Abstract
The fidelity of protein biosynthesis requires the aminoacylation of tRNA with its cognate amino acid catalyzed by aminoacyl-tRNA synthetase with high levels of accuracy and efficiency. Crucial bases in tRNALeu to aminoacylation or editing functions of leucyl-tRNA synthetase have been extensively studied mainly by in vitro methods. In the present study, we constructed two Saccharomyces cerevisiae tRNALeu knockout strains carrying deletions of the genes for tRNALeu(GAG) and tRNALeu(UAG). Disrupting the single gene encoding tRNALeu(GAG) had no phenotypic consequence when compared to the wild-type strain. While disrupting the three genes for tRNALeu(UAG) had a lethal effect on the yeast strain, indicating that tRNALeu(UAG) decoding capacity could not be compensated by another tRNALeu isoacceptor. Using the triple tRNA knockout strain and a randomly mutated library of tRNALeu(UAG), a selection to identify critical tRNALeu elements was performed. In this way, mutations inducing in vivo decreases of tRNA levels or aminoacylation or editing ability by leucyl-tRNA synthetase were identified. Overall, the data showed that the triple tRNA knockout strain is a suitable tool for in vivo studies and identification of essential nucleotides of the tRNA.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) are a family of key enzymes present in three kingdoms of life (1). They catalyze aminoacylation of their cognate tRNAs with amino acids to form correct aminoacyl-tRNAs, which are the substrates of protein biosynthesis in the ribosome. Generally, aminoacylation of tRNA is performed by a two-step reaction—activation of the amino acid with ATP and its subsequent transfer to the 3′-end of the tRNA (2). Based on structural features of the amino acid activation domain, aaRSs can be divided into two classes—I and II (3). The fidelity of the aminoacylation reaction for certain aaRSs is threatened by a limited number of non-cognate standard amino acids. However, some aaRSs have evolved proofreading mechanisms to hydrolyze misactivated amino acids or misacylated tRNAs, known as pre-transfer editing or post-transfer editing, respectively (4).
Leucyl-tRNA synthetase (LeuRS) belongs to class Ia aaRS, which is characterized by a Rossmann fold with two signature peptides, HIGH and KMSKS. Additionally, almost all LeuRSs contain a large insertion domain called connective peptide 1 (CP1) within the catalytic Rossmann fold domain. CP1 is defined as a classic editing domain, which can hydrolyze the aminoacyl bond of mischarged aa-tRNA to ensure the fidelity of the catalytic process (5,6).
The cognate substrate of LeuRS, tRNALeu, has multiple unique characteristics. First, it belongs to the class II tRNAs, along with tRNASer and tRNATyr, characterized by a long variable stem and loop while other tRNAs only harbor a short variable arm (7). Second, in the normal genetic code, leucine is specified by six different codons and at least four tRNALeu isoacceptors exist in vivo. Therefore, how LeuRS recognizes the isoacceptors exhibiting diverse sequences and structural heterogeneity remains an interesting question. Third, LeuRS contains both an amino acid activation domain and an editing domain. The 3′-end of tRNALeu should shuttle between the two active sites during aminoacylation and editing processes. Our recent studies have shown that translocation of the tRNALeu 3′-terminus is crucial for the enzymatic activity (8).
Our laboratory has investigated the interaction between LeuRS and tRNALeu from various species, including Escherichia coli, Aquifex aeolicus, Giardia lamblia and human (cytosolic and mitochondrial) (6,8–11). The above-mentioned tRNALeu could be purified after overexpression in E. coli or transcribed by T7 RNA polymerase in vitro. Several extensive in vitro studies have been carried out to clarify the recognition elements of tRNALeu from different species during the LeuRS-tRNALeu interaction. A73, which is absolutely conserved in all tRNALeu, was found to be the discriminator base. In E. coli, the amino acid-accepting end (CCA76) has been shown to be essential for both aminoacylation and editing (8). Similarly, the tRNA folding resulting from the tertiary interactions between the D-loop and the TΨC-loop is crucial for recognition by LeuRS during both steps (9). In the bacterium A. aeolicus, interactions between LeuRS and tRNALeu anticodon arm are essential for translocation of the tRNA from the synthetic to the editing site (10,12). However, nucleotides of tRNALeu that specifically control the editing reaction are unknown. In tRNAIle, the specific nucleotides from the D-loop are sufficient to trigger the editing response (13). In tRNAAla, both aminoacylation and editing have been shown to be sensitive to the G3•U70 base pair in the acceptor stem (14).
Despite considerable efforts, transcripts of tRNALeu from Saccharomyces cerevisiae (yeast tRNALeu) could not be synthesized in vitro with a reasonable accepting activity (15). In vitro transcription by T7 RNA polymerase is a powerful approach to study recognition elements of tRNAs; but it has some limitations. First, the point-by-point site-directed mutagenesis method is time-consuming if an exhaustive investigation is carried out. Second, in vitro transcribed tRNAs may be inactive due to the absence of critical base modifications (16). An alternative approach consists in using in vivo assays, which may overcome some of these limitations and approach the physiological conditions. Thus, in some conditions, it may become a useful substitute for the T7 RNA transcription approach. In vivo screening assays are often based on tRNA knockout strains. In E. coli, new approaches such as one-step inactivation of chromosomal genes and specific plasmids have been developed to delete a tRNA gene in order to investigate its structure and function in vivo (17,18). In eukaryotes, a yeast tRNA knockout strain was used to discriminate between active and inactive tRNAArg molecules (19). In mammals, a tRNASec knockout mouse was constructed, with the aim of studying the relationship between organic aging and selenocysteine metabolism (20,21).
Here, we constructed two knockout yeast strains for tRNALeu genes. In the first strain, the unique gene of tRNALeu(GAG) was deleted, and the three genes of tRNALeu(UAG) were deleted in the second. These strains were used to study the two tRNAs in vivo. The results showed that tRNALeu(UAG), but not tRNALeu(GAG), is essential for yeast growth. A genetic selection was performed to isolate lethal mutants of tRNALeu(UAG) unable to complement the yeast knockout strain. In addition, selection on plates containing the toxic analog norvaline (Nva) led to the identification of tRNA mutants unable to be edited by LeuRS (4,5,22). In this way, seven critical nucleotides for aminoacylation of yeast tRNALeu(UAG) and one for the editing function of yeast LeuRS were identified.
MATERIALS AND METHODS
Materials
Ethylenediaminetetraacetic (EDTA), PEG4000, lithium acetate, polyvinylpyrrolidone (PVP), bovine serum albumin (BSA), Ficoll 400, sodium citrate, potassium acetate, glucose, magnesium chloride, Tris–HCl and maleic acid were purchased from Sango (Shanghai, China). The yeast minimal synthetic defined (SD) base and dropout (DO) supplement base were purchased from Clontech (Mountain View, CA, USA). 5-fluoroorotic acid (5-FOA), restriction endonucleases and T4 DNA ligase were obtained from Fermentas (Burlington, Canada). Amino acids were purchased from Sigma (St Louis, MO, USA). The KOD plus DNA polymerase and dNTPs were purchased from TOYOBO (Osaka, Japan). Digoxigenin blocking reagent and anti-digoxigenin-AP were purchased from Roche (Basel, Switzerland). All the radioactive amino-acids and ATP are purchased from PerkinElmer (Waltham, MA, USA). T7 RNA polymerase was prepared in our laboratory as previously described. (23). Streptavidin Agarose Resin was purchased from Throme-Pierce (Rockford, IL, USA). Ni2+–NTA superflow column was purchased from Qiagen (Dusseldorf, Germany).
Plasmids pALR5 (tR4, URA, ADE3) (24), pRS313 (HIS3, CEN6, ARSH4), pRS314 (TRP1, CEN6, ARSH4), pRS315 (LEU2, CEN6, ARSH4) and pGPD415 (LEU2, CEN6, ARSH4, GPD promoter) (25,26) were from the Institut de Biologie Moléculaire et Cellulaire du CNRS (Strasbourg, France); pET28-ycleuS (KAN, ycleuS) and pET28-ycleuS-D419A (KAN, ycleuS-D419A) were constructed previously and stored in our laboratory (22); E. coli strain BL21(DE3) was purchased from Stratagene (La Jolla, CA, USA). pRS317 (LYS2, CEN6, ARSH4) and pRS412 (ADE2, CEN6, ARSH4), S. cerevisiae strains YPH499 (Mata ura3-52 lys2-801Amber ade2-101Ochre trp1-Δ63 his3-Δ200 leu2-Δ1) and YPH501 (ura3-52 lys2-801Amber ade2-101Ochre trp1-Δ63 his3-Δ200 leu2-Δ1) (26,27) were gifts from JinQiu Zhou in our institute. Digoxigenin-labeled RNA probes were synthesized by Takara (Otsu, Shiga, Japan).
Gene cloning and site-directed mutagenesis
The yeast tRNALeu(UAG) gene and its upstream and downstream DNA fragments were amplified from genome extracted from wild-type (WT) yeast strain YPH501 by PCR using forward primer 5′-AAAGAGCTCGACAAACGACTG-3′ (SacI site in italics) and reverse primer 5′-AAAGGATCCGTTGCGCCAAGA-3′ (BamHI site in italics), which introduced restriction sites for cloning. After digestion with SacI and BamHI, the PCR product was inserted into the same sites of pALR5 to give the rescuing plasmids pALR-T [tR4, URA, ADE3, TL(UAG)]. Similarly, the digested PCR product was inserted into pRS314 to give pRS314-R [TRP1, CEN6, ARSH4, TL(UAG)] which was used as a template for the random mutagenesis. The single-point mutants of tRNALeu(UAG) were constructed by PCR according to the protocol provided by the KOD-Plus-Mutagenesis Kit (TOYOBO, Japan) (28), using mutation-containing primers and pRS314-R as template.
