Abstract
Background
High throughput, brain-wide analysis of neural circuit connectivity is needed to understand brain function across species. Combining such tractography techniques with small animal models will allow more rapid integration of systems neuroscience with molecular genetic, behavioral, and cellular approaches.
Methods
We collected DTI and T2 scans on 3 series of 6 fixed mouse brains ex vivo in a 9.4 Tesla magnet. The DTI analysis of ten mouse brains focused on comparing prelimbic (PL) and Infralimbic (IL) probabilistic tractography. To validate the DTI results a preliminary set of 24 additional mice were injected with BDA into the IL and PL. The DTI results and preliminary BDA results were also compared to previously published rat connectivity.
Results
We focused our analyses on the connectivity of the mouse prelimbic (PL) vs. infralimbic (IL) cortices. We demonstrated that this DTI analysis is consistent across scanned mice, with prior analyses of rat IL/PL connectivity, and with mouse PL and IL projections using the BDA tracer.
Conclusions
High-throughput ex vivo DTI imaging in the mouse delineated both common and differential connectivity of the IL and PL cortex. The scanning methodology provided a balance of tissue contrast, signal-to-noise ratio, resolution and throughput. Our results are largely consistent with previously published anterograde staining techniques in rats, and the preliminary tracer study of the mouse IL and PL provided here.
Keywords: Diffusion Tensor Imaging (DTI), Infralimbic/Prelimbic, Mouse, Prefrontal Cortex, Amygdala
INTRODUCTION
An increasing focus has been placed on differential functions of the medial Prefrontal Cortex (mPFC) in mammals. This region is involved in a number of regulatory functions in humans, and in rodents has been most associated with regulation of behavior related to appetitive and aversive processing (Choi et al.; Corcoran and Quirk, 2007; LaLumiere et al.; Milad and Quirk, 2002; Milad et al., 2004; Peters et al., 2008). The processing of conditioned fear is among the most well-understood neural circuits related to a specific behavior, and recently the mPFC has been shown to intimately regulate the amygdala in the modulation of learned fear (Corcoran and Quirk, 2007; Vidal-Gonzalez et al., 2006). The prelimbic (PL) component of the mPFC in mice and rats has been associated with the learning and expression of conditioned fear, but with no effect on innate fear behavior (McLaughlin et al., 2002; Powell et al., 2001). In contrast, the infralimbic (IL) component of the mPFC appears to have an opposite function, with its requisite involvement in the inhibition of learned fear behavior (Milad and Quirk, 2002; Nieuwenhuis and Takashima, 2011; Quirk et al., 2000) . Similarly, with appetitive behaviors, the PL appears to drive drug seeking while the IL suppresses such behaviors (LaLumiere et al., 2012; Rocha, et al 2010). What is particularly interesting about the mPFC regions is that despite their apparently opposite functions in emotion regulation, they are adjacent, sharing a thin cortical region of only a few hundred microns within the rodent mPFC. Furthermore, although the PL/IL connectivity has been examined in the rat (Vertes, 2004), these connections have not been described in the mouse, which is increasingly used for understanding the molecular genetic mechanisms of brain function.
Therefore, it is critical to further understand the complementary and differential roles of these regions, as well as to better understand their connectivity relationships to other brain regions. However, classical anatomical techniques, while feasible and serving as the gold standard, are slow - often requiring weeks from injection to image generation, low throughput as limited by the number of stereotaxic setups and animal surgery time, are limited to a few injections of tracer in regions of interest in each animal, and are not easily applied in small animal models. For example (and in the case of this paper), when working with a mouse brain, the injection targets are significantly smaller (leading to a high “miss” rate) and the diffusion of the tracer may impact surrounding regions of interest unless the volume of tracer injection is reduced (also reducing the neural tracing fidelity).
