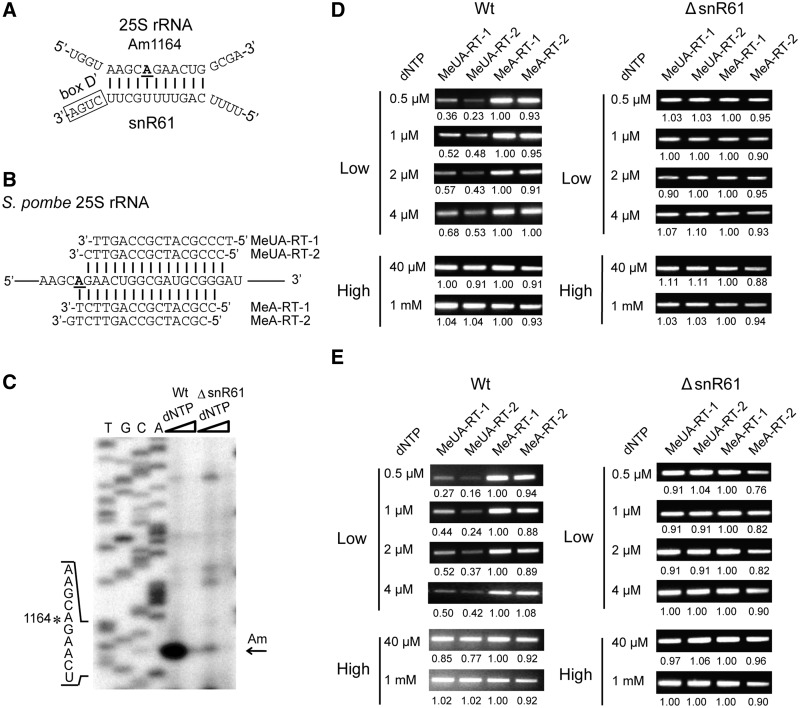
Precise mapping of a new 2′-O-methylated site in S. pombe rRNA by RTL-P
The 2′-O-methylation of Am1164 in the 25S rRNA of S. pombe was predicted to be guided by a novel box C/D snoRNA, snR61 (accession number AJ617330), which possesses an 11-nt-long antisense stretch complementary to the 25S rRNA (Figure 4A). To test whether this methylation was dependent on snR61, we designed a set of RT primers, including two MeUA-RT (MeUA-RT-1/2) and two MeA-RT (MeA-RT-1/2) primers as well as PCR primers to detect the modification site (Figure 4B). The experiment could have been performed using only the two RT primers (MeUA-RT-2 and MeA-RT-1) instead of the four RT primers; nevertheless, the use of the other two primers (MeUA-RT-1 and MeA-RT-2) proved useful for interpretation of the results. The RT primer extension was performed using either 20 ng (Figure 4D) or 2 ng (Figure 4E) total RNA per RT reaction, which was followed by PCR using the RT products as DNA templates. As expected, the MeUA-RT PCR products were obviously less than the MeA-RT products at low dNTP concentrations (<4 μM) in wild type S. pombe; however, the band intensities of the RT-PCR products were approximately equal when the RT products were generated using high dNTP concentrations (>40 μM). The same experiment was then performed on the ΔsnR61 S. pombe mutant strain, which is deficient for the Am1164 methylation of the 25S rRNA. As expected, no significantly different intensities of the RT-PCR products generated from the MeUA-RT or MeA-RT primers were found in the low or high dNTP lanes (Figure 4E). The traditional RT sequencing method was also performed to verify the methylation site in the 25S rRNA of S. pombe with 30 μg total RNA (Figure 4C). These results show that our novel method can be applied to accurately detect modification sites in rRNAs.
Figure 4.
The detection of the Am1164 2'-O-methylation in the 25S rRNA of S. pombe by RTL-P. (A) The potential base-pairing interaction between the snR61 snoRNA and its target region in the 25S rRNA of S. pombe. (B) A schematic diagram of the RT primer design. (C) An analysis of Am1164 by the traditional primer extension method in wild type (Wt) and mutant (△snR61) S. pombe. The primer extension was performed in the presence of increasing dNTP concentrations (4 μM and 0.15 mM). A subset of the rRNA sequence is indicated on the left. The position of the reverse transcriptase stop site is indicated by an arrow, and the methylated site is marked with an asterisk. T/G/C/A = the rDNA sequence ladders. (D and E) The detection of Am1164 by RTL-P at varying dNTP concentrations with 20 ng (D) and 2 ng total RNA (E) in each RT reaction. All assays were independently carried out at least three times, and the representative results of a single experiment are shown. The ratio of PCR signal intensity is shown beneath each lane.
