Abstract
RlmM (YgdE) catalyzes the S-adenosyl methionine (AdoMet)-dependent 2′O methylation of C2498 in 23S ribosomal RNA (rRNA) of Escherichia coli. Previous experiments have shown that RlmM is active on 23S rRNA from an RlmM knockout strain but not on mature 50S subunits from the same strain. Here, we demonstrate RlmM methyltransferase (MTase) activity on in vitro transcribed 23S rRNA and its domain V. We have solved crystal structures of E. coli RlmM at 1.9 Å resolution and of an RlmM–AdoMet complex at 2.6 Å resolution. RlmM consists of an N-terminal THUMP domain and a C-terminal catalytic Rossmann-like fold MTase domain in a novel arrangement. The catalytic domain of RlmM is closely related to YiiB, TlyA and fibrillarins, with the second K of the catalytic tetrad KDKE shifted by two residues at the C-terminal end of a beta strand compared with most 2′O MTases. The AdoMet-binding site is open and shallow, suggesting that RNA substrate binding may be required to form a conformation needed for catalysis. A continuous surface of conserved positive charge indicates that RlmM uses one side of the two domains and the inter-domain linker to recognize its RNA substrate.
INTRODUCTION
Ribosomal RNAs (rRNAs) form complex tertiary structures that in mature ribosomes are stabilized by numerous ribosomal proteins. Large rRNAs in all species carry post-transcriptional modifications, the number of which range from 14 in mycoplasma to more than a hundred in humans. In Escherichia coli, there are in total 36 rRNA modifications: C-, O- or N-methylated nucleosides, pseudouridines, dihydrouridine and hydroxycytidine (reviewed in (1–2), 3) clustered in functionally important regions such as contact areas with mRNA and tRNAs, the catalytic peptidyl transferase center (PTC), the peptide exit tunnel and the intersubunit bridges.
The functional importance of rRNA modifications has been studied using ribosomes containing in vitro–transcribed rRNA. Although functional 30S subunits can be assembled using unmodified 16S rRNA (4,5), 50S subunits containing unmodified 23S rRNA are catalytically inactive. For assembly of catalytically active 50S subunits, it is necessary that nucleotides 2445–2523 (E. coli numbering used throughout) in the PTC region of domain V of E. coli 23S rRNA (Figure 1A) carry all or a subset of the native modifications (6).
Figure 1.
Location of residue C2498 in E. coli 23S rRNA. (A) Secondary structure of the central loop region of domain V of E. coli 23S rRNA based on pdb 2qam (14). Modifications found in wild-type E. coli 23S rRNA are shown in green. The 2′O-methylcytosine 2498 is shown in red. An arrow indicates site of primer for extension analysis. (B). Structural surrounding of C2498 in the E. coli 70S ribosome (pdb 2qam (14)). The 2′O position of C2498 is marked with an asterisk. The 570 region of domain II is shown in pale green, the 2031 region in pale cyan, the 2454 region in wheat and the 2498 region in light yellow. Helix 89 extends toward the bottom right in the figure.
While eukaryotes and archaea use ribonucleoprotein (RNP) complexes to carry out guide RNA-mediated rRNA recognition and modification, bacteria use site-specific modification enzymes that, in general, recognize the structure of their targets rather than the sequence. Many rRNA modifications are not conserved between different bacteria and knockouts of individual rRNA-modifying enzymes are all viable and lead to mild or no phenotype (for example (7–9)). This suggests that the modifications in a certain region in combination with the rRNA sequence in that particular species may fine-tune the structure, stability and function of the ribosome.
The ribosomal PTC is the target for numerous antibiotics (10,11). Most likely because the sequence conservation in this region is functionally critical, resistance to these antibiotics often comes through post-transcriptional modifications, for example, C8 methylation of A2503 by the Cfr enzyme (12) leading to resistance to several classes of antibiotics. In addition, in some cases knockouts of native modifications in this region lead to higher sensitivity to antibiotics, suggesting an evolutionary link to intrinsic resistance to natural antibiotics (10,13). There are still many remaining questions regarding the structure and function of these enzymes.
RlmM, also called YgdE, was identified as the enzyme responsible for the stoichiometric 2′O-ribose methylation of C2498 (9). Nucleoside 2498 is located at the start of a conserved sequence CXUCGAU in the central loop of domain V of 23S rRNA that has four modification sites (Figure 1A). In high-resolution crystal structures of ribosomes from different bacteria (14–16), C2498 and U2500 are base paired to G2454 and A2453, respectively, at the end of helix 89, and the modification site is surrounded by the peptidyl transferase loop, the 570 region and the 2031 region of 23S rRNA (Figure 1B). The 2′O methylation adds hydrophobicity and has been suggested to stabilize the structure by filling a void in the packing between these regions (2). Sequence analysis showed that RlmM contains a C-terminal Rossmann-like fold methyltransferase (MTase) domain that uses S-adenosyl methionine (AdoMet) as a methyl donor. This class of enzymes contain a 2′O-ribose MTase-specific consensus K-D-K-E/H catalytic tetrad necessary to perform methyl transfer from AdoMet to the 2′OH of the target (17).
RlmM has been shown to be active on protein-free 23S rRNA from an rlmM knockout strain but not on 50S subunits or 70S ribosomes from the same strain, suggesting that it acts early in ribosome assembly (9). Furthermore, in analysis of ribosome assembly intermediates accumulated in vivo in the presence of the antibiotics erythromycin or chloramphenicol, the C2498 modification appeared in the intermediate steps of 50S assembly. Thus, RlmM in vivo must also be active on 23S substrates that have bound a subset of the ribosomal proteins (18). Detailed structural information regarding 50S assembly intermediates is lacking, but the modification site is not accessible in the mature 50S subunit (Figure 1B). The knockout of RlmM is viable but leads to lower fitness compared with wild type in competition assays (9).
In this work, we have solved the crystal structure of E. coli RlmM and its complex with AdoMet and demonstrated RlmM activity on in vitro-transcribed rRNA.
MATERIALS AND METHODS
In vitro transcription and RNA preparation
Unmodified 23S rRNA was prepared by T7 RNA polymerase transcription in vitro from pCW1 plasmid containing the E. coli rrlB 23S rRNA gene (plasmid constructed in (19)) cut with AflII (Fermentas). DNA template for domain V (nucleotides 2021–2625) of the 23S rRNA was obtained by polymerase chain reaction (PCR) from the pCW1 plasmid using the primers 5′-TAATACGACTCACTATAGGGAACTCGCTGTGAAGATGC-3′ and 5′-CGTATGCAGCTTAAGCCCACGGCAGATAGGGAC-3′. The resulting DNA was digested with AflII and gel purified (Qiagen). Transcription was carried out at 37°C for 6 h using 200 mM 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) pH 7.5, 4.4 mM each nucleotide triphosphate (NTP), 5 mM DL-Dithiothreitol, 2 mM spermidine, 30 mM MgCl2, 0.01% Triton X-100, 300 nM template DNA and 10 U of T7 RNA polymerase in 250 µl, followed by DNase I (Fermentas) treatment and phenol/chloroform extraction and ethanol precipitation. RNA was quantified by agarose gel electrophoresis. 23S rRNA from the wild-type BW25113 strain (20) was extracted using sodium dodecyl sulfate/phenol extraction and ethanol precipitation after cells were grown in Luria Broth (LB) at 37°C overnight.
