Abstract
Cell-cycle progression requires careful regulation to ensure accurate propagation of genetic material to the daughter cells. Although many cell-cycle regulators are evolutionarily conserved in the protozoan parasite Trypanosoma brucei, novel regulatory mechanisms seem to have evolved. Here, we analyse the function of the histone methyltransferase DOT1A during cell-cycle progression. Over-expression of DOT1A generates a population of cells with aneuploid nuclei as well as enucleated cells. Detailed analysis shows that DOT1A over-expression causes continuous replication of the nuclear DNA. In contrast, depletion of DOT1A by RNAi abolishes replication but does not prevent karyokinesis. As histone H3K76 methylation has never been associated with replication control in eukaryotes before, we have discovered a novel function of DOT1 enzymes, which might not be unique to trypanosomes.
INTRODUCTION
Trypanosoma brucei is a unicellular eukaryotic parasite that causes sleeping sickness in humans and the ‘nagana’ disease in African cattle. There has been substantial progress in recent years to identify and characterize regulators of the cell cycle in T. brucei. Although many orthologues of conserved eukaryotic cell-cycle regulators are present in trypanosomes, there is a considerable divergence of their functions and a number of key regulators are missing [reviewed in (1)]. Furthermore, trypanosomes appear to use different checkpoint control mechanisms compared to other eukaryotic model organisms. Considering that T. brucei diverged from the eukaryotic lineage a few hundred million years ago, it is conceivable that this parasite developed novel pathways to control its cell cycle.
The first evidence that histone-modifying enzymes are involved in cell-cycle progression in T. brucei was obtained when two members of the DOT1 histone methyltransferase family were characterized (2). DOT1 proteins are evolutionarily conserved histone H3 lysine 79 (H3K79) methyltransferases. H3K79 di-methylation (H3K79me2) is thought to prevent the formation of heterochromatin, as mutation of H3K79 increases the in vivo interaction of the silencing proteins Sir2p and Sir3p with chromatin in budding yeast (3). Furthermore, H3K79 methylation is enriched on H3.3, a histone variant found at transcriptionally active loci in Drosophila and mammals (4,5). Genome-wide analysis showed that di- and tri-methylation of H3K79 clearly correlate with actively transcribed chromatin in mammalian cells (6) and there are hints that H3K79 methylation is involved in chromosome segregation and cell-cycle regulation. San-Segundo and colleagues demonstrated that Dot1p is important for the meiotic ‘pachytene checkpoint’ in yeast (7). Finally, H3K79me2 but not H3K79me3 is enriched at some genes that are cell-cycle regulated (8). However, neither over-expression nor deletion of yeast Dot1p causes any obvious cell-cycle-related phenotype. Recently, Vos and colleagues presented the hypothesis that progressive histone methylation by Dot1 functions as a timer during the cell cycle in yeast (9).
Trypanosomes have two DOT1 homologues, which were termed DOT1A and DOT1B (2). DOT1A is essential and mediates mono- and di-methylation of H3K76 (H3K76me1/2) whereas DOT1B also catalyses H3K76 tri-methylation (H3K76 in T. brucei is homologous to H3K79 in other organisms). H3K76 tri-methylation is involved in different biological processes such as antigenic variation and developmental differentiation (10,11). The first indication that methylation of H3K76 is involved in cell-cycle regulation in T. brucei was discovered when the appearance of this modification was monitored during cell-cycle progression (2). H3K76me2 can be detected mainly during mitosis and cytokinesis, and depletion of DOT1A by RNAi causes severe cell-cycle defects including the emergence of cells with a reduced DNA content, suggesting that H3K76me2 plays an important role in accurate cell-cycle progression. In summary, various experiments described in the literature indicate that H3K79 methylation is involved in transcriptional regulation, the control of accurate chromosome segregation and possibly cell-cycle regulation. However, the function of H3K79 methylation in these processes in yeast or other organisms is still not well understood.
In this study, we aimed to unravel which step of the cell-cycle is disturbed in DOT1A mutants and how exactly DOT1A influences cell-cycle progression. First, we analysed the exact distribution of the different methylation states during cell-cycle progression in T. brucei. Mono-methylated H3K76 was first detectable in G2 phase cells and did not coincide with the replication marker proliferating cell nuclear antigen (PCNA) suggesting that newly incorporated H3 is unmethylated during S phase. Depletion of DOT1A decreased H3K76 mono- and di-methylation and generated cells with a reduced DNA content as observed previously. Using BrdU-incorporation, we could show that these cells originated from a complete inhibition of replication without preventing cell division. To further test if H3K76 methylation regulates replication in trypanosomes, we used a tetracycline-inducible system to over-express DOT1A. Interestingly, we observed premature H3K76 methylation already in S phase cells rather than a general increase of methylation levels. This temporal deregulation of H3K76 methylation caused continuous replication of nuclear DNA, which generated parasites with an increasing DNA content. In summary, genetic manipulation of DOT1A activity disturbs accurate replication, suggesting that carefully timed H3K76 methylation is essential for proper replication control in T. brucei.