An additional plasmid expressing a chimeric tRNA(GAG) was constructed to rescue the triple tRNALeu(UAG) knockout strain. The gene TL(GAG) was amplified by PCR as a 700-bp DNA fragment containing the natural promoter. It was subsequently cloned in pRS317 and the anticodon GAG was mutated to UAG. The resulting plasmid (pRS317-C) was expressing a chimeric tRNA exhibiting the main body of tRNALeu(GAG) and a UAG anticodon. The plasmid could efficiently complement the triple tRNALeu(UAG) knockout strain. It was particularly useful during the northern blot analyses of tRNALeu(UAG) mutants since it did not hybridize with the probe designed to recognize tRNALeu(UAG).
In situ disruption of the genes encoding tRNALeu(GAG) and tRNALeu(UAG)
Disruption of the gene for tRNALeu(GAG) was performed as follow. First, a DNA fragment carrying the 1.5-kb DNA fragment of HIS3 gene ligated to the flanking sequences of tRNALeu(GAG) gene was constructed. For this, two PCR amplifications were performed by LA-Taq enzyme on yeast genomic DNA. A 1-kb DNA fragment located upstream of the tRNA gene was amplified by forward primer 5′-AAAGGATCCTAGTGCTGATGGTATCCC-3′ (BamHI in italics) and reverse primer 5′-AAAGCGGCCGCAATTCATCTGTTGGGTAA-3′ (NotI in italics). A second 1-kb DNA fragment located downstream was amplified by forward primer 5′-AAATCTAGAGAATCTACGCACTTCA AT-3′ (XbaI in italics) and reverse primer 5′-AAAGGGCCCGAATCTTGGAACAGGTTG-3′ (ApaI in italics). The amplified upstream and downstream fragments were cleaved by BamHI + NotI and by XbaI + ApaI, respectively. A 1.5-kb marker gene HIS3 was amplified by PCR from pRS313 by using forward primer 5′-AAAGCGGCCGCTCGCGCGTTTCGGTGATG-3′ (NotI in italic) and reverse primer 5′-AAATCTAGAGATTTCGGCCTATTGGTT-3′ (XbaI in italic). The HIS3 gene was cleaved by NotI and XbaI. The three fragments above were ligated between the BamHI and ApaI of pcDNA3 to generate a recombinant plasmid (PCDNA-R) in which the recombinant DNA fragment insert [designated UHD or tl(gag)::HIS3] consisted of the HIS3 marker gene in between the upstream and downstream DNA fragments of the yeast tRNALeu(GAG) gene (Supplementary Figure S1). The recombined fragment was excised by BamHI and ApaI from pcDNA3-R, and the linear DNA fragment was introduced into the yeast diploid strain YPH501. Yeast transformants were grown on SD-His− plates at 30°C for 2–3 days. It was expected that the selected His+ colonies were exhibiting a tRNALeu(GAG) gene replaced by a HIS3 marker gene after homologous recombination with the UHD DNA fragment. To verify this hypothesis, sporulation of His+ colonies was induced on appropriate plates (1% potassium acetate, 0.1% yeast extract and 0.05% glucose). Tetrads were dissected and the resulting haploids were plated on YPDA plates (1% yeast extract, 2% peptone, 2% glucose, 0.004% adenine and appropriate agar) and further analyzed (described earlier). The yeast strain deleted for tRNALeu(GAG) gene was named tl(gag)-Δ1.
We used the same method to delete the three genes of tRNALeu(UAG), and the yeast strain was named tl(uag)-Δ1-3.
Construction of the random mutation library and screening by plasmid shuffling
Random mutagenesis of tRNALeu(UAG) was performed by hydroxylamine treatment. Plasmid pRS314-R (Trp+) was incubated in an appropriate volume of mutagenesis buffer (0.8 M hydroxylamine–HCl, 50 mM sodium phosphate, pH 6, 1 mM EDTA) at 37°C for 48 h to statistically obtain only one mutation per tRNA gene (29). Then, the DNA solution was extracted by phenol/chloroform, and precipitated with ethanol. The library was used to transformed tl(uag)-Δ1-3 strain in which the three genes for tRNALeu(UAG) were deleted. Transformants were grown at 30°C on plates of SD medium minus tryptophan (SD − Trp−) for 2–3 days. Then, transformants were replicated to SD − Trp− plates containing 5-FOA and uracil to allow shuffling of the pALR-T maintenance plasmid [containing URA3 gene whose product converts 5-FOA to toxic 5-fluorouridine monophosphate (5-FUMP)]. The lethal clones were selected. The DNA was extracted for sequencing of the tRNALeu(UAG) mutated gene in pRS314-R.
Complementation assays by site-directed mutated tRNALeu
Plasmids carrying the site-directed mutated tRNALeu(UAG) genes were introduced separately into the tRNA knockout strain and transformants were grown at 30°C and treated as above with the randomly mutated library. Transformants were subjected to drop-test on 5-FOA containing plates. The growth of the yeast cells was then observed and calculated (19,22).
Measurement of the growth rate
Yeast cells containing the different tRNA mutations were grown in liquid SD − Trp− medium. Each culture was diluted in liquid media to an initial A600 = 0.1. The diluted culture was shaken at 30°C. The A600 value was measured at various intervals, and the growth rate was calculated according to the logistic growth equation (30).
Analysis of in vivo aminoacylation by northern blot
The yeast gene knockout strains containing different mutations were grown in liquid SD − Trp− media to A600≈0.6, and then were harvested by centrifugation. Cell pellets were suspended in 1 ml of extraction buffer (0.3 M sodium acetate, pH 4.5, 10 mM EDTA) and extracted with one volume of phenol extraction (pH 4.5). The supernatant was then re-extracted with one volume of phenol/chloroform (pH 4.5). Three volumes of 100% ethanol were mixed with the supernatant to precipitate the RNA, and the final precipitate was dissolved in 50 μl of storage buffer (10 mM sodium acetate pH 4.5).
tRNAs were separated by acid 12% PAGE containing 8 M urea. Thirty micrograms of tRNA samples were loaded on a 0.4-mm thick gel in 0.1 M sodium acetate buffer pH 5. Electrophoresis was carried out at 4°C and constant 12 W (≈400 V) for 24 h in 0.1 M pH 4.5 sodium acetate buffer. The portion of the gel containing the tRNAs was transferred to a nylon membrane (Millipore, Bedford, MA, USA) by electrophoresis at constant 100 V for 30 min in 0.5× TBE buffer. The nylon membrane was then baked for 30 min at 80°C (19,31).
Pre-hybridization was performed at 65°C for 6 h in 15 ml of solution (10× Denhardt’s solution, 6× SSC and 0.5% SDS). Hybridization was performed at 65°C for 6 h in 15 ml of the same solution, in the presence of a tRNALeu(UAG) probe (5′-AGGGA TTCGA ACCCT TGCAT CCGAA GATAT-3′ for the mutation of the anticodon arm or 5′-CCUUG CAUCC GAAGA UAUCA GAGCC UAAAU-3′ for the other region), or a 5S RNA probe (5′-ACCCA CTACA CTACT CGGTC AGGCT CTTAC-3′ as an internal control) labeled with digoxigenin at each 5′-end. Membranes were washed at 65°C in a 2% SSPE, 0.5% SDS solution for 20 min, hybridized with anti-digoxigenin-AP and visualized with CDP-star, following standard procedures (32). Signals were quantified using a Fuji Bioimager Bas2000.
Preparation of proteins and tRNALeus
The ycLeuRS and its mutant ycLeuRS-D419A were purified as described (22). Escherichia coli strain BL21(DE3) was transformed with plasmids pET28-ycleuS and pET28-ycleuS-D419A which were constructed in our laboratory (22). Transformants were grown in 500 ml Luria–Bertani liquid medium (1% peptone, 0.5% yeast extract and 1% NaCl) containing 100 μg/ml kanamycin and grown at 37°C until the A260 reached 0.6. IPTG was added to a final concentration of 0.2 mM, and the incubation continued at 22°C for another 10 h. The cells were harvested by centrifugation at 4000g for 10 min. The cell pellets were then sonicated and centrifuged at 34 000g for 1 h. The resulting supernatants were applied to Ni–NTA Superflow columns and incubated for 1 h. The columns were washed with 20 ml wash buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl and 20 mM imidazole) and eluted with 5 ml elution buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl and 250 mM imidazole). ycLeuRS and ycLeuRS-D419A were then pooled and concentrated by dialysis against 10 mM potassium phosphate buffer (pH 7.5) containing 50% glycerol.
The gene-encoding yeast elongation factor 1α was PCR-amplified from yeast genomic DNA and cloned into plasmid pGPD425 after the 6 His-encoding sequence, giving an N-terminal 6 His-tagged protein (33). The recombinant plasmid was transformed into yeast strain YPH501. Transformed yeast cells were grown at 30°C for 24 h. His-tagged EF-1α protein was purified by Ni–NTA affinity chromatography as described above.
Yeast tRNALeu(UAG) WT and its mutants were prepared as described (34). The genes encoding mutated tRNALeus were transformed into our constructed yeast strain tl(uag)-Δ1-3, respectively. The transformants were cultured in 500 ml liquid YPDA medium and grown at 30°C for 24 h. Total tRNA was extracted by phenol from the harvested yeast cells as described above. Then 0.1 mg 5′-biotin labeled DNA probe with the sequence 5′-AGGGA TTCGA ACCCT TGCAT CCGAA GATAT-3′, a sequence complementary to the regions from the anticodon arm to the TΨC-loop of S. cerevisiae tRNALeu, bound to 300 μl Streptavidin Agarose Resin, and 2.5 mg of yeast total RNA were mixed and incubated at room temperature for 1 h. The mixture was washed with 1 ml of NTE buffer (5 mM Tris–HCl pH 8.0, 2.5 mM EDTA and 200 mM NaCl) at room temperature, and tRNA was eluted with 1 ml 0.1× NTE buffer at 65°C. The tRNA was further purified by electrophoresis on a 10% polyacrylamide/8 M urea gel, followed by passive elution in 10 mM Tris–HCl (pH 7.5) containing 0.3 M NaCl and 0.5 mM EDTA, and precipitation by ethanol after phenol–chloroform extraction. Eluted tRNALeu(UAG)s were renatured by heating at 80°C for 5 min and slow cooling to room temperature (10).
