Abstract
Furvina®, also denominated G1 (MW 297), is a synthetic nitrovinylfuran [2-bromo-5-(2-bromo-2-nitrovinyl)-furan] antibiotic with a broad antimicrobial spectrum. An ointment (Dermofural®) containing G1 as the only active principle is currently marketed in Cuba and successfully used to treat dermatological infections. Here we describe the molecular target and mechanism of action of G1 in bacteria and demonstrate that in vivo G1 preferentially inhibits protein synthesis over RNA, DNA and cell wall synthesis. Furthermore, we demonstrate that G1 targets the small ribosomal subunit, binds at or near the P-decoding site and inhibits its function interfering with the ribosomal binding of fMet-tRNA during 30S initiation complex (IC) formation ultimately inhibiting translation. Notably, this G1 inhibition displays a bias for the nature (purine vs. pyrimidine) of the 3′-base of the codon, occurring efficiently only when the mRNA directing 30S IC formation and translation contains the canonical AUG initiation triplet or the rarely found AUA triplet, but hardly occurs when the mRNA start codon is either one of the non-canonical triplets AUU or AUC. This codon discrimination by G1 is reminiscent, though of opposite type of that displayed by IF3 in its fidelity function, and remarkably does not occur in the absence of this factor.
INTRODUCTION
New antimicrobial compounds, targeting un- or under-exploited targets, having novel modes of action and therefore not recognized by existing resistance mechanisms, are urgently needed to cope with the spread of almost untreatable multidrug-resistant pathogens which are causing fatal outbreaks worldwide (1,2).
In the course of a screening program aimed at the discovery of new antimicrobial agents, promising data were obtained with a synthetic nitrovinylfuran, which displayed a broad spectrum of antimicrobial activity. This compound, denominated Furvina® (also known and referred to as G1 in this article), is 2-bromo-5-(2-bromo-2-nitrovinyl)-furan (MW 297), a furylethylenic derivative in which the nitro group is not attached to the furan ring (Figure 1A insert) and is obtained in two synthetic steps in high yields with >99.8% purity (3,4).
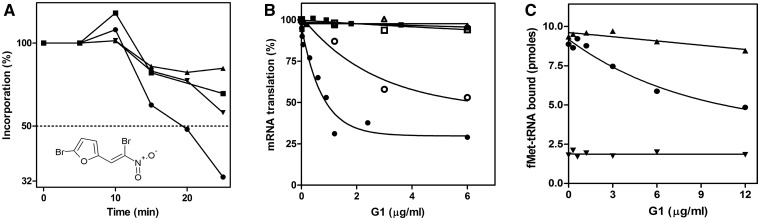
Figure 1.
Inhibitory effects of G1 in vivo and in vitro. (A) Effect of G1 (whose chemical structure is presented in the insert) on the in vivo incorporation of [3H] thymidine (filled triangle), [3H] uridine (filled inverted triangle), [35S] methionine (filled circle) and [3H] N-acetylglucosamine (filled square) by Escherichia coli MRE600 cells. The antibiotic (3 µg/ml) was offered at 5 min; (B) effect of the indicated amounts of G1 on the in vitro translation of the universal 027IF2Cp(A) mRNA (filled circle) or on the poly(U)-dependent poly(Phe) synthesis (filled square). To identify the G1 target within the translational apparatus, 027IF2Cp(A) mRNA was translated in an E. coli system reconstituted using 30 S (open circle) or 50 S (open square) ribosomal subunits centrifuged after 10 min incubation at 37°C without (open triangle) or with the indicated amounts of G1 (open circle, open square) as described in the text. The E. coli cell-free extracts were incubated at 37°C for 15 min as described in (9), and the synthesized products were quantified by determining the amount of radioactive precursor incorporated following the hot TCA procedure. Further details can be found in the ‘Materials and Methods’ section. (C) Effect of increasing concentrations of G1 on the formation of 30 S (filled circle) and 70S (filled triangle) initiation complexes as determined from the amount of ribosome-bound f[35S] Met-tRNA in response to mRNA detected by nitrocellulose filtration. Binding of f[35S] Met-tRNA to non-coded 30S subunits (filled inverted triangle).
G1 displays a significant antimicrobial effect, comparable to that of some other antimicrobial agents currently in clinical use, being active against bacteria, yeast and filamentous fungi with minimal inhibitory concentration (MIC) values ranging from 0.5 to 32 μg/ml (5–7). Because of these and other features, such as very low levels of toxicity, this antibiotic was approved for topic therapy in Cuba where it is marketed as Dermofural®, an ointment used to treat dermatological infections of both bacterial and fungal origin.
However, so far nothing is known concerning the target and mechanism of action of G1, and this study is aimed at filling this gap in our knowledge. Indeed, it is important to obtain information about these functional aspects of G1, not only in the perspective of a possible use of this antibiotic for the systemic treatment of diseases caused by G1-sensitive microorganisms but also in light of the interesting finding that this antibiotic is active against both bacteria and mycetes which belong to two different kingdoms of life and display important structural and biological differences.
Our results indicate that in bacteria G1 preferentially inhibits protein synthesis, both in vivo and in vitro and shows that it binds to the 30S ribosomal subunit and inhibits P-site decoding thereby blocking translation initiation, which represents an underexploited antibiotic target (8). Finally, our data show that G1 displays the remarkable property of discriminating between initiation triplets, being active against mRNAs bearing the canonical AUG or the rare AUA start codon, but being rather ineffective against initiation directed by the non-canonical codons AUU and AUC bearing a pyrimidine in the 3′-position.