MATERIALS AND METHODS
Trypanosome cell lines and plasmid construction
Trypanosoma brucei procyclic forms (strain 427) and bloodstream forms (MITat 1.2, clone 221a) were cultured in modified SDM-79 medium (12) at 27°C and in HMI-9 medium (13) at 37°C, respectively. Transfection and drug selection were described previously (14).To generate the inducible DOT1A over-expressing cell line, the DOT1A open reading frame was PCR-amplified with a primer pair, which included the sequence of a TY-epitope and cloned into the pLew100 plasmid (15). The resulting plasmid was linearized with NotI and transfected by electroporation into the procyclic cell line 29–13 as described previously (15). Expression was induced with 500 ng/ml tetracycline. To generate the DOT1A mutant cell line, the open reading frame of the DOT1A-G138R mutant was PCR-amplified from plasmid pFF20 (16) and cloned into the pLew100 plasmid. Over-expression was carried out as for wild-type DOT1A. A PCR-based method was used for C-terminal in vivo tagging of TbPCNA in 29–13 (17). The DOT1A RNAi cell line used in this work was generated using p2T7TA RNAi vectors as described in details elsewhere (2). The additional control RNAi cell lines were generated using a next generation hairpin vector system. Briefly, two fragments of the DOT1A ORF (nucleotide position 142–602) or a GFP ORF were cloned head-to-tail downstream of a tetracycline-inducible PARP promoter. Both constructs were digested with NotI prior to transfection. RNAi was induced by adding 100 ng/ml tetracycline to the cell culture.
Western blot analysis and antibodies
Total protein extracts were separated by 15% SDS–PAGE and transferred onto a PVDF membrane. Polyclonal antibodies specific for H3K76me2 or H3K76me3 were described elsewhere (2). The polyclonal antibody specific for H3K76me1 was raised by immunizing rabbits with KLH-conjugated peptide VSGAQK[Me1]EGLRFC. Antisera were affinity-purified using the same peptide immobilized to a SulfoLink coupling gel (Pierce). Specificity was evaluated by peptide competition assays. A polyclonal antibody specific for histone H3 was raised by immunizing rabbits with recombinant H3 protein. The TY-antibody (BB2) and the PFR-antibody were kindly provided by Keith Gull. Primary antibodies were detected with Alexa Fluor 680- and IRdye 800-coupled antibodies.
Immunofluorescence analysis
Immunofluorescence analysis (IFA) was carried out as previously described (2). Primary antibodies were visualized with Alexa Fluor 488- and 594-coupled antibodies. Vertical stacks (0.2 µm steps) were captured using personal DV (Applied Precision) deconvolution (softWoRx Software) microscopy. The average Z-projection of three images is shown.
Fluorescence in situ hybridization
The α/β-tubulin gene cluster on chromosome 1 was used as a marker for a large chromosome. A DNA probe was prepared by labelling a PCR product with the Biotin-nick Translation Mix (Roche Diagnostics). Average probe length was ∼150 bp. PCF (5 × 106) were fixed in 3.6% formaldehyde for 15 min at room temperature (RT), washed twice in PBS and allowed to settle on silanized coverslips for 30 min at RT before permeabilization with 0.1% NP-40/PBS for 5 min. Biotin-labelled probe was mixed with herring sperm DNA (0.2 mg/ml) and yeast tRNA (0.2 mg/ml) in hybridization mix. Final hybridization was carried out, after denaturation at 95°C for 5 min, at 37°C for 16 h in water saturated environment. The samples were washed with 50% formamide, 2× SSC for 30 min at 37°C, 10 min in 2× SSC at 50°C, 60 min in 0.2× SSC at 50°C and 10 min in 4× SSC at RT. Cells were incubated with Cy3-conjugated streptavidin (Sigma) in 1% BAS/PBS for 1 h at RT for detection of the biotinylated DNA.
DNA labelling
5-Ethynyl-2-deoxyuridine (EdU) and/or 5-Bromo-2-deoxyuridine (BrdU) were added to cell cultures at concentrations of 300 and 200 µM, respectively. In addition, 100 µM 2-deoxycytidine was provided. Cells were fixed in 1% formaldehyde and allowed to settle on silanized glass cover slips. After permeabilization with 0.5% Triton-X-100 in PBS, DNA was denatured with 2 M HCl for 30 min followed by neutralization with 0.1 M sodium borate. Detection of EdU was performed as described in the manufacturer’s protocol (Click-iTEdU Imaging Kit, Invitrogen). BrdU was detected with a monoclonal anti-BrdU antibody (Caltag, JU-4) followed by incubation with an Alexa 488 antibody. RNAi of DOT1A was induced for 12 h before adding BrdU to cells for a further 7 h. Cells were fixed with ethanol for flow cytometry analysis and sorted with a FacsAriaII cell sorter (Becton Dickinson). Sorted cells were allowed to settle on glass cover slips and detection of BrdU was performed as described above.
Flow cytometry
Cells were prepared as described previously (2) and were analysed with the FACSCalibur (BD Biosciences). Acquired data were analysed with FlowJo software (Tree Star).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described previously (18) using antibodies specific for H3K76me1, H3K76me2 and H3. Precipitated DNA of three independent replicates was amplified using a whole-genome amplification kit (Sigma) before hybridization on a custom-made DNA microarray (Roche NimbleGen). Data from the microarrays (.pair format) was read and analysed using R (http://www.r-project.org) and Bioconductor (http://www.bioconductor.org). All functions were used with default parameters unless stated otherwise. The raw probe signals were transformed into log2-values and then adjusted with a scale normalization as described previously (19).
Mass spectrometry
Histones were separated by 15% SDS–PAGE, stained by Comassie blue and excised from the gel. Isolated bands were diced into smaller pieces, destained, chemically modified and digested with trypsin. The resulting peptides were desalted and analysed by LC–MS/MS. All fragment ion spectra were recorded in the LTQ part of the instrument. Spectra were analysed with the Xcalibur and Proteome Discoverer software packages and quantified by extracting the XIC area under the peak that was verified to contain the corresponding peptide by at least one MS/MS spectrum (Thermo Scientific).