Aminoacylation, deacylation and AMP formation
The aminoacylation assay of WT yctRNALeu(UAG) and its mutants were determined at 30°C in a reaction mixture containing 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 2 mM dithiothreitol, 4 mM ATP, 20 μM [3H]leucine, 2 μM tRNALeu and 1 nM ycLeuRS. Kinetics constants for WT yctRNALeu(UAG) and its mutants were determined in the reaction mixture mentioned above, in the presence of various concentrations (from 0.05 to 2 μM) of the relevant yctRNALeu(UAG) substrates.
To determine the effect of EF-1α on leucine charging activity, aminoacylation was performed in the presence of a mix of 0.1 mg/ml EF-1α and 4 mM GTP (pre-incubated with pyruvate kinase for 30 min). In parallel, a negative control was performed in which an equal concentration of BSA and GTP was added.
Aminoacylated [3H]Leu-tRNALeu(UAG) WT and mutants were obtained by charging the corresponding tRNAs with Leu in the presence of WT ycLeuRS. The spontaneous hydrolysis levels of Leu-tRNALeu(UAG)s were measured in 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 1 μM [3H]Leu-tRNALeu(UAG)s, with 0.1 mg/ml EF-1α and 4 mM GTP or with equal concentration of BSA and GTP at 30°C. Aliquots (10 μl) of reaction solution were quenched on Whatman filter pads at different times. Radioactivity was quantified in a scintillation counter (Beckman, Pasadena, CA, USA).
Misacylated Ile-tRNALeu(UAG) WT and mutants were obtained by charging with Ile in the presence of ycLeuRS-D419A (22). The editing activities of ycLeuRS toward mischarged [3H]Ile-tRNALeu and mutants were measured at 30°C in 60 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 2 mM dithiothreitol and 1 μM [3H]Ile-tRNALeu and mutants. The reactions were initiated with 1 nM ycLeuRS.
Because the editing reaction consumes ATP, it can be measured using the breakdown of ATP (to AMP and PPi) in the presence of non-cognate amino acids (4,6). AMP formation was measured in reaction mixtures containing 60 mM Tris–HCl (pH7.5), 10 mM MgCl2, 5 mM DTT, 3 mM ATP, 20 nM [α-32P]ATP, 15 mM Nva, 5 U/ml pyrophosphatase, in the presence or absence of 5 μM tRNA. The reactions were incubated at 30°C and initiated by 1 μM ycLeuRS. Aliquots (1.5 μl) were quenched in 6 μl of 200 mM sodium acetate (pH 5.0). Quenched aliquots (1.5 μl) were dripped into a TLC plate. Nva-[32P]AMP, [32P]AMP and [32P]ATP were separated in 0.1 M ammonium acetate and 5% acetic acid. The plates were visualized by phosphor imaging and the data were further analyzed (5).
RESULTS
Deletion of the gene for tRNALeu(GAG) does not affect cell viability
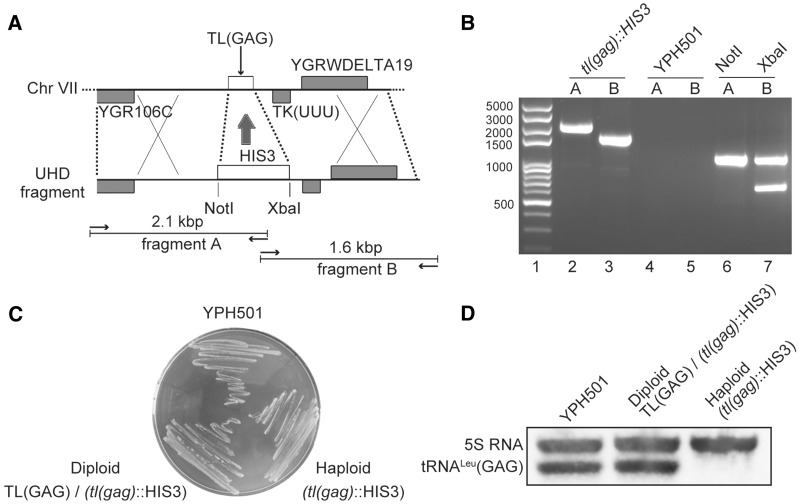
The genome of S. cerevisiae contains 21 tRNALeu genes distributed as 10 tRNALeu(CAA) genes, seven tRNALeu(UAA) genes, three tRNALeu(UAG) genes and one tRNALeu(GAG) gene [TL(GAG)]. Based on the assumption that the single-copy TL(GAG) for tRNALeu(GAG) should be essential according to the decoding capacity of the anticodon GAG (35), we first deleted the TL(GAG) gene in the diploid YPH501 strain. The TL(GAG) gene is located between positions 700 679 and 700 760 in chromosome VII and is flanked by a tRNALys(UUU) gene and two genes YGR106C and YGRWDELTA19 (Figure 1A) (35). By the method described above, plasmid pcDNA3-R was constructed carrying HIS3 flanked by the upstream and downstream sequences of TL(GAG) (so called UHD fragment). The linear BamHI-ApaI UHD fragment was introduced into the yeast strain YPH501. In this way, the 0.1-kb TL(GAG) gene was substituted by the 1.5-kb HIS3 gene, keeping the recombination flanking sequences intact. Transformants were grown on SD/His− plates in order to select recombination events. To confirm that the diploid yeast transformants had lost TL(GAG) as expected, we examined the corresponding region of the chromosome by PCR amplification and sequencing (Invitrogen, Shanghai, China). Two DNA fragments were PCR-amplified from primers hybridizing to the flanking sequences on one side and to HIS3 on the other side (Figure 1A). PCR products of 2.1-kb and 1.6-kb sizes were amplified as expected from the strains carrying the UHD gene modification. Additionally, the presence of NotI and XbaI sites resulting from the insertion of HIS3 was verified by digestion, producing 1.05-kb and 1 + 0.6-kb fragments, respectively (Figure 1B). These data confirmed that the UHD fragment was inserted into the yeast strain. However, after dissecting the tetrads obtained after incubation of the diploid strain on sporulation medium, two of the spores were His− as WT haploids, while the other two haploids which lacked TL(GAG) were exhibiting a His+ phenotype. We found that the haploids knockout strain could grow perfectly on YPDA plates (Figure 1C). These data suggested that tRNALeu(GAG) may not be an essential tRNA in yeast and that its decoding capacity overlaps with another tRNALeu. To verify that tRNALeu(GAG) was not produced any more in the knockout strain, we performed a northern blot analysis on the haploid His+ strain and indeed no such tRNA could be detected (Figure 1D). In addition, we compared the growth rate of the knockout strain [named tl(gag)-Δ1] and WT YPH499 strain in different conditions. At 23°C, 30°C and 37°C, on YPDA medium, similar growth rates were observed (Supplementary Figure S2). When grown on YPDA with excess NaCl or glycerol, no difference was observed between the strains (data not shown) (36). Altogether, the data showed that the gene TL(GAG) of tRNALeu(GAG) may be a dispensable gene and thus it is not a suitable gene to construct a strain dedicated to genetic selection of tRNA mutants.
Figure 1.
Strategy of construction and characterization of the tRNALeu(GAG) knockout strain. (A) Schematic map of the chromosomal TL(GAG) region and of the UHD DNA fragment constructed to exchange the TL(GAG) gene. Arrows symbolize the PCR primers used to verify the knockout experiment. Predicated sizes and restriction sites are indicated. (B) Agarose gel of the PCR products of YPH501 recipient strain and tl(gag)::HIS3 strain. Lane 1: 100 bp plus DNA ladder (Transgene, Beijing, China); lanes 2 and 3: PCR products amplified with tl(gag)::HIS3 genomic DNA. Lanes 4 and 5: PCR products amplified from the recipient YPH501 yeast strain. Lane 6: PCR product from lane 2 digested with NotI. Lane 7: PCR product from lane 3 digested with XbaI. (C) Growth of strains on YPDA plate. Parental strain YPH501 [a/α TL(GAG) +/+], the knockout diploid strain [a/α TL(GAG)/tl(gag)::HIS3] and the haploid knockout strain [tl(gag)::HIS3] are equally growing on YPDA. (D) Northern blot analysis on the RNA content of the yeast strains. 5S RNA was detected as an internal control. The tRNA probe was the following 5′-CCCGC GCCUC CGAAG AGAUC AGGAC CUCGA-3′.