Cloning, protein expression and purification
ASKA clone(–) JW2777 containing the E. coli rlmM/ygdE gene was obtained from the ASKA clone collection (21). The rlmM gene was amplified by PCR using the primers 5′-ATGAATAAGGTTGTATTGCTGTGCCG-3′ and 5′-GGATCGCTCGTCGCGACGAC-3′ and subcloned into pEXP5-CT/TOPO (Invitrogen, USA) to generate a C-terminally His-tagged construct. The resulting plasmid, pAP01-rlmm, was verified by sequencing and used to transform E. coli BL21(DE3) cells. The cells were cultured in LB with 100 µg/ml ampicillin at 37°C to A600 of 0.5. Protein expression was induced with 0.5 mM isopropyl-β-thiogalactopyranoside at 18°C for 20 h. The cells were harvested by centrifugation, washed in 20 mM Tris–HCl, pH 7.5, 200 mM NaCl and stored at −20°C. All purification steps were performed at room temperature. The cells were re-suspended in buffer A (20 mM Tris–H2SO4, pH 8.5, 300 mM Na2SO4, 5% glycerol, 10 mM β-mercaptoethanol) supplemented with 0.1% Triton X-100, 5 mg lysozyme and complete protease inhibitor (Roche, Germany) and lysed using a cell disruptor (Constant Systems Ltd, UK). The lysate was cleared by centrifugation and loaded into a Bio-Rad Econo-Pac gravity flow column containing Ni-Sepharose™ (GE Healthcare, Sweden) pre-equilibrated with buffer A containing 10 mM imidazole. After incubation for 1 h, the column was washed with 40 mM imidazole in buffer A, and the protein was eluted with 250 mM imidazole in buffer A. RlmM was further purified through a HiLoad™ 16/60 Superdex™ 75 prep grade column (GE Healthcare) equilibrated in buffer A. Peak fractions eluting at monomer size were pooled and concentrated to 10 mg/ml using a Vivaspin 30 kDa cut-off concentrator (Sartorius Stedim Biotech, Germany). Optimal buffer composition for the protein purification was identified by differential scanning fluorimetry (22), where Na2SO4 was identified as the salt that gave the highest thermal stability to RlmM.
In vitro methylation assay
Prior to methylation reactions, 200 nM rRNA (23S in vitro transcript or transcribed domain V) was suspended in 40 mM HEPES pH 7.6, 100 mM NH4Cl, 10 mM MgCl2, 12 mM β-mercaptoethanol, 20% glycerol and 0.1% Triton X-100 and warmed for 10 min at 50°C followed by 15 min at 37°C (9). A final concentration of 1 mM AdoMet was then added as the methyl donor followed by addition of Ni-purified RlmM to a final concentration of 100 nM. The reaction was carried out at 37°C for 90 min or as noted otherwise. Reactions were quenched in 50:50 phenol:chloroform followed by ethanol precipitation. Precipitated RNA was dissolved in H2O prior to primer extension. For time course analysis, 10 µl aliquots were quenched in 50:50 phenol:chloroform at 0 s (before enzyme addition), 30 s, 1, 2, 4, 8, 20, 45 and 90 min. Primer extension analysis was used to detect the modification.
Primer extension
300 ng of oligodeoxyribonucleotide complimentary to the E. coli 23S rRNA nucleotide sequence 2540–2556 (5′-CUUGUCGGUAUGGGAAC-3′ (9)) was 32P-5′-labeled by incubation with 15 U polynucleotide kinase (Fermentas) and 20 µCi γ-[32P]-adenosine triphosphate (ATP; 3000 Ci/mmol stock concentration; PerkinElmer) at 37°C for 45 min followed by inactivation by incubation at 95°C for 10 min (23). The labeled primer mix was purified using an Illustra G-50 spin column (GE Healthcare). Five nanograms of labeled primers were annealed to the rRNA in hybridization buffer (56 mM HEPES-KOH, pH 7 and 111 mM KCl) by heating to 85°C for 3 min followed by slow cooling to 37°C over 15 min and then put directly at 4°C (24). The hybridization mix was combined with 1U Avian Myeloblastosis Virus reverse transcriptase (NEB; 10-fold diluted in 50 mM Tris–HCl, pH 8.4; 2 mM DTE and 50% (v/v) glycerol (24)) and extension mix (100 µM deoxy-ATP, deoxycytidine triphosphate, deoxythymidine triphosphate; 116 mM Tris–HCl, pH 8.4; 9 mM MgCl2; 9 mM DTE; (24)) and 0.5 µM deoxyguanosine triphosphate (dGTP) (9) and incubated at 37°C for 45 min to generate cDNA complimentary to the rRNA. The reaction mixture was then ethanol precipitated with 30 µl 300 mM NaOAc, 1 mM ethylenediaminetetraacetic acid and 90 µl 100% ethanol (24). Dideoxy sequencing products were generated from pCW1 using DNA polymerase (Sequenase Version 2.0, USB). DNAs were separated on a 20 × 40 cm× 0.75 mm 6% polyacrylamide/7 M urea gel run at 30 W until the bromophenol blue dye reached the bottom. The gel was dried at 80°C under vacuum and analyzed by phosphoimaging (Bio-Rad FX).
Quantity One software (Bio-Rad) was used to quantitate the band intensities corresponding to stops at nucleotides C2499, G2502, G2508 and U2511. Percent reaction of C2498 in the time course assay was estimated by taking the ratio of the intensity of the C2499 band against average intensity of G2502, G2508 and U2511 of the same lane.
Crystallization and data collection
In initial crystallization trials, needle clusters were observed in 0.2 M ammonium tartrate dibasic pH 7.0, 20% PEG 3350 (PEG/Ion screen, Hampton Research). After optimization and streak seeding, clusters of rod-shaped crystals grew in 1 week at 20°C in sitting drop vapor diffusion experiments containing 1.5 μl RlmM (10 mg/ml) and 1.5 μl reservoir solution (0.3 M ammonium tartrate, pH 7.0 and 20% PEG 3350). Crystals were briefly soaked in reservoir solution containing 15% glycerol and vitrified in liquid nitrogen. All data were collected at 100 K at MAX-Lab, Lund, Sweden on MarCCD detectors. 1.9 Å diffraction data were collected at beamline I911-2 and processed in space group P31. Attempts to solve the structure by molecular replacement failed. A second crystal form obtained by streak seeding into drops containing 3.0% dextran sulfate sodium salt as an additive produced single crystals that grew to 100 × 100 × 50 µm in 1 week. Native data to 1.95 Å were collected on the same beamline and processed in space group P3121. These crystals were soaked in cryoprotectant solution containing 5 mM K2PtCl4 for 20 min and flash frozen in liquid nitrogen for collection of single-wavelength anomalous dispersion (SAD) data to 2.2 Å resolution at beamline I911-3. RlmM was co-crystallized with 5 mM AdoMet in the P3121 condition, and a 2.6 Å dataset was collected at beamline I911-2. All diffraction data were processed and scaled using XDS (25). Data statistics are summarized in Table 1.
Table 1.
Summary of crystallographic data and refinement statistics
| Dataset | Apo | Apo | AdoMet | K2PtCl4 |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P31 | P3121 | P3121 | P3121 |
| Unit-cell parameters | ||||
| a, b, c (Å) | 132.8, 132.8, 41.5 | 92.1, 92.1, 83.1 | 92.3, 92.3, 83.6 | 92.9, 92.9, 86.4 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution range (Å)a | 20.1–1.9 (2.0–1.9) | 30.0–1.95 (2.05–1.95) | 30.0–2.6 (2.7–2.6) | 30.0–2.2 (2.3–2.2) |
| Wavelength (Å) | 1.041 | 1.041 | 1.041 | 1.072 |
| Unique reflections | 64 294 | 30 029 | 12 983 | 42 258 |
| Completeness (%) | 99.8 (98.2) | 99.8 (98.0) | 99.8 (99.9) | 99.8 (99.5) |
| Redundancy | 4.9 (4.6) | 7.3 (6.8) | 7.4 (7.5) | 11.9 (11.7) |
| Rmeas (%) | 7.1 (49.0) | 6.1 (61.0) | 15.7 (71.3) | 22.3 (71.5) |
| I/σ (I) | 19.7 (4.4) | 25.0 (4.3) | 16.4 (3.3) | 19.4 (3.3) |
| Refinement | ||||
| Resolution range (Å) | 20.1–1.9 | 28.8–1.95 | 28.9–2.6 | |
| Reflections (test set) | 61 052 (3212) | 28 520 (1502) | 12 333 (650) | |
| Number of atoms | ||||
| Protein | 5808 | 2916 | 2888 | |
| Water | 552 | 195 | 87 | |
| Other | 157 | 134 | 103 | |
| Rwork/Rfree (%) | 16.4/20.6 | 16.9/20.6 | 16.3/19.6 | |
| Average B-factor (Å2) | ||||
| Protein | 21.4 | 33.0 | 28.6 | |
| Water | 28.0 | 39.1 | 26.8 | |
| Other | 30.7 | 52.1 | 49.7 | |
| RMSD from ideal bond length (Å) | 0.016 | 0.008 | 0.008 | |
| RMSD from ideal bond angle (°) | 1.567 | 1.072 | 1.134 | |
| Ramachandran plot | ||||
| Preferred (%) | 98.3 | 98.3 | 97.2 | |
| Allowed (%) | 1.53 | 1.66 | 2.80 | |
| Outliers (%) | 0.14 | 0.00 | 0.00 | |
| PDB ID | 4AUK | 4ATN | 4B17 | |
aValues within parenthesis represent the highest resolution bin.