RESULTS
Cell-cycle-regulated H3K76 methylation levels
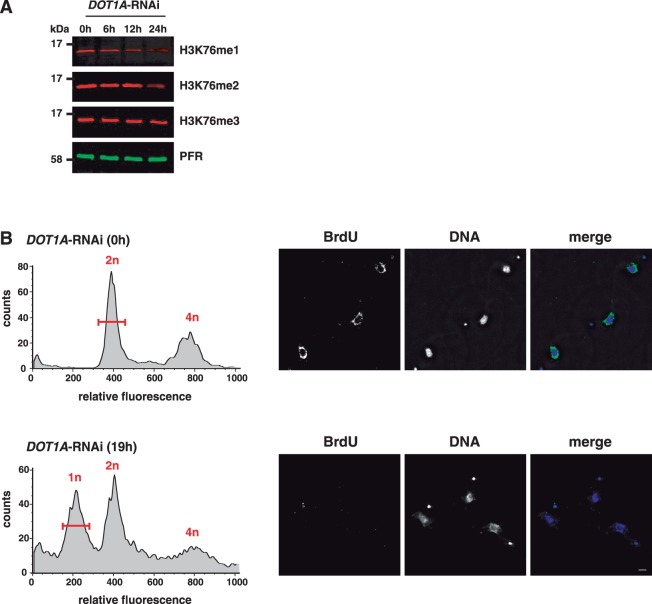
We reported previously that H3K76me2 is mainly restricted to mitosis and cytokinesis (2). To obtain a complete picture of the H3K76 methylation pattern during the cell cycle, we generated an antibody specific for H3K76 mono-methylation. The specificity of the anti-H3K76me1 antibody was confirmed by peptide competition assays (Supplementary Figure S1). To determine the position of the parasite in the cell cycle, we stained the nucleus (N) and the single mitochondrial genome (kinetoplast, K) with DAPI. Replication and segregation of the kinetoplasts precede those of the nuclei, therefore they can be used as markers for the position of an individual cell in the cell cycle as indicated in Figure 1B (20,21). Interestingly, H3K76me1 was detected mainly in G2 and M phase cells (Figure 1A). H3K76me2 could be detected mainly during mitosis and cytokinesis and H3K76me3 is not cell-cycle regulated, as described previously (2). As S phase is not defined precisely by the nucleus and kinetoplast configuration, we used the PCNA, a conserved marker for replication foci (22), to analyse if H3K76me1 and replication coincide. Distinguishable foci of TY-tagged PCNA were clearly detectable in >90% of cells with an elongated kinetoplast (Figure 1B, 1NeK, S phase), which were always negative for mono- or di-methylated H3K76, suggesting that replication and de novo H3K76 methylation exclude each other.
Figure 1.
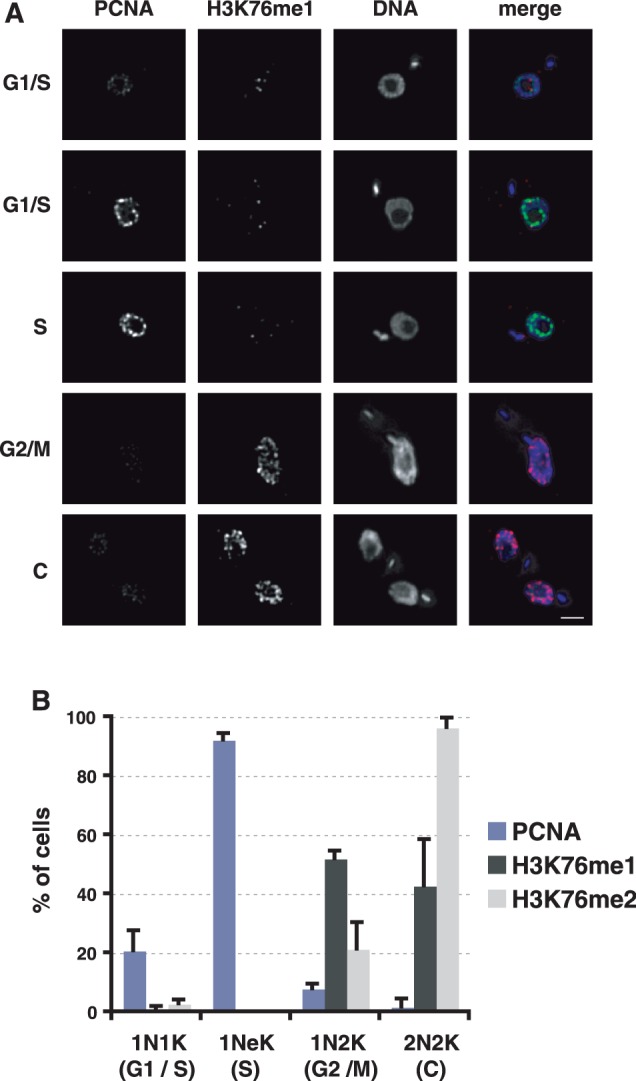
H3K76me1 is undetectable in S phase cells. (A) Indirect IFA of different cell-cycle stages in PCF expressing PCNA-TY (green) and H3K76me1 (red). DNA was stained with DAPI. Scale bar, 2 µm. (B) Quantification of cell cycle-dependent expression of PCNA-TY, H3K76me1 and H3K76me2. Three independent experiments were analysed.