MATERIALS AND METHODS
Preparation of biological materials
Escherichia coli 70S, 30S and 50S ribosomal subunits, S30 cell extracts, S100 post-ribosomal supernatant, f[35S] Met-tRNAfMet, IF1, IF2 and IF3166Alexa488, two types of model mRNAs (022mRNA and 027IF2Cp(A) mRNA) and fMet-tRNAfMet carrying a quencher (QSY35) at position 8 were prepared as described (9–11). In vivo effect of G1 on macromolecular syntheses was assessed as described in (9) and in Supplementary Data.
In vitro translation
Universal 027IF2Cp(A) mRNA translation and poly(U)-dependent poly-phenylalanine (poly-Phe) synthesis with E. coli cell-free extracts were performed as described, but for the omission of reducing agents such as DTT or β-mercaptoethanol (9,11).
30S initiation complex formation
Binding of fMet-tRNA to 30S ribosomal subunits programmed with 022 mRNA (with the indicated initiation codons) was determined (9,10) by filtration through nitrocellulose discs (as described in Supplementary Data) and by fluorescence stopped flow analysis performed in a Kintek SF-2004 stopped-flow apparatus. Syringe 1 was filled with 20 mM Tris–HCl (pH 7.7) buffer containing 7 mM Mg acetate, 60 mM NH4Cl, 0.5 mM guanosine 5′-triphosphate (GTP), 0.2 µM E. coli 30S subunits, 0.2 µM IF1, IF2 and IF3166Alexa488, 0.4 µM 022 mRNA and the indicated amounts of G1; syringe 2 was filled with the same buffer containing 0.4 µM fMet-tRNA carrying a quencher (QSY35) at position 8. The signal generated upon mixing equal volumes (20 µl) of the two solutions was measured upon excitation at 488 nm after passing a 515-nm cutoff filter as described (10).
Kinetics of 30S IC-50S ribosomal subunit association
For the light scattering measurements, equal volumes (20 µl) of 30S initiation complex (IC) (0.1 µM), prepared as mentioned earlier in the text (Syringe 1) and 50S (0.3 µM; Syringe 2), were allowed to mix at 20°C in a stopped flow apparatus; changes of light scattering were recorded exciting at 430 nm and measuring the scattered light at 90° with respect to the incident beam without a filter (12).
Antibiotic binding to ribosomal subunits
30S (1 µM) or 50 S (1 µM) E. coli ribosomal subunits were incubated for 10 min at 20°C in the presence of increasing concentrations of G1 in 200 µl of buffer containing 20 mM Tris–HCl pH 7.7, 10 mM Mg acetate, 60 mM NH4Cl and supplemented with 20% sucrose. The samples were centrifuged for 1 h at 100 K rpm at 4°C in a Sorvall S100-AT3 rotor. The pellets obtained were then re-suspended in 20 µl of buffer containing 20 mM Tris–HCl pH 7.7, 10 mM Mg acetate and 60 mM NH4Cl. The antibiotic-exposed subunits were then tested, in combination with stoichiometrically equivalent amounts of the complementary non-G1-exposed subunits, for their capacity to support 027IF2Cp(A)mRNA-directed in vitro translation in an E. coli cell-free system.
Probing the ribosomal-binding site of G1 by hydroxyl radical cleavage and primer extension analysis
This was carried out essentially as described in (13) and in Supplementary Data.
RESULTS
Inhibitory effects of G1 in vivo and in vitro
As mentioned above, G1 (or Furvina®) inhibits with comparable efficiency (i.e. with similar MIC) the growth of several Gram-positive and Gram-negative pathogenic bacteria, as well as that of the pathogenic yeast Candida albicans. Furthermore, with the exception of Enterococcus faecalis, for which the MIC and the minimal bactericidal concentration (MBC) values are clearly divergent, in all other cases MIC and MBC have almost identical values, indicating that G1 is endowed with both bacteriostatic and bactericidal properties (Table S1 supplementary Data). Although the mechanism of action of G1 in pathogenic yeasts will be the subject of a forthcoming study, in this work we have investigated the mechanism of action of G1 in bacteria.
The first approach that we used toward the elucidation of the mechanism of action of G1 was to identify the target of this antibiotic by following the in vivo synthesis of different bio-macromolecules in E. coli cells exposed to a G1 concentration of 3 µg/ml, which is just below the MIC value in this organism (4–8 µg/ml). The results obtained in this experiment indicate that G1 preferentially inhibits protein synthesis compared to RNA, DNA and cell wall syntheses (Figure 1A); this finding is fully supported by the results of an in vitro mRNA translation test, which shows that G1 is a powerful inhibitor of protein synthesis in a bacterial cell-free extract (Figure 1B). However, comparison of the effect of G1 on translation programmed by a natural-like model mRNA and by poly-uridylic acid shows that, unlike mRNA-directed protein synthesis, poly-Phe synthesis is not affected by the antibiotic (Figure 1B). Although indicating that elongation functions (i.e. ribosomal A-site, peptidyl transferase, translocation and peptide channel) are unlikely to be the targets of G1 inhibition, this result suggests that this antibiotic affects the initiation or the termination phase of protein synthesis or the function of one or more aminoacyl-tRNA synthetases, with the exception of that responsible for charging tRNAphe. The nature of the product synthesized by the bacterial translational system in the presence of G1 was also investigated immunologically with a monoclonal antibody directed against the protein encoded by the 027IF2Cp(A) mRNA template used to program the cell-free system. The results of this experiment indicate that, unlike with the aminoglycoside streptomycin, protein synthesis inhibition by G1 is not accompanied by mRNA misreading. In fact, streptomycin has a more severe effect on the production of the correct translational product (as immunologically determined by enzyme-linked immunosorbent assay) compared with the incorporation of a radioactive precursor into a hot-trichloroacetic acid insoluble product, whereas G1 causes the same level of inhibition regardless of the method used to detect the product (Supplementary Figure S1). Under mild alkaline conditions in aqueous solution, upon water attack on the exocyclic double bond, G1 undergoes a rather rapid degradation, yielding 5-Br-furfural and Br-nitromethane. However, unlike G1, neither isolated 5-Br-furfural nor Br-nitromethane, nor a mixture of the two molecules was found to have any effect on protein synthesis (Supplementary Figure S2); this result leads to the conclusion that in our experimental system translation inhibition is exclusively a property of intact G1.