The detection of a 2′-O-methylated site in the 3′-end of small RNAs
The 3′-end of small RNAs, including animal piRNAs, endo-siRNAs and plant miRNAs, are fully methylated. To test the capability of RTL-P to detect methylation on endo-siRNAs, we assayed three individual mouse piRNAs (piRNA-1, piRNA-2 and piRNA-3) as well as two miRNAs from mouse testis as controls (miR-449 and miR-34 b). The mouse piRNA-1 [corresponding to piR-131190 (22)] and the other piRNAs have been verified to be 2′-O-methylated at their 3′-terminal nucleotide (23,24). The experimental RTL-P procedure was modified slightly to allow for RT-primer binding (Figure 5A). The small RNAs analyzed were first ligated to a RNA adapter, which provided a binding site for the RT primer. The results of the modified RTL-P showed the piRNAs were methylated at the 3′-termini (Figure 5B). In contrast, RTL-P did not detect the modification at the 3′-termini of the control mouse miRNAs, miR-449 and miR-34 b (Figure 5B), which is consistent with the data that demonstrates animal miRNAs may not be modified unlike their counterparts in plants (25).
Figure 5.
The detection of 2'-O-methylation at the 3'-ends of small RNAs. (A) An experimental scheme for the detection of 2′-O-methylation by RTL-P. (B) The detection of 2'-O-methylation at the 3'-ends of piRNAs and miRNAs. The 3' modification of small RNAs (piRNAs) is identified by the difference in the signal intensities of the RT-PCR products produced with an unanchored (①) or anchored RT primer (②) in the RT reaction at a low concentration of dNTPs (0.4 μM). In contrast, no significant differences in the signal intensities of the RT-PCR product are observed for 3' non-modified small RNAs (miRNAs) regardless of the dNTP concentration or primer (i.e. anchored or unanchored) used in the RT reaction. The ratio of PCR signal intensity is shown beneath each lane. Each assay was performed at least three times and the results of a representative assay are shown. N = any nucleotide base.
The detection of partially modified RNA species
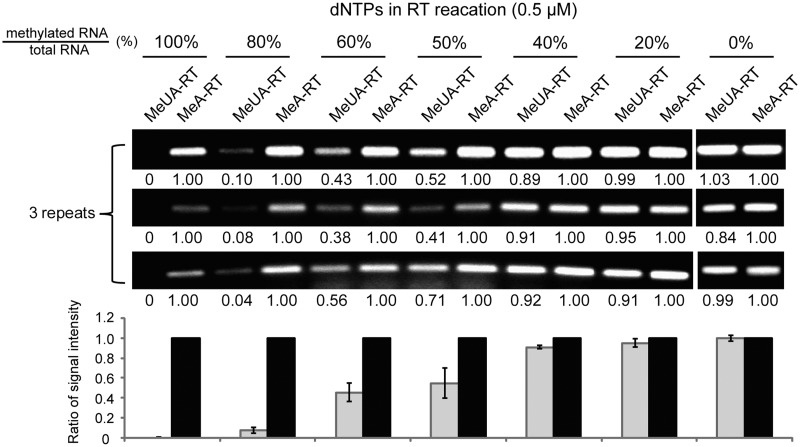
Generally, the rRNAs and piRNAs are fully 2′-O-methylated. To test whether RTL-P can be used for partially modified RNA species, we examined the sensitivity of RTL-P by using in vitro synthesized RNA substrates containing a 2′-O-methylation site but diluted with increasing amounts of the identical but unmodified RNA sequences. Our result demonstrated that RTL-P can effectively detect the partially modified RNA species at least with 50% of 2′-O-methylation at a given position (Figure 6).
Figure 6.
The detection of partially 2'-O-methylated sites in RNA species. The percentage of RNA substrates with a 2'-O-methylation site in total RNA detected is shown above each lane; the MeUA-RT/MeA-RT signal intensity ratio is indicated beneath each lane.
