Abstract
The RNA degradosome is a multi-enzyme assembly that contributes to key processes of RNA metabolism, and it engages numerous partners in serving its varied functional roles. Small domains within the assembly recognize collectively a diverse range of macromolecules, including the core protein components, the cytoplasmic lipid membrane, mRNAs, non-coding regulatory RNAs and precursors of structured RNAs. We present evidence that the degradosome can form a stable complex with the 70S ribosome and polysomes, and we demonstrate the proximity in vivo of ribosomal proteins and the scaffold of the degradosome, RNase E. The principal interactions are mapped to two, independent, RNA-binding domains from RNase E. RhlB, the RNA helicase component of the degradosome, also contributes to ribosome binding, and this is favoured through an activating interaction with RNase E. The catalytic activity of RNase E for processing 9S RNA (the ribosomal 5S RNA precursor) is repressed in the presence of the ribosome, whereas there is little affect on the cleavage of single-stranded substrates mediated by non-coding RNA, suggestings that the enzyme retains capacity to cleave unstructured substrates when associated with the ribosome. We propose that polysomes may act as antennae that enhance the rates of capture of the limited number of degradosomes, so that they become recruited to sites of active translation to act on mRNAs as they become exposed or tagged for degradation.
INTRODUCTION
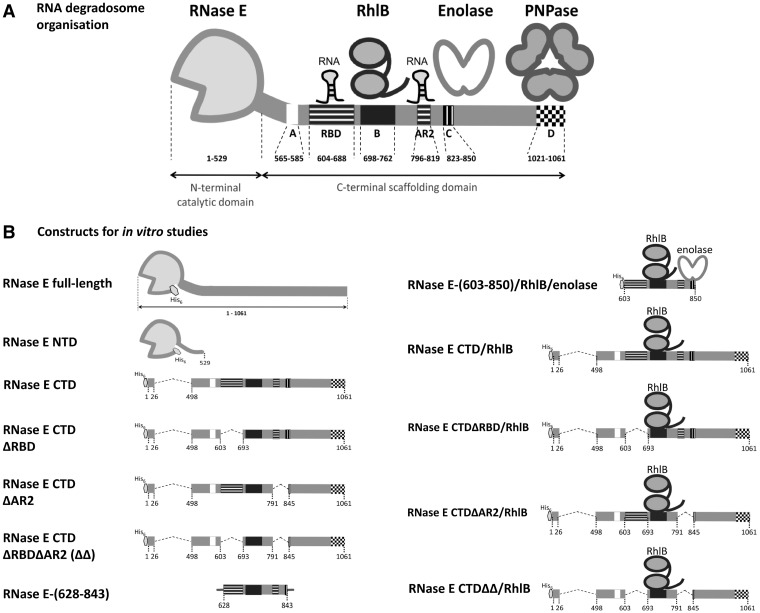
The endoribonuclease RNase E, a key enzyme of RNA turnover and processing in Escherichia coli and many other proteobacteria, forms a multi-enzyme machine of RNA metabolism, known as the RNA degradosome (1–3) (Figure 1A). The C-terminal half of RNase E acts as the scaffold for the RNA degradosome assembly and recruits its core constituents, which are the exoribonuclease polynucleotide phosphorylase (PNPase), the glycolytic enzyme enolase and the RNA helicase RhlB, a member of the DEAD-box ATP-dependent helicase family (3,4). These, and other functional components, are gained or lost according to environmental conditions and growth stage (5–7). Auxiliary degradosome components include the RNA chaperone Hfq, which facilitates the activity of small regulatory RNAs (sRNA), and poly(A) polymerase, which adds poly(A) tails at the 3′-end of RNAs to favour their degradation (5,8,9). While the RNase E C-terminal domain (CTD) is not essential for cell viability, deletion of this domain diminishes organism fitness (10), affects messenger RNA (mRNA) profiles and increases median mRNA half-lives (11). Analogous or homologous degradosome-like assemblies are found in divergent bacterial lineages (12,13), which also indicates a likely functional importance for such assemblies. The degradosome shares certain functional and structural analogies with the eukaryotic and archaeal exosomes (14).
Figure 1.
Organization of the RNA degradosome and its component constructs. (A) Schematic representation of the RNA degradosome. RNase E CTD organizes the degradosome and is punctuated by small recognition elements: segment A is the membrane-binding amphipathic helix corresponding to RNase E residues 565–585; segment B is the RhlB binding site; an enolase dimer binds segment C and a PNPase trimer binds segment D. RBD and AR2 are the two RNA binding sites of RNase E CTD. (B) Recombinant constructs used in this study. The hexagon represents the hexa-histidine tag and the grey lines in the RNase E-(628-843) diagram represent the flanking non-RNase E sequences derived from its expression vector (a 13-residue head and a 20-residue tail).
Accumulating evidence suggests that ribosome constituents are potential partners for the RNA degradosome. Ribosomal RNA co-purifies with cell-extracted degradosome preparations (15), which is in accord with the known roles of RNase E and other RNA degradosome components in the genesis and quality control of ribosomal RNA (16–18). The canonical RNA degradosome components have been identified as binding partners of ribosomal proteins from the 30S subunit (S5, S10, S13) and the 50S subunit (L4 and L23) (19). The functional role of these interactions has not been established, but it has been observed that the L4 protein from the 50S subunit inhibits the mRNA degradation activity of RNase E (20), leading to the proposal that L4 released from the ribosome during stress conditions impedes RNase E activity and consequently stabilizes transcripts encoding stress–response proteins (21). Furthermore, RNase E has been identified as the second strongest correlative factor in optimal ribosome synthesis, suggesting that the enzyme affects a key control point (22). Although it is not exactly clear how RNase E might play that role, a picture is emerging in which the enzyme is deeply networked in regulatory processes involving the RNA and protein components of the ribosome.
Whereas ribosomal RNA and protein constituents are known to interact with RNase E, there is presently no indication whether the complete ribosomal 70S assembly or its 30S and 50S subunits are partners of the ribonuclease or the RNA degradosome assembly. We have studied the interactions of the degradosome and its components with the ribosome (recombinant constructs are illustrated in Figure 1B), and our data from in vivo and in vitro experiments show that the degradosome can form a stable complex with the isolated ribosome and translating polysomes, and that RNase E and ribosomal proteins are in physical proximity in vivo. Interactions with the ribosome impede RNase E activities for complex, processed substrates, but not for simpler single-stranded substrates, suggesting that the enzyme could cleave unstructured substrates when associated with the ribosome. Based on our experimental findings, we suggest that the potential association of RNase E and the degradosome with ribosomes may facilitate the turnover of translated mRNAs and aid non-coding regulatory RNAs to terminate translation efficiently.
MATERIALS AND METHODS
Two-stage purification of RNase E-70S ribosome complex from cell cytoplasmic extract
Escherichia coli strain JE28 expressing histidine-tagged 70S (23) was grown in 2X YT media at 37°C and harvested at mid-log phase by centrifugation (1800g for 15 min at 4°C). Cell pellets were resuspended in lysis buffer (20 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 30 mM NH4Cl, 4 mM β-mercaptoethanol, one protease inhibitor tablet (Roche) per 50 ml of buffer) supplemented with 0.75 mg/ml of lysozyme, then twice shock-frozen in liquid nitrogen and thawed. Deoxycholate was added to 0.25% v/v and the lysate was left for 5 min on ice, and then clarified by centrifugation (22 000g for 20 min at 4°C). About 12.1 A260 U of lysate were layered onto a gradient of 10–50% sucrose and 0.02–0.1% v/v glutaraldehyde in 20 mM HEPES pH 7.5, 10 mM MgCl2, 100 mM KCl, 4 mM β-mercaptoethanol (24). Samples were spun at 256 136g at 4°C for 2 h with a SW40 rotor (Beckman) in a Beckman LE-80 K Ultracentrifuge and then fractionated. Fractions containing ribosome and RNase E (identified by western blot using anti-ribosomal protein and anti-RNase E antibodies) were pooled, exchanged into HS buffer (20 mM HEPES pH 7.5, 30 mM NH4Cl, 150 mM KCl, 10 mM MgCl2, 1 mM β-mercaptoethanol) and concentrated using a 10-kDa MWCO concentrator (Sartorius Stedim Biotech). The concentrated sample was applied to a His-select spin column (Sigma) pre-equilibrated with HS buffer, washed with HS buffer supplemented with 10 mM imidazole, and proteins were eluted with HS buffer supplemented with 300 mM imidazole. The eluted fractions were pooled, concentrated using a 5-kDa MWCO concentrator (Sartorius Stedim Biotech) and assessed by western blot.
In vivo cross-linking and immunoprecipitation of His-tagged 70S ribosome
Escherichia coli strain JE28 (23) encoding His-tagged 70S ribosome was also used for in vivo cross-linking. Cells were grown in 2X YT media at 37°C, pelleted at mid-log phase by centrifugation (1800g for 15 min at 4°C), washed once with ice-cold buffer (100 mM HEPES pH 8.0, 150 mM NaCl, 20 mM MgCl2) and resuspended in the same buffer to a final cell density of 3 × 1010 cells/ml (25). Dimethyl suberimidate (DMS, Pierce Biotechnology) was added to the resuspended cell culture to a final concentration of 16 mM. After 1 h at 4°C, the reaction was quenched by addition of Tris–HCl (pH 8.0) to a final concentration of 20 mM. The cross-linked cells were collected by centrifugation (3000g for 20 min at 4°C). Immunoprecipitation products of His-tagged 70S ribosome were obtained by the protocol described for 70S ribosome purification (23).