Besides classical tracing are other methods including Magnetic Resonance Imaging (MRI), which provides an excellent and powerful set of customizable methodologies to allow non-invasive in vivo and ex vivo analysis of the mouse brain. A number of different MRI methods have been developed and implemented to allow high-throughput ex vivo imaging of fixed tissue or in vivo samples. Several groups have developed sophisticated workflows that allow rapid in vivo analysis in mice, although this often requires the construction of custom enclosures, receiver coils, or modifications to the underlying hardware. For anatomical scans and morphometric analysis of brain structures, many of the studies focus on T2 and T2* weighted pulse sequences, which provide a good balance of tissue contrast, signal-to-noise ratio (SNR) and resolution. The choice of optimal scan parameters of course remains a tradeoff, as there is inherently a tradeoff between SNR, image resolution, scan time, and sample preparation which can be a particular problem in diffusion tensor imaging which requires multiple acquisition volumes.
Diffusion Tensor Imaging (DTI) is a MRI technique that generates its signal and contrast primarily based on the degree and direction that water diffuses along neuronal axons (Zhang et al., 2003). DTI has had some preliminary success when used to characterize the developing mouse embryo (Zhang et al., 2003). DTI has also been extensively used in both human (Douaud et al., 2011; Gutman et al., 2009; Wedeen et al., 2008) and primate studies (Li et al., 2010; Rilling et al., 2008; Wedeen et al., 2008) to compare and contrast the structural connectivity of brain regions between the model species imaged. Using computation techniques, it is subsequently possible to reconstruct putative connections in silico via various computational algorithms. A few studies have also directly compared the results of tractography to the gold standard tracer-injection studies in the porcine model and have generally shown a high concordance between the histologically defined pathways and those defined based on MRI Tractography results (Dyrby et al., 2011; Dyrby et al., 2007). Furthermore, one study has compared the DTI analysis of the tracts of the murine olfactory bulb to corresponding MRI based tracing conducted using MEMRI (Gutman et al., 2012).
The generation of high-throughput MRI DTI presents a number of challenges in mice—largely related to the small size of the mouse brain. For example, the resolution required to gather isotropic voxels less than a fifth of a millimeter (or about the size of the smaller nuclei in the mouse brain) makes a highfield magnet (e.g 7 or greater teslas) with a complimentary gradient and RF coil set ideal. Additionally, the higher resolution increases the length of the scan tremendously. Furthermore, in order to compute probabilistic tractography, a minimum of 30 gradient directions is recommended (although technically only 6 are required to compute a ‘tensor’), necessitating the collection of at least 31 separate volumes (when including at least one B0), which dictates and even longer scanning session. Considering both the resolution and number of gradients such work is often not feasible with in vivo models. Importantly, the addition of 30 or more directions and higher resolution are imperative as it allows estimations of a more sophisticated tensor model that allows tracking through regions of “crossing fibers” (Behrens et al., 2007; Behrens et al., 2003; Johansen-Berg and Behrens, 2006).
In this study, we report our implementation of a high-throughput ex vivo diffusion tensor imaging protocol that allows for both high-resolution and a high number of diffusion weighted directions. For these studies, groups of fixed ex vivo mice brains were scanned in a single acquisition session. We then tested the feasibility and fidelity of using probabilistic tractography in the mouse by analyzing the connectivity of the IL and PL cortices (areas separated by less than a millimeter) with the results validated against previous tract-tracing experiments in rats, and also against a preliminary BDA tracing of the mouse PL and IL projections. Comparisons to rat anatomy are important a major theme of this paper as there is an assumed direct and corresponding homology. Furthermore, comparisons to the classical tracing within mice provides a validation of the DTI technique. Specifically, the mPFC region was chosen as it has received both increasing interest within the neuroscience community (Maddux and Holland, 2011; Nieuwenhuis and Takashima, 2011), as well as provides some of the technical challenges in terms of proximity.
Our DTI and classical tracing data suggest that IL and PL share a majority of projections but also have unique connectivity as well. More importantly, it demonstrates that these methods can be utilized to examine differential connectivity of mPFC (and other regions of interest) in genetically modified models as well as pre- and post-experimental manipulation, e.g. following stress, fear conditioning, drug sensitization, etc. In this paper, we demonstrate the feasibility of tracking complicated pathways in the mouse through the use of probabilistic tractography, and we validate these results using a comparative anatomy approach of known anatomical connections of the rat IL/PL (Vertes, 2004) and also using a preliminary tracing study of the mouse IL/PL. Finally, our data provides a preliminary comparison of rat and mouse IL/PL connectivity, often assumed to be homologous.