Construction of triple knockout strain of the tRNALeu(UAG) genes
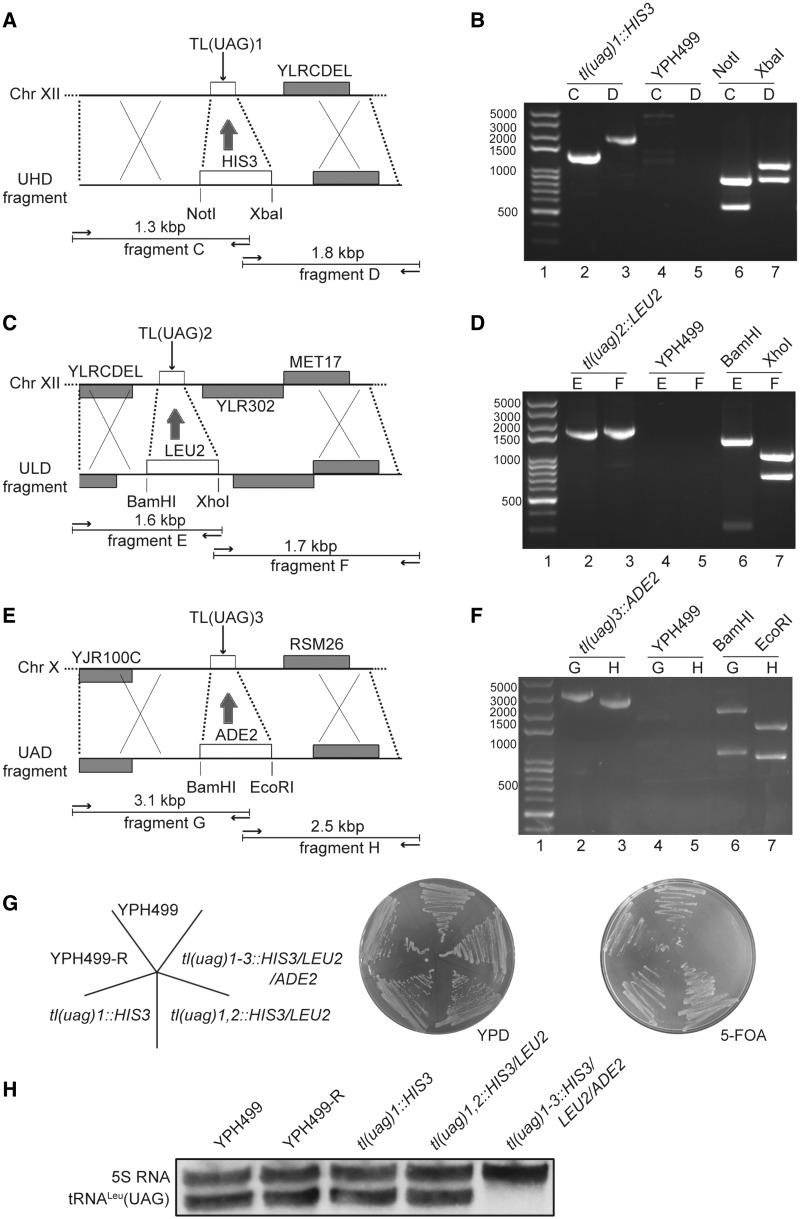
The triple deletion of tRNALeu(UAG) genes was performed in the haploid strain YPH499 [which was chosen due to the simple genetic manipulations with having fewer spores to dissect (37)]. tRNALeu(UAG) is transcribed from three TL(UAG) copies, two of them located on chromosome XII and one on chromosome X. Considering that TL(UAG) might be essential, a rescuing plasmid pALR-T [tR4, URA, ADE3, TL(UAG)] was first introduced into YPH499. The deletion of the three copies of TL(UAG) was carried out successively as described above, adding sequentially the HIS3, LEU2 and ADE2 genes. PCR- and sequencing analysis confirmed that the genes were deleted and replaced by the auxotrophic markers (Figure 2A–F). The deleted strains were named tl(uag)-Δ1, tl(uag)-Δ1,2 and tl(uag)-Δ1-3. A plasmid-shuffling assay was performed on plates containing 5-FOA to check if the rescuing plasmid pALR-T could be shuffled from these strains. On 5-FOA plates, strains tl(uag)-Δ1 and tl(uag)-Δ1,2 were perfectly growing as the WT strain indicating that the rescuing plasmid was not essential. After deletion of the third tRNALeu gene [tl(uag)-Δ1-3], the strain was unable to grow on 5-FOA-containing medium, indicating that the rescuing plasmid could not be lost (and thus 5-FOA was metabolized in 5-FUMP toxic for cells) (Figure 2G). This strongly suggests that the triple knockout experiment was successful and that tRNALeu(UAG) is an essential tRNALeu which has a decoding capacity that cannot not be covered by another tRNALeu. Northern blot analysis further confirmed that the tl(uag)-Δ1-3 strain was not containing any tRNALeu(UAG) and that the triple knockout was successful (Figure 2H). Altogether, the data showed that the knockout of the three tRNALeu(UAG) genes is lethal for the yeast cells. The triple mutant can be complemented by a plasmid copy of the rescuing tRNA and a shuffle assay can be set up to select and test mutated tRNALeu(UAG).
Figure 2.
Construction of the tRNALeu(UAG) triple knockout yeast strain. (A, C and E) Schematic map of the chromosomal regions containing the three tRNALeu(UAG) genes and of the homologous recombination DNA fragment constructed to delete the TL(UAG) genes. Arrows symbolize the PCR primers used to verify the knockout experiment. Predicated sizes and restriction sites are indicated. (B, D and F) Agarose gels of the PCR products of YPH499 strain and tRNALeu deleted strains. Lanes 1: DNA ladder; lanes 2 and 3: PCR products amplified with genomic DNAs from deleted strains. Lanes 4 and 5: PCR products amplified from the recipient YPH499 yeast strain. Lanes 6: PCR products from lanes 2 digested with NotI or BamHI. Lanes 7: PCR products from lanes 3 digested with XbaI, XhoI or EcoRI. (G) Parental strain (YPH499), derivative strain WT-R (transformed pALR-T into YPH499), single knockout strain tl(uag)1::HIS3, double knockout strain tl(uag)1,2::HIS3/LEU2 and triple knockout strain tl(uag)1-3::HIS3/LEU2/ADE2 were grown on a normal YPDA medium and then spread over on a plate containing 5-FOA. (H) Northern blot analysis on the RNA content of the three knockout strains. 5S RNA was detected as an internal control. The tRNA probe was the following 5′-CCUUG CAUCC GAAGA UAUCA GAGCC UAAAU-3′.
Site-directed mutagenesis to test the triple tRNA knockout strain
We mutated several well-defined tRNALeu identity elements to validate the use of strain tl(uag)-Δ1-3 for in vivo studies. The discriminator base A73, a positive identity determinant for recognition by LeuRS, and the crucial G18-Ψ55 base pair for maintaining the tertiary structure of tRNALeu were chosen (9,10). These two elements of tRNALeu are necessary for aminoacylation in most species. Three mutations, A73C, G18U and Ψ55G were constructed and the mutated genes was introduced into strain tl(uag)-Δ1-3. Then, the three transformants were plated on 5-FOA-containing medium and no one could grow, confirming that the mutations induced lethality of the cells. Another mutant, Ψ55C, which allowed formation of the tertiary base pair with G18 could grow normally at the presence of the 5-FOA (Figure 3 and Supplementary Figure S3). These results matched those of former in vitro data and strongly supported that strain tl(uag)-Δ1-3 as an efficient system for in vivo investigations on tRNALeu identity determinants.
Figure 3.
Scheme and diagram showing the mutations studied and the effect on cell growth. (A) Cloverleaf structure of tRNALeu(UAG) indicating the mutations (underlined) performed by random selected (in bold) or performed by side-directed mutagenesis (in italics). Putative impacts of the mutations are suggested. (B) Relative growth rate of yeast strains harboring a mutant tRNALeu measuring in liquid medium. The error bars are standard deviations from three independent measurements.
Essential nucleotides revealed by screening of a randomly mutated library
To identify the essential nucleotides of tRNALeu, a randomly mutated library of plasmid pRS314-R [Trp2, TL(UAG)] was generated by chemical modification by hydroxylamine that preferentially induces G→U or C→A transversions (29). The library was introduced into strain tl(uag)-Δ1-3 rescued by pALR-T and the transformants were grown in SD − TRP− plates. Five thousand colonies were obtained. They were replicated on 5-FOA-containing medium in order to induce pALR-T shuffle. A total of 11 clones did not grow on 5-FOA-containing medium, meaning that the shuffle could not occur because pRS314-R contained a TL(UAG) gene unable to sustain cell growth or grew slowly. Plasmids from the 11 clones were extracted. The phenotypes were confirmed by a second transformation and plating on 5-FOA, then the plasmid DNA was sequenced.
Among the 11 mutations, 9 appeared within TL(UAG) whereas the 2 others were located in another part of the plasmid outside of the tRNA gene. Among the lethal mutations were G18U in the D-loop, G36U and G37U (two occurrences) in the anticodon loop, C61A and C62A (two occurrences) in the T-stem. Among the mutants that grew slowly, one was harboring a G2U mutation in the acceptor stem (70% of WT of the growth rate) and the second one a mutation G30U in the anticodon stem (inducing a 45% growth decrease) (Figure 3A). We grew yeast cells containing the mutants of tRNALeu on both solid and liquid media to further confirm the phenotype. On solid medium, lethal or slow growth phenotype was observed directly (Supplementary Figure S3). In liquid medium, the A600 value of the culture was measured at various intervals, and the growth rate was calculated (Figure 3B).
Surprisingly, tRNALeu(UAG) harbor a long variable arm carrying six G or C nucleotides that could have been mutated by hydroxylamine, however, no lethal mutation was selected in the variable arm. It is known that this tRNA subdomain is a crucial element for proper tRNA-enzyme recognition, which involves more structural constraints than sequence-specific interactions with the synthetase (38). Therefore, single–site mutations may not disrupt or affect the LeuRS:tRNALeu complex network of constraints.
Effects of the mutations on the steady state and aminoacylation levels of tRNALeu
Mutations in the tRNA primary sequences may impact several processes, such as transcription, stability, bases modification, aminoacylation, editing and interaction with the translation factors and ribosome (39). Some tRNAs are also involved in signal transduction as cellular factors (40). To identify the mutations that affect the steady state and aminoacylation status of tRNALeu, we performed northern blots analyses. By extracting the tRNAs in acidic conditions, we could preserve the aminoacyl-ester bond and analyze the levels of in vivo aminoacylation (31).
To rescue the cells harboring a lethal tRNALeu mutants, we complemented the cells with a chimeric tRNALeu consisting of the body of tRNALeu(GAG) in which the codon was changed to (UAG). This tRNA was expressed from the rescuing plasmid pRS317-C. In that way, we could rescue the cells and analyze the mutated tRNALeu(UAG) content by northern blot without hybridizing the rescuing plasmid. Total tRNAs extracts were prepared and the tRNALeu(UAG) levels were analyzed (19).