To identify the molecular target of G1 within the translational apparatus, isolated ribosomal subunits were pre-incubated with increasing amounts of G1, spun down by centrifugation, re-suspended in buffer and tested for their capacity to support mRNA translation in combination with the complementary untreated subunits. A clear reduction of the synthetic activity was observed in the translation systems containing G1-treated 30S subunits, whereas the control subunits that were processed in the same way but incubated without G1, as well as the G1-treated 50S subunits did not display any activity loss. Furthermore, the extent of inhibition suffered by the G1-exposed 30S subunits clearly increases as a function of the G1 concentration used in the pre-incubation (Figure 1B). These results indicate that G1 binds to the small ribosomal subunit and remains bound to this particle at a concentration high enough to cause translational inhibition. On the other hand, the large subunit does not display the same behavior and remains fully active in translation after incubation with G1 (Figure 1B), indicating either that this molecule does not bind to the 50S subunit or that the 50S-G1 interaction, if it exists, is too weak to withstand the centrifugation step.
In light of the evidence that G1 binds to the 30S subunit and of the possibility that it may inhibit translation initiation (Figure 1B), the effect of this antibiotic on the formation of the 30S IC and 70S IC was tested by measuring the amount of fMet-tRNA bound to mRNA-programed ribosomes. The results of these ‘classic’ nitrocellulose-binding studies demonstrate that although fMet-tRNA binding to the 70S monomers was not or only marginally affected, formation of the 30S IC was found to be severely inhibited by G1 (Figure 1C). Furthermore, since fMet-tRNA can bind to non-coded ribosomes and occupy a puromycin-reactive position provided that both IF2 and GTP are present (14), an additional experiment was carried out to determine if G1 can also inhibit this type of binding. The results of this experiment show that this antibiotic does not inhibit IF2-GTP-assisted binding of fMet-tRNA to non-coded 30S and highlight the importance of P-site codon–anticodon interaction for the inhibition by G1 (Figure 1C).
Additional experiments aimed at determining whether the inhibition is influenced by the order of addition of the antibiotic with respect to the other ribosomal ligands demonstrated that G1 is effective in inhibiting 30S IC formation only if offered to the small subunit before fMet-tRNA, whereas even large amounts of G1 cannot displace 30S-bound fMet-tRNA (not shown). These findings and the lack of inhibition of 70S IC formation (for which the 30S IC is an intermediate) suggest that fMet-tRNA and G1 compete for the same binding site and that, once 30S-bound, fMet-tRNA prevents, either directly or allosterically, G1 binding.
Effect of G1 on 30S IC formation
As already mentioned, the above experiments were carried out according to a standard protocol (i.e. by nitrocellulose filtration after a fairly long incubation) that measures the level of ribosome-bound fMet-tRNA at equilibrium. Thus, deeper insight into the mechanism of G1 inhibition was obtained by analyzing the real-time kinetics of 30S IC formation and of 50S subunits docking to the 30S IC to yield a 70S IC. Binding of fMet-tRNA to the 30S subunits in the presence of increasing concentrations of G1 was followed in a stopped-flow apparatus by measuring the quenching of the emission of a fluorophore covalently linked to 30S-bound IF3 by a quencher (QSY35) placed at position 8 of an approaching fMet-tRNA molecule (10). Joining of the 50S subunit to the 30S IC was followed by monitoring the light scattering increase due to the increased mass of the 70S particles produced in the process. Both curves describing fMet-tRNA binding to yield a 30S IC (Figure 2A) and subunit joining to yield a 70S IC (Figure 2B) can be fitted by two-exponential equations and increasing concentrations of G1 cause a progressive reduction of the level of both fMet-tRNA-binding and subunit joining (Figure 2C). However, only the apparent rate (kapp) of the fast binding step in 30S IC formation was progressively reduced as a function of increasing concentrations of the antibiotic (Figure 2D); by contrast, G1 does not cause any reduction of the kapp of subunit joining (Figure 2D).
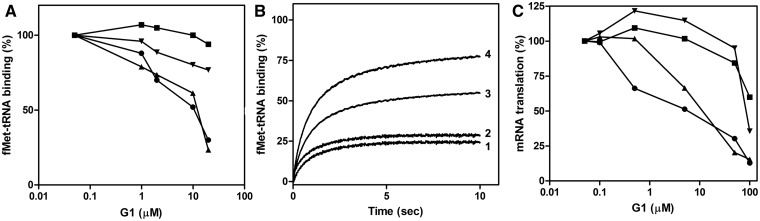
Figure 2.