METHODS
Animals
All animals used in this study were treated humanely following a procedure approved by the Institutional Animal Care and Use Committee at Emory University. A total of 18 C57BL6 mice (Jackson Labs) were used for the MRI imaging presented in the study. 10 Brains were of sufficient quality with the entire brain through cerebellum visualized to be included in final analyses. Animals were ~ 100 days at old at time of fixation. A total of 24 C57BL6 mice (Jackson Labs) were used to conduct a preliminary anterograde tracing study of IL and PL.
Fixation Procedure
Mice were euthanized using pentobarbital, and brains were perfused with 10 mls Phosphate Buffered Saline (PH 7.4) followed by 10 mls 4% paraformaldehyde (PF)/PBS. The brains were then extracted from the skull (to ensure a close interface between the brain and embedding matrix as discussed below) and were post-fixed (for 24 hours) in the same 4% PF/PBS, then rinsed in PBS and stored at 4 C in PBS until they were embedded in an agarose-gadolinium (III) oxide matrix for imaging. No exogenous contrast agent was used for this study.
Mounting procedure
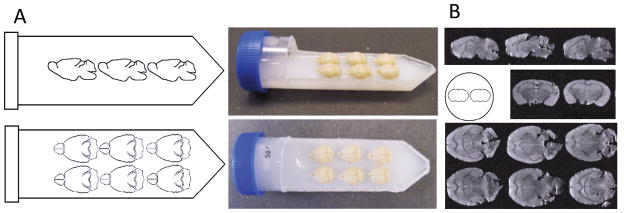
Following at least 24 hours of post-fixation, the brains were embedded in a 50 ml falcon tube (Fisher Scientific), with an approximate outer diameter of 3.0 cm and a length of 11.5 cm from cap to tip, arranged in a 2 x 3 matrix (see Figure 1). Brains were embedded in 2% agarose (Phenix Research Products Low EEO, Molecular Biology Grade Agarose) doped with an insoluble mixture of 2 mM gadolinium (III) oxide (Acros Organics Fisher Scientific). Of note, several iterations of T2 scans with different gadolinium concentrations were conducted with the 2x3 configuration to optimize integration with the Bruker coil, minimizing signal drop-off and maximizing SNR. Doping with gadolinium (III) oxide serves to robustly reduce the T1 relaxation time in the agarose/water solution, providing better separation of the brains from the background.
Figure 1. High throughput layout utilized to scan 6 brains ex vivo.
A) Shows the dimensions and the scale used to maximize the volume of sensitivity of the gradient coil. B) Shows the corresponding images that are acquired with optimized T2 weight parameters.