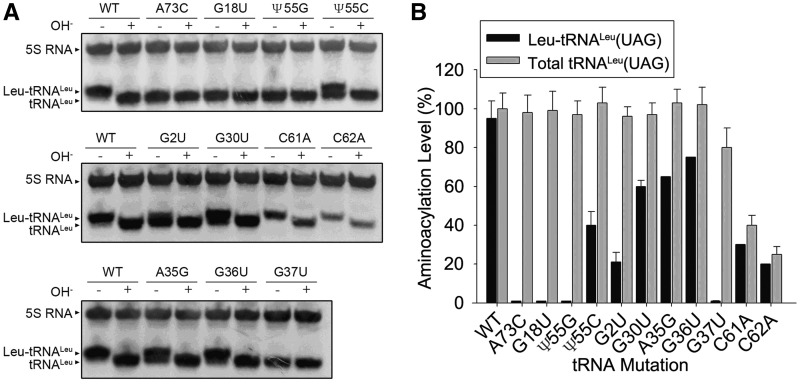
Almost all of the 11 mutations from site-directed and random mutagenesis affected the aminoacylation levels of tRNALeu(UAG). Mutant A73C was totally uncharged in vivo, which is consistent with in vitro results (10,34). In vitro, the long-distance interaction between G18 and Ψ55 is important to maintain the tertiary structure and the aminoacylation activity. The present work shows that preventing the interaction with G18U or Ψ55G mutations led to total uncharging of the tRNA by LeuRS (Figure 4). Mutation Ψ55C, which theoretically allowed formation of hydrogen bonds between bases 18 and 55, restored partial aminoacylation of tRNA (Figure 4), suggesting that the structure, the specific nucleotides or their modifications play a role during the aminoacylation of tRNA.
Figure 4.
Northern blot analysis of tRNALeu samples. (A) Total RNA samples from lethal mutants were extracted under acidic conditions, electrophoresed on a 6.5% polyacrylamide gel containing 8 M urea at pH 5.0, 4°C, and transferred to a Hybond membrane. The blot was probed with an oligonucleotide complementary to 5S RNA (control for RNA content) and tRNALeu. ‘+’ and ‘–’ marks refer to alkaline treatment (OH−) used to deacylate the RNA. (B) Grey scale scan and quantization of the amount of Leu-tRNALeu. The amounts were normalized according to 5S RNA. The error bars are standard deviations from three independent measurements.
The two mutations of G2U and G30U of yeast tRNALeu(UAG) exhibiting a reduced growth rate in the triple tRNA knockout strain were only partially aminoacylated in vivo (Figure 4). The G2U mutation significantly reduced the aminoacylation level (∼40% compared of WT), suggesting that the modification of the geometry of the acceptor arm by changing the original G:U wobble base pair is responsible of the aminoacylation defect. In addition, we cannot exclude that G2 is a determinant of the leucine identity. On the other hand, G30U was ∼70% as aminoacylated as the WT tRNALeu, suggesting that the mutation more likely impacted the translation process (Figure 4).
The lethal mutations C61A and C62A located in the T-loop led to drastically reduced tRNALeu(UAG) levels (Figure 4), suggesting that the transcription process was deficient or the stability of the transcripts reduced. Indeed, tRNAs are RNA polymerase III transcripts and the ICR (internal control region) box is located into the T-loop (41), thus we cannot exclude that mutations C61A and C62A disturb the transcription process.
Although the G36U mutation changed the anticodon of tRNALeu(UAG) from leucine anticodon UAG to serine anticodon UAU (thus switching the decoding property of the tRNA), the mutant of tRNALeu could still be charged with an amino acid by LeuRS to a significant level (Figure 3A). However, the mutation was lethal as expected due to the swap of decoding properties from leucine to serine codons. Surprisingly, mutation G37U totally abolished aminoacylation, indicating that G37 residue is essential for the aminoacylation of tRNALeu (34) (Figure 4).
It has been reported that A35 of yeast tRNALeu(UAG) is crucial for aminoacylation of tRNA. Introducing a A35G was found to reduced the kcat and Km of tRNALeu(UAG) dramatically in vitro (34). In the present investigation we did not select any mutant of A35, probably because adenine residues are poorly reactive to hydroxylamine. Thus, we constructed the A35G mutation and introduced it into the triple tRNA-knockout strain. The mutation induced a lethal phenotype on 5-FOA plate (Figure 3 and Supplementary Figure S3) as expected since the decoding property was shifted from Leu to Pro. We analyzed the in vivo aminoacylation level of the mutant and found that the aminoacylation level was decreased by about 30% (Figure 4), which is an effect significantly different from the in vitro reported data (total abolished aminoacylation) (34). Thus, the anticodon mutations affected aminoacylation slightly; indicating that A35 or G36 are not critical elements of the aminoacylation reaction, which is the universal recognition model between LeuRS and tRNALeu from various species in many in vitro studies (9,10).
The anticodon stem is involved in editing function of LeuRS
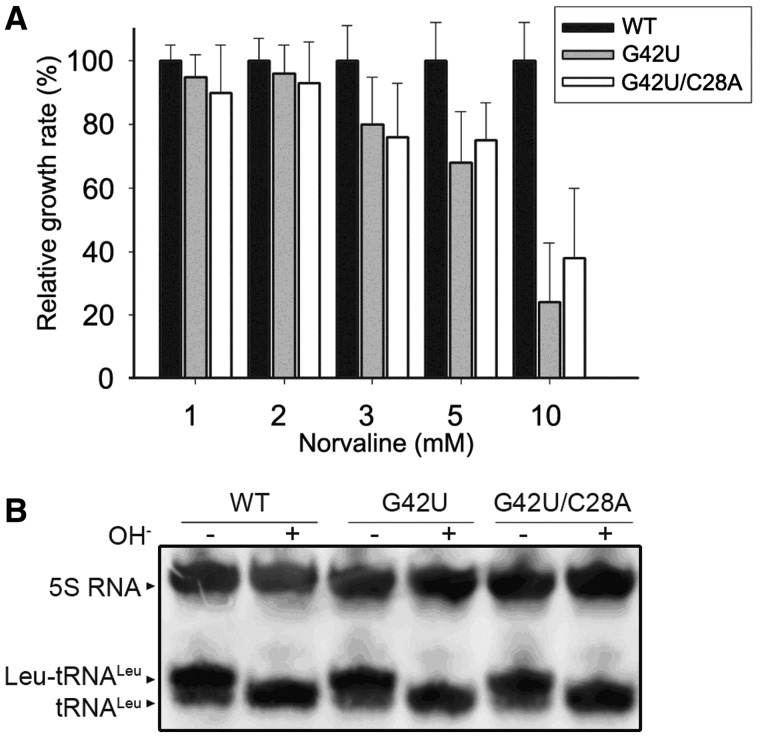
LeuRS contains an editing domain that deacylates misacyl-tRNA that could alter the fidelity of the translation process. Under usual growth conditions, yeast cells are not exposed to an excess of non-cognate amino acids and the editing function is hardly observable. However, when yeast cells are exposed to an excess of non-cognate amino acids, mis-aminoacylation of tRNA occurs and editing becomes essential for the viability (5,22). To assay the editing properties, we replicated the previous 5000 transformants of the triple knockout strain tl(uag)-Δ1-3 on plates containing 10 mM norvaline (Nva), an analog of leucine (22). We identified a single clone showing a reduced growth rate on 5-FOA plates. The tRNALeu gene was exhibiting a G42U mutation (Figure 5A and Supplementary Figure S4). A dose-dependent assay was set up in the present of various concentrations of Nva to further confirm that G42U mutant was sensitive to 5-FOA (Figure 5A and Supplementary Figure S4). In the presence of 3 mM Nva, the yeast strain harboring the G42U mutation grew slower than the WT in the same conditions (Supplementary Figure S4). When northern blots were performed on tRNA extracts prepared in acidic conditions, a complete aminoacylation of G42U mutant was observed (Figure 5B). However, Leu-tRNALeu and Nva-tRNALeu are very similar molecules that cannot be distinguished by PAGE. Thus, we concluded that that G42U mutation does not prevent the aminocylation by Nva or Leu but more likely affect the editing of Nva-tRNALeu, therefore leading to growth rate defects on 5-FOA plates.
Figure 5.
Toxic effect of Nva observed with mutants of the anticodon stem. (A) The growth rate of yeast strains expressing WT tRNALeu, G42U and G42U/C28A mutant tRNALeu in Nva-containing liquid medium. The growth rates of the mutants are compared to the WT rate that is normalized to 100%. (B) Northern blot analysis of the WT, G42U and G42U/C28A mutants. Probes were directed against 5S RNA (internal control) and tRNALeu. The error bars are standard deviations from three independent measurements.
Because G42 forms a strong G:C base pair with C28, it might strengthen the stability of the anticodon stem which may be critical for the editing activity. To test this hypothesis, we further constructed a double mutant C28A/G42U exhibiting a weaker A:U base pair. The in vivo aminoacylation level of the double mutant was not affected and was similar to the single G42U mutant (Figure 5B) while the growth effect recovered to some extent as compared with the G42U single point mutation tRNALeu(UAG) (Figure 5A and Supplementary Figure S4).
Altogether, the results suggest that the anticodon stem plays a role during the editing reaction. A perfect base pairing for a correct geometry and/or a high thermodynamic affinity seem to be needed in the anticodon stem of tRNALeu for an optimal editing activity catalyzed by LeuRS.