Effect of G1 on 30S initiation complex formation. (A) Effect of G1 on the kinetics of 30S IC formation. Binding of fMet-tRNA to 30S subunits programmed with 022AUGmRNA in the presence of the indicated amounts of G1 was determined in a fluorescence stopped-flow apparatus monitoring the quenching of the IF3166Alexa488 fluorescence by fMet-tRNA labeled at the position 8 with QSY35 quencher. Additional details concerning the experimental conditions and procedure can be found in Materials and Methods and in ref (10). (B) Effect of G1 on the rate of 70S IC formation from 30S IC. Kinetics of 50S subunit docking to the 30S IC in the presence of the indicated amounts of G1. The experiment was carried out by stopped-flow kinetics using the light scattering change as a measurable. (C) level of 30S-bound fMet-tRNA and of 50S docking to the 30S IC measured from the amplitude of the fluorescence (filled circle) and light scattering (filled triangle) signal changes observed as a function of increasing concentrations of G1; (D) apparent rate (kapp) of fMet-tRNA binding to the 30S subunits (filled circle) and of 50S docking to 30S IC (filled triangle) as a function of increasing concentrations of G1. Further details can be found in Supplementary data.
Initiation codon bias of G1 inhibition
Since codon–anticodon interaction in the P-site is the target of G1 inhibition, the influence of the nature of the initiation codon on the fMet-tRNA-binding kinetics in the presence of G1 was investigated. The results of this experiment show that G1 inhibits initiator tRNA binding to 30S subunits programmed with an mRNA bearing the canonical AUG, as well as with the rarely used AUA (15,16) start codon (Figure 3A) but not to those programmed with an mRNA containing the non-canonical AUU and AUC initiation codons (Figure 3B). This somewhat surprising result prompted us to test and compare the effect of G1 on the translation of four mRNAs identical but for the nature of the initiation triplet, namely, 022AUGmRNA, 022AUAmRNA, 022AUUmRNA and 022AUCmRNA (17). The results obtained confirm the indications derived from the analysis of the initiation codon-dependence of G1 inhibition of 30S IC formation. Indeed, translation of the mRNA bearing the non-canonical AUU and AUC start codon proved to be much less sensitive to G1 inhibition than translation of an mRNA beginning with the canonical AUG or with the rare AUA initiation triplet.
Figure 3.
Initiation codon bias of G1 inhibition. (A) Effect of G1 on f[35S]Met-tRNA binding to the 30S ribosomal subunits programmed with 022AUGmRNA (filled circle), 022AUAmRNA (filled triangle), 022AUCmRNA (filled inverted triangle) and 022AUUmRNA (filled square) in the presence of the indicated amounts of G1; (B) Effect of G1 on the kinetics of fMet-tRNA binding to the 30S ribosomal subunits programmed with 022AUGmRNA (tracing 1), 022AUAmRNA (tracing 2), 022AUCmRNA (tracing 3) and 022AUUmRNA (tracing 4) in the presence of 6 µg/ml G1. Formation of the initiation complex was followed by fluorescence stopped-flow analysis monitoring the quenching of the IF3166Alexa488 fluorescence caused by fMet-tRNA QSY35. (C) Inhibition of in vitro translation of an E. coli cell-free system programmed with 022AUGmRNA (filled circle) 022AUAmRNA (filled triangle), 022AUCmRNA (filled inverted triangle) and 022AUUmRNA (filled square) in the presence of the indicated amounts of G1.
Localization of G1 binding site on the 30S ribosomal subunit
The topographical localization of G1 on the 30S ribosomal subunit was then studied by looking at the effects of this antibiotic on the in situ cleavage of 16S rRNA by hydroxyl radicals generated by Fe(II)-ethylenediaminetetraacetic acid (EDTA). To minimize effects due to possible non-specific, weak interactions of G1 with the 30S subunit, the cleavage reaction was carried out on ribosomal subunits pre-incubated with G1 and then subjected to centrifugation as described above.
Primer extension analysis of the cleavage sites within the 16S rRNA was carried out using oligonucleotide primers chosen to cover the entire length of the 16S rRNA molecule. These analyses gave a clear indication that G1 mainly affects bases at or near the P-site. Indeed, bases 1399 and 1404–1406, adjacent to the mRNA initiation codon, and bases 1336–1339, which constitute one side of the P/E gate (Figure 4D and E), are protected by the antibiotic (Figure 4A upper and lower panels and B, C and E). On the other hand, bases 1387–1389, in the P-site decoding region (Figure 4B, C and E) and implicated in codon discrimination by IF3 (18,19), as well as bases 1394,1396 and 1398, adjacent to the 3′-side of the mRNA initiation codon, become more exposed upon G1 binding (Figure 4A lower panel and B, C and E). Likewise, bases 1227 and 1229, which are near the N-terminal domain of S13 (Figure 4D and E), are more exposed in the presence of the antibiotic (Figure 4A middle panel and B, C and E). An additional protection effect by G1 is seen between bases 1211 and 1215 (Figure 4A middle panel); since these bases are somewhat distant from the P-site, it is not clear whether this represents a secondary G1-binding site or a long-distance conformational effect caused by the occupation of a single binding site by this antibiotic. It should be mentioned here that when protection from hydroxyl radical cleavage was performed on 30S subunits not subjected to pre-incubation with G1 and centrifugation (see above), the same protection pattern of the P-decoding site by G1 was observed (data not shown).