Structure determination and refinement
Using the Pt-SAD data in P3121, eight platinum sites were located using PHENIX (26). Heavy atom refinement, phasing and density modification were performed in PHENIX. The figure of merit was 0.36 (0.70 after density modification). The initial model was built using PHENIX AutoBuild (26) and improved by cycles of manual building in Coot (27) and refinement against the native RlmM data in PHENIX. Refinement statistics are presented in Table 1. The P31 structure was solved by molecular replacement in Phaser (28), using the separate domains of the P3121 structure as search models. The AdoMet complex structure was solved by the same method, using the complete P3121 structure as search model. The models were manually rebuilt in Coot and refined in PHENIX. Refinement statistics are presented in Table 1. The quality of the structures was assessed using MolProbity (29). The refined coordinates and the structure factors have been deposited in the protein data bank (PDB) with accession numbers 4ATN (P3121), 4AUK (P31) and 4B17 (P3121 AdoMet complex). Structure figures were prepared using PyMOL (The PyMOL Molecular Graphics System, Version 1.2r1, Schrödinger, LLC).
Sequence analysis
BLAST was used to identify full-length homologues of E. coli RlmM. Multiple sequence alignment was performed in CLUSTALW2 (30).
RESULTS
RlmM modifies in vitro transcribed 23S rRNA at C2498
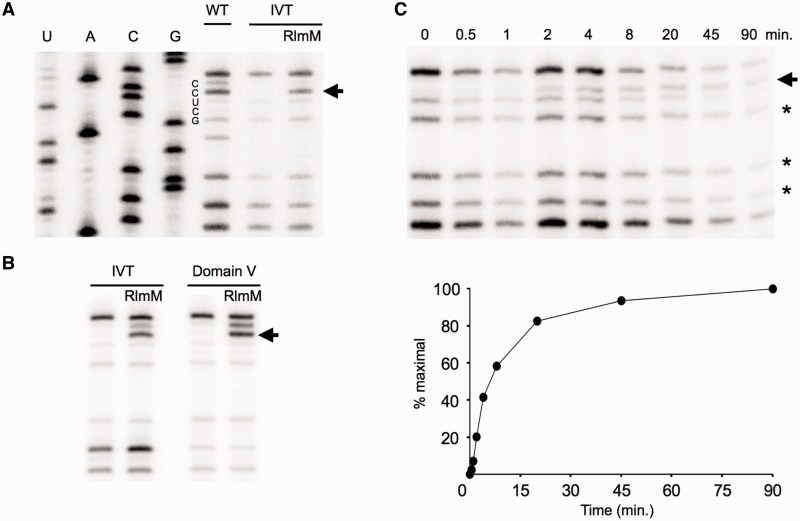
To investigate whether RlmM can modify unmodified in vitro transcripts of full length 23S rRNA (IVT), we tested our purified, recombinant RlmM in the presence of excess AdoMet. Most RNA modifications, such as 2′O methylation, can be detected directly using primer extension by reverse transcriptase and gel electrophoresis (24). Low dNTP concentrations cause reverse transcriptase to pause one nucleotide before a 2′O methylation site (31) and the optimal dGTP concentration for Cm2498 detection reported by Purta et al. was also used here. A prominent C2499 band, induced by methylation on C2498, appeared after incubation of IVT with RlmM, showing RlmM does not rely on prior modifications to the 23S rRNA for activity (Figure 2A). Smaller fragments were also detected at G2502, G2508 and U2511 in all lanes, presumably due to strong secondary structure and limiting dGTP for the templated C that follows in each case. A reverse transcriptase stop on U2504 corresponding to wild-type methylation on A2503 can also be seen in the WT lane and not in the IVT lane, as expected (Figure 2A). Standardizing the intensity of the C2499 band to the average of band intensities of these smaller fragments allowed comparisons between lanes. On the basis of band intensity, the extent of modification in the in vitro reaction is nearly the same as found in the E. coli wild-type 23S rRNA (Figure 2A), which was previously determined to be stoichiometric on 23S rRNA (32). In addition, RlmM is capable of modifying unmodified 23S rRNA domain V alone to the same extent as full length IVT (Figure 2B). This demonstrates that RlmM recognition is neither dependent on other domains of the 23S rRNA nor other rRNA modifications.
Figure 2.
Primer extension analysis of in vitro methylation of C2498. Arrows denote stops at the nucleotide before Cm2498. (A) Recombinant RlmM modifies C2498 of IVT based on primer extension analysis. WT: 23S rRNA purified from wild-type E. coli; IVT: in vitro transcript of 23S rRNA; RlmM: IVT incubated with RlmM enzyme. U, A, C and G correspond to DNA sequencing lanes. (B) RlmM modifies C2498 of unmodified 23S rRNA domain V alone. Other labels as in A. (C) Time course of IVT methylation. Top: stars indicate bands used for comparing band intensities between lanes. Bottom: plot of modification time course. Intensity of the C2499 position at time 0 was subtracted from each time point.
Although the primer extension assay is not highly quantitative, it was possible to use it to measure methylation of IVT by RlmM over time, showing >80% reaction completion after 45 min with a rudimentary initial velocity (Figure 2C; Supplementary Figure S1) similar to other MTases under similar in vitro conditions (33–34).
Structure determination of RlmM
E. coli RlmM with a C-terminal His-tag was crystallized with polyethylene glycol and ammonium tartrate in two crystal forms belonging to space group P31 with two molecules per asymmetric unit and space group P3121 with one molecule in the asymmetric unit, respectively (Table 1). Molecular replacement trials with other Rossmann-like fold MTase domains failed to provide sufficient phase information to build the structure. The P3121 structure was solved by SAD phasing using a K2PtCl4 derivative and refined against the native data to 1.95 Å resolution. The resulting model was used to solve the 1.9 Å P31 structure by molecular replacement. The final refined structures of RlmM contain residues 1–357. The C-terminal residues 358–366 and the His-tag were disordered in all structures. RlmM was also co-crystallized with AdoMet in the P3121 crystal form and the resulting structure will be described below. On the basis of its profile on size exclusion chromatography, we believe RlmM to be a monomer in vivo.
Overall structure
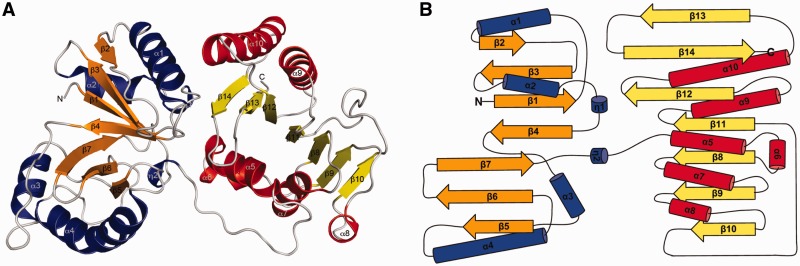
RlmM forms a bi-lobed structure with the approximate dimensions of 62 × 41 × 33 Å3 (Figure 3A). The structure is organized into an N-terminal THUMP domain and a C-terminal Rossmann-like MTase domain (Figure 3B). The two domains are connected by a long loop containing a 310 helix. The loop forms hydrophobic interactions with residues from the C-terminal domain and the relative orientation of the two domains is stabilized by inter-domain hydrogen bonds and hydrophobic interactions. The buried surface area in the inter-domain interaction is roughly 900 Å2, corresponding to 10% of the area of the N-terminal domain.
Figure 3.
The overall structure of E. coli RlmM. (A) Cartoon representation showing the N-terminal THUMP domain with α-helices in blue and β-strands in orange and the C-terminal Rossmann-like MTase domain with α-helices in red and β-strands in yellow. (B) Topology diagram of E. coli RlmM, colored as in A.
The structures of RlmM in the two crystal forms are close to identical. In the P31 structure, molecules A and B superpose with a root mean square deviation (RMSD) of 0.55 Å for 355 Cα atoms, and the P3121 structure superposes onto molecules A and B of the P31 structure with RMSDs of 0.72 Å and 0.77 Å, respectively, for 355 Cα atoms. Due to crystal packing, there are conformational differences in the loop preceding β11 and the β4-α3 loop. However, when the P31 structures are superposed on the N-terminal domain of the P3121 structure, there is a minor rotational movement of the C-terminal domain, resulting in shifts of up to 3.4 Å at the outer edge of the C-terminal domain without affecting the inter-domain packing (Supplementary Figure S2). Below, unless stated otherwise, the P3121 structure will be described.