The mounting procedure is simple and straightforward providing both accurate and precise ex vivo imaging. The lid is cut from a 50mL Falcon Tube with a Dremel Tool using a Dremel Plastic Cutting Wheel (Model 4000 and EZ476 respectively, Bosch Tools). The first cut is made along the length of the falcon tube at the point of maximum diameter connecting each cut on the side at the tip of the tube, a perpendicular cut is made through the long axis near the cap (Figure 1A). The lid section then has two 0.5 cm holes cut into it using a Dremel high speed cutting bit (192, Bosch Tools). Concurrently, a solution of deionized water containing 2% (by weight) agarose and 2 mM Gadolinum (III) Oxide (note: Gadolinium III Oxide is not soluble and needs to be maintained in suspension) is mixed on a stir plate until homogenous and then heated in a microwave. Following heating, the solution is placed back onto the stir plate, to ensure homogeneity; the solution is then poured into the tube, until the level of the solution is approximately 2 mm below the edge created when cutting out the top (replace the beaker onto the stirplate). The tube containing the agarose/Gadolinum III oxide is then placed into a 4°C freezer (to accelerate the setting of the agarose before the gadolinium forms a concentration gradient due to gravity), until the agarose is firm. Next, a thin layer of the agarose/gadolinium mixture is formed by filling the remaining space of the tube (up to the edge, ~2 mm), and the perfused brains are placed into the layer immediately (this layer is necessary to ensure the mixture can get into the ventral spaces at the interface of the cerebellum, brainstem, and forebrain else air pockets form creating an artifact when imaging; it is also possible to submerge the brains in the mixture then place them onto the hardened layer). Once that layer has firmed, the lid is then hot glued into it original orientation, creating a water tight seal. Upon the glue hardening, the remaining Agarose Gad (III) oxide solution is used to fill the remaining volume of the tube using the two 0.5 cm holes previously drilled (one serving as a point of pipette tip insertion, and the other as an air escape). The whole tube is then allowed to set. The two holes are then covered with hot glue to prevent dehydration of the agarose/gadolinium gel. Since the contrast in DTI is based on the diffusion rate of water, the tube is then allowed to equilibrate with ambient room temperature for a minimum of six hours prior to scanning.
Imaging Parameters
High resolution DTI with multiple gradient directions, can result in a scanning sequence that lasts several days. Therefore the ability to image multiple brains in a single session is the crucial feature that makes this experiment practical. All imaging was performed on a 9.4 T 20 cm horizontal bore scanner (Bruker, Billerica, MA) interfaced with an ACANCE console (Bruker, Billerica, MA). To obtain maximum resolution for the DTI/tractography experiments, a 6cm gradient insert (150 G/cm) was used with a 35 mm transmit/receive volume coil. The data were collected in the axial orientation with the read-out direction oriented to the long axis of the tube. For each tube of brains, T2-weighted images were first acquired at 100 micron isotropic resolution (10 averages, TE = 26ms, matrix 256 x 512, 20 averages, scan time ~ 16 hours). Diffusion-weighted images of the 6 mouse brains were then acquired using a 2-D spin-echo based sequence with 161 micron isotropic resolution. (1 average, TE = 26.9 ms, TR=10000 ms, matrix size of 256 x 128, ~ 55–60 axial slices collected/tube, 60 gradient directions with a diffusion weighting b=2000 s/mm2, and 3 b=0 s/mm2 images, scan time ~ 60 hrs). The Massachusetts General Hospital (MGH) 60 direction DTI gradient sequence was used for data acquisition; the calculated tensors were verified using the built-in gradient tables in trackvis.org (Wang et al., 2007; Wedeen et al., 2008).
ROI Selection
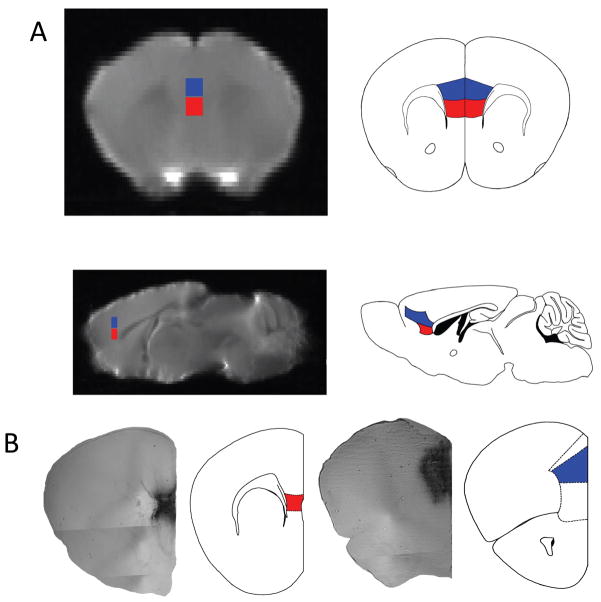
For each T2 high resolution brain scan, two 10 x 4 x 5 voxel cubed (1.0 mm x 0.4 mm x 0.5 mm) ROIs were drawn, and were only inclusive of the IL and PL volumes, respectively, [See Figure 2] based on the atlas of Paxinos and Franklin (Paxinos and Franklin, 2004).