Figure 4.
Effect of G1 on the in situ hydroxyl radical cleavage of 16S rRNA. (A) Primer extension analysis of the cleavage sites by hydroxyl radicals generated by Fe-EDTA oxidation; from top to bottom three electrophoretic separations covering the 1200–1400 region of 16S rRNA are presented. The bases whose accessibility is decreased (magenta) or increased (blue) in the presence of 1, 2 and 3 µM G1 (last three lanes on the right) are indicated on side of the gels. Lanes G and A contain the sequencing reactions, while16S rRNA untreated and cleaved in the absence of G1 is presented in lanes K and C, respectively. In the middle panel, it is possible to see that G1 protects U1211–G1215. This protected site, being far away from the P sites where the bases affected by G1 are located is not indicated in the structures of panels B and C, but it is discussed in the text. The bases whose exposure is affected by G1 binding are shown, using the same color code of panel (A), in the background of the 3D structure of the upper portion of the 30S subunit (light gray) (B) containing bound mRNA (red) or (C) containing both mRNA (red) and P-site-bound tRNA (green). (D) close-up of the P-site codon-anticodon base-pairing (red) highlighting the bases (cyan) contributing to the stability of the canonical base-pairing (5′-AUG 3′–3′-UAC 5′) and involved in the IF3-dependent discrimination against non-canonical initiation triplets as described in the text; (E) this is the same image presented in panel (D) in which the 30S components whose exposure to hydroxyl radical cleavage is either decreased (magenta) or increased (blue) upon G1 binding are shown in space filling. Further experimental details are given in the ‘Materials and Methods’ section.
Regardless of the actual reason for the finding that G1 affects two areas of the 30S subunit, a riddle that can be solved only by X-ray crystallographic analysis, the chemical probing results are fully consistent with the premise that G1 binds or at least affects the structure of the P-site and with its effect on P-site binding of fMet-tRNA.
Finally, a relationship between the initiation codon discriminations operated by G1 and IF3 is highlighted by the finding that the bias displayed by G1 disappears in the absence of IF3. In fact, when 30S IC is allowed to form in the absence of the factor, fMet-tRNA binding to the 30S subunit in response to AUG, AUA, AUU or AUC start codons is inhibited to the same extent by G1 (Figure 5).
Figure 5.
Initiation codon discrimination in G1 requires the presence of IF3. Binding of f[35S]Met-tRNA to 30S subunits programmed with 022 mRNAs bearing the indicated initiation triplets was performed in the presence (white bars) or in the absence (gray bars) of IF3 (at a 1:1 stoichiometric ratio to the 30S subunits). The incubation was carried out for 5 min at 37°C as described in Supplementary Data. Each bar represents the average of three independent experiments.
DISCUSSION
G1 (or Furvina®) is a bactericidal antibiotic endowed with the interesting property of inhibiting with comparable efficiency the growth of both bacteria and yeast. In spite of the fact that G1 is already in therapeutic use, essentially nothing was known concerning its mechanism of action and this study was aimed at filling this gap.
Here we demonstrate that G1 inhibits bacterial growth by inhibiting protein synthesis, both in vivo and in vitro; furthermore, since G1 did not inhibit poly(U)-dependent poly(Phe) synthesis and did not cause misreading, we were able to rule out that G1 inhibits or interferes with elongation functions such as A-site decoding, peptidyl transferase activity, translocation, and with the function of the peptide exit channel of the ribosome. Instead, our data indicate that G1 targets the P-site of the 30S subunit and inhibits one of the earliest steps of the translation initiation pathway, namely, initiator fMet-tRNA binding to the 30S subunit to yield a 30S IC. Fully consistent with this type of inhibition is the evidence of a stable and specific interaction of G1 with the 30S subunit and the topographical localization of this antibiotic at and near the P-site decoding center. In addition, analyses by fast kinetics of initiator tRNA binding to the 30S subunit showed that both apparent rate (kapp) and level of fMet-tRNA binding decreased in the presence of G1. Furthermore, also the level of 30S IC–50S subunit association is lower in the presence of G1. However, since the kapp of 30S IC–50S docking is not influenced by G1, it seems safe to conclude that the antibiotic does not directly interfere with this process and that the reduced level of subunits association, as measured by the increase in the light scattering signal, is a likely consequence of the reduced level of productive 30S IC generated in the presence of G1.
As to the mechanism by which G1 inhibits fMet-tRNA binding, the finding that non-coded, IF2-GTP-assisted, binding of fMet-tRNA to the 30S subunit is not affected by the antibiotic suggests that codon–anticodon pairing in the P-site plays a central role as a target of the inhibition; this premise is further underlined by the surprising finding that G1 discriminates between canonical/non-canonical (e.g. AUG vs. AUU or AUC) initiation codons, being able to inhibit both 30S IC formation and mRNA translation only if the mRNA contains an initiation triplet bearing a purine in the 3′-position, as in the case of the canonical AUG codon or of the AUA triplet occasionally found in nature as a result of a mutation (15) or used under special circumstances (16). Thus, G1 displays an opposite bias with respect to that exercised by initiation factor IF3 whose fidelity function consists in the discrimination against non-canonical start codons both in vitro and in vivo (17,20–22).