A DALI search (35) for similar 3D structures identified several homologues to the N-and C-terminal domains, respectively (see subsequent sections), but did not identify the complete domain arrangement of RlmM in any other structure in the PDB.
Sequence analysis
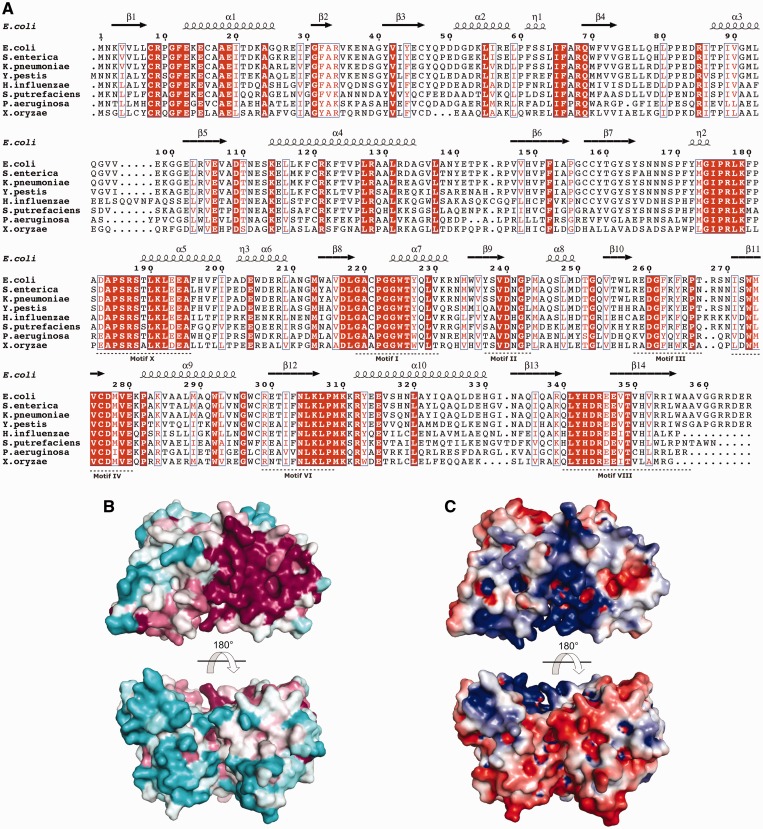
RlmM was aligned to a representative set of full-length orthologs from gammaproteobacteria and betaproteo-bacteria, and the resulting gammaproteobacterial sequence alignment was used to identify conserved residues (Figure 4A). The sequence identity to E. coli RlmM ranges from 44% for the Xanthomonas oryzae protein to 94% for the Salmonella enterica protein.
Figure 4.
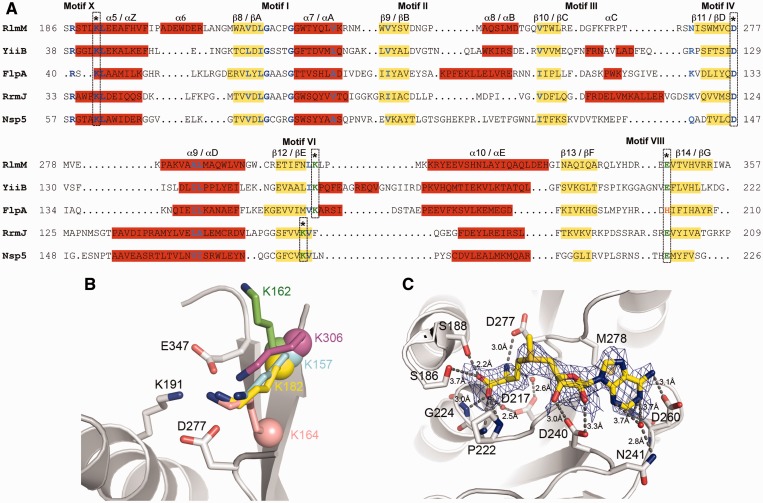
(A) Multiple sequence alignment of E. coli RlmM with seven gammaproteobacterial sequences of the RlmM family (image generated using ESPript (66)). Conserved residues are shown in white on red background and conservative substitutions are shown in red on white background. NCBI accession numbers: Escherichia coli (NP_417286.1), Salmonella enterica (NP_457375.1), Klebsiella pneumoniae (YP_002236829.1), Yersinia pestis (NP_670451.1), Haemophilus influenzae (YP_005179378.1), Shewanella putrefaciens (ADV53762.1), Pseudomonas aeruginosa (NP_250254.1), Xanthomonas oryzae (YP_452543.1). Motifs I–IV, VI, VIII and X in the MTase domain (17) are indicated below the alignment. (B) Surface of RlmM colored according to sequence conservation using ConSurf (36). The color spectrum ranges from magenta (highest conservation) to cyan (lowest conservation). Orientation of top view as in Figure 3A. (C) Electrostatic surface potential of RlmM. The color spectrum ranges from deep red (–4 kT) to deep blue (+4 kT).
Mapping of the degree of sequence conservation in all full-length homologs onto the RlmM structure using ConSurf (36) revealed a large conserved patch on one side of the MTase domain that extends toward the long helix α4 in the THUMP domain (Figure 4A and B). Many of these conserved or conservatively substituted residues are arginines and lysines, resulting in a strong patch of positive charge that overlaps with the conserved surface (Figure 4C).
N-terminal domain
The N-terminal domain (amino acids 1–176) is a THUMP domain according to the classification in the SCOP database. It contains a seven-stranded mostly anti-parallel β-sheet (β5 and β6 are parallel) forming a half-barrel with one short and one longer helix on each side (Figure 3A and B). It can be further divided into a ferredoxin-like domain (FLD, amino acid 1–73) and a C-terminal ‘minimal THUMP domain’ (amino acid 74–164) corresponding to the two sides of the half-barrel. The minimal THUMP domain is an RNA-binding fold containing a four-stranded mixed β-sheet that was identified on the sequence level and named after its predicted occurrence in thiouridine synthases, RNA methylases and pseudouridine synthases of all kingdoms of life (37). The minimal THUMP domain was first observed in the structure of the tRNA thiouridine synthase ThiI (38), where it is similarly fused to an N-terminal FLD. This arrangement was subsequently observed in several structures and is now classified as the THUMP domain family in the SCOP database. The ‘minimal THUMP’ has also been observed fused to an N-terminal helical domain in the pseudouridine synthase Pus10 (39).
In a DALI (35) search with the N-terminal domain, the most similar structures were the two tRNA:m2G6 MTases Trm14 from Pyrococcus furiosus (pdb 3tm5, RMSD 2.7 Å for 145 Cα atoms) and TrmN from Thermus thermophilus (pdb 3tma, RMSD 3.3 Å for 141 Cα atoms) (40) and the N-terminal part of the rRNA m2G2445 MTase RlmKL (pdb 3v8v, RMSD 2.8 Å for 135 Cα atoms) (41). Interestingly, in both of these enzymes, the THUMP domains are followed by catalytic Rossmann-like fold MTase domains, but in a different orientation compared with RlmM (further discussed in the subsequent sections). The same is true for a putative methylase from Clostridium difficile. THUMP domains only share 10–15% sequence identity, explaining why these domains were not detected in sequence analysis. The THUMP domain also occurs in combination with other catalytic domains; an N-terminal cytidine deaminase domain in the archaeal tRNA C8 deaminase CDAT8 (42) and a C-terminal PP-loop pyrophosphatase domain in ThiI (38). In TAN1, a protein essential for the ac4C modification in tRNA, a tRNA-binding THUMP domain occurs by itself and needs an un-identified catalytic subunit for modification activity (37,43). Thus, the THUMP domain has the role of RNA recognition in several different enzymes that act on rRNA or tRNA.