Polysome preparation and electron microscopy
Plasmid pZA11 was prepared encoding ompD in which the stop codon is replaced with a 51-bp SecM stalling sequence (TTC AGC ACG CCC GTC TGG ATA AGC CAG GCG CAA GGC ATC CGT GCT GGC CCT) under the control of the T7 promoter. Escherichia coli BL21(DE3) was transformed with the plasmid, induced during exponential growth at mid-log phase with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and harvested after 1 h. The clarified cell lysate was mixed with purified recombinant RNA degradosome and applied to a sucrose gradient (10–50% sucrose in 20 mM HEPES pH 7.5, 10 mM MgCl2, 100 mM KCl, 4 mM β-mercaptoethanol). Samples were spun at 256 136g at 4°C for 2 h with a SW40 rotor (Beckman) in a Beckman LE-80K Ultracentrifuge. Recombinant RNA degradosome was applied under the same condition as a control. Specimens from the fractionated gradient were probed with antibodies and blotted onto glow-discharge carbon coated copper grids and stained with 1% uranyl acetate. Images were collected with a Tecnai G2 electron microscope.
Preparation of E. coli strains encoding fluorescent fusion protein
Bacterial strains encoding genomically tagged fluorescent protein are summarized in Supplementary Table S1. Genomic rpsQ-PhiYFP (encoding S17-fused to Phialidium yellow fluorescent protein, hereafter PhiYFP) and rpsP-PhiYFP (encoding S16-PhiYFP fusion protein) were constructed in E. coli LEC strain [a DY329 derivative (26), with genotype W3110 ΔlacU169 nadA::Tn10gal490 pglΔ8[λ cI857 Δcro bio A] rne::rne-6xGly-cfp, encoding RNase E-CFP fusion protein) by homologous recombination, employing PCR amplified recombining cassettes (26). The strain LYLY was constructed by homologous recombination of the E. coli DY329 strain to express genomically encoded PhiYFP protein (DY329 tetA::pT5/lacO-PhiYFP). Genetic engineering of E. coli DY329 chromosome by homologous recombination was performed as previously published with minor modifications (27). Each DNA cassette contains the homology region H1 for the target ribosomal protein, a six-Glycine hinge, the full cistron sequence of a fluorescent protein [cyan fluorescent protein (CFP) or PhiYFP], and a chloramphenicol resistance marker, followed by the homology region H2 for each target gene. The cells had no visible aggregates when viewed by fluorescence microscopy.
Analysis of in vivo protein–protein proximity with donor (CFP-tagged RNase E) and acceptor (PhiYFP-tagged ribosomal proteins)
In the E. coli LEC strain, RNase E is genomically tagged with CFP. To measure the fluorescence resonance energy transfer (FRET) signal in vivo, two fusion genes of rpsP-PhiYFP and rpsQ-PhiYFP expressing PhiYFP-tagged 30S ribosomal subunit proteins S16 and S17, respectively, were constructed in the genome of E. coli LEC strain by homologous recombination. Fluorescent signal measurement was made in bacterial cultures grown overnight in LB media. Cells were pelleted, washed and resuspended in phosphate buffered saline (pH 7.4) at 2.0 A600. Samples were analysed at the following wavelengths: CFP channel, excitation at 433 nm or 458 nm, emission at 505 nm; PhiYFP channel, excitation at 514 nm, emission at 537 nm; FRET channel, excitation at 433 nm or 458 nm, emission at 537 nm under agitation at room temperature using a Synergy™ H4 Hybrid Microplate Reader spectrofluorimeter (BioTek Instruments Inc). The apparent FRET (FRETapp) was calculated as R − R0, where R is the ratio of the subtraction of the crosstalk C to the signal B2 in the FRET channel, to the signal A2 in the acceptor channel of strains containing both acceptor and donor fusions [R =(B2 − C)/A2]. The crosstalk C of the donor is calculated as the ratio of the signal C1 in the FRET channel and the signal C2 in the donor channel of the strain containing only the donor fusion (LEC strain). This ratio C1/C2 is then multiplied to the signal C3 in the donor channel of strains containing both acceptor and donor fusions. This gives C = (C1/C2) × C3. On the other hand, R0 is given as the ratio of the signal B1 in the FRET channel and the signal A1 in the acceptor channel of strains containing only the acceptor fusion (LYLY strain); therefore, R0 = B1/A1. Finally, we have FRETapp = [(B2 − C)/A2] − B1/A1]. The calculations assume that the fluorescence spectrum of YFP is the same whether it is free in the cytoplasm or conjugated to other proteins, based on our observation that the fluorescence spectrum of YFP is the same whether it is free in the cytoplasm or conjugated to the ribosomal proteins S17 or S16 in the strains co-expression RNase E-CFP (Supplementary Figure S2G and H).
Recombinant protein preparation
Recombinant degradosome was prepared as described previously (7). The N-terminal catalytic domain of RNase E (residues 1–529, RNase E NTD) was purified as described previously (28). RNase E C-terminal domain (CTD) and its deletion constructs were overexpressed and purified as described by Callaghan et al. (29). RhlB was prepared as described by Worrall et al. (30). Reconstitution of RNase E CTD constructs/RhlB subassemblies was based on a previous protocol (31) and the subassemblies were further purified by size exclusion chromatography [Sephacryl S-200 HR column (GE Healthcare) with 20 mM sodium potassium phosphate buffer, pH 7.7, 150 mM NaCl]. Full-length RNase E with a hexa-His-tag at the N-terminal end was prepared from E. coli strain BL21(DE3) harbouring pET15b-rne (A.J. Carpousis, CNRS Toulouse). The expression and purification of the recombinant RNase E was based on several reported protocols (32–34).
The 70S ribosome and its 50S and 30S subunits were prepared as previously described from E. coli strain JE28 encoding His6-tagged 70S ribosome (23) with additional purification steps. After eluting 70S ribosome from an affinity column (23), the samples were pooled and dialysed against 70S-buffer A (100 mM Tris pH 7.5, 150 mM KCl, 30 mM NH4Cl, 10 mM MgCl2 and one EDTA-free protease inhibitor tablet, Roche) with 0.5 mM fresh DTT for 2.5 h with an 8 kDa MWCO dialysis membrane. The dialysed samples were recovered and spun at 100 000g for 2 h at 4°C (Optima Max, Beckman). In total, 200 µl of 70S-buffer A with 3 mM fresh DTT was layered over each pellet and left on ice for 24 h before resuspension. Concentrated 70S ribosome was subsequently applied to a Sephacryl S-500 HR column (GE Healthcare) equilibrated with 70S-buffer A supplemented with 500 mM NaCl. Fractions containing pure 70S ribosome were pooled and concentrated by a 100 kDa MWCO concentrator (Millipore).
Electrophoretic mobility shift assays
For recombinant degradosome (RNase E F575E) and 70S ribosome analysis, 5 nM of recombinant degradosome was mixed with 0.5, 1.25, 2.5, 12.5, 25, 50, 100 and 250 nM of 70S ribosome to a total reaction volume of 20 μl. A casting of 0.5% agarose gel was made with 0.5X TB buffer supplemented with 10 mM MgCl2, 10 mM DTT and 0.01% β-dodecylmaltoside. Electrophoresis was performed at 50V for 4 h. For RNase E CTD constructs and 70S ribosome analysis, 2.4 μM of CTD constructs was mixed with 0.3 μM of 70S. The total volume was 10 μl. A casting of 0.5% agarose gel was made with 0.5X TB buffer supplemented with 10 mM MgCl2 and 0.01% Triton X-100 and electrophoresis was performed at 30V for 5–6 h. For RNase E CTD constructs and ribosome subunits (50S and 30S) analysis, 2.4 μM of CTD constructs was mixed with 2.4 μM of 50S or 30S, whereas RNase E-(628-843) was used at 3.1 μM. The total volume was 10 μl. A casting of 0.6% agarose gel was made with TG buffer (24 mM Tris–HCl, 190 mM glycine, pH 8.3 supplemented with 10 mM MgCl2).
Proteins in the native agarose gel were stained with Coomassie Blue (0.025% Coomassie Blue R-250, 40% methanol, 7% acetic acid) and destained with 50% methanol and 7% acetic acid, followed with Milli-Q water or subsequently electrotransferred onto polyvinylidene fluoride (PVDF) membrane for western blot. For liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of the 70S/degradosome sample, the shifted band was excised from the Coomassie Blue-stained agarose gel, treated with trypsin, and analysed by the Cambridge Centre for Proteomics using liquid chromatography and tandem mass spectrometry with an orbitrap spectrometer.
Sucrose gradient co-sedimentation
A 10–50% (w/v) sucrose gradient was prepared in 20 mM HEPES pH 7.5, 10 mM MgCl2, 100 mM NH4Cl, 4 mM β-mercaptoethanol with polyallomer tubes (14 ml, 14 x 95 mm, Beckman Coulter) (35). RNase E CTD constructs and 70S ribosome were mixed at 16:1 molar ratio, and their final concentrations were as follows: 11.2 μM CTD and 0.7 μM 70S to a total of 712.7 μl; 43.8 μM CTDΔRBD and 2.8 μM 70S to a total of 165 μl; 60 μM CTDΔAR2 and 3.8 μM 70S to a total of 121 μl; 38.5 μM CTDΔΔ and 2.4 μM 70S to a total of 167 μl; 35 μM CTDΔΔ/RhlB and 2.2 μM 70S to a total of 183 μl. Samples were spun in a SW40 rotor (Beckman) and spun at 256 136g at 4°C for 2 h in a Beckman LE-80K Ultracentrifuge and then fractionated.