Figure 2. IL and PL MRI seeds for DTI tractrography and PL BDI injection site.
Each brain is placed in a standard space that is also matched (same dimensions, with the ability to overlay the same rulers place on the atlas, to stereotactically determine regions of interest) to the Paxinos and Franklin Mouse Atlas used to determine areas of connectivity from the IL (shown in red) and Pl (shown in blue) seeds. A). Presented is a representation of the MRI images matched to slices in the atlas, as determined by specific anatomical landmarks (particularly the white matter tracts of the corpus callosum, the lateral and third ventricles and the aqueduct to the 4th ventricle, the internal capsule, and the hippocampus). B) Shows the injection site of the BDA/ABC/DAB in the PL and IL area at 4x magnification using phase contrast. The representative atlas slice is shown to the right with IL highlighted in red and Pl highlighted in blue, for the PL injection the cingulate outlined more dorsally and the medial orbital cortex outlined below.
DTI registration
Given that each raw image (both T2 weighted and diffusion tensor image (DTI) modalities) from the scanner contained a 2 x 3 matrix of brains, the initial processing step was the parsing of the raw data into individual brain volumes via manual segmentation using fslroi (FSL, Oxford, UK). In accordance with the FSL DTI pipeline, individual masks were generated for each mouse brain in the matrix using the Brain Extraction Toolkit (BET) (FSL, www.fmrib.ox.ac.uk/fsl) to isolate the brain from the background. Since BET is intended to extract the brain from the skull and the mouse skulls were not present (see Fixation Procedure), the fractional intensity threshold for extraction was determined empirically (a reduction in the threshold by 0.05 recursively till the extraction successfully isolated the brain from background) to be 0.05 from the default 0.5. Masks were subsequently edited manually to correct any remaining errors in the masking process. A study-specific high-resolution mouse template was generated by nonlinearly registering each mouse brain to a single reference image. The default FSL non-linear parameters were used, with the exception that the reference mask was based on the selected reference brain. The mouse brain with the best orientation, signal to noise, and contrast between white and grey matter was used for this purpose. Non-linear registration was validated by checking for correct alignment of the surface of the brain and internal alignment of the anterior commissure, corpus callosum, and cerebellum. An averaged image of the registered brains was then constructed and used as the reference image for subsequent analysis (FNIRT and FSLMATHS, respectively, www.fmrib.ox.ac.uk/fsl), and transformation matrices were generated between the reference image and individual mouse brain data sets using FNIRT (FSL, www.fmrib.ox.ac.uk/fsl). In this study results from 10 mice are presented of the 18 scanned. The initial two tubes of 6 brains had two problems that cost 4 brains from each tube. The first problem was that brain volumes were clipped or aliased (as a byproduct of trying to use a minimal FOV to cut down the imaging time), the solution was to reduce the distance between the brains measuring the embedding matrix to ensure it was smaller than the selected field of view. The second problem that arose was an issue with centering the tube in the coil, which led to a few brains with notable signal loss in the cerebellum which compromised the registration algorithm. The final tube, which had the brains placed in a tighter 2 x 6 matrix, and which was accurately centered with an iterative flashtripilot approach and a 3 plane positional scan resulted in all six brains being used in the analysis. Overall, this methodology is quite similar to those commonly employed in group level MRI imaging as described here (Lerch et al., 2011)
Probabilistic tractography
Diffusion-based probabilistic tractography was performed using the FSL imaging suite on a Linux workstation. As part of the standard FSL pipeline, the IL and PL ROIs drawn on T2 weighted high resolution space were transformed into each individual mouse's DTI space. Tractography was then computed for each voxel within the seed mask (using n=5000 streamline fibers/voxel and curvature threshold of 0.2) and then back-transformed into the high resolution mouse-standard space. For all tractography, a multi-fiber reconstruction algorithm was employed allowing the reconstruction of more geometrically complex pathways, including regions of crossing fibers (Behrens et al., 2007).