Unlike in the A-site, where 3′ wobbling is a common event, in the P-site the 3′-nucleotide of the initiation codon is rather stringently decoded and only codons having a 3′-G are recognized as ‘canonical’ starts. As shown in Figure 4D, stability of the canonical pairing is given by hydrogen bonding between C34 of tRNA anticodon and the 3′G of mRNA codon and by stacking interactions provided by G966 and C1400 of 16S rRNA (23). The discrimination of canonical vs non-canonical initiation triplets also relies on the existence of a molecular gate separating P-site and E-site (23). This gate is constituted by G1338/A1339, which bind to one side of the anticodon stem loop of P-site bound fMet-tRNA and A790, which binds to the opposite side (Figure 4D). A large body of evidence, including biochemical and mutational studies, has clearly implicated G1338, A1339 and A790 in the IF3-dependent initiation codon discrimination and indicated that IF3 controls the conformation of this gate preventing/allowing the drifting of the P-site-bound tRNA (fMet-tRNA or an elongator aa-tRNA) into the E-site depending on the nature of the initiation triplet (18,19,24). Furthermore, bases 1387–1389 of h28 and the entire h44, the A1413–G1487 base pairing and base A1408, in particular, have been implicated in non-canonical codon discrimination (19,25,26). In fact, in vivo translation from a non-canonical initiation codon increases as a result of a C1389U substitution, whereas the h44 conformation around A1408 changes under the influence of IF1, oscillating from an initiation-unfavorable to an initiation-favorable conformation depending upon the occurrence or not of a canonical codon–anticodon pairing in the P site (25). Finally, IF3 could also destabilize the incorrect complexes through an effect mediated by S13, a protein that contacts the P-site-bound tRNA (Figure 4 D and E) and is a close neighbor of IF3 (23,27).
Analysis of the 16S rRNA cleavage patterns clearly shows that essentially all the aforementioned bases implicated in P-site decoding and in IF3-dependent discrimination against non-canonical start codons are affected by G1 binding. In fact, bases 1399 and 1404–1406, adjacent to the mRNA initiation codon, and bases 1336–1339, which constitute one side of the P/E gate (Figure 4D and E), are protected by the antibiotic (Figure 4A upper and lower panels and B, C and E). On the other hand, bases 1387–1389, which have been implicated in codon discrimination by IF3 (18,19) and 1394,1396 and 1398 adjacent to the 3′-side of the mRNA initiation codon, become more exposed upon G1 binding (Figure 4A lower panel and B,C and E). Finally, also bases 1227 and 1229, which are near the N-terminal domain of S13 (Figure 4D and E), are more exposed by the antibiotic (Figure 4A middle panel and B, C and E). Taken together, the finding that several nucleotides are protected and others become more exposed to hydroxyl radical cleavage in the presence of G1 gives a clear indication that, in addition to possibly direct shielding effects, G1 also induces conformational changes at specific 16S rRNA sites affecting the overall structure of the P-decoding region of the 30S ribosomal subunit. Thus, it may be surmised that, as with IF3 (24,28,29), also in the case of G1 a ligand-induced conformational change of the 30S subunit is at the root of initiation codon discrimination. Beyond the ‘opposite’ nature of the discriminated codons, the large overlap between the 16S rRNA bases implicated in P-site decoding and IF3 fidelity function and those protected/exposed by G1 underlies the similarity of the mechanisms determining initiation codon bias. The existence of a causal link between the bias operated by IF3 and G1 is also clearly suggested by the finding that initiation codon discrimination by G1 is suppressed in the absence of IF3 (Figure 5).
Although the majority of the anti-bacterial drugs target the translational apparatus, there are components and steps of protein synthesis, such as translation initiation, which represent unexploited or underexploited antibiotic targets. Concerning translation initiation, aside from the inhibitors of methionyl-tRNA synthetase (30) and tetrahydrofolate synthesis and consequently of initiator Met-tRNA formylation (31), only thiostrepton, evernimicin and a handful of P-site inhibitors (edeine, kasugamycin and GE81112) have been found to inhibit this early step of translation. However, although both thiostrepton and evernimicin interfere with the function of IF2 (32–34), they cannot be considered specific IF2 inhibitors since they also inhibit elongation factors EF-G and EF4 (35,36). On the other hand, among the P-site inhibitors only GE81112 can be regarded as a bacterial- and P-site-specific inhibitor (37,38), the others being either active against other targets and/or also active in eukaryotic systems (37,39).
Thus, the present discovery that the antibiotic G1 is a specific inhibitor of the 30S P-site, which blocks the formation of the 30S IC, promises to increase the potential weapons capable of selectively blocking the initial steps of bacterial protein synthesis. In light of the excellent knowledge of the 3-D structure of the 30S ribosomal subunit (e.g. 40), which represents the G1 target and of the fairly small size (MW 297) of this antibiotic, G1 can be also regarded as a potentially interesting pharmacophore in the perspective of developing, through computational chemistry, rational design or fragment-based drug design a variety of efficient tools to fight bacteria, in particular, those which have acquired multiple resistance to drugs targeting more common steps of translation (41–44).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1 and 2 and Supplementary Methods.
FUNDING
Cuban government and a Ministero dell’Istruzione, dell’Università e della Ricerca Futuro in Ricerca grant [RBFR 10X4YN_001 to D.P.]. Funding for open access charge: Ministero dell’Istruzione, dell’Università e della Ricerca.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
A.F., L.B., C.L.P. and C.O.G. wish to thank all the colleagues and friends around the world who have made this research possible with their generous gift of chemicals, fine biochemicals, precursors, etc. thereby compensating for the lack of financial support offered by the Italian government to their research.