Surface plasmon resonance
70S ribosome was biotinylated with N-hydroxysuccinimidobiotin (NHS-biotin, Pierce Biotechnology, Inc.) in 20 mM HEPES pH 7.5, 150 mM KCl, 10 mM MgCl2 according to the manufacturer’s instructions. The SA sensorchip surface was immobilized with 150 RUs of biotinylated 70S ribosome in running buffer HBS-P, 50 μM EDTA, 10 mM MgCl2, 50 mM KCl and 1 mM β-mercaptoethanol. Purified RNase E CTD at a concentration range of 0–250 nM was injected at a 60 μl/min constant flow rate. Under the same conditions, RNase E CTDΔRBD was at 0–250 nM, RNase E CTDΔAR2 at 0–250 nM and RNase E CTDΔΔ at 0–600 nM. The analyte contact time was 60s and the dissociation time was 5 min. The surface was regenerated with running buffer containing 1 M NaCl and the regeneration period lasted five cycles of 60s injection. The interactions were tested in triplicate. Data were evaluated by Biacore T100 evaluation software and fit to the 1:1 kinetic binding model to give the KD of each interaction tested.
For subassemblies of RNase E CTD constructs and RhlB binding to 70S ribosome, the SA sensorchip surface was immobilized with 4000 RUs of biotinylated 70S ribosome in running buffer HBS-P, 50 μM EDTA, 10 mM MgCl2, 50 mM KCl and 1 mM β-mercaptoethanol. Purified RNase E CTD/RhlB, CTDΔRBD/RhlB, CTDΔAR2/RhlB and CTDΔRBDΔAR2/RhlB at 300 nM were injected at a 30 μl/min constant flow rate. Purified RNase E NTD was injected at the same concentration (300 nM) to compare the association/dissociation curves. The analyte contact time was 60s and the dissociation time was 3 min. The surface was regenerated with running buffer containing 1 M NaCl and the regeneration period lasted five cycles of 60s injection. Data were evaluated by Biacore T100 evaluation software.
RNA preparation and RNase E activity assays
9S RNA was produced and utilized for RNA processing assay by RNase E constructs as previously described (7,36). The preparation of ompD mRNA and sRNA MicC were described by Pfeiffer et al. (37). Proteins (RNase E full-length and RNase E NTD) and 70S ribosome samples were prepared at 2 µM with 2X reaction buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 100 mM KCl, 20 mM MgCl2, 2 mM DTT and 1U/µl RNaseOUT) and RNA samples were prepared at 2 µM in Milli-Q water. For the ompD, MicC and Hfq mixture, 2 μM of ompD and MicC were pre-incubated separately at 50°C for 2 min and cooled to room temperature on the bench. The two RNAs were then mixed with reaction buffer and incubated at 37°C for 5 min before incubating with 2 μM of Hfq. After 10 min of incubation with Hfq at the same temperature, the RNA substrate mixture was ready for degradation assay. The mixtures of RNase E NTD:70S and RNase E:70S at 1:0.25, 1:0.5 and 1:1 molar ratio were prepared on ice following by pre-incubation at 37°C for 10 min. After pre-incubation, all the mixtures were left on ice for 1 min before adding RNA substrate (9S or ompD mRNA or Hfq-MicC-ompD ternary complex). The reaction mixture (10 μl) was incubated at 37°C for 30 min and then quenched by 10 µl of PK mix (0.5 mg/ml Protease K in 100 mM Tris–HCl pH 7.5, 12.5 mM EDTA, 150 mM NaCl and 1% w/v SDS) at 50°C for 20 min. The samples with 10 µl of 2X RNA loading dye (Fermentas) were heated at 90°C for 3 min before loading into denaturing polyacrylamide gel. Denaturing gel electrophoresis, nucleic acids staining and analyses were conducted as previously described (36).
RESULTS
RNase E co-purifies with 70S ribosome
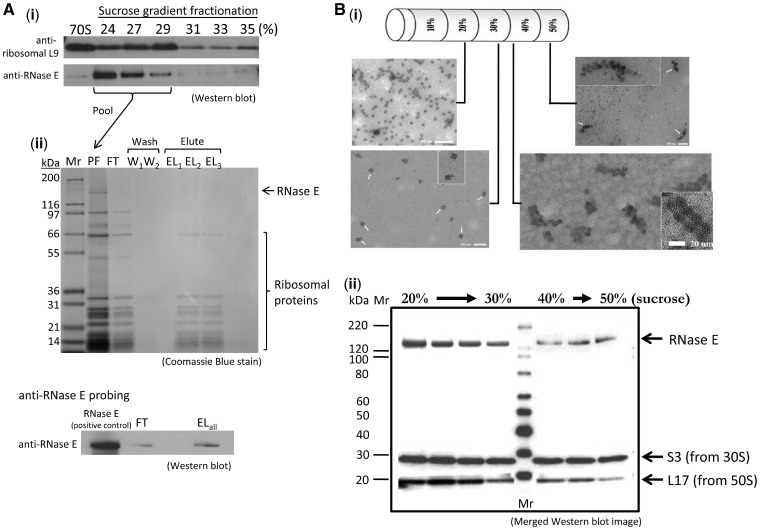
An E. coli strain expressing genomically encoded histidine-tagged ribosomal protein L12 (23) was used to isolate potential binding partners from whole cell extracts (strain JE28). Soluble whole cell extracts from the JE28 strain were resolved by ultracentrifugation in a 10–50% sucrose gradient that included a 0.02–0.1% (v/v) crosslinker glutaraldehyde gradient to aid complex stability (38). Both RNase E and ribosomal proteins were found at 24–29% sucrose by western blotting of the fractionated gradient [Figure 2A (i)]. These fractions were pooled and applied to a His-select spin column for the second stage affinity purification, and a signal for RNase E was detected for the pooled elution fractions [Figure 2A (ii)]. A trace of RNase E was detected in the flow through fraction due to column overloading, as ribosomes are present in the flow through fraction as well [lower panel, Figure 2A (ii)]. Although a proportion of the material had been lost during the course of preparation, the results suggest that the complex of RNase E and ribosome could be isolated after the two-stage purification (flowchart illustrated in Supplementary Figure S1).
Figure 2.
Isolation of RNase E-70S ribosome and RNase E-polysome complexes from E. coli cell extracts. (A) An RNase E-70S ribosome complex can be isolated from E. coli cell extracts with a two-step procedure. (i) Initial purification using sucrose gradient ultracentrifugation with chemical crosslinker in the gradient. Escherichia coli JE28 cell cytoplasmic extract was resolved in a 10–50% sucrose gradient containing glutaraldehyde. Fractions from the sucrose gradient were resolved on a denaturing gel and detected using antibodies against RNase E and ribosomal protein L9, which gave representative signal for the antibody detection of the 70S particle. Signals of both RNase E and L9 indicate that the two proteins co-migrated at 24–29% sucrose solution. (ii) The fractions enriched for RNase E and L9 were pooled and analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE), showing that ribosomal proteins from both 50S and 30S subunits are present, as well as a band corresponding to RNase E (lane PF). This pooled material was used for affinity purification with a His-select spin column. The flow through fraction (FT), two aliquots of washing step (W1, W2) and three aliquots of eluted fractions (EL1 − EL3) were analysed by 4–12% SDS–PAGE (upper panel). Western blot analysis of FT fraction and pooled elutions (ELall) was performed to detect RNase E in these samples (lower panel). After probing with anti-RNase E antibody, a faint band appeared in the FT fraction (due to column overloading as a portion of ribosomes also appeared in FT) and a more defined signal of RNase E was present in the EL fraction. (B) RNase E co-sediments with polysomes in the dense sucrose gradient fractions when overexpressing the artificially paused transcript ompD-SecM in E. coli. The stop codon of the Salmonella ompD transcript is replaced by a sequence, SecM, known to stall ribosomes during translation. (i) Electron microscopy images of sucrose gradient fractionation samples. The specimens were visualized by uranyl acetate negative staining at a magnification of 29 000. (ii) The mixture of endogenous polysome and recombinant degradosome were resolved on a sucrose gradient. Samples from different fractions containing 70S ribosomes, tetrasomes and polysomes were probed with antibodies against ribosomal proteins and RNase E. The western blot image is merged after separately probing with anti-RNase E, anti-S3 and anti-L17 antibodies. The recombinant degradosome itself localizes in the lighter fractions (results not shown).
Identification of 70S in vivo partners by in situ chemical crosslinking
A culture of the JE28 strain expressing histidine-tagged ribosomal protein L12 was treated with bifunctional chemical crosslinker (dimethyl suberimidate), and ribosomes were enriched from the cell extracts by step-elution from a Nickel chelating column and then pelleted by ultracentrifugation at 150 000g. Compared with controls, several new cross-linked species in the range of 90–150 kDa were observed by denaturing gels (not shown). Mass spectrometry analysis of these bands from the gels identified RNase E and its degradosome partners RhlB and PNPase (Supplementary Table S2). Interestingly, the dominating species identified was the beta-subunit of RNA polymerase, consistent with close coupling of translation and transcription. Many tRNA synthetases were also observed, suggesting perhaps that these enzymes might be compartmentalized with translating ribosomes.