Although H3K79 methylation was associated with cell-cycle-regulated gene expression in yeast (8), there is no clear cell-cycle-related phenotype after genetic manipulation of DOT1 (neither after deletion nor over-expression). In trypanosomes, cells showed a severe cell-cycle defect after depletion of DOT1A and a population of cells with a potentially haploid DNA content was generated (2). To specify the observed phenotype we tested if a replication defect is the source of these aberrant cells.
RNAi-mediated depletion of DOT1A abolishes replication of nuclear DNA
Depletion of DOT1A by RNAi causes a reduction of H3K76me2 and H3K76me1 as expected (Figure 2A). To unravel if replication is affected by depletion of DOT1A we used BrdU-labelling of newly synthesized DNA. After induction of DOT1A-RNAi a population with 1n DNA content was detectable by propidium iodide (PI) staining followed by FACS analysis as described previously (Figure 2B, lower panel). An additional RNAi cell line with different DOT1A target sequences showed basically an identical phenotype whereas a GFP knockdown caused no cell-cycle-related phenotype (Supplementary Figure S2). Twelve hours post-induction, cells were labelled with BrdU for 7 h and the 1n population was sorted for indirect IFA with a BrdU-specific antibody (Figure 2B, lower panel, bracket). Surprisingly, none of these cells showed BrdU-incorporation suggesting that these cells progressed through mitosis and cytokinesis without previous DNA replication. As a control, uninduced cells with 2n DNA content (G1 phase cells) were sorted and analysed (Figure 2B, upper panel, bracket). Almost 100% of this population showed BrdU incorporation, suggesting that depletion of DOT1A abolishes replication completely but does not prevent karyokinesis, which leads to cells with a reduced DNA content. Karyokinesis without preceding replication has not been described before in trypanosomes. Thus, it is unclear if chromosomes have been segregated accurately in these cells. This will be the subject of future analysis. Next, we wanted to test whether over-expression of DOT1A also has an effect on replication in trypanosomes.
Figure 2.
RNAi-mediated depletion of DOT1A inhibits replication. (A) Western blot analysis of different H3K76 methylation states in DOT1A-depleted cells. Two major proteins of the paraflagellar rod (PFR) were used as loading control. (B) Flow cytometry analysis of PI-stained non-induced BSF cells (upper left panel) and cells 19 h post-induction of DOT1A-RNAi (lower left panel). Non-induced cells with a DNA content of 2n and DOT1A-depleted cells with a DNA content of 1n (both indicated by a red bar) were sorted and incorporation of BrdU (green) was analysed (right panels). Cells were incubated with BrdU for 7 h before fixation. DNA was stained with DAPI. Scale bars, 2 µm.
DOT1A over-expression is lethal and generates aneuploid cells
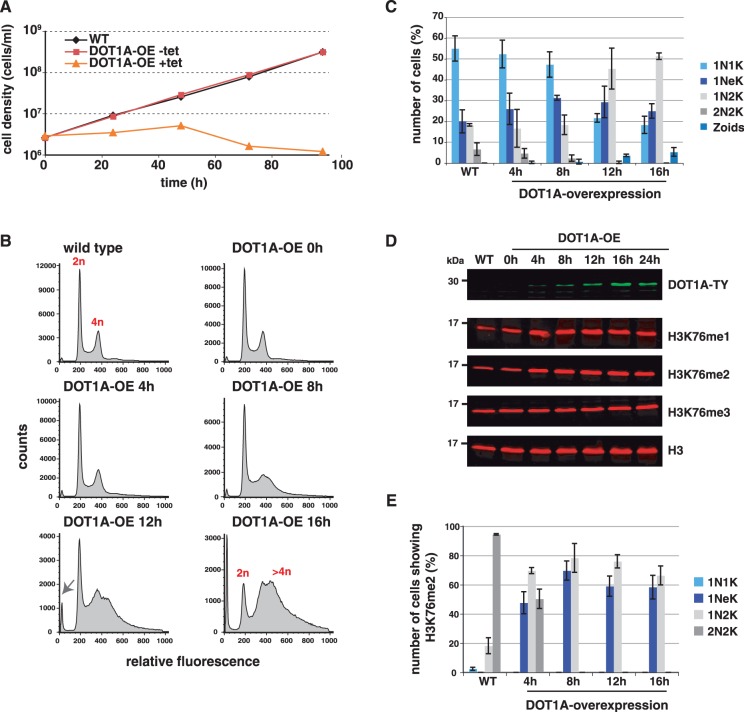
To analyse the effects of DOT1A over-expression, we employed an inducible T7 RNA polymerase-driven expression system (15). We expressed a peptide-tagged DOT1A-TY fusion protein, as a specific antibody for trypanosome DOT1A is not available. In contrast to yeast, DOT1A over-expression was lethal in T. brucei (Figure 3A). Within 24 h post-induction growth of the population ceased almost completely. Next, we analysed whether the growth defect is associated with cell-cycle-related phenotypes. To this end, we stained tetracycline-induced cells with PI to monitor cell-cycle profiles by flow cytometry (Figure 3B). Cell-cycle profiles of uninduced cells (Figure 3B, 0 h) were indistinguishable from those of wild-type cells with ∼45% of the cells in G1 phase, 11% in S phase and ∼28% in G2/M phase. After induction of DOT1A over-expression two additional populations were detected: enucleated cells (so called ‘zoids’, Figure 3B, lower panel, arrow) and a population with an apparent aneuploid DNA content (Figure 3B, >4n). Emergence of enucleated cells has been observed in trypanosomes before. Trypanosomes are able to enter cytokinesis without preceding karyokinesis, due to the lack of certain checkpoints (23). As a result, one daughter cell lacks a nucleus and the other has a nucleus with 4N DNA content. Sixteen-hours post-induction we saw 7% zoids and a dramatic increase of aneuploid cells to ∼50% of the population while the G1 phase population decreased (Figure 3B).To demonstrate that the development of the replication phenotype is dependent on methyltransferase activity of DOT1A, we also over-expressed a mutant (DOT1A-G183R) with only residual activity (Gülcin Dindar, unpublished data). Although expression levels were comparable to over-expressed wild-type DOT1A, we could detect only minor changes of the cell-cycle profiles at later time points after induction (Supplementary Figure S3B).