DISCUSSION
We have developed a novel approach, RTL-P, to detect 2′-O-methylated sites in RNA molecules. RTL-P does not require expensive reagents, sophisticated procedures or dangerous radioactive labeling and can be performed in any molecular biology laboratory with standard equipment. By designing specific RT primers, the approach can be applied not only to screen transcripts to detect the presence or absence of methylated sites but also to detect the exact location of the modified sites in the RNAs of interest.
Compared with previously reported methods, RTL-P is easier to perform and more sensitive. Although traditional RT-based methods enhance their sensitivity by utilizing radiolabeled probes, large amounts of starting material (∼2–20 μg total RNA) is usually required for the primer extension reaction depending on the concentration of the RNA of interest (1). In contrast, RTL-P employs PCR to amplify the target cDNAs to increase the detection sensitivity. With the RTL-P method, the cDNA product generated from 2 ng of total RNA was enough to detect the 2′-O-methylated residue in the human 18 S rRNA (Figure 2). This result demonstrated that the sensitivity of RTL-P is nearly 1000 times greater than traditional RT-based methods. Compared with two other RT based approaches (12,13) that apply fluorescent primers or fluorescent dye such as SYBR green to increase the detection sensitivity, RTL-P needs no sophisticated procedures such as capillary electrophoresis or expensive reagents including fluorescent dye. Moreover, RTL-P can be used to effectively detect the partially 2′-O-methylated site at a given position (Figure 6). Our data revealed that the extent of methylation has a direct impact on the efficiency of RT reaction. Partial modification with more than 50% of 2′-O-methylation at a given position of RNA species can significantly impede the RT reaction at a low dNTP concentration.
In the RTL-P assay, different PCR products (i.e. R/Fu and R/FD products in the Figure 2D) mixed together before the ratio of their intensity signals were analyzed by gel electrophoresis. Alternatively, it might render the RTL-P approach easier and reduce the experiment error if the multiplex PCR was performed in a single tube [Reverse Transcription at Low dNTP concentrations followed by Multiplex PCR (RTL-MP)] in which different PCR primers were mixed together (Supplementary Figure S1). However, multiple primers used in a single tube increase the possibility of primer–dimer formation. Researchers have to perform the ‘pre-PCR’ to mitigate this problem by a laborious primer design and validation process before using RTL-MP.
RTL-P serves as a powerful tool for the localization of modified residues in low-abundance cellular RNAs, including small non-coding RNAs. 2′-O-methylation is an important characteristic for certain small RNAs, including animal piRNAs, endo-siRNAs and plant miRNAs. Therefore, identification of 2′-O-methyl groups in the 3-RNA termini helps to distinguish the different types of small RNAs. However, modifications located too close to the RNA termini, including RNAs shorter than 40 nt, such as piRNAs and miRNAs, are difficult to detect with traditional RT-based approaches, which require annealing an oligonucleotide primer to the RNA 3′-termini. In this study, we solved this issue by ligating a RNA adapter to the 3′-end of the small RNAs, which allowed the successful application of RTL-P to detect the modified nucleotides at the 3′-end of piRNAs. These results suggest that RTL-P can be applied to the other small RNA species for the detection of 2′-O-methylation. Additionally, with this ligation strategy, the RNA adapter could also serve as a universal binding site for the RT primer allowing for the use of RTL-P for the high-throughput analysis of 2′-O-methylation in small RNAs.
Similar to traditional RT-based approaches, RTL-P requires that the region of the RNA targeted by the RT primers is accessible and not hidden within a stable secondary structure, such as a stem loop. These secondary structures and some sequences can also induce RT pauses, which could cause the identification of false positive modifications. In some cases, performing the RT reaction at ≥42°C temperatures may serve as a solution to this problem. Therefore, other reverse transcriptases, such as the avian Mieloblastosis virus (AMV) reverse transcriptase, would be more applicable than the Moloney murine leukemia virus (M-MLV) reverse transcriptase used in this study due to its higher thermostability (up to 70°C) (26) and decreased sensitivity to RNA secondary structures (27). It is noted that the number of PCR cycles greatly influences the RT-PCR profile. In general, the less starting templates are used for subsequent PCR, the more PCR cycles will be required for an optimal ratio of PCR signal intensities (Figure 2D). Although the ratio of PCR signal intensity correlated with the PCR cycles (Figure 2E), the linear range of signal ratios versus cycle number is limited. No differences in product quantity can be detected if the PCR reaction is allowed to reach the plateau phase during later cycles. Therefore, an optimum range of cycle number should be carefully determined by performing PCR assays with a serial of cycles to find the optimal ratio. Comparing the quantity of the RT-PCR products generated under different experimental conditions, such as varied dNTP concentrations in the RT reaction and different cycles of PCR, generally yields quite reliable results. However, it should be noted that several 2′-O-methylation sites in rRNAs do not cause any pauses (19), which cannot be detected by the RT-based methods or by RTL-P. The use of other complementary RNA analysis techniques, including OH- cleavage or 2′-OH reactivity, LC/MS and 2D-TLC, is useful to confirm the modifications detected by the RTL-P method and to detect novel methylated sites.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figure 1.