Aminoacylation, deacylation and EF1-α protection assays on tRNALeu(UAG)s
Yeast tRNALeu(UAG) WT and three representative mutants (A35G, G37U, G42U) were purified from yeast transformants containing their genes using biotin-Streptavidin Agarose Resin. The partially purified tRNALeu(UAG)s were further separated on a denaturing gel. The final tRNAs looked homogeneous in the gel (Supplementary Figure S5). The plateau value of WT and its two mutants A35U and G42U reached 1600 pmole/A260, however, the mutant G37U could not be aminoacylated (Supplementary Table S1). This is consistent with the undetectable in vivo growth rate (Figures 3, 4 and 5B). Kinetic constants of ycLeuRS for WT, A35U and G42U mutants of tRNALeu(UAG) were assayed, which showed that the catalytic efficiency of ycLeuRS for these tRNALeus was not obviously changed. However, leucylation of G37U mutant was abolished completely (Table 1) which corroborates the in vivo results (Figures 3, 4A and 5B). Because modified G37 in many tRNALeus is crucial to their leucylation (42), G37U mutation must change the modification of G37, and our results further indicated the importance of modified G37 in tRNALeu.
Table 1.
Kinetic constants of ycLeuRS for yctRNALeu(UAG) WT and mutants in the aminoacylation reaction
| Mutants of tRNA | Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) | kcat/Km (relative) |
|---|---|---|---|---|
| WT | 0.34 ± 0.04 | 0.24 ± 0.04 | 0.72 | 1.00 |
| A35G | 0.95 ± 0.18 | 0.42 ± 0.08 | 0.44 | 0.83 |
| G37U | NM | NM | NM | NM |
| G42U | 0.55 ± 0.11 | 0.33 ± 0.05 | 0.60 | 0.61 |
Each parameter was determined from a Lineweaver-Burk plot. NM means not measurable. All rates represent the average of three trials with the standard deviations indicated.
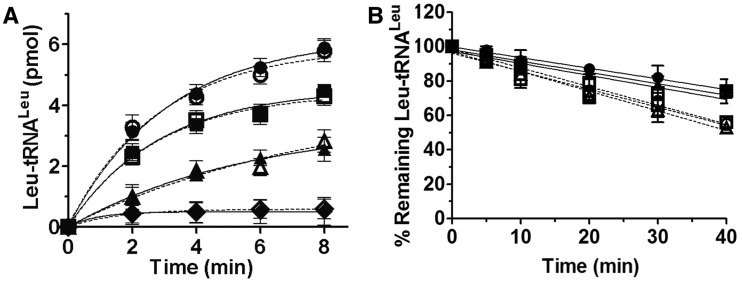
In vivo, the ester linkage between the amino acid and tRNA is labile and is readily hydrolyzed if the aa-tRNA is not recognized and bound by elongation factor. To eliminate the possibility that some mutated tRNAs have lost their binding capacity to elongation factor in vivo, we cloned the gene encoding yeast EF-1α (33), purified the protein and carry out charging and protection assays. In the presence or absence of EF-1α, leucylation of tRNALeu (UAG) WT and its three mutants A35G, G37U and G42U were assayed. The data showed that their accepting activities were not improved by the presence of EF-1α (Figure 6A). Moreover, we assayed the protection of aminoacylated Leu-tRNALeu by EF-1α from hydrolysis in water in the absence of ycLeuRS. Under physiological conditions (pH 7.5), the ester linkage between Leu and tRNALeu(UAG) was moderately hydrolyzed and the presence of EF-1α only slightly improved the stability of the ester linkage (Figure 6B). No obvious difference was observed between WT tRNALeu(UAG), A35G and G42U mutants indicating that the lethal phenotype of the mutants is not due to a loss of protection by EF-1α in vivo.
Figure 6.
Effect of EF-1α on the aminoacylation of tRNALeu(UAG)s and stability of the ester linkage. (A) Aminoacylation of tRNALeu(UAG)s by ycLeuRS (1 nM). Reactions were performed in the presence of EF-1α (solid lines) of tRNALeu -WT (filled circle), -A35G (filled triangle), -G37U (filled diamond) and -G42U (filled square). or in the presence of BSA (dashed lines) of tRNALeu -WT (unfilled circle), -A35G(unfilled triangle), -G37U (unfilled diamond) and -G42U (unfilled square). (B) Spontaneous hydrolysis of Leu-tRNALeu(UAG)s in the presence of EF-1α (solid lines): tRNALeu -WT (filled circle), -A35G (filled triangle) and -G42U (filled square) or in the presence of BSA (dashed lines): tRNALeu -WT (unfilled circle), -A35G(unfilled triangle) and -G42U (unfilled square). The error bars are standard deviations from three independent measurements.
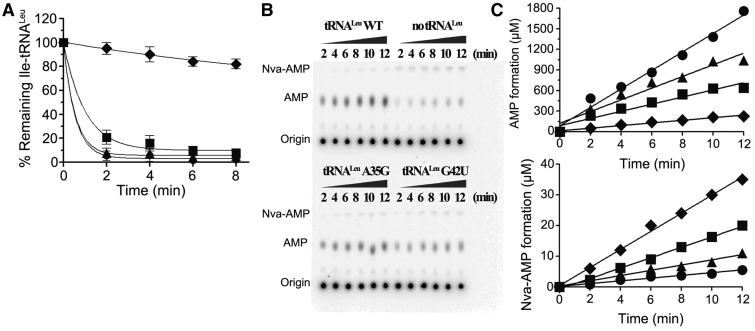
Our in vivo experiment reveals that excess Nva decreases the growth rate of yeast cells harboring the tRNALeu(UAG)-G42U mutant. Our previous work also elucidated that the Nva could be mischarged and incorrect proteins were generated (22). An in vitro editing assay was performed to investigate whether Nva could be charged to tRNALeu(UAG)-G42U. Although no Nva-tRNALeu(UAG)-G42U was accumulated when catalyzed by ycLeuRS (data not shown); the hydrolytic activity of ycLeuRS to mischarge Ile-tRNALeu(UAG)-G42U was reduced (Figure 7A), and in the presence of tRNALeu(UAG)-G42U and Nva, AMP formation was decreased (Table 2, Figure 7B and C) Additionally, Nva-AMP formation was increased (Figure 7B and C) as compared to tRNALeu(UAG) WT. Further, tRNALeu(UAG)-A35G revealed a slight decrease in editing activity. The in vivo and in vitro data both indicated that A35 and G42 at the anticodon stem and loop are important for editing.
Figure 7.
In vitro editing the mischarged tRNALeu(UAG) WT and mutants by ycLeuRS and the AMP and Nva-AMP formations with the corresponding tRNAs. (A) Deacylation assays catalyzed by ycLeuRS (1 nM) of 1 μM of Ile-tRNALeu WT (filled circle), -A35G (filled triangle), -G42U (filled square), reaction without enzyme (filled diamond) was performed as spontaneous hydrolysis control. (B) TLC plates showing the [32P]AMP and Nva-[32P]AMP formation catalyzed by ycLeuRS (1 μM). (C) Quantification of the AMP or Nva-AMP formation with tRNALeu(UAG) -WT (filled circle), -A35G (filled triangle), -G42U (filled square) and without tRNA (filled diamond), respectively. kobs values of AMP and Nva-AMP formations are calculated from the slopes and reported in Table 2.
Table 2.
Observed steady-state constants of ycLeuRS in AMP and Nva-AMP synthesis reaction in the presence of Nva and yctRNALeu(UAG) WT or its mutants
| Mutants of tRNA | AMP formation kobs (s−1) | AMP formation (relative) | Nva-AMP formation kobs (s−1) | Nva-AMP formation (relative) |
|---|---|---|---|---|
| WT | 1.93 ± 0.23 | 1.00 | (7.20 ± 0.56) × 10−3 | 1.00 |
| A35G | 1.41 ± 0.08 | 0.73 | (1.25 ± 0.13) × 10−2 | 1.74 |
| G42U | 0.74 ± 0.10 | 0.38 | (2.32 ± 0.65) × 10−2 | 3.22 |
| No tRNA | 0.30 ± 0.03 | 0.16 | (5.16 ± 1.71) × 10−2 | 7.17 |
All rates represent the average of three trials with the standard deviations indicated.
DISCUSSION
Saccharomyces cerevisiae is using superwobbling to read leucine codons
The anticodon of a tRNA molecule pairs by hydrogen bonding with a codon in mRNA. Pairing between the second and third positions of anticodons with the second and first positions of codons follows the usual rules of base pairing proposed by Crick (43), with some play or ‘wobble’ between the first anticodon base and the third codon base during pairing in order to expand the coding capacity in protein synthesis. For example, the base pair between base 34 of tRNA and the third base of triplet codon of mRNA may be Watson–Crick pairing G34–C3, U34–A3 and wobble base pairing G34–U3, U34–G3. Since the original hypothesis of Crick, more and more wobble base pairings between anticodon and codon have been identified and they differ in various organisms and systems. Several modifications of the first anticodon base also participate in pairing (44)—For instance, in E. coli, tRNALeu(UAG) translates CUA, CUU, CUG, but not CUC in vitro due to a uridine-5-oxyacetic acid modification (45). Modification of bases may also have opposite effects and expand the decoding capacity. tRNAVal(UAC) of E. coli and tRNAPro(UGG) of Salmonella, contain a 5-oxyacetic acid modified U34 which helps to decode a degenerate codon box (46). The structural basis for the expansion of the decoding capacity of tRNA by modification of uridines was recently solved (47). In addition it is now evident that unmodified U34 pairs with all four bases in third position of codons (in UNN family boxes). Thus, a single anticodon with a general formula UNN is potentially able to translate the four codons in its family box. The phenomenon, called ‘superwobbling’ represents one possible mechanism of reduction of in number of tRNAs (48). This form of translation takes place in Mycoplasma and some mitochondria and chloroplasts (49). However, if superwobbling facilitates translation with a reduced set of tRNAs, it has to pay the price of a reduced translation efficiency compared to most organisms that prefer more than one anticodon per family box (48).