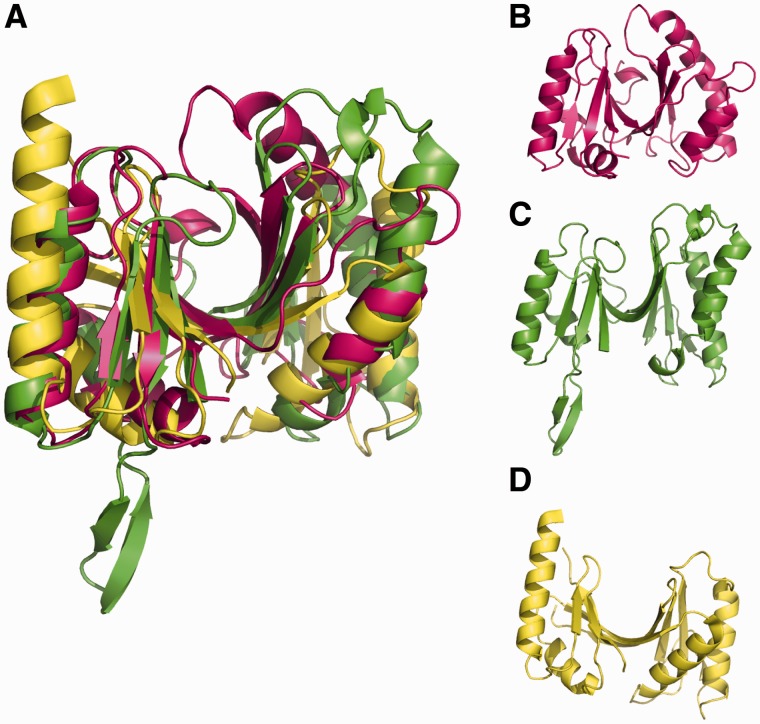
In comparison with previously observed THUMP domains, RlmM has a few unique features (Figure 5A–D). The strand β4 is shorter than in the other structures, making only three hydrogen bonds to β7 and thus making the half-barrel into two nearly separate β-sheets. The lengths of β4 and β7 at the center of the sheet affect the shape of the half-barrel, so that RlmM has more of a V-shape, whereas ThiI where both strands participate in the two sheets forms a shallow U and Trm14, TrmN and RlmKL, which have only β4 bridging the two sheets are intermediate. In RlmM, helix α4 is also longer than in the other structures in the direction of the barrel opening, which further contributes to the closing of the opening by pushing β4 inwards. RlmM also contains an extra 310 helix that forms hydrophobic interactions with the center of the barrel. Each of the compared THUMP domains have unique structural features, such as an extra β-hairpin in Trm14 and a longer α1 in ThiI, suggesting that the ancient THUMP domain (37) has evolved to form RNA-binding modules of different shape (Figure 5B–D).
Figure 5.
The THUMP domain of RlmM. (A) Overlay of the THUMP domain of RlmM (pink) with THUMP domains from Trm14 (pdb 3tm5, (40), green) and ThiI (pdb 2c5s (38), yellow). (B). THUMP domain of RlmM. (C) THUMP domain of Trm14. (D) THUMP domain of ThiI.
C-terminal domain
The C-terminal domain of RlmM (residues 186–356) adopts a Rossmann-like fold belonging to the Class I AdoMet-dependent MTases (44). This catalytic domain consists of a mixed seven-stranded β-sheet flanked by four helices on one side and two helices on the opposite side (Figure 3A and B). The sequence alignment revealed that conserved residues in RlmM cluster in and around the functional motifs I–VIII and X in AdoMet-dependent MTases (Figure 4A) (45). Motifs I–III are important for AdoMet binding, whereas motifs IV, VI, VIII and X form the 2′O-ribose MTase-specific K-D-K-E/H catalytic tetrad (46). The second Lys is thought to act as a general base by deprotonating the ribose 2′OH nucleophile that attacks the reactive methyl group of AdoMet, whereas the Asp stabilizes the positive charge on the sulfur (46) (Supplementary Figure S3).
In a DALI (35) search with the C-terminal domain, the most similar structures were bacterial RNA 2′O-ribose MTases YiiB and TlyA annotated as putative hemolysins (pdb 3opn, RMSD 2.3 Å for 154 Cα atoms and pdb 3hp7, RMSD 2.5 Å for 156 Cα atoms). The same structures were found in a BLAST search of the pdb with the RlmM sequence prior to solving the structure. Other DALI hits were archaeal fibrillarin FlpA (pdb 1nt2, RMSD 2.3 Å for 152 Cα atoms (47)) and the bacterial FtsJ/RrmJ (pdb 1eiz, RMSD 2.5 Å for 148 Cα atoms (48)). The DALI results also included rRNA base methylating enzymes with lower structural similarity such as ErmC′ (49) and KsgA (50) and the flaviviral non-structural protein 5 (Nsp5) that performs ribose as well as base methylation on an mRNA substrate (51). The RlmM MTase domain shared 12–23% sequence identity with the top hits over 140–156 residues.
Structures of the 2′O-ribose MTases YiiB (pdb 3opn), FlpA (pdb 1nt2 (47)), RrmJ (pdb 1eiz (48)) and Nsp5 (pdb 2wa2 (51)) were superposed onto the RlmM MTase domain. Structure-based sequence alignment (Figure 6A) and structural superposition confirmed that the catalytic tetrad K191-D277-K306-E347 is present in the RlmM MTase domain. A closer look revealed that while residues K191, D277 and E347 of RlmM superimpose on the conserved K, D and E/H residues of all the 2′O-ribose MTases, residue K306 of the RlmM MTase domain is shifted forward by two residues relative to RrmJ and Nsp5 and superimposes onto the corresponding lysine residues of FlpA and YiiB (Figure 6B). This shift in RlmM differs from the alignment reported based on the primary sequence, where the FXXK motif in RlmM was aligned to the same motif in RrmJ and Nsp5 (9). A previous bioinformatic and phylogenetic study identified the two-residue shift of the second catalytic K in fibrillarin (52), but the shift in YiiB/TlyA (Figure 6A and B) has not been reported before.
Figure 6.
Comparison of the RlmM MTase domain with homologues identified by the DALI search. (A) Structure-based sequence alignment of the RlmM MTase domain with YiiB from Lactococcus lactis (pdb 3opn), FlpA from Archaeoglobus fulgidus (pdb 1nt2 (47)), RrmJ from E. coli (pdb 1eiz (48)) and Nsp5 from Modoc virus (pdb 2wa2 (51)). α-helices (red background) and β-strands (yellow background) are labeled according to the RlmM MTase domain (numbers) and the consensus AdoMet-dependent MTases (letters). Strictly conserved residues are shown in deep-blue and similar residues in sky-blue. Residues forming the catalytic tetrad K-D-K-E/H are marked with an asterisk. The two-residue shift of the second catalytic K between RlmM, YiiB, FlpA and RrmJ & Nsp5 is indicated. The difference between FlpA and the other proteins in the fourth catalytic residue is highlighted. (B) Comparison of the second Lys of the catalytic tetrad in superposed structures of MTase domains of RlmM, FlpA, YiiB and RrmJ. RlmM is shown in grey. K306 in RlmM (pink), K157 in YiiB (cyan), K162 (green) and K182 (yellow) in FlpA exhibit a two-residue shift of the second lysine relative to K164 in RrmJ (salmon). The Cα atoms of the lysines are shown as spheres. In FlpA, the side chain of K162 (green) moves to the orientation of K182 (yellow) when bound to substrate RNA. (C) The AdoMet-binding site in RlmM. The simulated annealing Fo–Fc omit map of AdoMet is contoured at 2.0 σ. AdoMet is shown in yellow and the interacting residues are shown in grey and colored by atom type. Waters are shown as red spheres and hydrogen bonds as dotted lines.
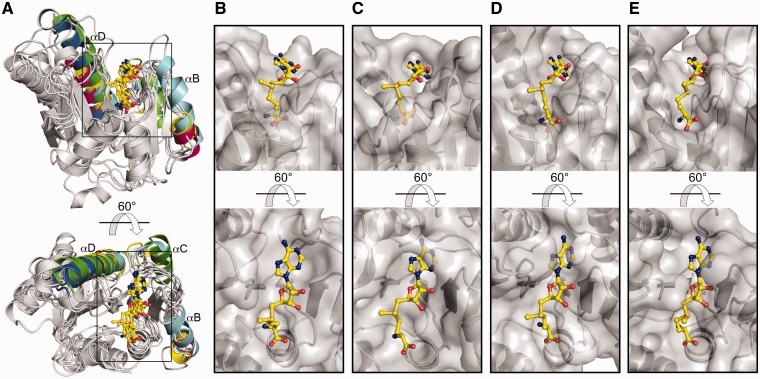
Other differences between the MTase domain in RlmM and other 2′O-ribose MTases can be seen in the relative lengths of helices αB, αC and αD that surround the AdoMet-binding site (Figure 6A and 7A). As discussed in the subsequent sections, these helices influence the shape of the AdoMet-binding pocket.