RNase E co-sediments with polysomes in gradient ultracentrifugation
To test if RNase E can also interact with polysomes in vivo, we enriched for stalled polysomes by overexpressing an artificial transcript in which the stop codon of ompD is replaced with a translation pausing sequence, namely the SecM signal (39). Cells expressing the plasmid were also treated with chloramphenicol before harvesting to stop translation. The stalled ribosomes accumulated on the artificially paused transcript and could be separated from 70S particles by ultracentrifugation in sucrose gradients. Negative stain electron microscopy images of the sucrose gradient fractions revealed that 70S ribosomes were distributed at ∼20% sucrose, whereas polysomes were observed at 40–50% sucrose [Figure 2B (i)]. Fractions containing 70S ribosomes, tetrasomes and polysomes were probed with antibodies against ribosomal proteins and RNase E [Figure 2B (ii)]. The merged western blot shows that RNase E is present in both ribosome and polysome fractions. Purified recombinant degradosome sediments at earlier fractions (10–20% sucrose; data not shown), suggesting that the co-sedimentation of polysomes and RNase E is due to a physical interaction.
Protein–protein interaction between RNase E and 30S ribosomal proteins by FRET in vivo
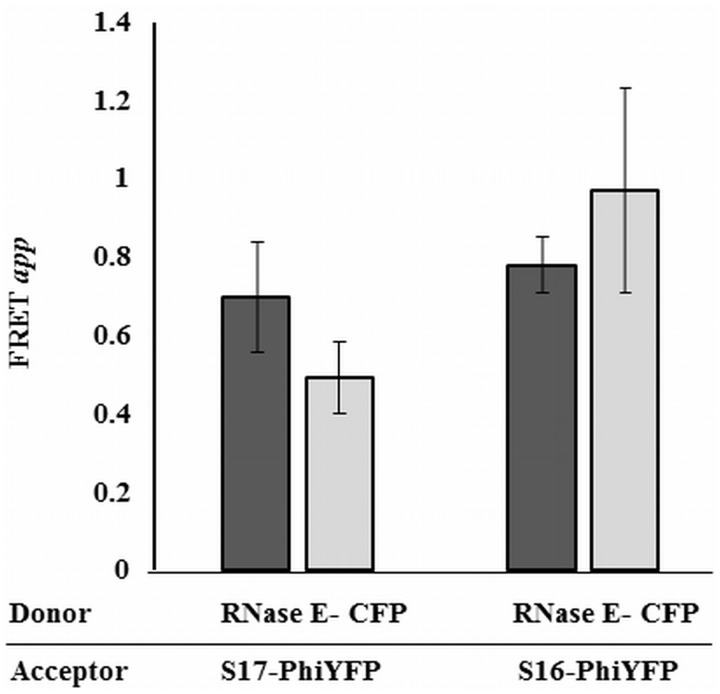
The chromosomal genes encoding small subunit ribosomal proteins S16 and S17 were fused with PhiYFP in two separate engineered constructs. These two constructs were selected for their surface accessibility in the 70S particle, and they each gave a good fluorescent signal. The two fusions were also introduced into a strain that carries an RNase E- CFP fusion protein. The proximity between the selected ribosomal proteins and RNase E was determined in vivo by measuring the FRET signal between the PhiYFP-tagged ribosomal proteins (acceptors) and the CFP-tagged RNase E (donor) by end-point fluorimetry (Figure 3 and Supplementary Figure S2). The apparent FRET (FRETapp) was calculated as described in the ‘Materials and Methods’ section, and the calculations assume that the fluorescence spectrum of PhiYFP is the same, whether it is free in the cytoplasm or conjugated to other proteins. The strength of the signal is consistent with a separation of <100 Å between the fluorophores, suggesting that the selected protein pairs (S16/RNase E and S17/RNase E) are in close proximity, and these most likely would be part of the 30S complex rather than free proteins. These observations are consistent with the hypothesis of an in vivo interaction between the 30S particle and RNase E.
Figure 3.
Evidence for proximity between RNase E and small subunit ribosomal proteins in vivo. The apparent fluorescence resonance energy transfer (FRETapp) between RNase E-CFP donor and PhiYFP acceptor fusions of S16 or S17 from the 30S, was measured by end point fluorimetry as described in ‘Materials and Methods’ section. Cells co-expressing chromosomal RNase E-CFP and the fluorescent 30S ribosomal proteins were excited at 433 nm (dark grey bars) and at 458 nm (light grey bars) and the fluorescence at 537 nm was recorded. Fluorescence from E. coli LEC cells with PhiYFP and RNase E-CFP were used as background corrections to estimate the FRET signal (Supplementary Figure S2 for details). Standard error is indicated on top of each histogram.
In vitro analysis of degradosome/ribosome interactions
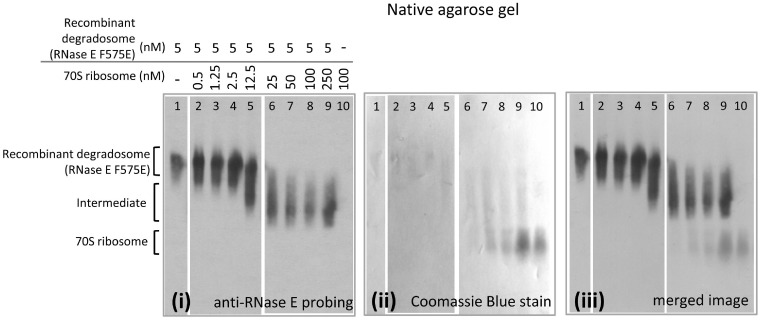
To explore the potential interactions of the degradosome (RNase E/RhlB/enolase/PNPase) and ribosome in vitro, binding was evaluated using agarose gel electrophoresis under native conditions (Figure 4). Native gel shifts were also evaluated by western blot using anti-RNase E antibody to probe the degradosome complex position, and this permitted work at much lower concentrations. For these experiments, the substitution F575E was introduced to improve the protein solubility and overcome a problem with aggregation of RNase E during purification (M. Górna, unpublished data). F575E is in RNase E Segment A (Figure 1A) (29), corresponding to residues 565–585, forms an amphipathic α-helix and is involved in cytoplasmic membrane association (29,40). The recombinant RNA degradosome (with the mutant RNase E F575E) was mixed with 70S ribosome in different molar ratios and resolved by native agarose gel electrophoresis. A diffuse, mobility-shifted species was identified as containing RNase E by western blotting [Figure 4 (i)] and total protein visualized by staining with Coomassie Blue [Figure 4 (ii)]. Shifted species are seen at the molar ratio of 1:2.5 (RNA degradosome:70S ribosome) and increase with an increasing ratio of 70S ribosome to RNA degradosome, but then remains the same for samples above molar ratios at 1:10, implying that there is a binding saturation point. Excess free 70S ribosome can be seen on the merged image of Coomassie Blue stain and western blot [Figure 4 (iii)]. It is expected that in vivo ribosomes will be ≥100-fold excess over RNase E and the degradosome.
Figure 4.
Native gel electrophoretic mobility shift assay of degradosome and 70S ribosome. Electrophoretic mobility shift of purified recombinant degradosome with purified His-tagged 70S ribosome in a native agarose gel. Purified recombinant RNA degradosome with a mutation in the membrane association segment A (F575E) and 70S ribosome were mixed in different ratios: 1:0.1, 1:0.25, 1:0.5, 1:2.5, 1:5, 1:10, 1:20 and 1:50 (degradosome:ribosome). The proteins were electrotransferred onto PVDF membrane and probed with anti-RNase E antibody (i) and then total protein visualized with Coomassie Blue (ii). With an increasing ratio of ribosome to degradosome, RNase E is seen to migrate to a position between that identified for free degradosome and free 70S ribosome, as seen on the merged image (iii).
70S ribosomes stripped of the S1 protein using polyU-sepharose also bound to the recombinant degradosome with the native agarose gel assay, suggesting that the putative interaction of the two assemblies is not mediated through the S1 domain (data not shown).
The interaction of wild-type recombinant degradosome (prepared with wild-type RNase E) and 70S ribosome was also observed using native agarose gel electrophoresis and the mobility shift was similar to the binding of mutant recombinant degradosome (RNase E F575E) and 70S ribosome (data not shown). The intermediate band was excised and analysed by tandem mass spectrometry, identifying peptides from both the small and large ribosomal subunits and from the canonical degradosome components: RNase E, PNPase, enolase and helicase (Supplementary Table S3). These data suggest that the degradosome and ribosome can form a stable complex, and that the canonical components of the degradosome are retained in the complex. The mass spectrometry data also identified minor components that co-purified with the degradosome or ribosome. These include the DEAD-box helicases SrmB, CsdA and RhlE, which are involved in ribosome assembly (41). Other degradosome-associated proteins of note were the RNA chaperone Hfq and poly(A) polymerase (5). The western blot analysis using anti-Hfq antibodies indicates that Hfq co-purifies with His-tagged 70S ribosome (results not shown). These observations are consistent with findings that Hfq is associated with polysome fractions (42). Pre-treatment of the 70S ribosome with RNase I under conditions that degrades naked RNA had little effect on the binding of the degradosome, suggesting that the interaction is not dependent on exposed transcript (Supplementary Figure S3).