Figure 3.
DOT1A over-expression generates aneuploid cells and disturbs the H3K76 methylation pattern. (A) Cumulative growth curves of wild-type, non-induced and induced DOT1A over-expressing cell lines. (B) Flow cytometry analysis of PI-stained DOT1A over-expressing cells. Relative amount of DNA content is indicated. The population of enucleated cells (‘zoids’) is marked by an arrow. (C) Quantitative analysis of the number of nuclei (N) and kinetoplasts (K) in wild-type and DOT1A over-expressing cells. Mean values of three independent experiments are presented with standard deviation (n > 100). (D) Western blot analysis of DOT1A expression and different H3K76 methylation states in DOT1A over-expressing cells. Histone H3 was used as loading control. (E) Percentage of H3K76me2 positive cells in different cell cycle stages. Mean values of three independent experiments are presented with standard deviation (n > 70 for each cell cycle phase). 2N2K cells were only analysed in wild-type cells and 4 h after induction because of low amounts of 2N2K cells at later time points.
The numbers of nuclei and kinetoplasts per cell were determined up to 16 h post-induction (Figure 3C). In agreement with the flow cytometry analysis a decrease of 1N1K cells (mainly G1 phase) could be detected (reduced to 20%). Simultaneously, the percentage of 1N2K (G2 phase) cells increased to 50%, while the proportion of 1NeK cells (S-phase) did not change significantly which points to inhibition of chromosome segregation or karyokinesis. The amount of 2N2K cells (post-mitotic) decreased (none detectable 16 h post-induction). At this time point 5% of enucleated zoids were observed in accordance with the flow cytometry data. Strikingly, none of the cells show multiple nuclei/kinetoplasts. This strongly supports the hypothesis that karyokinesis is inhibited and the DNA content increases constantly in individual nuclei leading to the generation of cells with aneuploid nuclei.
To better understand the effect of DOT1A over-expression on H3K76 methylation levels, we performed quantitative western blot analysis using antibodies specific for H3K76 mono-, di- and tri-methylation (Figure 3D). Surprisingly, H3K76me1 and H3K76me2 levels of whole cell lysates did not increase although over-expression of DOT1A-TY could be detected (Figure 3D, upper panel). To understand the replication phenotype in DOT1A over-expressing cells, we monitored the temporal appearance of the H3K76 methylation pattern of single cells during the cell cycle. Immunofluorescence analyses of wild-type populations revealed that ∼2% of 1N1K cells, 20% of 1N2K cells and 95% of 2N2K cells are positive for H3K76me2 (Figure 3E). 1NeK cells (S phase) never show H3K76me2. In contrast, 70% of 1NeK cells were positive for H3K76me2 6 h post-induction. The proportion of 1N2K cells showing H3K76me2 increased to 80% 8 h post-induction. The amount of 2N2K cells was very low and could not be analysed. These results showed that H3K76me2 appeared earlier in the cell cycle after DOT1A over-expression and was, in contrast to wild-type cells, abundant in many S phase cells shortly after induction. In accordance with the shifted H3K76me2 pattern, H3K76me1 also occurred earlier in the cell cycle after induction (data not shown).
In summary, the temporal occurrence of H3K76me2 shifted in the cell cycle after DOT1A over-expression and is likely responsible for the appearance of aneuploid cells.