Figure 7.
RlmM has an unusually open AdoMet-binding pocket. (A) Cartoon representation of the structural superposition of the AdoMet complexes of related MTase domains. Helices αB, αC and αD of the different structures are colored according to the respective structure: RlmM (pink), YiiB (yellow), FlpA (cyan), RrmJ (green) and Nsp5 (blue). (B–E) AdoMet-binding site in the MTase domains. AdoMet is shown in yellow and the MTase domains in partially transparent surface representation. (B) RlmM. (C) FlpA. (D) RrmJ. (E) Nsp5.
Structure of the RlmM–AdoMet complex
Trials to soak AdoMet into pre-formed RlmM crystals gave only partial density for the ligand, indicating low occupancy and/or multiple conformations of the ligand. However, using co-crystallization in the P3121 crystal form, electron density for the complete AdoMet was observed at 2.6 Å resolution (Figure 6C). The RlmM AdoMet complex structure superposed on the apo structure with an RMSD of 0.19 Å for the entire protein (357 Cα atoms) as well as for the individual domains, showing that no global conformational change takes place upon AdoMet binding.
AdoMet binds in extended conformation to a shallow negatively charged pocket formed by the conserved residues in motifs I–IV of the MTase domain (Figure 6C and Supplementary Figure S4). Comparison of the AdoMet-binding pocket in RlmM with AdoMet complex structures of FlpA (pdb 1nt2), RrmJ (pdb 1eiz) and Nsp5 (pdb 2wa2) (Figure 7A) reveals some differences. The shape of the AdoMet-binding pocket depends on the presence and length of helices αB, αC and αD that position the motif II and III loops to surround the adenine nucleobase and ribose moiety of AdoMet. Due to the short αB and αD and the missing αC, the AdoMet-binding pocket in RlmM is open and shallow (Figure 7B). In contrast, helices αB and αC in RrmJ, FlpA, YiiB and Nsp5 contribute to deeper AdoMet-binding pockets with narrow openings (Figure 7C–E). In Nsp5, another monomer stabilizes the long motif II loop to form an enclosed AdoMet-binding pocket.
There are also differences in the motif II and III loops that interact with the base of AdoMet. In the RlmM–AdoMet complex, the base binds to a hydrophobic surface formed by M278 in motif IV interacting with G261 in motif III, as in most related complex structures (Supplementary Figure S5). However, the base is normally partially or completely covered by a ‘lid’ residue from the motif II loop that narrows the opening of the AdoMet-binding pocket. In RlmM, the motif II loop (D240–G242) has an open conformation in all our structures. In the P3121 structure, the motif II loop is involved in crystal packing, but there is no evidence for conformational changes in this loop upon AdoMet binding to other 2′O-ribose MTases. The motif II loop would need to re-orient by 3.1 Å for N241 to be positioned to cover the adenine ring from the top as the corresponding Leu in RrmJ and Nsp5 (Supplementary Figure S5) and the Tyr in FlpA. There is no structure of YiiB or TlyA in complex with AdoMet, but in the apo YiiB structure, the Val of the motif II loop seems positioned to reach the AdoMet-binding site.
Furthermore, in the adenine nucleobase, the N6 amino group makes a hydrogen bond interaction with the β-carboxyl group of D260, the N1 atom is oriented to make polar interactions with the β-carboxyl group and the backbone amide of G261 and the N3 atom makes a water-mediated hydrogen bond to N241 (Figure 6C and Supplementary Figure S4). The interactions of the ribose and the methionine moiety of AdoMet are similar to what is seen in other Class-I MTases. The ribose of AdoMet is stabilized by hydrogen bonds of the 2′OH and 3′OH with the conserved D240. The carboxylate group of the methionine moiety interacts with the side chains of S186 and S188 and the backbone amides of P222, G223 and G224. The amino group of the methionine moiety forms a hydrogen bond with the catalytic D277 in motif IV. At the C-terminal end of β8, D217 extends to make water-mediated hydrogen bond to the carboxylate and amino groups of the methionine moiety.
On AdoMet binding, there is a slight shift of the catalytic tetrad so that K191 moves away from D277 and comes closer to E347 (Supplementary Figure S6). According to the reaction mechanism proposed for RrmJ (46), for catalysis to be possible, D277 would need to be close enough to stabilize the positive charge on the AdoMet sulfur (currently 4.64 Å) and on K306 which will deprotonate the substrate 2′OH (Supplementary Figure S3). This would be possible if motif II-mediated closure of the AdoMet-binding pocket would push the ligand closer to the catalytic tetrad. Since no such change is observed upon AdoMet binding, it is possible that RlmM cannot do this by itself.
RNA-binding site of RlmM
The THUMP domain contains conserved surface-exposed residues in the β1-α1 loop, the N-terminal end of helix α1 and the loop before β4 (Figure 4A and B). Furthermore, there are three conserved basic residues in helix α4 and another three in the linker to the catalytic domain. Together, the basic residues form a continuous positively charged surface (helix α4, continuing with the β1-α1 loop, the N-terminus of α1, the loop before β4 and the linker between the N- and C-terminal domain; Figure 4C) connected to the conserved active site surface of the catalytic domain. The positively charged residues overlap with the conserved region, indicating their functional importance and suggesting that this surface is where RlmM binds to 23S rRNA.
Trm14/TrmN and RlmKL also contain N-terminal THUMP domains followed by a Rossmann-like fold MTase domains (40,41). These two structures have close to identical packing between the two domains. In comparison, the catalytic domain in RlmM is rotated by ∼90° (Figure 8). Surface conservation of these structures (Figure 4B; (40)) suggest that both domain arrangements have evolved to bind to the RNA substrate and position it next to the active site on the catalytic domain, but using different parts of the THUMP domain.
Figure 8.
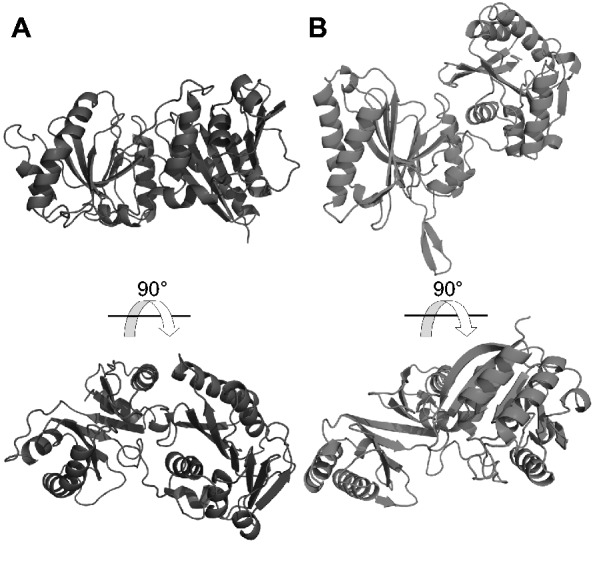
RlmM displays a novel orientation between the THUMP domain and the MTase domain. RlmM (A) and Trm14 (pdb 3tm5 (40), B) were superposed based on the N-terminal THUMP domain (left) and shown side by side. The Rossmann-like fold MTase domain of RlmM displays a 90° rotation relative to the equivalent domain of Trm14. The conserved, putative RNA-binding surface of RlmM is visible in the bottom view, while the putative RNA-binding surface of Trm14 is visible in the top view (40).
The only available structure of a ribose 2′O MTase in complex with RNA is the fibrillarin subunit in the complete archaeal box C/D RNP in complex with substrate RNA and guide RNA (53). Superposition of the fibrillarin–RNA complex from this structure (pdb 3pla) onto the RlmM MTase domain suggests that RlmM could make a minor groove interaction with an RNA double-helix placed across the catalytic domain. However, the comparison does not provide any clues regarding how the RNA potentially could influence the AdoMet-binding site.
DISCUSSION
Structural relatives of RlmM
We were initially puzzled by seeing that the catalytic domain of RlmM was more similar to YiiB and TlyA, annotated as putative hemolysins, than to other known rRNA-modifying enzymes. However, TlyA from Mycobacterium tuberculosis has proven 2′O-ribose MTase activity on C1409 of 16S rRNA and C1920 of 23S rRNA (54,55). In contrast, the suggested hemolytic activity is controversial (56,57).