The data here presented showed that the gene for tRNALeu(GAG) could be deleted without impairing yeast growth, whereas the knockout of the gene of tRNALeu(UAG) was lethal. This suggests that CU(C/U) codons were decoded by another tRNA which should obviously be tRNALeu(UAG). Thus, the ability of yeast tRNALeu(UAG) to translate the four codons might imply a superwobble mechanism as observed with tRNAs carrying unmodified U34 (50). Indeed, the spectrophotometric investigations performed on yeast tRNALeu(UAG) showed that U34 was not modified and was even able to translate the six Leu codons in vitro (50). Thus, our finding that protein synthesis occurs in the absence of tRNALeu(GAG) demonstrates that a single tRNA species with an unmodified U in the wobble position can accurately decode triplets with all four nucleotides in the third codon position, therefore supporting the superwobble hypothesis to sustain protein biosynthesis. Based on protein synthesis defects, it was proposed that superwobbling is less efficient than standard and wobble base-pairing, presumably because codon–anticodon recognition is delayed (48). Here, we did not observe any growth defect of the tRNALeu(GAG) knockout cells, however, we did not thoroughly analyze growth rates in different conditions, thus, we cannot exclude that superwobbling is less efficient than standard and wobble base-pairing.
An intriguing question is whether the capacity for superwobbling of tRNALeu(UAG) carrying an unmodified U is used spontaneously in WT yeast cells or only in tRNALeu(GAG) deleted cells. The capacity of reading may be latent in the WT cells and revealed by the tRNA deletion.
The six Leu codons are spread in a four-codon box (CUN) and a two-codon box (UUR). In S. cerevisiae, the usage of codons CUU, CUC, CUG, CUA, UUG and UUA is 12.9%, 5.8%, 11.2%, 14.2%, 27.8% and 28.1%, respectively (51). At the tRNA gene level, we can see that four tRNA genes are in charge of decoding the four-codon box and 17 tRNA genes for the two-codon box. Of interest, we can notice that CU(C/U) codons that are theoretically decoded by tRNALeu(GAG) represent 18.7% (12.9 + 5.8%) of the total number of Leu codons, which is a high value for a tRNA transcript from a single gene copy. For instance, tRNALeu(UAG) decoding CU(G/A) is transcribed from three genes and represents 24.4% (11.2 + 14.2%) of Leu codons. The extreme example is tRNALeu(CAA) which decodes UUG codons (27.8% of total Leu codons) from 10 copies of gene. Altogether, the data indicate that the ratio of codons to read per tRNA isoacceptor gene is the highest for tRNALeu(GAG), suggesting that superwobbling by tRNALeu(UAG) might occur in vivo as soon as a deficit of charged tRNALeu(GAG) appears. However, it remains unclear whether superwobbling happens in normal growth conditions or only extreme conditions.
tRNALeu(UAG) triple knockout strain as a tool to select tRNALeu mutants in vivo
After construction of the triple tRNALeu(UAG) knockout strain, three loss-of-function mutations (A73U, G18U and Ψ55G) were constructed to test whether the strain could be used to isolate lethal mutations. The three mutations were chosen because they modified the main identity element (A73) or the tertiary structure of the tRNA (G18, Ψ55) that has been shown to be essential for Leu identity in almost all tRNALeu isoacceptors tested. In our assay, the triple knockout strain transformed with the mutated tRNA genes was unable to grow or survive because the cells could not shuffle the rescuing plasmid carrying the WT tRNALeu gene. The result of this first assay was lethality in agreement with the in vitro data that showed critical decreases of the aminoacylation efficiency of the three tRNA mutants (9,10). The result confirmed that the triple knockout strain and the rescuing plasmid could be used to detect tRNA mutations critical for aminoacylation or other functions of the tRNA.
We constructed a library of tRNALeu genes randomly mutated by hydroxylamine and used it to isolate tRNA mutations that induced lethality of the yeast knockout strain. Five thousand transformants were plated on minimal medium supplemented by the required amino acids and then replicated on plates containing 5-FOA. Clones those were unable to grow on the later medium contained lethal tRNALeu genes. Similarly, the 5000 clones were replicated on medium containing an excess of norvaline (Nva) in order to isolate tRNALeu mutants inducing a toxic effect in the presence of Nva. Nva is an analog of Leu that is misactivated and mischarged on tRNALeu by LeuRS and then proofread by the editing activity of the same enzyme (5,22). From this selection, it was expected to isolate tRNALeu mutants that impact the tRNA-dependent editing activity of LeuRS, leading to the incorporation of higher amounts of Nva into the proteins and then to toxicity.
As a result from the selections, we isolated tRNALeu mutants that induced effects that could be classified in three distinct groups as follows.
(i) The first group contained the two lethal mutations located in the T-arm: C61A and C62A. Northern blot analysis showed that the two tRNAs were perfectly aminoacylated, however the steady-state levels of the two tRNAs were severely decreased, to ∼30% and 25% of the WT level for C61A and C62A, respectively. These low levels might result from a transcription defect since transcription by RNA polymerase III (41) is using an internal promoter called internal control region (ICR) located in both A box (nt 8–20) and B box (nt 51–63) (52). Thus, lethal mutations in positions 61 and 62 might affect transcription as already proposed for equivalent mutations isolated in tRNAArg and tRNASer in yeast (19,53). Alternatively, the low levels of tRNAs could result from a rapid degradation due to unstable tRNA structures. Indeed, the two mutations C61A and C62A introduced mismatches in the T-arm that might destabilize the tRNALeu structures and target them to rapid degradation.
(ii) The second group contained tRNALeu mutations inducing decreases of the in vivo aminoacylation levels: G2U, G30U, A35G, G36U and G37U. The G2U mutation probably destabilized the structure of the acceptor arm by changing the G:U wobble base pair to a U–U. G2U mutation caused a decrease in the aminoacylation level and a decrease in growth of the yeast cells. Due to its location, G2U might influence the CCA acceptor end conformation during the aminoacylation reaction and explain the decrease of aminoacylation level. The decrease of the aminoacylation level of mutant G30U is even lower compared to G2U, however, it impacts growth rate of yeast cells as G2U mutant. Therefore, this moderate effect can hardly explain the in vivo growth defect. As G30U introduced a U30–C40 mismatch in the anticodon-arm, one might speculate that the mutation impacts protein biosynthesis after aminoacylation, during codon reading on the ribosome, for instance.
Yeast cells harbor four distinct tRNALeu isoacceptors decoding the six Leu codons. The variety of anticodons suggests that the anticodon triplet is not crucial for LeuRS recognition. Indeed, our data showed that A35G and G36U mutation caused only moderate decreases of the aminoacylation levels of ∼40% and 20%, respectively. This indicates that nucleotides from the anticodon loop are involved in some way in the aminoacylation process as previously proposed (34). However, in our case, the two mutations were lethal because they induced a loss of Leu-decoding capacity of the yeast cells and not because of the reduced aminoacylation.
Nucleotide 37 was also isolated as a lethal mutation G37U. Nucleotide 37, which is often a modified nucleotide (54), is involved in the reading frame maintenance, ensuring accurate translation on the ribosome (42,54). For instance, a previously isolated G37U mutation in yeast tRNATyr was shown to be lethal despite normal transcription and aminoacylation levels (55), suggesting a defect at the translation level. Similarly, mammalian tRNALys lacking A37 modification caused type II diabetes (56). Surprisingly, our data showed that G37U mutation in yeast tRNALeu totally abolished aminoacylation in vivo. We further purified G37U mutated tRNA from yeast and found that its aminoacylation was also abolished in vitro. Therefore, the lethal effect was first due to a defect of aminoacylation and revealed that G37 is an essential determinant for LeuRS recognition.
(iii) The third group of mutations contained G42U and derivative G42U/C28A, both located in the anticodon arm. Both induced a growth inhibition when the yeast cells were grown on medium containing high amounts of Nva, an analog of Leu, which is usually edited by LeuRS to prevent its incorporation into proteins. Editing of tRNA by LeuRS is dependent on the translocation of the 3′ CCA end from the synthetic site to the editing site (8,57). Different nucleotides from the tRNA are involved in the process. In E. coli tRNAVal, a mutation of A76U specifically abolished the editing response without impairing the aminoacylation (58). In tRNALeu from A. aeolicus, discrete editing determinants in the anticodon arm were found to stabilize the conformation of the post-transfer editing state. In the present work, we identified a G42U mutation that reduced tolerance of yeast to excess of Nva, suggesting some toxic effect due to decrease of the editing activity of LeuRS. The data also showed that a strong C28:G42 base pair is strictly required and cannot be replaced by a A:U pair. In both cases, the aminoacylation of tRNALeu was kept intact. Here the data show that yeast growth was considerably reduced despite standard aminoacylation and tRNA levels. This strongly suggests that the toxic effect was due to the inability to clear mischarged products that appeared in the presence of elevated concentrations of Nva. This hypothesis was further confirmed by showing that the editing activity of purified G42U mutant was reduced, leading to Nva-adenylate accumulation that could be later charged on tRNALeu and incorporated into proteins.
tRNALeu(UAG) triple knockout strain as a tool to obtain tRNALeu mutants
Transcripts from S. cerevisiae tRNALeu (both GAG and UAG isoacceptors) are very poor substrates for LeuRS and hardly used for biochemical investigations (15). The study on the interaction between yeast tRNALeu and LeuRS is difficult. We could not obtain yeast tRNALeu with high enough accepting activity from E. coli transformants containing its gene or by T7 polymerase transcription. Using the knockout strains tl(uag)-Δ1-3, we could not only investigate the function of tRNALeu by the growth rate of yeast cells in vivo but also obtain any tRNALeu(UAG) mutant with the modified bases from yeast tl(uag)-Δ1-3 transformants containing the mutant’s gene. In vitro the studies on effect of a series of interesting individual nucleotide of yeast tRNALeu(UAG) on its function becomes possible. We will use the yeast knockout strains to study the effect of more nucleotides of tRNALeu(UAG) on aminoacylation and editing of ycLeuRS by site-directed mutagenesis. Further studies are in progress to find new selection tools and perform more extensive analysis of the tRNA properties.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
FUNDING
Funding for open access charge: The Natural Science Foundation of China [30930022 and 31130 064]; National Key Basic Research Foundation of China [2012CB911000 and 2012CB911001].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Jin-Qiu Zhou in our Institute for supplying the yeast strains and plasmids and Andrew Flagg from Department of Biology in Case Western Reserve University for his helping in preparing and improving the manuscript.