The scaffold of the degradosome, RNase E, interacts with ribosomes through two RNA binding sites in its CTD
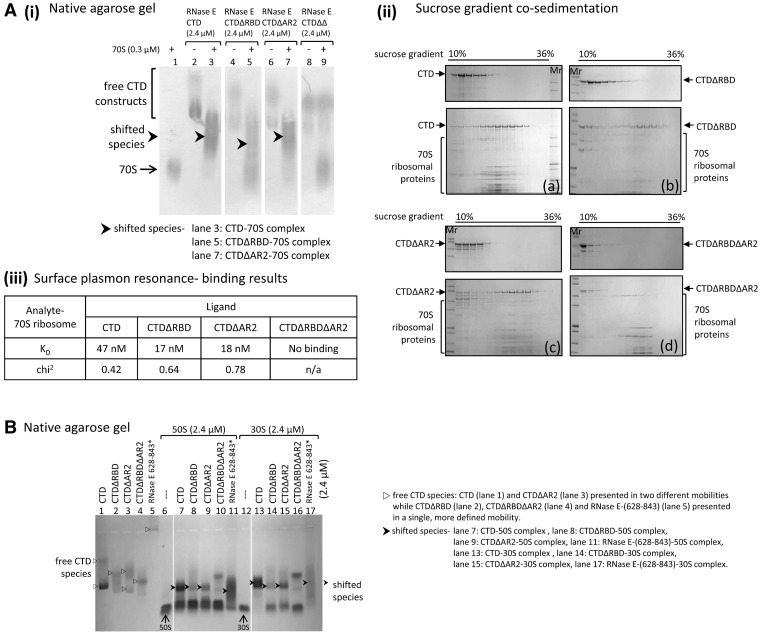
In addition to the protein binding sites required for degradosome assembly, the RNase E CTD also harbours two sites for RNA binding (Figure 1A) (3). With native agarose gel electrophoresis, purified CTD shifted mobility in the presence of 70S ribosome, and similar shifts were also observed with purified CTD constructs lacking either of the RNA-binding sites [CTDΔRBD and CTDΔAR2; Figure 1B and Figure 5A (i), lanes 1–7]. A CTD construct lacking both of the RNA binding sites, CTDΔRBDΔAR2 (or CTDΔΔ), failed to shift mobility in the presence of 70S ribosome [Figure 5A (i), lane 9 compared with lanes 8 and 1].
Figure 5.
RNase E CTD constructs containing at least one RNA-binding site interact with 70S ribosome and its subunits, 50S and 30S. (A) Interactions between RNase E CTD constructs and 70S ribosome in vitro. (i) Electrophoretic mobility shift assay of RNase E CTD constructs and 70S ribosome. Prior to electrophoresis, protein samples were mixed at 8:1 (CTD constructs:70S ribosome) molar ratio. Protein samples were stained with Coomassie Blue. (ii) Gradient co-sedimentation of RNase E CTD construct (a), CTDΔRBD (b), CTDΔAR2 (c) and CTDΔRBDΔAR2 (d) with (lower gel) and without (upper gel) 70S ribosome. Before layering protein samples on a 10–50% sucrose gradient, the samples of CTD constructs with 70S ribosome were mixed at a 16:1 molar ratio. The fractionated samples were analysed using 4–12% SDS–PAGE and the protein samples were stained with Coomassie Blue. (iii) Data for dissociation constants of RNase E CTD constructs and 70S ribosome were estimated by kinetic analyses. Biotinylated ribosomes were immobilized on a SA chip while RNase E CTD constructs were injected as analytes. (B) Native gel electrophoresis of RNase E constructs and ribosome subunits (50S and 30S). Prior to electrophoresis, RNase E constructs were mixed with 50S and 30S ribosomal subunits, respectively at 1:1 molar ratio, except the RNase E-(628–843) peptide was mixed with the subunits at 1.3:1 peptide:subunit ratio (asterisk). The protein samples were stained with Coomassie Blue.
RNase E CTD can avidly bind a 27-mer RNA with fluorescent 5′-end label that is predicted to have no secondary structure (compare lanes 2 and 5 in panel iii, Supplementary Figure S4). Upon introducing 70S ribosome to pre-mixed RNase E CTD-27 mer RNA complex, the 27-mer RNA is displaced, and a shifted species of RNase E CTD and 70S ribosome forms (Supplementary Figure S4, lanes 1–4). The 70S ribosome also interacts with the peptide RNase E-(628-843) and the degradosome subassembly RNase E-(603-850)/RhlB/enolase, which contains RhlB and enolase reconstituted on a part of RNase E CTD (Figure 1B and Supplementary Figure S5).
The interactions of the CTD with the 70S ribosome could also be detected using ultracentrifugation with sucrose gradients. The CTD constructs distributed at the 10–18% sucrose concentration range, whereas 70S ribosome distributed at 18–32% sucrose, as expected, based on the relative masses of the two samples. When mixed with 70S ribosome, the CTD constructs containing at least one of the RNA binding domains co-migrated with 70S at 20–30% sucrose. However, CTDΔΔ and 70S ribosome still migrated separately [Figure 5A (ii)]. The co-sedimentation results are consistent with native gel shifts and suggest that RBD and/or AR2 RNA binding sites on RNase E CTD are the 70S ribosome contacting regions.
The strengths of the association between the RNase E CTD constructs and 70S ribosome were measured by surface plasmon resonance [Figure 5A (iii) and Supplementary Figure S6]. The KD of the interaction of CTD, CTDΔRBD and CTDΔAR2 with 70S ribosome is in the nanomolar range, indicative of tight binding. The binding of the CTD is less strong compared with either of the constructs lacking one of the two RNA binding sites. Whether this indicates negative cooperativity between the two binding sites is not presently clear. RNase E NTD, which encompasses RNase E residues 1–529 and is the catalytic domain, did not have an obvious or specific interaction with 70S ribosome in the native agarose gel electrophoresis (Supplementary Figure S7A), and only weakly co-sedimented with the ribosome in gradient centrifugation (Supplementary Figure S7B).
Interactions of RNase E with the isolated 30S and 50S subunits
RNase E domains were tested for binding to the isolated 50S and 30S ribosomal subunits. Native gel results suggest that RNase E binds 30S ribosomal subunits and, to a lesser extent, the 50S subunits. At a ratio of 1:1, and a concentration of 2.4 µM, the RNase E CTD shifted all of the free 30S into a new complex (Figure 5B, compare lanes 1, 12 and 13). The CTD may also bind 50S, but the results suggest substantially weaker binding, because free 50S remains in the mixture of the components (Figure 5B, lane 7). Surface plasmon resonance data suggest that the dissociation constant of CTD-30S is in the same order as CTD-70S, whereas that of CTD-50S is ∼10-times weaker (data not shown). RNase E CTD variants containing one of the RNA binding sites showed some mobility shift in the presence of 30S and weaker binding to 50S (Figure 5B, lanes 14 and 15; lanes 8 and 9, respectively), whereas CTDΔΔ did not shift (Figure 5B, lanes 10 and 16). 50S and 30S subunits also caused a drastic mobility shift of RNase E-(628-843) peptide (Figure 5B, lanes 11 and 17). The recombinant degradosome forms a gel-shifted species with the isolated 30S subunit in native agarose gels (Supplementary Figure S3, lanes 7–10). The data taken together indicate that the two RNA binding sites on RNase E CTD are involved in interaction with the isolated ribosome subunits, and that the stronger contacts may be made to the 30S subunit.
RhlB interaction with ribosomes requires the degradosome scaffolding domain of RNase E
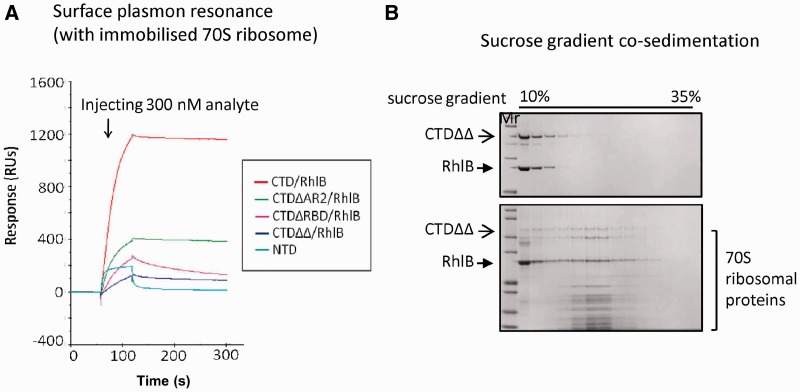
The RNase E CTD constructs were able to form a subassembly with RhlB. In fact, the RNase E CTD/RhlB, RNase E CTDΔRBD/RhlB and RNase E CTDΔAR2/RhlB complexes also interact with 70S ribosome, and the interactions were observed consistently by sucrose gradient co-sedimentation (data not shown) and surface plasmon resonance (Figure 6A). One result to emphasize is the interaction of RNase E CTDΔΔ/RhlB with 70S. In isolation, neither CTDΔΔ (lacking both of the RNA binding sites) nor RhlB bound to the 70S ribosome (Figure 5A, data not shown). However, combined CTDΔΔ and RhlB co-sedimented with 70S in sucrose gradient (Figure 6B). Earlier studies reported that RhlB requires a minimum binding peptide on RNase E CTD (residues 696–762) to activate its ATPase activity, indicating that interactions with RNase E affect the helicase conformation (30). Taken together, these results suggest that RhlB may adapt its conformation in the context of the degradosome assembly so that it becomes capable of ribosome binding. It is also possible that RhlB helps to pre-organize the RNA-binding sites in the RNase E CTD to bind better to the 70S ribosome.