Of the other similar structures, archaebacterial fibrillarins take part in guide-mediated ribose methylation in rRNA and tRNA, RrmJ is a bacterial 23S rRNA 2′O-ribose MTase and Nsp5 is involved in capping of viral mRNAs. Thus, similar structures are used for ribose methylation of a variety of substrates in viruses, bacteria and archaea using different mechanisms of substrate recognition.
A conserved catalytic site with a migrated Lys residue
We observed that RlmM and YiiB/TlyA, like fibrillarin have a two-residue shift forward of the second catalytic lysine compared with most 2′O MTases (Figure 6A and B). Although key catalytic residues are normally conserved among functionally related proteins, there are several examples of long- and short-distance migration of essential residues in the primary sequence (58). In these cases, although a residue is shifted in the sequence and structure, the positions of the atoms involved in catalysis are preserved. In 2′O-ribose MTases, the position of the ε-amino group of the second catalytic lysine is thought to be crucial for deprotonation of the ribose 2′OH, enabling its nucleophilic attack on the AdoMet methyl group (46,59) (Supplementary Figure S3). In FlpA, binding of the substrate RNA re-orients the Lys so that its ε-amino group can make the proper interaction with the 2′OH of the ribose, whereas RlmM and YiiB without RNA and AdoMet adopt conformations where the side chains of K306 and K157 are positioned similarly to the FlpA complex with substrate RNA (Figure 6B). This suggests that two optimized variants of the catalytic site for 2′O-ribose methylation have evolved and that further analysis of the evolutionary relationship between RlmM, FlpA, YiiB and RrmJ would be of interest.
Is RlmM dependent on rRNA substrate for closing the AdoMet pocket?
We managed to co-crystallize RlmM with AdoMet in the P3121 crystal form, but not in the P31 crystal form. Perhaps crystal contacts of the open motif II loop in the P3121 crystal form have a stabilizing effect on the AdoMet-binding site. Another possibility is that the crystallization additive dextran sulfate affects binding of AdoMet. The AdoMet-binding site is open and shallow compared with other 2′O MTases (Figure 7B–E), and the ligand does not make the interactions with the catalytic tetrad that would be needed for catalysis. Therefore, we hypothesize that RlmM may be a partner-dependent MTase that forms the correct interactions with AdoMet only in presence of RNA substrate. As exemplified in the subsequent sections, there are several examples of RNA MTases that only bind AdoMet in optimal position for methyl transfer in the presence of an auxiliary protein or the RNA substrate.
Protein-mediated stabilization occurs in the archaebacterial fibrillarin-Nop5p complex. Nop5p stabilizes the motif II and III regions through interactions with αB and β3 (Figure 6A and B), leading to a pre-ordered binding site for, and increased affinity to, AdoMet (60). Along the same line, the stimulatory subunit Nsp10 is essential for AdoMet binding and MTase activity of the SARS coronavirus-encoded Nsp16. Nsp16 has a flexible AdoMet-binding pocket surrounded by long loops that Nsp10 stabilizes through interactions with the motif II loop (61).
RNA-mediated stabilization has been observed in the antibiotic resistance protein KsgA that dimethylates the bases of A1518 and A1519 in E. coli 16S rRNA. A recent crystal structure revealed that substrate binding triggers conformational changes that stabilize motifs I and VIII in conformations essential for efficient AdoMet binding (62).
Activity assays with pure recombinant protein excludes the possibility that RlmM needs another protein for activity, suggesting that binding of the 23S rRNA substrate might be needed to induce the structural changes in the AdoMet-binding pocket necessary for methyl transfer. This hypothesis will be tested in future experiments.
RlmM interaction with its RNA substrate
Mapping of conserved surface residues and charge distribution demonstrate that RlmM in the crystal structures displays a conformation where the RNA-binding surface is nicely aligned with the catalytic site of the MTase domain (Figure 4B and C). Thus, we have no indication that global conformational changes in RlmM would be needed for RNA binding. The THUMP domains seem to have evolved different shapes from a common structural scaffold and distinct surfaces for RNA recognition in the different enzymes. In ThiI and RlmKL, a ‘channel’ formed by the THUMP beta half-barrel has been suggested to be used for RNA binding (38,41), but in RlmM, the corresponding groove is filled. Similar to proposals for Trm14/TrmN (40), the likely RNA-binding surface of RlmM involves both domains and a positively charged inter-domain linker; but in our model, RlmM would use a distinct surface of the THUMP domain for RNA binding compared with Trm14/TrmN and RlmKL. Thus, at least two different architectures using the THUMP domain together with the Rossmann-like fold MTase domain have arisen in evolution.
The conserved basic residues in RlmM form a large surface; from the methyl group on AdoMet, the distance to K114 at the N-terminus of the long helix α4 is roughly 25 Å and the distance along this helix to R128 is another 20 Å (Figure 4). Perhaps, RlmM uses a large binding surface to be able to recognize its target site in a flexible RNA or partly assembled ribosome.
The available examples of THUMP domains display low sequence similarity and we postulate that more examples of THUMP domains are likely to be identified in structures of other enzymes acting on RNA. There is so far no structure of a THUMP domain in complex with RNA. Still, the available THUMP structures suggest that THUMP domains can bind RNA in several ways. Thus, we cannot at present predict why this particular domain has proven useful for recognition of structured RNA in all kingdoms of life.
The RNA substrate of RlmM
Our assays show that RlmM is active on in vitro transcribed 23S rRNA substrate as well as on the in vitro transcribed 604 nt domain V construct. Thus, we can conclude that RlmM activity does not depend on prior modifications to the substrate, as has been found for other rRNA MTases such as RlmD, RlmKL and RlmN (33,34,63). In addition, since RlmM can modify the domain V construct, the interaction between domain V and domain II right at the modification site is not needed for substrate recognition. Due to these interactions between domain V and domain II, C2498 is not accessible in the conformation of the crystallized ribosome structures. Thus, these interactions are likely to form after modification, suggesting that RlmM in vivo may act on a substrate where 23S domain V is folded but not yet making tertiary interactions with the other domains.
The m8A2503 modification by Cfr in vivo leads to a decreased level of modification of C2498 by RlmM (64,65). Thus, Cfr acts prior or simultaneously with RlmM, and the modification has been suggested to hinder the interaction of RlmM with its target (9). In the structure of domain V in the context of the ribosome, the modification sites are 21 Å apart, and the most likely explanation for the interplay between the two modifications is that domain V in presence of the Cfr-catalyzed methylation prefers a conformation where the 2′O of C2498 is no longer accessible for RlmM. In the mature ribosome, A2503 stacks between A2059 and G2061 at the opposite side of the peptidyl transferase loop, and possibly the methylation, apart from sterically hindering antibiotic binding, also increases the stability of the stacked structure.
The RNA recognition mechanism used by RlmM remains to be clarified. The local RNA structure may change upon binding to the enzyme, but it would also be possible that RlmM acts on an available 50S assembly intermediate where the 2′-oxygen of C2498 is accessible.
ACCESSION NUMBERS
Pdb entries 4auk, 4atn, 4b17.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
Wenner-Gren Foundation Postdoctoral Fellowship (to T.R.S.); W. M. Keck Foundation (to A.C.F.); Swedish Foundation for Strategic Research; the Swedish Research Council (project grant and URRC Linneaus center); KAW (RiboCORE); Hagbergs Foundation (to M.S.). Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Yang Chen for comments on the manuscript and Marek Kwiatkowski for discussions and expert technical assistance.