DOT1A over-expression causes continuous replication of nuclear DNA
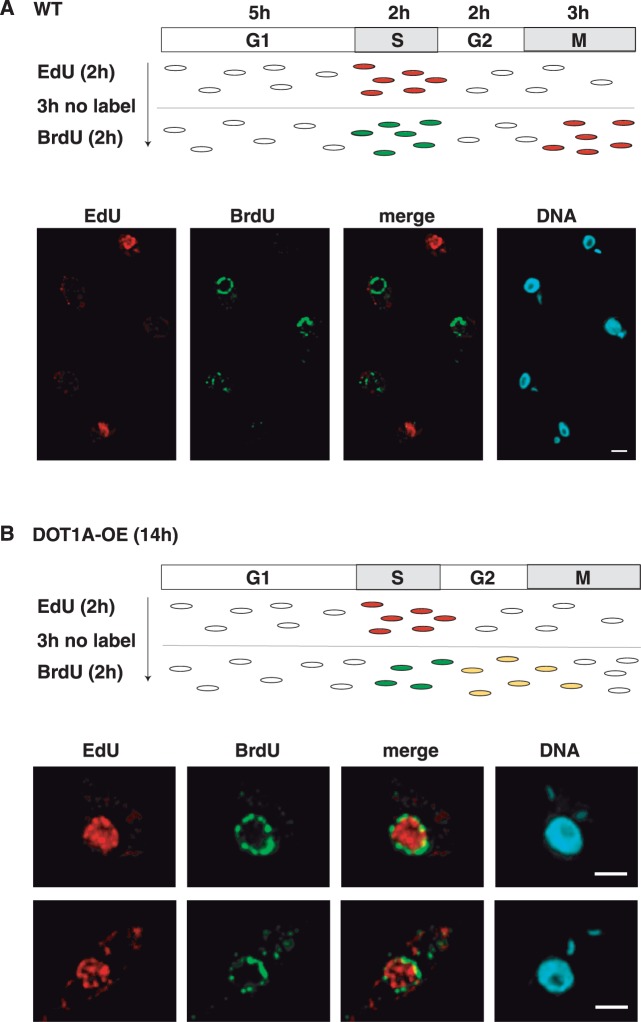
We did not observe a sharp peak in the FACS profile of the aneuploid cells; therefore it is unlikely that the DNA content was accurately doubled in an additional round of replication. The broad profile rather suggests a continuous replication of DNA without proper chromosome segregation and karyokinesis. To test this hypothesis, we established a protocol that allows the detection of DNA synthesis at distinct time points of the cell cycle by incorporation of differently labelled nucleotides. We first labelled newly synthesized DNA with EdU 7 h post-induction for 2 h (Figure 4A). We then removed EdU from the cell culture supernatant, waited for 3 h to ensure that all labelled cells exit S phase (which requires ∼2.5 h in our laboratory strain) and labelled again with BrdU for 2 h. We then analysed the cells by fluorescence microscopy (Figure 4). In a wild-type population cells could be labelled with either EdU or BrdU but never with both (Figure 4A). The distribution of the incorporated BrdU and EdU was different, which may have been caused by differences in mobility of an antibody compared to a small molecule. In DOT1A over-expressing cells we could detect cells that were clearly doubly labelled with EdU and BrdU, indicating that replication continued although cellular markers suggest that the cells were not in S phase anymore (Figure 4B, 1N2K cell). In summary, these data support our hypothesis that DOT1A over-expression causes continuous replication, which generates nuclei with increasing DNA content and prevents chromosome segregation and karyokinesis. To characterize the aneuploid cells in more detail, we sorted cells of this population and performed fluorescence in situ hybridization (FISH) with a probe specific for the tubulin gene array (Figure 5). Sorted cells of G1 and G2 phase were analysed as control and showed distinguishable signals for the tubulin gene locus as expected (Figure 5A, top panels). Two signals were detectable in most G1 phase cells and G2 phase cells showed either two or four signals. The lower panel of Figure 5A shows four distinguishable signals representing segregated chromosomes in a cell that nearly finished karyokinesis.
Figure 4.
DOT1A over-expression causes continuous replication of nuclear DNA. Incorporation analysis of EdU and BrdU in replicated DNA during S phase of (A) wild-type and (B) DOT1A over-expressing cells (14 h post-induction). Top panels show a schematic representation of the experimental design. Labelled cells are shown as coloured dots. Bottom panels show IFA of labelled parasites. Detection of EdU (red) is based on a ‘click reaction’, BrdU (green) was detected by an antibody. DNA was stained with DAPI (blue). Scale bars, 2 µm.
Figure 5.
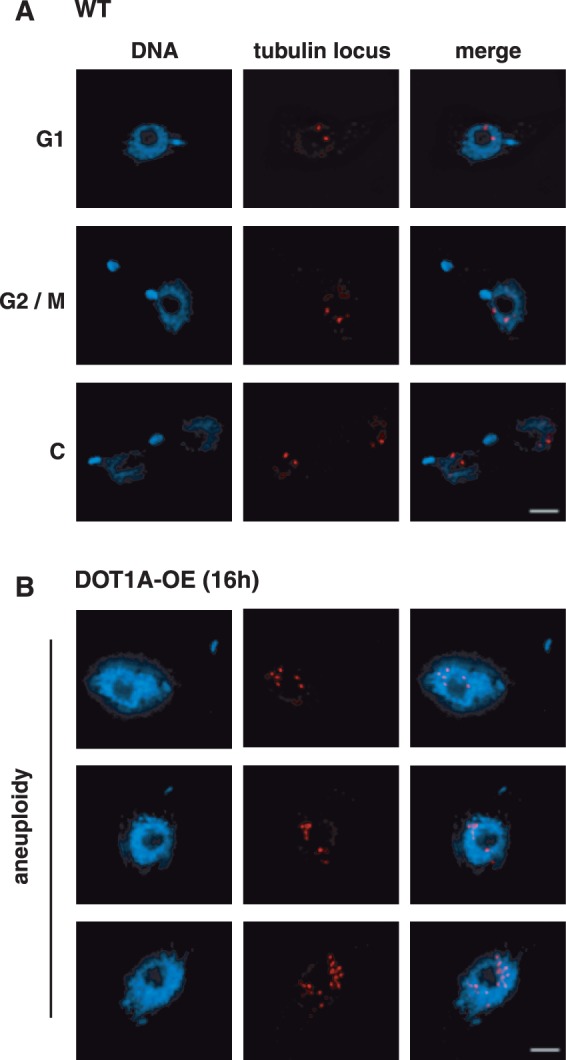
DOT1A over-expression causes re-initiation of replication. FISH of (A) wild-type and (B) DOT1A over-expressing cells. A probe specific for the tubulin gene array (red) was used for hybridization. DNA was stained with DAPI. Aneuploid nuclei show several distinguishable signals of the tubulin locus. Scale bars, 2 µm.