Figure 6.
Subassemblies of RNase E CTD constructs with RhlB, interact with 70S ribosome. (A) Surface plasmon resonance analysis. Equal amount of subassemblies of RNase E CTD constructs/RhlB or NTD were injected over immobilized 70S ribosome and their association/dissociation curves compared. The dissociation curves for the subassemblies indicate a stable interaction between analytes and ligands (with weaker stability for RNase E CTDΔRBD/RhlB). The interaction between RNase E NTD and 70S ribosome is considered to be weak or non-specific. (B) Co-sedimentation analysis of RNase E CTDΔRBDΔAR2/RhlB complex and 70S ribosome. The CTDΔRBDΔAR2/RhlB complex migrated with 70S ribosome to 23–24% sucrose (lower gel), whereas the subassembly itself distributed at 10–20% sucrose (upper gel).
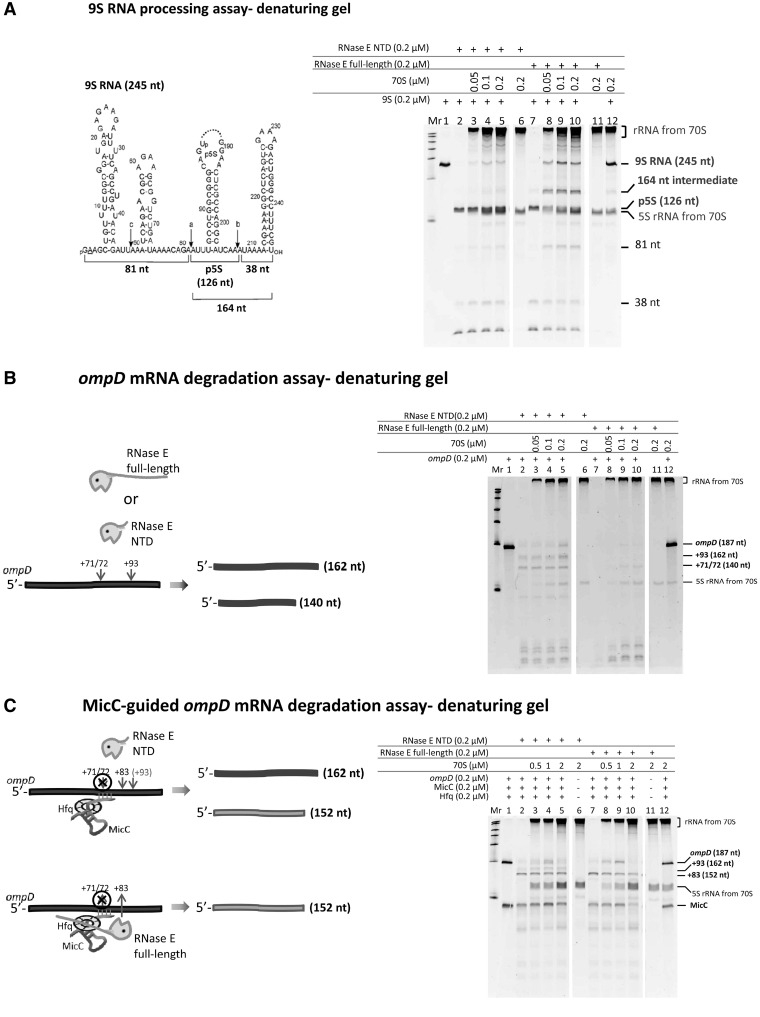
70S ribosome affects RNase E activity for processing, but not for degradation
The question arises whether the interaction with 70S particles and polysomes affects the catalytic activity of RNase E. To address this, we tested whether RNase E could cleave substrates in vitro in the presence of 70S ribosome. RNase E processing activity was assessed by the well-established 9S RNA processing assay (7,43,44). After in vitro treatment with purified recombinant RNase E, the 9S RNA was cleaved into three segments: the p5S (precursor of 5S rRNA, 126 nt), 81 and 38 nucleotides (Figure 7A, lanes 1, 2 and 7). Purified recombinant, full length RNase E (without the other degradosome components) and its N-terminal catalytic domain (RNase E-NTD) showed similar activities on processing 9S RNA after 30 min incubation. RNase E-NTD and full length RNase E were pre-incubated with 70S ribosome at 1:0.25, 1:0.5 and 1:1 molar ratio at 37°C for 10 min to allow the enzymes to bind to the ribosomes before addition of substrate. Comparing the amount of 9S remaining in each reaction to the standard degradation of 9S RNA, it could be seen that the RNase E-NTD and RNase E activities were inhibited by 70S ribosome, with a maximal inhibition at 1:0.5 enzyme:70S ribosome molar ratio (Figure 7A, compare lanes 2 and 5; 7 and 10). The 70S ribosome showed greater inhibition of the processing activity of full-length RNase E compared with the isolated catalytic domain (NTD); furthermore, a 164-nt intermediate was observed that was absent from the controls (Figure 7A, lanes 8–10). Somehow, the presence of the 70S affects the interactions of the RNase E-CTD with the structured substrate, and so affects its access by the RNase E catalytic domain.
Figure 7.
The presence of 70S ribosome decreases RNase E activity for processing the 9S RNA substrate but does not affect RNase E activity for degrading ompD mRNA. (A) Assay of 9S processing activity by full length RNase E and its isolated catalytic domain (NTD) in the presence of 70S ribosome. Lane 1 shows the initial 9S substrate amount provided in each reaction. NTD processed 98.7% of 9S (lane 2) and RNase E processed 98.5% (lane 7). 70S ribosome inhibits NTD activity with increasing concentration by 4.5, 10.9 and 9% and inhibits RNase E activity by 24.1, 42.3 and 30.2% (lanes 3–5 and lanes 8–10). In control assays, neither NTD nor RNase E digests 70S ribosome (lanes 6 and 11). The 9S processing reaction products were resolved on an 8% denaturing polyacrylamide gel. Reactant quantities are indicated in the panel above the gel. The amount of the remaining 9S substrate was quantified using GeneTools software (Syngene). (B) In ompD degradation analysis, lane 1 shows the amount of ompD provided in each reaction. In the absence of ribosome, RNase E and its isolated catalytic domain digested almost all the substrates (lanes 7 and 2). In the presence of 70S, RNase E and the catalytic domain retain most of their activity (lanes 8–10 and lanes 3–5). (C) In the degradation analysis of the Hfq–MicC–ompD ternary complex, lane 1 shows the equivalent amount of MicC and ompD provided in each reaction. Full length RNase E is as active as its isolated catalytic domain in the absence of 70S and the sRNA-induced cleavage (lanes 7 and 2). Increasing the amount of 70S slightly inhibits RNase E NTD and RNase E activity (lanes 3–5 and lanes 8–10). Inhibition was not observed with RNase E:70S at a 1:1 ratio (lane 10). In control assays, neither RNase E NTD nor RNase E digested 70S ribosome (lanes 6 and 11 in A, B and C).
The 9S substrate and 70S, both bind to RNase E-CTD, but poorly to RNase E-NTD (Supplementary Figure S8). One possible explanation for why RNase E activity for processing 9S RNA is inhibited by the presence of 70S ribosome, is that ribosome competes with 9S RNA for binding to the two RNA-binding domains of RNase E CTD and consequently blocks the presentation of 9S RNA to the RNase E active site. This could explain why 70S ribosome shows a greater impact on the processing activity of full length RNase E than on the activity of the catalytic domain of RNase E. Similarly, it accounts for the appearance of the 164-nt intermediate seen for the degradation of 9S RNA by full length RNase E in the presence of 70S ribosome (Figure 7A, lanes 8–10).
Effects of 70S on sRNA-mediated activity of RNase E
ompD is an mRNA encoding a porin membrane protein and a 187-nt segment of ompD mRNA (−69 to +118) is known to be cleaved by RNase E (37). The cleavage pattern of the 187-nt segment of the transcript by RNase E is similar in the presence and absence of 70S ribosome (Figure 7B). The sRNA MicC is a regulator of ompD expression, and it functions together with the RNA chaperone Hfq to trigger targeted mRNA degradation by RNase E (37,45). In the presence of Hfq, MicC and ompD, the full-length RNase E and the catalytic domain digested ompD by preferential cleavage at position + 83, which appears to be a stable product (Figure 7C, lanes 7 and 2). When the reaction mixture also includes 70S ribosome, RNase E and its catalytic domain activities to cleave ompD were modestly inhibited, with the greatest extent of inhibition seen at 1:0.5 molar ratio of enzyme:70S (Figure 7C, lanes 9 and 4). It is interesting to note that 70S here shows more impact on the full-length RNase E. These results could arise from 70S occupying the sites of the RNase E CTD that might bind Hfq and form a stable RNase E–Hfq–MicC complex.