REFERENCES
- 1.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Castañedo NR, Goizueta RD, González O, Pérez JA, González J, Silveira EA, Estrada E, Martínez A, Cuesta M, Lugo E, et al. Inventors Procedure for the obtention of 1-(5-bromofur-2-il)-2-bromo-2-nitroeteno and its action as microcide. 1999 Canada Patent 2,147,594, Japan Patent 2,875,969. [Google Scholar]
- 4.Castañedo NR, Goizueta RD, González O, Pérez JA, González J, Silveira EA, Estrada E, Martínez A, Cuesta M, Lugo E, et al. Inventors Procedimiento de obtencion del 1-(5-bromofur-2-il)-2-bromo-2-nitroeteno su accion como microcida. 2000 Certificado N 22446 Resolucion N 2190/1996. ONIITEM. Ciudad de La Habana. Cuba. [Google Scholar]
- 5.Blondeau JM, Castanedo N, Gonzalez O, Mendina R, Silveira E. In vitro evaluation of G1: a novel antimicrobial compound. Int. J. Antimicrob. Agents. 1999;11:163–166. doi: 10.1016/s0924-8579(98)00086-7. [DOI] [PubMed] [Google Scholar]
- 6.García Bernal M, Medina Marrero R, Hidalgo Llanes P, Machado Pérez R, Pérez Acosta Y, Vergara Salazar L. Comparación del efecto postantifúngico del G-1 y Anfotericina B frente a cepas de Candida albicans. Acta Farmacéutica Bonaerense. 2003;22:5–9. [Google Scholar]
- 7.García Bernal M, Medina Marrero R, Toraño Peraza G, Gutiérrez Ferrás LM. Comparación del efecto postantibiótico del G-1 y la Gentamicina frente a cepas de Staphylococcus aureus y Escherichia coli. Acta Farmacéutica Bonaerense. 2000;19:225–230. [Google Scholar]
- 8.Brandi L, Fabbretti A, Pon CL, Dahlberg AE, Gualerzi CO. Initiation of protein synthesis: a target for antimicrobials. Expert Opin. Ther. Targets. 2008;12:519–534. doi: 10.1517/14728222.12.5.519. [DOI] [PubMed] [Google Scholar]
- 9.Brandi L, Fabbretti A, Milon P, Carotti M, Pon CL, Gualerzi CO. Methods for identifying compounds that specifically target translation. Meth. Enzymol. 2007;431:229–267. doi: 10.1016/S0076-6879(07)31012-4. [DOI] [PubMed] [Google Scholar]
- 10.Milon P, Konevega AL, Peske F, Fabbretti A, Gualerzi CO, Rodnina MV. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Meth. Enzymol. 2007;430:1–30. doi: 10.1016/S0076-6879(07)30001-3. [DOI] [PubMed] [Google Scholar]
- 11.Brandi L, Dresios J, Gualerzi CO. Assays for the identification of inhibitors targeting specific translational steps. Meth. Mol. Med. 2008;142:87–105. doi: 10.1007/978-1-59745-246-5_8. [DOI] [PubMed] [Google Scholar]
- 12.Milon P, Konevega AL, Gualerzi CO, Rodnina MV. Kinetic checkpoint at a late step in translation initiation. Mol. Cell. 2008;30:712–720. doi: 10.1016/j.molcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Fabbretti A, Milon P, Giuliodori AM, Gualerzi CO, Pon CL. Real-time dynamics of ribosome-ligand interaction by time-resolved chemical probing methods. Meth. Enzymol. 2007;430:45–58. doi: 10.1016/S0076-6879(07)30003-7. [DOI] [PubMed] [Google Scholar]
- 14.La Teana A, Pon CL, Gualerzi CO. Late events in translation initiation. Adjustment of fMet-tRNA in the ribosomal P-site. J. Mol. Biol. 1996;256:667–675. doi: 10.1006/jmbi.1996.0116. [DOI] [PubMed] [Google Scholar]
- 15.Belin D, Hedgpeth J, Selzer JB, Epstein RH. Temperature-sensitive mutation in the initiation codon of the rIIB gene of bacteriophage T4. Proc. Natl Acad. Sci. USA. 1979;76:700–704. doi: 10.1073/pnas.76.2.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connor M, Gregory ST, Rajbhandary UL, Dahlberg AE. Altered discrimination of start codons and initiator tRNAs by mutant initiation factor 3. RNA. 2001;7:969–978. doi: 10.1017/s1355838201010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Teana A, Pon CL, Gualerzi CO. Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc. Natl Acad. Sci. USA. 1993;90:4161–4165. doi: 10.1073/pnas.90.9.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lancaster L, Noller HF. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Qin D, Abdi NM, Fredrick K. Characterization of 16S rRNA mutations that decrease the fidelity of translation initiation. RNA. 2007;13:2348–2355. doi: 10.1261/rna.715307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Risuleo G, Gualerzi CO, Pon CL. Specificity and properties of the destabilization, induced by initation factor IF3, of ternary complexes of 30S, aminoacyl-tRNA and polynucleotides. Eur. J. Biochem. 1976;67:603–613. doi: 10.1111/j.1432-1033.1976.tb10726.x. [DOI] [PubMed] [Google Scholar]
- 21.Sacerdot C, Chiaruttini C, Engst K, Graffe M, Millet M, Mathy N, Dondon J, Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
- 22.Sussman JK, Simons EL, Simons RW. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 1996;21:347–360. doi: 10.1046/j.1365-2958.1996.6371354.x. [DOI] [PubMed] [Google Scholar]