In contrast, cells of aneuploid populations contained enlarged nuclei with several clearly distinguishable signals (Figure 5B) 16 h after induction of DOT1A over-expression, which suggests multiple re-initiation events in the same locus within one S phase.
Our data clearly indicate that DOT1A is involved in replication control in T. brucei and suggest that coordinated regulation of the methyltransferase activity is important for accurate cell-cycle progression. How the different methylation levels are generated and maintained during the cell cycle will be the subject of further experiments.
Genome-wide distribution of H3K76 mono- and di-methylation
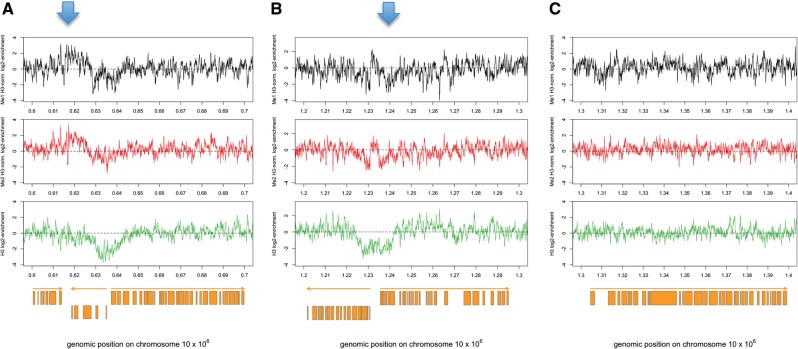
How does H3K76 methylation influence replication? The simplest explanation is that H3K76 methylation marks the loci in the genome where replication is initiated. These origins of replication are not well defined in most eukaryotes and have not been described in trypanosomes, yet. Therefore, we wanted to know if H3K76 mono- and di-methylation is associated with distinguishable domains in the genome or with specific DNA sequences. To address this question, we employed ChIP with specific antibodies for mono- or di-methylated H3K76 in combination with a custom-made microarray to find chromatin domains enriched with H3K76 methylation. After genome-wide analysis (data not shown), we could not define any sequence-specific enrichment of H3K76 methylation. However, we observed moderate enrichment in some functional domains (Figure 6). The genome of trypanosomes is organized in large polycistronic transcription units, which are bordered by well-defined transcription start sites (TSS) and transcription termination sites [TTS, (18)]. TSS and TTS are characterized by incorporation of histone variants into the nucleosomes, partial nucleosome depletion and enrichment of specific histone PTMs. Interestingly, we found a clear correlation of H3K76 methylation and TTS. For example, on chromosome 10 we could detect 27 areas of H3K76me1 enrichment using a 2-fold cut off and a peak width of at least five adjacent probes (Supplementary Figure S4 and Figure 6). Nine peaks correlated with convergent TTS, 10 peaks with internal TTS and two with divergent TSS. Six peaks were not associated with any functional domains. The distribution of H3K76me2 showed a very similar distribution (eight peaks at convergent TTS, seven peaks at internal TTS and two peaks within a polycistronic unit).
Figure 6.
Distribution of H3K76 mono- and di-methylation. Representative regions of chromosome 10 showing the distribution of H3K76me1 (black, upper panels), H3K76me2 (red, lower panels) and histone H3 (green, lower panels) are displayed. The distribution of H3K76 methylation is normalized with histone H3 distribution. Orange boxes represent ORFs and orange arrows indicate direction of transcription. Displayed are examples of (A) a convergent transcription termination site (TTS, marked by an arrow), (B) a divergent transcription start site (TSS, marked by an arrow) and (C) an internal part of a polycistronic unit.
In summary, these data suggest that some but not all TTS or TSS might function as origins of replication in Trypanosoma brucei. The hypothesis that accurate timing of de novo H3K76 methylation prevents premature initiation of replication will be in the focus of future research in our laboratory.
DISCUSSION
In eukaryotes replication origins are ‘licensed’ in order to guarantee only one round of replication per cell cycle during S phase [reviewed in (24)]. The ‘licensing model’ of replication control suggests three consecutive steps: first the pre-replicative complex (pre-RC) is found at origins during G1 phase, second the initiation of the replication displaces the Mcm (mini chromosome maintenance) complex from origins and third the license is re-established by stably binding the origin recognition complex (ORC) and Mcm complex proteins during mitosis or G1 phase. The regulation of licensing in metazoans is very complex and involves carefully coordinated interactions of cyclin-dependent kinases (CDKs) and licensing factors such as Cdt1. The licensing system is regulated rather indirectly, for example by CDK-dependent degradation of Cdt1 or by prohibiting Cdt1 activity by inhibitors such as geminin [reviewed in (25)].
There is clear evidence that the cell cycle is also regulated by cyclins and CDKs in trypanosomes [reviewed in (1)], the mechanisms of replication regulation are, however, completely unknown. Database searches suggest that geminin and Cdt1 homologues are absent in T. brucei, but Orc1/Cdc6 homologues have been identified (26,27). Therefore, it is unlikely that the trypanosome regulatory system is similar to that of metazoans. Although components of the pre-initiation complex such as Orc1 have been identified and characterized, their function in replication control remains elusive. Neither expression, localization, nor chromatin binding of Orc1 appear to be the mechanisms of replication regulation in T. brucei (26). How could replication be regulated in trypanosomes?