DISCUSSION
Our data from in vivo and in vitro experiments show that RNase E and the RNase E-based degradosome assembly can form a stable complex with the isolated ribosome. Immunoprecipitation and sucrose gradient co-sedimentation identify the RNase E-70S ribosome and polysome complexes from cell extracts. FRET measurements from bacterial cultures indicate a close proximity between RNase E and two proteins (S16 and S17) of the 30S subunit in vivo. The complex of recombinant degradosomes and ribosomes can be resolved by native electrophoresis, and the interactions of subassemblies of the degradosome have been evaluated by surface plasmon resonance. Using these approaches, the recognition sites of the degradosome for the ribosome have been mapped to the two RNA binding domains in the C-terminal scaffolding region of RNase E. The binding data also indicate that the DEAD-box helicase RhlB interacts with the ribosome, but this interaction requires allosteric activation of the helicase by RNase E. The ribosome binding sites in RNase E can function independently, although their physical proximity likely favours cooperativity through a chelate effect. These domains are likely to interact with the RNA in the ribosome, but it is also possible that they may contact the ribosomal proteins.
Estimates of the dissociation constant (KD) for the binary association of the RNase E CTD and 70S ribosome are in the nanomolar range by surface plasmon resonance. Gel mobility assays of the recombinant degradosome and 70S are more consistent with KD in the micromolar to submicromolar range. We note that it is estimated that there are roughly 250 RNase E tetramers in E. coli and other bacteria, such as Caulobacter crescentus (46). In contrast, the number of ribosomes is about 70 000 for exponential growth (47), so there is an excess of approximately 300 ribosomes for each degradosome, corresponding to perhaps 20 polysomes for every degradosome. Because RNase E is anticipated to be a tetramer in vivo and the degradosome may have up to three tetramers in principle, it is likely that each of the CTD domains and associated RhlB could contribute independently to 70S ribosome interactions in the context of a polysome. In this way, polysomes may effectively act as antenna for degradosomes, so that they are recruited to sites of active translation.
RNase E is membrane-associated (40), and in the context of the full assembly, its interactions with the ribosome may be constrained or enhanced by interactions with the membrane and with canonical components. RhlB and the two RNA binding sites on RNase E CTD that interact with the 70S ribosome were also identified in an earlier study as the interaction sites of RraA, the putative endonuclease inhibitor (36). Evidence indicates that the association of RraA with RNase E CTD remodels the degradosome assembly and limits the accessibility of RNA substrates for cleavage. Potentially, RraA may influence a dynamic equilibrium between the ribosome and the degradosome, but this hypothesis awaits experimental verification.
One important aspect of the RNase E CTD is its contribution to the mechanism of Hfq/sRNA-mediated mRNA degradation. This domain recruits the RNA chaperone Hfq (45) and sRNA. The association of the Hfq/sRNA complex with RNase E may direct cleavage of a transcript near the pairing site (37). The presence of 70S ribosome does not interfere with the sRNA-guided mRNA degradation (Figure 7C) or the turnover of a short single-stranded RNA oligomer (data not shown). However, the processing activity of RNase E for more complex folded RNA, namely the 9S precursor of ribosomal 5S RNA, is modestly inhibited in the presence of 70S ribosome. This inhibition might be mediated by the CTD, because the RNase E catalytic domain interacts only weakly with 70S ribosome.
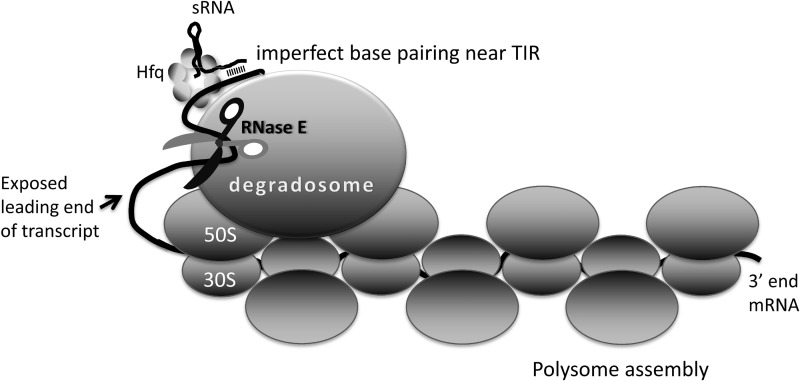
A proposed functional model for degradosome–polysome interactions
Under what circumstances might the degradosome-70S ribosome macromolecular complex be expected to occur? One possibility is that the degradosome serves as an assembly factory during the biogenesis of ribosomes, or a recycling centre during ribosome catabolism. We suggest another model in which RNase E bound to 70S ribosome may serve a regulatory role on certain translated transcripts in E. coli, where it may cleave an mRNA at a guided site when triggered by sRNA binding or other activating signal (Figure 8). We envisage that the degradosome may rest passively on a ribosome, moving from one to the next as they roll along in a polysome assembly. Cryo-electron microscopy studies of E. coli polysomes suggest that between 40 and 52 nucleotides bridge the adjacent mRNA exit and entry points (48), but this would be in a sequestered core that might protect against endoribonuclease cleavage. If a sRNA/Hfq binary complex forms on the emerging 5′-end of the transcript, it could be bound by the passive degradosome assembly. In this position, it is poised to trigger cleavage of transcripts as they issue from the end of the polysome (Figure 8). Our data from LC-MS/MS identifies a minor signal for Hfq in the degradosome/70S ribosome complex, and our western blot analyses using anti-Hfq antibodies, identifies Hfq as co-purifying with His-tagged 70S. Furthermore, Hfq and the sRNA RyhB are associated with polysome fractions (42). The proposed mode of operation depicted in Figure 8 can be viewed as a ‘graceful’ translation exit, which ensures that synthesis of nascent peptides is not stopped prematurely.
Figure 8.
A hypothetical model for the activity of the degradosome on polysomes. Taken together, the available data give a picture of the degradosome as a regulator of post-transcriptional gene expression, waiting passively on a translating polysome for stochastic encounter with the transcript, or for an activating signal. RNA on the polysome is likely to be protected from RNase E attack, judging from structural data (48). If the degradosome remains on the polysome, then it might attack, in a stochastic manner, the spooling RNA as it emerges from the polysome assembly through the catalytic domain of RNase E (shown as scissors in the diagram). It is envisaged that a sRNA could recognize and associate with the 5′ translation initiation region (TIR) as it emerges from the most recently loaded 70S ribosome. This binary complex can associate with RNase E, and increase the rate of encounter with the free end of the transcript for cleavage. The oligomeric degradosome has multiple RNA-binding sites, enabling simultaneous binding to both sRNA/Hfq/transcript and to the 70S ribosomes in a polysome.
The interactions of the degradosome with polysomes may account for the finding that RNase E can be activated by the base-pairing of a sRNA with mRNA to cleave the transcript distant from the site of RNA–RNA pairing. The sRNA RyhB base-pairs with the target sodB mRNA (encoding superoxide dismutase) at the 5′-UTR (untranslated region), and this induces RNase E to cleave within the coding region during active translation (49). The mechanism also involves the RNA chaperone, Hfq. The model interpreting this behaviour proposes that the binding of RyhB at the 5′-UTR triggers two processes in sequence: first, prevention of a new round of translation, by simply occluding the ribosome binding sites; second, triggering cleavage downstream only after any translating ribosome have read past the distal cleavage site. This two-stage mechanism avoids the fate of accumulating ribosomes that are trapped on a cleaved, stop-less transcript (49). If the degradosome remains associated with the sRNA/Hfq/5′-UTR region, as in the model shown in Figure 8, then the emerging transcript might spool until a structural signal is recognized by a component of the degradosome. This model is consistent with the observation that the degradosome assembly is required for this action at a distance (49) and for other cases of sRNA-mediated gene silencing (50,51).
It seems paradoxical that nature might have evolved a ribonuclease that can associate closely with the RNA-based machinery of translation, especially considering that the instructions for the machine are vulnerable to irreversible inactivation by inadvertent cleavage. However, there is precedent for other ribonucleases to interact with the ribosome with functional consequences. For example, genetic data define a role of ribonucleases in ribosome turnover in starvation response, and ribonuclease toxins of toxin–antitoxin pairs terminate translation by cleaving transcripts at the A site of translating ribosomes (52,53). Other studies suggest that endoribonucleases play a role in rescuing stalled ribosome from faulty transcripts (52,54–56). In eukaryotes, ribonucleases interact with ribosomes as part of nonsense mediated RNA decay, and in response to transcripts lacking stop codons (57,58). Our results suggest that in E. coli, the RNA degradosome can associate with ribosomes and polysomes, and we speculate that this may cause cessation of translation, either through stochastic exposure of the emerging transcript, or through the guidance of sRNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–8 and Supplementary Materials and Methods.