Determining Connectivity of the IL/Pl
Connectivity, based on the probabilistic tractography results, was determined in two separate but complimentary ways. First, a single mouse was chosen as a representative sample from the group (middle column of figures 3A/B), and that individual’s tractography was transformed into the high resolution standard mouse space. The raw tractography was thresholded to exclude voxels which received less than 10 total tracts from the seed ROI. Using anatomical landmarks, including the corpus collosum, the ventricles (lateral, 3rd and 4th ventricles, plus the aqueduct) and the hippocampus, it was possible to review MRI slices and assign the voxels with tractography results to a particular area of the brain labeled in the Paxinos & Franklin atlas.
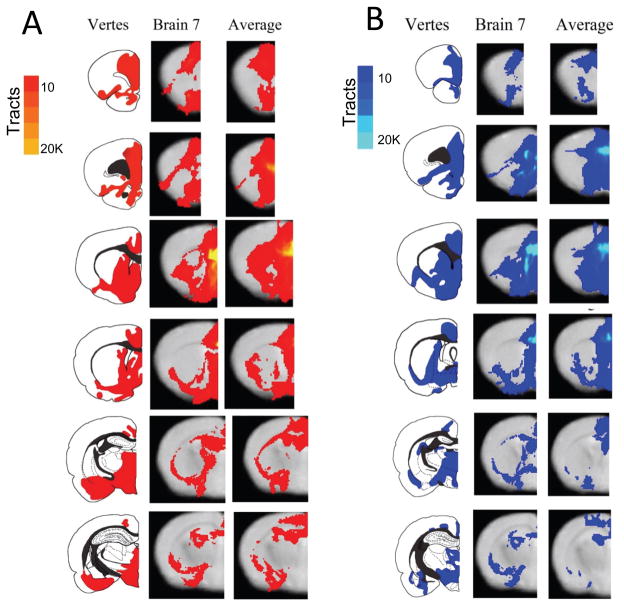
Figure 3.
Figure 3A. Comparison of infralimbic cortex results between the Vertes retrograde tracing study using a rat, the probtrack results from a representative brain, and the probtrack results from all 10 brains placed into standard space and averaged. The images for the probtrack results presented in the right two columns are thresholded to at least 10 tracts passing through a voxel to help ameliorate results from voxels with few passing tracts in only one or two brains. Notice there is considerable overlap between the rat and mouse connectivity results, and also between the representative mouse and all the mice averaged together.
Figure 3B. Comparisons of prelimbic cortex results of the connectivity between the Vertes Retrograde tracing study using a rat, the probtrack results from a representative brain, and the probtrack results from all 10 brains placed into standard space and averaged. The images for the probtrack results presented in the right two columns are thresholded to at least 10 tracts passing through a voxel to help reduce results from voxels with few passing tracts in only one or two brains. Notice there is considerable overlap between the rat and mouse connectivity results, and also between the representative mouse and all the mice averaged together.
In another analysis, the tracts from all 10 mice were placed in the standard space analogous to the methodology describe above, and then were averaged (right hand most column of Figure 3A/3B for IL and PL respectively) together yielding an average number of tracts passing through the voxels in the standard mouse space, which could be similarly assigned anatomic labels by referencing the Paxinos & Franklin Atlas.
Intra-mice comparison of IL/PL connectivity
To disambiguate regions of overlapping connectivity from the IL/PL seeds, a strategy to exploit the relative probability of connectivity between IL and PL was employed. Conceptually, for each individual brain the tractography results of both the IL and PL were compared for each voxel. If the number of tracts from the IL seed were greater in a voxel relative to those from the PL seed, that voxel was classified as IL dominant, and vice versa. For each animal, a binarized map of where the IL or the PL connectivity was dominant was generated. By subsequently merging the individual binarized maps, a map indicating the proportion of animals which showed tracts passing through that voxel that were dominant for either the IL or the PL (Figure 6).
Figure 6. Relative IL to PL connectivity.