Recently, Tardat and colleagues described that H4K20 methylation by histone methyltransferase PR-Set7 is involved in replication control in mammalian cells (27). Similarly to H3K76 methylation, H4K20me1 accumulates during G2 phase by cell-cycle-regulated expression of PR-Set7. Expression of a degradation-resistant PR-Set7 mutant or forced over-expression of wild-type PR-Set7 resulted in re-replication of DNA in G2 phase-arrested cells, suggesting that methylated H4K20 functions in replication licensing (28).
Although alternative targets of DOT1 methyltransferases cannot be completely excluded, there could be a very similar function of histone H3K76 methylation in T. brucei. The regulation of H3K76 methylation appears to be mediated by careful coordination of DOT1A and DOT1B activity. As shown in other organisms, H3K76 methylation is only possible in a nucleosomal context (2). Hence, accumulation of H3K76 mono-methylation in G2 phase and H3K76 di-methylation during mitosis and cytokinesis could be mediated by DOT1A after new histones are incorporated into chromatin. H3K76 mono- or di-methylation might be essential in G2 or M phase to license origins of replication. H3K76 is fully tri-methylated shortly after cytokinesis (A. Gassen, unpublished data), which requires a massive up-regulation of DOT1B activity and might prevent replication initiation outside S phase.
Our data suggest that H3K76 methylation is involved in the activation of origin licensing in trypanosomes most likely at TTS at the end of polycistronic units. An indirect effect on H4K18 methylation (the homologous residue to mammal H4K20 in trypanosomes) can be excluded because H4K18 methylation levels do not change after manipulation of DOT1A expression (Supplementary Figure S5). To fully unravel the mechanisms of replication regulation, origins of replication have to be characterized in T. brucei and the influence of H3K76 methylation on pre-RC assembly has to be analysed. Furthermore, we have to learn how the different methylation activities of DOT1A and DOT1B are regulated during the cell cycle and how specific chromatin domains are targeted by these methyltransferases. This may unravel a mechanism that is not unique to trypanosomes but might be active in other protozoan parasites or even in higher eukaryotes as a redundant pathway for replication regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [JA1013/2-1 and the Sonderforschungsbereich (collaborative research network) (SFB) TR5]. Funding for open access charge: DFG collaborative research center TR5.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank all members of the Janzen laboratory, Daniela Tonn, Nicola Jones, Tim Krueger and Sandra Hake for carefully reading the manuscript and Michael Boshart for valuable discussions.
REFERENCES
- 1.Hammarton TC. Cell cycle regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;153:1–8. doi: 10.1016/j.molbiopara.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janzen CJ, Hake SB, Lowell JE, Cross GA. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 3.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 4.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 6.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.San-Segundo PA, Roeder GS. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell. 2009;11:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Vos D, Frederiks F, Terweij M, van Welsem T, Verzijlbergen KF, Iachina E, de Graaf EL, Altelaar AF, Oudgenoeg G, Heck AJ, et al. Progressive methylation of ageing histones by Dot1 functions as a timer. EMBO Rep. 2011;12:956–962. doi: 10.1038/embor.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figueiredo LM, Janzen CJ, Cross GA. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6:e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figueiredo LM, Cross GA, Janzen CJ. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat. Rev. Microbiol. 2009;7:504–513. doi: 10.1038/nrmicro2149. [DOI] [PubMed] [Google Scholar]
- 12.Brun R, Schonenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 13.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 14.Burkard G, Fragoso CM, Roditi I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 2007;153:220–223. doi: 10.1016/j.molbiopara.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for dominant negative approaches in Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 16.Alibu VP, Storm L, Haile S, Clayton C, Horn D. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 2005;139:75–82. doi: 10.1016/j.molbiopara.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Oberholzer M, Morand S, Kunz S, Seebeck T. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 2006;145:117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- 21.Siegel TN, Hekstra DR, Cross GA. Analysis of the Trypanosoma brucei cell cycle by quantitative DAPI imaging. Mol. Biochem. Parasitol. 2008;160:171–174. doi: 10.1016/j.molbiopara.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufmann D, Gassen A, Maiser A, Leonhardt H, Janzen CJ. Regulation and spatial organization of PCNA in Trypanosoma brucei. Biochem. Biophys. Res. Commun. 2012;23:698–702. doi: 10.1016/j.bbrc.2012.02.082. [DOI] [PubMed] [Google Scholar]
- 23.Ploubidou A, Robinson DR, Docherty RC, Ogbadoyi EO, Gull K. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci. 1999;112:4641–4650. doi: 10.1242/jcs.112.24.4641. [DOI] [PubMed] [Google Scholar]
- 24.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 25.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell. Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Godoy PD, Nogueira-Junior LA, Paes LS, Cornejo A, Martins RM, Silber AM, Schenkman S, Elias MC. Trypanosome prereplication machinery contains a single functional orc1/cdc6 protein, which is typical of archaea. Eukaryot. Cell. 2009;8:1592–1603. doi: 10.1128/EC.00161-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang HQ, Li Z. The Cdc45.Mcm2-7.GINS protein complex in trypanosomes regulates DNA replication and interacts with two Orc1-like proteins in the origin recognition complex. J. Biol. Chem. 2011;286:32424–32435. doi: 10.1074/jbc.M111.240143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- 29.Frederiks F, van Welsem T, Oudgenoeg G, Heck AJ, Janzen CJ, van Leeuwen F. Heterologous expression reveals distinct enzymatic activities of two DOT1 histone methyltransferases of Trypanosoma brucei. J. Cell Sci. 2010;123:4019–4023. doi: 10.1242/jcs.073882. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.