REFERENCES
- 1.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sergiev PV, Golovina AY, Prokhorova IV, Sergeeva OV, Osterman IA, Nesterchuk MV, Burakovsky DE, Bogdanov AA, Dontsova OA. Modifications of ribosomal RNA: From enzymes to function. In: Rodnina MV, Wintermeyer W, Green R, editors. Ribosomes Structure, Function, and Dynamics. Vienna: Springer Verlag GmbH; 2011. pp. 97–110. [Google Scholar]
- 3.Havelund JF, Giessing AM, Hansen T, Rasmussen A, Scott LG, Kirpekar F. Identification of 5-hydroxycytidine at position 2501 concludes characterization of modified nucleotides in E. coli 23S rRNA. J. Mol. Biol. 2011;411:529–536. doi: 10.1016/j.jmb.2011.06.036. [DOI] [PubMed] [Google Scholar]
- 4.Krzyzosiak W, Denman R, Nurse K, Hellmann W, Boublik M, Gehrke CW, Agris PF, Ofengand J. In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into a functional 30S ribosome. Biochemistry. 1987;26:2353–2364. doi: 10.1021/bi00382a042. [DOI] [PubMed] [Google Scholar]
- 5.Melancon P, Gravel M, Boileau G, Brakier-Gingras L. Reassembly of active 30S ribosomal subunits with an unmethylated in vitro transcribed 16S rRNA. Biochem. Cell Biol. 1987;65:1022–1030. doi: 10.1139/o87-134. [DOI] [PubMed] [Google Scholar]
- 6.Green R, Noller HF. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 7.Bjork GR, Isaksson LA. Isolation of mutants of Escherichia coli lac king 5-methyluracil in transfer ribonucleic acid or 1-methylguanine in ribosomal RNA. J. Mol. Biol. 1970;51:83–100. doi: 10.1016/0022-2836(70)90272-x. [DOI] [PubMed] [Google Scholar]
- 8.Huang L, Ku J, Pookanjanatavip M, Gu X, Wang D, Greene PJ, Santi DV. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry. 1998;37:15951–15957. doi: 10.1021/bi981002n. [DOI] [PubMed] [Google Scholar]
- 9.Purta E, O'Connor M, Bujnicki JM, Douthwaite S. YgdE is the 2′-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol. Microbiol. 2009;72:1147–1158. doi: 10.1111/j.1365-2958.2009.06709.x. [DOI] [PubMed] [Google Scholar]
- 10.McCusker KP, Fujimori DG. The chemistry of peptidyltransferase center-targeted antibiotics: enzymatic resistance and approaches to countering resistance. ACS Chem. Biol. 2012;7:64–72. doi: 10.1021/cb200418f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polacek N, Mankin AS. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. Biol. 2005;40:285–311. doi: 10.1080/10409230500326334. [DOI] [PubMed] [Google Scholar]
- 12.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toh SM, Mankin AS. An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors. J. Mol. Biol. 2008;380:593–597. doi: 10.1016/j.jmb.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 15.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 16.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 17.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siibak T, Remme J. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA. 2010;16:2023–2032. doi: 10.1261/rna.2160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzmann CJ, Cunningham PR, Ofengand J. Cloning, in vitro transcription, and biological activity of Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 1990;18:3515–3520. doi: 10.1093/nar/18.12.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 22.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 23.Toh SM, Xiong L, Bae T, Mankin AS. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA. 2008;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 31.Maden BE, Corbett ME, Heeney PA, Pugh K, Ajuh PM. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic ribosomal RNA. Biochimie. 1995;77:22–29. doi: 10.1016/0300-9084(96)88100-4. [DOI] [PubMed] [Google Scholar]
- 32.Smith JE, Cooperman BS, Mitchell P. Methylation sites in Escherichia coli ribosomal RNA: localization and identification of four new sites of methylation in 23S rRNA. Biochemistry. 1992;31:10825–10834. doi: 10.1021/bi00159a025. [DOI] [PubMed] [Google Scholar]
- 33.Agarwalla S, Kealey JT, Santi DV, Stroud RM. Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J. Biol. Chem. 2002;277:8835–8840. doi: 10.1074/jbc.M111825200. [DOI] [PubMed] [Google Scholar]
- 34.Kimura S, Ikeuchi Y, Kitahara K, Sakaguchi Y, Suzuki T. Base methylations in the double-stranded RNA by a fused methyltransferase bearing unwinding activity. Nucleic Acids Res. 2012;40:4071–4085. doi: 10.1093/nar/gkr1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aravind L, Koonin EV. THUMP—a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001;26:215–217. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- 38.Waterman DG, Ortiz-Lombardia M, Fogg MJ, Koonin EV, Antson AA. Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 2006;356:97–110. doi: 10.1016/j.jmb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 39.McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J. Mol. Biol. 2007;373:1243–1254. doi: 10.1016/j.jmb.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 40.Fislage M, Roovers M, Tuszynska I, Bujnicki JM, Droogmans L, Versees W. Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life. Nucleic Acids. Res. 2012;40:5149–5161. doi: 10.1093/nar/gks163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang KT, Desmolaize B, Nan J, Zhang XW, Li LF, Douthwaite S, Su XD. Structure of the bifunctional methyltransferase YcbY (RlmKL) that adds the m7G2069 and m2G2445 modifications in Escherichia coli 23S rRNA. Nucleic Acids Res. 2012;40:5138–5148. doi: 10.1093/nar/gks160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randau L, Stanley BJ, Kohlway A, Mechta S, Xiong Y, Soll D. A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science. 2009;324:657–659. doi: 10.1126/science.1170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johansson MJO, Bystrom AS. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA. 2004;10:712–719. doi: 10.1261/rna.5198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 46.Hager J, Staker BL, Bugl H, Jakob U. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 2002;277:41978–41986. doi: 10.1074/jbc.M205423200. [DOI] [PubMed] [Google Scholar]
- 47.Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 2003;10:256–263. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- 48.Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U. RNA methylation under heat shock control. Mol. Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- 49.Schluckebier G, Zhong P, Stewart KD, Kavanaugh TJ, Abad-Zapatero C. The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J. Mol. Biol. 1999;289:277–291. doi: 10.1006/jmbi.1999.2788. [DOI] [PubMed] [Google Scholar]
- 50.O'Farrell HC, Scarsdale JN, Rife JP. Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli. J. Mol. Biol. 2004;339:337–353. doi: 10.1016/j.jmb.2004.02.068. [DOI] [PubMed] [Google Scholar]
- 51.Jansson AM, Jakobsson E, Johansson P, Lantez V, Coutard B, de Lamballerie X, Unge T, Jones TA. Structure of the methyltransferase domain from the Modoc virus, a flavivirus with no known vector. Acta Crystallogr. D Biol. Crystallogr. 2009;65:796–803. doi: 10.1107/S0907444909017260. [DOI] [PubMed] [Google Scholar]
- 52.Feder M, Pas J, Wyrwicz LS, Bujnicki JM. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2'-O-methyltransferases. Gene. 2003;302:129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- 53.Lin J, Lai S, Jia R, Xu A, Zhang L, Lu J, Ye K. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature. 2011;469:559–563. doi: 10.1038/nature09688. [DOI] [PubMed] [Google Scholar]
- 54.Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2'-O-methylations in 16S and 23S rRNAs. Mol. Cell. 2006;23:173–182. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 55.Rahman A, Srivastava SS, Sneh A, Ahmed N, Krishnasastry MV. Molecular characterization of tlyA gene product, Rv1694 of Mycobacterium tuberculosis: a non-conventional hemolysin and a ribosomal RNA methyl transferase. BMC Biochem. 2010;11:35. doi: 10.1186/1471-2091-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones CM, Niederweis M. Mycobacterium tuberculosis can utilize heme as an iron source. J. Bacteriol. 2011;193:1767–1770. doi: 10.1128/JB.01312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arenas NE, Salazar LM, Soto CY, Vizcaino C, Patarroyo ME, Patarroyo MA, Gomez A. Molecular modeling and in silico characterization of Mycobacterium tuberculosis TlyA: possible misannotation of this tubercle bacilli-hemolysin. BMC Struct. Biol. 2011;11:16. doi: 10.1186/1472-6807-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Todd AE, Orengo CA, Thornton JM. Plasticity of enzyme active sites. Trends Biochem. Sci. 2002;27:419–426. doi: 10.1016/s0968-0004(02)02158-8. [DOI] [PubMed] [Google Scholar]
- 59.Liu L, Dong H, Chen H, Zhang J, Ling H, Li Z, Shi PY, Li H. Flavivirus RNA cap methyltransferase: structure, function, and inhibition. Front Biol. 2010;5:286–303. doi: 10.1007/s11515-010-0660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aittaleb M, Visone T, Fenley MO, Li H. Structural and thermodynamic evidence for a stabilizing role of Nop5p in S-adenosyl-L-methionine binding to fibrillarin. J. Biol. Chem. 2004;279:41822–41829. doi: 10.1074/jbc.M406209200. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tu C, Tropea JE, Austin BP, Court DL, Waugh DS, Ji X. Structural basis for binding of RNA and cofactor by a KsgA methyltransferase. Structure. 2009;17:374–385. doi: 10.1016/j.str.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J. Am. Chem. Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 65.Giessing AM, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.