Given the large amount of overlap of probtrack results, the data from the average of the 10 mice were categorized as either Pl or IL dominant, by comparing the magnitude of tracts per voxel in the IL and the PL results, if the number of tracts passing through a voxel was greater for the probtrack results from the IL seed (Red), compared to the PL seed (Blue), the voxel was classified as an IL seed (and vice versa). The images above display the classification within the same standard space to directly compare the relative strengths of the connections. Most notable is the rather dominant character of the IL connectivity when compared to the PL.
Anterograde Tracer Injection of BDA into the Mouse IL/PL
To have a within species classical tracing study for comparison to the DTI, we stereotactically (Kopf, Tujunga, Ca) injected with a pump (World Precision Instruments, Inc. Sarasota, Fl) either 0.25 μl of BDA (10,000 MW) into the PL or 0.15 μl of BDA into the IL (a smaller target) of the mouse anesthetized with 0.15 ml of Ketamine/Medetomidine (Domitor), using a 5 μl Hamilton syringe (30 gauge beveled needle). The coordinates were determined using the Paxinos and Franklin mouse atlas (PL, 1.7 mm anterior of bregma, 0.3 mm lateral, and 3 mm ventral; IL 1.6 mm anterior of bregma, 0.3 mm lateral, and 3.3 mm ventral). Fifteen minutes after the delivery of the BDA, the syringe was removed over the course of 2 minutes to minimize the drag of DBA through the injection site. The mice were allowed to recover for 5 days before they were perfused, analogous to those prepared for MRI. After 24 hours post fixation, the brains were placed in 30% wt sucrose solution for cryoprotection. The following day the brains were then sectioned (35 μM) using a freezing microtome and the free-floating sections were rinsed with PBS-T (2% Tween). Then sections were incubated in 1% hydrogen peroxide (H2O2) in PBS-T for 30 minutes to quench peroxidase activity. The sections were then subsequently incubated in avidin-biotin-peroxidase complex (Vector Elite ABC kit) for 4 hours. Color development was then conducted with 3,3’-diaminobenzidine (DAB, Dako, Carpinteria, Ca) for 30 minutes. The sections were then mounted onto poly-Lysine-coated coverslips, allowed to dry overnight, and were dehydrated and cover-slipped for dark field and phase analysis via a light microscope (Nikon Eclipse, E800).
RESULTS
Probabilistic Tractography (PT)
The probabilistic tractography results from seeding the infralimbic and the prelimbic areas are presented in Figure 3A and 3B. A tabulation of the areas that are covered by the probabilistic tracts, sorted by IL/PL, is presented in Table 1. In Figure 3, connectivity is compared between the results described in Vertes (Vertes, 2004) using anterograde HRP injections into IL/PL into the rat brain, the results of computed tractography from a single example mouse brain [middle], along with a tractography map generated by identifying patterns of consistent connectivity demonstrated between at least 7 of the 10 mice at the voxel level [right].
Table 1.