REFERENCES
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu. Rev. Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 3.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 4.Tan M, Zhu B, Zhou XL, He R, Chen X, Eriani G, Wang ED. tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase. J. Biol. Chem. 2010;285:3235–3244. doi: 10.1074/jbc.M109.060616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Ma JJ, Tan M, Yao P, Hu QH, Eriani G, Wang ED. Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase. Nucleic Acids Res. 2011;39:235–247. doi: 10.1093/nar/gkq763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cusack S, Yaremchuk A, Tukalo M. The 2 A crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinberg S, Misch A, Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993;21:3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou XL, Du DH, Tan M, Lei HY, Ruan LL, Eriani G, Wang ED. Role of tRNA amino acid-accepting end in aminoacylation and its quality control. Nucleic Acids Res. 2011;39:8857–8868. doi: 10.1093/nar/gkr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du X, Wang ED. Tertiary structure base pairs between D- and TpsiC-loops of Escherichia coli tRNA(Leu) play important roles in both aminoacylation and editing. Nucleic Acids Res. 2003;31:2865–2872. doi: 10.1093/nar/gkg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao P, Zhu B, Jaeger S, Eriani G, Wang ED. Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps. Nucleic Acids Res. 2008;36:2728–2738. doi: 10.1093/nar/gkn028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao R, Zhao MW, Hao ZX, Yao YN, Wang ED. A T-stem slip in human mitochondrial tRNALeu(CUN) governs its charging capacity. Nucleic Acids Res. 2005;33:3606–3613. doi: 10.1093/nar/gki677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu B, Zhao MW, Eriani G, Wang ED. A present-day aminoacyl-tRNA synthetase with ancestral editing properties. RNA. 2007;13:15–21. doi: 10.1261/rna.228707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale SP, Auld DS, Schmidt E, Schimmel P. Discrete determinants in transfer RNA for editing and aminoacylation. Science. 1997;276:1250–1252. doi: 10.1126/science.276.5316.1250. [DOI] [PubMed] [Google Scholar]
- 14.Guo M, Chong YE, Beebe K, Shapiro R, Yang XL, Schimmel P. The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science. 2009;325:744–747. doi: 10.1126/science.1174343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lincecum TL, Jr, Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Grotli M, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 16.Pütz J, Florentz C, Benseler F, Giegé R. A single methyl group prevents the mischarging of a tRNA. Nat. Struct. Biol. 1994;1:580–582. doi: 10.1038/nsb0994-580. [DOI] [PubMed] [Google Scholar]
- 17.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriel K, McClain WH. Plasmid systems to study RNA function in Escherichia coli. J. Mol. Biol. 2001;310:543–548. doi: 10.1006/jmbi.2001.4786. [DOI] [PubMed] [Google Scholar]
- 19.Geslain R, Martin F, Camasses A, Eriani G. A yeast knockout strain to discriminate between active and inactive tRNA molecules. Nucleic Acids Res. 2003;31:4729–4737. doi: 10.1093/nar/gkg685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc. Natl Acad. Sci. USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sengupta A, Lichti UF, Carlson BA, Ryscavage AO, Gladyshev VN, Yuspa SH, Hatfield DL. Selenoproteins are essential for proper keratinocyte function and skin development. PLoS One. 2010;5:e12249. doi: 10.1371/journal.pone.0012249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao P, Zhou XL, He R, Xue MQ, Zheng YG, Wang YF, Wang ED. Unique residues crucial for optimal editing in yeast cytoplasmic Leucyl-tRNA synthetase are revealed by using a novel knockout yeast strain. J. Biol. Chem. 2008;283:22591–22600. doi: 10.1074/jbc.M801181200. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Chen J, Wang E, Wang Y. T7 RNA polymerase transcription of Escherichia coli isoacceptors tRNA(Leu) Sci. China C Life Sci. 1999;42:185–190. doi: 10.1007/BF02880055. [DOI] [PubMed] [Google Scholar]
- 24.Geslain R, Martin F, Delagoutte B, Cavarelli J, Gangloff J, Eriani G. In vivo selection of lethal mutations reveals two functional domains in arginyl-tRNA synthetase. RNA. 2000;6:434–448. doi: 10.1017/s1355838200992331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mumberg D, Müller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhou XL, Zhu B, Wang ED. The CP2 domain of leucyl-tRNA synthetase is crucial for amino acid activation and post-transfer editing. J. Biol. Chem. 2008;283:36608–36616. doi: 10.1074/jbc.M806745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isackson PJ, Bertrand KP. Dominant negative mutations in the Tn10 tet repressor: evidence for use of the conserved helix-turn-helix motif in DNA binding. Proc. Natl Acad. Sci. USA. 1985;82:6226–6230. doi: 10.1073/pnas.82.18.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bacher JM, de Crécy-Lagard V, Schimmel PR. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc. Natl Acad. Sci. USA. 2005;102:1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Köhrer C, Rajbhandary UL. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods. 2008;44:129–138. doi: 10.1016/j.ymeth.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SW, Li Z, Moore PS, Monaghan AP, Chang Y, Nichols M, John B. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010;38:e98. doi: 10.1093/nar/gkp1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visweswaraiah J, Lageix S, Castilho BA, Izotova L, Kinzy TG, Hinnebusch AG, Sattlegger E. Evidence that eukaryotic translation elongation factor 1A (eEF1A) binds the Gcn2 protein C terminus and inhibits Gcn2 activity. J. Biol. Chem. 2011;286:36568–36579. doi: 10.1074/jbc.M111.248898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soma A, Kumagai R, Nishikawa K, Himeno H. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNA(Leu) J. Mol. Biol. 1996;263:707–714. doi: 10.1006/jmbi.1996.0610. [DOI] [PubMed] [Google Scholar]
- 35.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mori S, Kajita T, Endo T, Yoshihisa T. The intron of tRNA-TrpCCA is dispensable for growth and translation of Saccharomyces cerevisiae. RNA. 2011;17:1760–1769. doi: 10.1261/rna.2851411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Séraphin B, Kretzner L, Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palencia A, Crépin T, Vu MT, Lincecum TL, Jr, Martinis SA, Cusack S. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 19:677–684. doi: 10.1038/nsmb.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoagland M. Enter transfer RNA. Nature. 2004;431:249. doi: 10.1038/431249a. [DOI] [PubMed] [Google Scholar]
- 40.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 41.Stewart TS, Söll D, Sharp S. Point mutations in the 5′ ICR and anticodon region of a Drosophila tRNAArg gene decrease in vitro transcription. Nucleic Acids Res. 1985;13:435–447. doi: 10.1093/nar/13.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waas WF, Druzina Z, Hanan M, Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J. Biol. Chem. 2007;282:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]
- 43.Crick FH. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 44.Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 45.Sørensen MA, Elf J, Bouakaz E, Tenson T, Sanyal S, Björk GR, Ehrenberg M. Over expression of a tRNA(Leu) isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J. Mol. Biol. 2005;354:16–24. doi: 10.1016/j.jmb.2005.08.076. [DOI] [PubMed] [Google Scholar]
- 46.Nasvall SJ, Chen P, Björk GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weixlbaumer A, Murphy FVt, Dziergowska A, Malkiewicz A, Vendeix FA, Agris PF, Ramakrishnan V. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogalski M, Karcher D, Bock R. Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 2008;15:192–198. doi: 10.1038/nsmb.1370. [DOI] [PubMed] [Google Scholar]
- 49.Andachi Y, Yamao F, Iwami M, Muto A, Osawa S. Occurrence of unmodified adenine and uracil at the first position of anticodon in threonine tRNAs in Mycoplasma capricolum. Proc. Natl Acad. Sci. USA. 1987;84:7398–7402. doi: 10.1073/pnas.84.21.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weissenbach J, Dirheimer G, Falcoff R, Sanceau J, Falcoff E. Yeast tRNALeu (anticodon U–A–G) translates all six leucine codons in extracts from interferon treated cells. FEBS Lett. 1977;82:71–76. doi: 10.1016/0014-5793(77)80888-0. [DOI] [PubMed] [Google Scholar]
- 51.Percudani R, Pavesi A, Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- 52.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat. Rev. Genet. 2011;12: 459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 53.Nichols M, Bell J, Klekamp MS, Weil PA, Söll D. Multiple mutations of the first gene of a dimeric tRNA gene abolish in vitro tRNA gene transcription. J. Biol. Chem. 1989;264:17084–17090. [PubMed] [Google Scholar]
- 54.Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat. Struct. Mol. Biol. 2009;16:1109–1115. doi: 10.1038/nsmb.1653. [DOI] [PubMed] [Google Scholar]
- 55.Wang SS, Hopper AK. Isolation of a yeast gene involved in species-specific pre-tRNA processing. Mol. Cell. Biol. 1988;8:5140–5149. doi: 10.1128/mcb.8.12.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei FY, Tomizawa K. Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes [Review] Endocr. J. 2011;58:819–825. doi: 10.1507/endocrj.ej11-0099. [DOI] [PubMed] [Google Scholar]
- 57.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 58.Tardif KD, Liu M, Vitseva O, Hou YM, Horowitz J. Misacylation and editing by Escherichia coli valyl-tRNA synthetase: evidence for two tRNA binding sites. Biochemistry. 2001;40:8118–8125. doi: 10.1021/bi0103213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.