FUNDING
Istituto Pasteur Fondazione Cenci Bolognetti Fellowship (to D.D.); BBSRC [BB/F013140/1 to A.J.C.]; CONACyT [CB-2010-01-152857 to J.G.M.]; Wellcome Trust funding (to B.F.L.). Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank A.J. Carpousis, Kenny McDowall, Zbyszek Pietras, Kasia Bandyra, Jörg Vogel, Kai Pappenfort, Kathrin Fröhlich, Venki Ramakrishnan, Hilde De Reuse, and Maja Górna for critical comments and helpful advice. We thank Jörg Vogel, Kasia Bandyra and Nelly Said for the MicC and ompD expression constructs. We thank Isabella Moll for generously providing antibodies against the large and small ribosomal subunit proteins and Hfq, A.J. Carpousis for antibodies against RNase E and many different expression vectors, S. Sanyal for the E. coli strain expressing His-tagged 70S, and Stan Cohen for providing monoclonal antibody against RNase E. We thank Len Packman, Svenja Hester, Julia Howard, Kathryn Lilley and Mike Deery (Cambridge Centre for Proteomics) for mass spectrometry analyses. We thank Jeremy Skepper for advice in recording EM images.
REFERENCES
- 1.Jain C, Deana A, Belasco JG. Consequences of RNase E scarcity in Escherichia coli. Mol. Microbiol. 2002;43:1053–1064. doi: 10.1046/j.1365-2958.2002.02808.x. [DOI] [PubMed] [Google Scholar]
- 2.Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J. Mol. Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- 3.Carpousis AJ. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 4.Górna MW, Carpousis AJ, Luisi BF. From conformational chaos to robust regulation: The structure and function of the multi-enzyme RNA degradosome. Q. Rev. Biophys. 2011;45:105–145. doi: 10.1017/S003358351100014X. [DOI] [PubMed] [Google Scholar]
- 5.Carabetta VJ, Silhavy TJ, Cristea IM. The response regulator SprE (RssB) is required for maintaining Poly(A) polymerase I-degradosome association during stationary phase. J. Bacteriol. 2010;192:3713–3721. doi: 10.1128/JB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prud’homme-Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: evidence for a ‘cold shock degradosome’. Mol. Microbiol. 2004;54:1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 7.Worrall JAR, Górna M, Crump NT, Phillips LG, Tuck AC, Price AJ, Bavro VN, Luisi BF. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J. Mol. Biol. 2008;382:870–883. doi: 10.1016/j.jmb.2008.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 2011;79:419–432. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 9.Morita T, Kawamoto H, Mizota T, Inada T, Aiba H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 2004;54:1063–1075. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- 10.Leroy A, Vanzo NF, Sousa S, Dreyfus M, Carpousis AJ. Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol. Microbiol. 2002;45:1231–1243. doi: 10.1046/j.1365-2958.2002.03104.x. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein JA, Lin P-H, Cohen SN, Lin-Chao S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Natl Acad. Sci. USA. 2004;101:2758–2763. doi: 10.1073/pnas.0308747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcaida MJ, DePristo MA, Chandran V, Carpousis AJ, Luisi BF. The RNA degradosome: life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Carpousis AJ, Luisi BF, McDowall KJ. Endonucleolytic Initiation of mRNA Decay in Escherichia coli. Prog. Mol. Biol. Transl. Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- 14.Symmons MF, Williams MG, Luisi BF, Jones GH, Carpousis AJ. Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- 15.Bessarab DA, Kaberdin VR, Wei C-L, Liou G-G, Lin-Chao S. RNA components of Escherichia coli degradosome: Evidence for rRNA decay. Proc. Natl Acad. Sci. USA. 1998;95:3157–3161. doi: 10.1073/pnas.95.6.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghora BK, Apirion D. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- 17.Misra TK, Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J. Biol. Chem. 1979;254:11 154–11159. [PubMed] [Google Scholar]
- 18.Cheng Z-F, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl Acad. Sci. USA. 2003;100:6388–6393. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 20.Singh D, Chang S-J, Lin P-H, Averina OV, Kaberdin VR, Lin-Chao S. Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc. Natl Acad. Sci. 2009;106:864–869. doi: 10.1073/pnas.0810205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaberdin VR, Lin-Chao S. Unraveling new roles for minor components of the E. coli RNA degradosome. RNA Biol. 2009;6:402–405. doi: 10.4161/rna.6.4.9320. [DOI] [PubMed] [Google Scholar]
- 22.Thiele I, Fleming RMT, Bordbar A, Schellenberger J, Palsson B. Functional characterization of alternate optimal solutions of Escherichia coli's transcriptional and translational machinery. Biophys. J. 2010;98:2072–2081. doi: 10.1016/j.bpj.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ederth J, Mandava CS, Dasgupta S, Sanyal S. A single-step method for purification of active His-tagged ribosomes from a genetically engineered Escherichia coli. Nucleic Acids Res. 2009;37:e15. doi: 10.1093/nar/gkn992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H-W, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009;16:1148–1153. doi: 10.1038/nsmb.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palva ET, Randall LL. Nearest-neighbor analysis of Escherichia coli Outer membrane proteins, using cleavable cross-links. J. Bacteriol. 1976;127:1558–1560. doi: 10.1128/jb.127.3.1558-1560.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu D, Ellis HM, Lee E-C, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Court DL, Sawitzke JA, Thomason LC. Genetic engineering using homologous recombination. Annu. Rev. Genet. 2002;36:361–388. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- 28.Callaghan AJ, Grossmann JG, Redko YU, Ilag LL, Moncrieffe MC, Symmons MF, Robinson CV, McDowall KJ, Luisi BF. Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain. Biochemistry. 2003;42:13 848–13855. doi: 10.1021/bi0351099. [DOI] [PubMed] [Google Scholar]
- 29.Callaghan AJ, Aurikko JP, Ilag LL, Günter Grossmann J, Chandran V, Kühnel K, Poljak L, Carpousis AJ, Robinson CV, Symmons MF, et al. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 2004;340:965–979. doi: 10.1016/j.jmb.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 30.Worrall JAR, Howe FS, McKay AR, Robinson CV, Luisi BF. Allosteric activation of the ATPase activity of the Escherichia coli RhlB RNA helicase. J. Biol. Chem. 2008;283:5567–5576. doi: 10.1074/jbc.M708620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khemici V, Toesca I, Poljak L, Vanzo NF, Carpousis AJ. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: Functional replacement of RhlB by RhlE. Mol. Microbiol. 2004;54:1422–1430. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- 32.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNase E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 33.Mackie GA, Coburn GA, Miao X, Briant DJ, Prud'homme-Généreux A, Stickney LM, Hankins JS, Lynne EM, Cecilia MA. 2008. Preparation of the Escherichia coli RNase E Protein and Reconstitution of the RNA Degradosome. Methods in Enzymology. Academic Press, Vol. 447, pp. 199–213. [DOI] [PubMed] [Google Scholar]
- 34.Miczak A, Kaberdin VR, Wei CL, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl Acad. Sci. USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasouly A, Schonbrun M, Shenhar Y, Ron EZ. YbeY, a heat shock protein involved in translation in Escherichia coli. J. Bacteriol. 2009;191:2649–2655. doi: 10.1128/JB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Górna MW, Pietras Z, Tsai Y-C, Callaghan AJ, Hernández H, Robinson CV, Luisi BF. The regulatory protein RraA modulates RNA-binding and helicase activities of the E. coli RNA degradosome. RNA. 2010;16:553–562. doi: 10.1261/rna.1858010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeiffer V, Papenfort K, Lucchini S, Hinton JCD, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 38.Stark H, Grant JJ. GraFix: Stabilization of fragile macromolecular complexes for single particle Cryo-EM. Methods Enzymol. 2010;481:109–126. doi: 10.1016/S0076-6879(10)81005-5. [DOI] [PubMed] [Google Scholar]
- 39.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 40.Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 2008;70:799–813. doi: 10.1111/j.1365-2958.2008.06454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trubetskoy D, Proux F, Allemand F, Dreyfus M, Iost I. SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo. Nucleic Acids Res. 2009;37:6540–6549. doi: 10.1093/nar/gkp685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Argaman L, Elgrably-Weiss M, Hershko T, Vogel J, Altuvia S. RelA protein stimulates the activity of RyhB small RNA by acting on RNA-binding protein Hfq. Proc. Natl Acad. Sci. USA. 2012;109:4621–626. doi: 10.1073/pnas.1113113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cormack RS, Mackie GA. Structural requirements for the processing of Escherichia coli 5 S ribosomal RNA by RNase E in vitro. J. Mol. Biol. 1992;228:1078–1090. doi: 10.1016/0022-2836(92)90316-c. [DOI] [PubMed] [Google Scholar]
- 44.Carpousis AJ, Leroy A, Vanzo N, Khemici V. 2001. Escherichia coli RNA degradosome. Methods in Enzymology. Academic Press, Vol. 342, pp. 333–345. [DOI] [PubMed] [Google Scholar]
- 45.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardwick SW, Chan VSY, Broadhurst RW, Luisi BF. An RNA degradosome assembly in Caulobacter crescentus. Nucleic Acids Res. 2011;39:1449–1459. doi: 10.1093/nar/gkq928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bremer H, Dennix PP. Modulation of Chemical Composition and Other Parameters of the Cell by Growth Rate. 2nd edn. Washington DC: American Society for Microbiology; 1996. [Google Scholar]
- 48.Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W. The native 3D organization of bacterial polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 49.Prévost K, Desnoyers G, Jacques J-F, Lavoie F, Massé E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 53.Neubauer C, Gao Y-G, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayes CS, Bose B, Sauer RT. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 2002;277:33825–33832. doi: 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- 55.Hayes CS, Sauer RT. Cleavage of the A Site mRNA Codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 56.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J. Biol. Chem. 2004;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 57.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 58.Becker T, Armache J-P, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34 –Hbs bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]