- 23.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelly AC, Weir RJ, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 24.Fabbretti A, Pon CL, Hennelly SP, Hill WE, Lodmell JS, Gualerzi CO. The real-time path of translation factor IF3 onto and off the ribosome. Mol. Cell. 2007;25:285–296. doi: 10.1016/j.molcel.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Qin D, Fredrick K. Control of translation initiation involves a factor-induced rearrangement of helix 44 of 16S ribosomal RNA. Mol. Microbiol. 2009;71:1239–1249. doi: 10.1111/j.1365-2958.2009.06598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin D, Liu Q, Devaraj A, Fredrick K. Role of helix 44 of 16S rRNA in the fidelity of translation initiation. RNA. 2012;18:485–495. doi: 10.1261/rna.031203.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pon CL, Pawlik RT, Gualerzi C. The topographical localization of IF3 on E. coli 30S ribosomal subunits as a clue to its way of functioning. FEBS Lett. 1982;137:163–167. doi: 10.1016/0014-5793(82)80339-6. [DOI] [PubMed] [Google Scholar]
- 28.Pon CL, Gualerzi CO. Effect of initiation factor 3 binding on the 30S ribosomal subunits of Escherichia coli. Proc. Natl Acad. Sci. USA. 1974;71:4950–4954. doi: 10.1073/pnas.71.12.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrelli D, La Teana A, Galofaro C, Spurio R, Pon CL, Gualerzi CO. Translation initiation factor IF3: two domains, five functions one mechanism. EMBO J. 2001;20:4560–4569. doi: 10.1093/emboj/20.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochsner UA, Young CL, Stone KC, Dean FB, Janjic N, Critchley IA. Mode of action and biochemical characterization of REP8839, a novel inhibitor of methionyl-tRNA synthetase. Antimicrob. Agents Chemother. 2005;49:4253–4262. doi: 10.1128/AAC.49.10.4253-4262.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Achari A, Somers DO, Champness JN, Bryant PK, Rosemond J, Stammers DK. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 1997;4:490–497. doi: 10.1038/nsb0697-490. [DOI] [PubMed] [Google Scholar]
- 32.Laue H, Valensise T, Seguin A, Lociuro S, Islam K, Hawser S. In vitro bactericidal activity of iclaprim in human plasma. Antimicrob. Agents Chemother. 2009;53:4542–4544. doi: 10.1128/AAC.00766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandi L, Marzi S, Fabbretti A, Fleischer C, Hill WE, Gualerzi CO, Lodmell JS. The translation initiation functions of IF2: targets for thiostrepton inhibition. J. Mol. Biol. 2004;335:881–894. doi: 10.1016/j.jmb.2003.10.067. [DOI] [PubMed] [Google Scholar]
- 34.Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. A quantitative kinetic scheme for 70S translation initiation complex formation. J. Mol. Biol. 2007;373:562–572. doi: 10.1016/j.jmb.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikolajka A, Liu H, Chen Y, Starosta AL, Márquez V, Ivanova M, Cooperman BS, Wilson DN. Differential effects of thiopeptide and orthosomycin antibiotics on translational GTPases. Chem. Biol. 2011;18:589–600. doi: 10.1016/j.chembiol.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gale EF, Cundliffe E, Reynolds PE, Richmond MH, Waring MJ. Molecular Basis of Antibiotic Action. London: Wiley; 1981. [Google Scholar]
- 37.Brandi L, Fabbretti A, La Teana A, Abbondi M, Losi D, Donadio S, Gualerzi CO. Specific, efficient, and selective inhibition of prokaryotic translation initiation by a novel peptide antibiotic. Proc. Natl. Acad. Sci. USA. 2006;103:39–44. doi: 10.1073/pnas.0507740102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandi L, Lazzarini A, Cavaletti L, Abbondi M, Corti E, Ciciliato I, Gastaldo L, Marazzi A, Feroggio M, Fabbretti A, et al. Novel tetrapeptide inhibitors of bacterial protein synthesis produced by a Streptomyces sp. Biochemistry. 2006;45:3700–3710. doi: 10.1021/bi052540k. [DOI] [PubMed] [Google Scholar]
- 39.Wilson DN. The A-Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 40.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 41.Mochalkin I, Miller JR, Narasimhan L, Thanabal V, Erdman P, Cox PB, Prasad JV, Lightle S, Huband MD, Stover CK. Discovery of antibacterial biotin carboxylase inhibitors by virtual screening and fragment based approaches. ACS Chem. Biol. 2009;4:473–483. doi: 10.1021/cb9000102. [DOI] [PubMed] [Google Scholar]
- 42.Rawls KA, Lang PT, Takeuchi J, Imamura S, Baguley TD, Grundner C, Alber T, Ellman JA. Fragment-based discovery of selective inhibitors of the Mycobacterium tuberculosis protein tyrosine phosphatase PtpA. Bioorg. Med. Chem. Lett. 2009;19:6851–6854. doi: 10.1016/j.bmcl.2009.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheich C, Puetter V, Schade M. Novel small molecule inhibitors of MDR Mycobacterium tuberculosis by NMR fragment screening of antigen 85C. J. Med. Chem. 2010;53:8362–8367. doi: 10.1021/jm100993z. [DOI] [PubMed] [Google Scholar]
- 44.Fabbretti A, Brandi L, Gualerzi CO. How to cope with the quest for new antibiotics. FEBS Lett. 2011;585:1673–1681. doi: 10.1016/j.febslet.2011.04.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.