FUNDING
Funding for open access charge: National Basic Research Program of China [No. 2011CB811300]; National Natural Science Foundation of China [No. 30830066, 31000571, 30900249 and 30870530]; Guangdong Provincial Natural Science Foundation of China [No. 10451027501005659 and No. 9451027501002892]; Ph.D. Programs Foundation of Ministry of Education of China [No.20090171120032].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 2.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 3.Maden BE. Mapping 2′-O-methyl groups in ribosomal RNA. Methods. 2001;25:374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- 4.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 5.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douthwaite S, Kirpekar F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007;425:1–20. doi: 10.1016/S0076-6879(07)25001-3. [DOI] [PubMed] [Google Scholar]
- 10.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 11.Grosjean H, Keith G, Droogmans L. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol. Biol. 2004;265:357–391. doi: 10.1385/1-59259-775-0:357. [DOI] [PubMed] [Google Scholar]
- 12.Deryusheva S, Gall JG. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol. Biol. Cell. 2009;20:5250–5259. doi: 10.1091/mbc.E09-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belin S, Beghin A, Solano-Gonzalez E, Bezin L, Brunet-Manquat S, Textoris J, Prats AC, Mertani HC, Dumontet C, Diaz JJ. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One. 2009;4:e7147. doi: 10.1371/journal.pone.0007147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu YT, Shu MD, Steitz JA. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 15.Lapham J, Yu YT, Shu MD, Steitz JA, Crothers DM. The position of site-directed cleavage of RNA using RNase H and 2′-O-methyl oligonucleotides is dependent on the enzyme source. RNA. 1997;3:950–951. [PMC free article] [PubMed] [Google Scholar]
- 16.Saikia M, Dai Q, Decatur WA, Fournier MJ, Piccirilli JA, Pan T. A systematic, ligation-based approach to study RNA modifications. RNA. 2006;12:2025–2033. doi: 10.1261/rna.208906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchhaupt M, Peifer C, Entian KD. Analysis of 2′-O-methylated nucleosides and pseudouridines in ribosomal RNAs using DNAzymes. Anal. Biochem. 2007;361:102–108. doi: 10.1016/j.ab.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat. Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- 19.Maden BE, Corbett ME, Heeney PA, Pugh K, Ajuh PM. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995;77:22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- 20.Zhou H, Chen YQ, Du YP, Qu LH. The Schizosaccharomyces pombe mgU6-47 gene is required for 2′-O-methylation of U6 snRNA at A41. Nucleic Acids Res. 2002;30:894–902. doi: 10.1093/nar/30.4.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu HL, Michot B, Bachellerie JP. Improved methods for structure probing in large RNAs: a rapid 'heterologous' sequencing approach is coupled to the direct mapping of nuclease accessible sites. Application to the 5′-terminal domain of eukaryotic 28S rRNA. Nucleic Acids Res. 1983;11:5903–5920. doi: 10.1093/nar/11.17.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 23.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′-termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- 24.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3′-termini of mouse Piwi-interacting RNAs are 2'-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- 25.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs B, Zhang K, Rock MG, Bolander ME, Sarkar G. High temperature cDNA synthesis by AMV reverse transcriptase improves the specificity of PCR. Mol. Biotechnol. 1999;12:237–240. doi: 10.1385/MB:12:3:237. [DOI] [PubMed] [Google Scholar]
- 27.Otsuka Y, Schoenberg DR. Approaches for studying PMR1 endonuclease-mediated mRNA decay. Methods Enzymol. 2008;448:241–263. doi: 10.1016/S0076-6879(08)02613-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.