| Structure | Vertes IL | Brain 17 IL | Subtraction IL | Vertes PL | Brain 17 PL | Subtraction PL |
|---|---|---|---|---|---|---|
| Telencephalon | ||||||
| Cortex | ||||||
| Cingulate | +++ | ✓ | ✓+ | +++ | ✓ | ✓ |
| Ectorhinal | + | -- | ✓ | -- | -- | -- |
| Entorhinal | ++ | ✓ | ✓ | ++ | ✓ | -- |
| Frontal | ||||||
| Medial | +++ | ✓ | ✓+ | +++ | ✓ | ✓ |
| Lateral | -- | - | - | -- | - | - |
| Infralimbic | +++ | ✓ | ✓ | +++ | ✓ | ✓ |
| Insular | ||||||
| Agranular | + | ✓ | ✓ | +++ | ✓ | ✓ |
| Dysgranular | ++ | ✓ | ✓ | +++ | -- | ✓ |
| Granular | + | ✓ | -- | -- | -- | -- |
| Lateral Agranular (Motor) | -- | -- | -- | -- | -- | -- |
| Medial Agranular (Motor) | + | ✓ | ✓+ | + | ✓ | ✓ |
| Occipital | -- | -- | -- | -- | -- | -- |
| Orbital | ||||||
| Lateral | + | ✓ | ✓ | + | ✓ | ✓ |
| Medial | +++ | ✓ | ✓+ | +++ | ✓ | ✓ |
| Ventral | + | ✓ | ✓ | + | ✓ | ✓ |
| Ventrolateral | + | ✓ | ✓ | + | ✓ | ✓ |
| Perirhinal | + | ✓ | ✓ | ++ | ✓ | ✓ |
| Piriform | ||||||
| Anterior | -- | ✓ | ✓ | + | ✓ | ✓ |
| Posterior | ++ | ✓ | ✓ | -- | ✓ | ✓ |
| Prelimbic | +++ | ✓ | ✓ | +++ | ✓ | ✓ |
| Retrosplenial | + | ✓ | ✓ | + | ✓ | ✓ |
| Somatosensory I | -- | -- | ✓ | -- | -- | -- |
| Somatosensory II | -- | -- | ✓ | -- | -- | -- |
| Temporal | -- | -- | -- | -- | -- | -- |
| Nucleus Accumbens | ||||||
| Shell | + | ✓ | ✓+ | +++ | ✓ | ✓ |
| Core | + | ✓ | ✓ | +++ | ✓ | ✓+ |
| Amygdala | ||||||
| Anterior | ++ | ✓ | ✓+ | -- | ✓ | -- |
| Basolateral | + | ✓ | ✓+ | +++ | ✓ | ✓ |
| Basomedial | +++ | ✓ | ✓+ | + | ✓ | ✓ |
| Central | +++ | ✓ | ✓+ | +++ | ✓ | ✓ |
| Cortical | ++ | -- | ||||
| Medial | +++ | ✓ | ✓+ | -- | ✓ | ✓ |
| Lateral | + | ✓ | ✓+ | + | ✓ | ✓ |
| Posterior | + | -- | ||||
| Anterior Olfactory Nucleus | +++ | ✓ | ✓ | +++ | ✓ | ✓+ |
| Bed Nucleus of the Stria Terminalis | +++ | ✓ | ✓ | -- | ✓ | ✓ |
| Caudate-Putamen | ++ | ✓ | ✓ | ++ | ✓ | ✓ |
| Claustrum | -- | ✓ | ✓ | +++ | ✓ | -- |
| Diagonal Band Nucleus | ||||||
| Horizontal | +++ | ✓ | ✓+ | + | ✓ | ✓ |
| Vertical | ++ | ✓ | ✓ | -- | ✓ | -- |
| Endopiriform Nucleus | ++ | ✓ | ✓+ | -- | ✓ | -- |
| Globus Pallidus | -- | -- | ✓ | -- | -- | ✓+ |
| Hippocampus | ||||||
| Ammon’s Horn | -- | -- | ✓+ | -- | -- | ✓ |
| Dentate Gyrus | -- | -- | ✓+ | -- | -- | ✓ |
| Subiculum | -- | -- | ✓+ | -- | -- | ✓ |
| Lateral Septum | ||||||
| Dorsal Nucleus | ++ | ✓ | ✓+ | -- | ✓ | ✓ |
| Intermediate Nucleus | + | ✓ | ✓+ | -- | ✓ | ✓ |
| Ventral Nucleus | +++ | ✓ | ✓+ | + | ✓ | ✓ |
| Lateral Preoptic Area | +++ | ✓ | ✓+ | + | ✓ | ✓ |
| Magnocellular Preoptic Area | + | ✓ | ✓+ | + | ✓ | ✓ |
| Medial Preoptic Area | +++ | ✓ | ✓+ | + | ✓ | ✓ |
| Median Preoptic Nucleus | ++ | -- | ||||
| Medial Septal Nucleus | + | ✓ | ✓ | -- | -- | -- |
| Olfactory Tubercle | ++ | ✓ | ✓ | +++ | ✓ | ✓+ |
| Septofimbrial Nucleus | -- | ✓ | ✓+ | -- | ✓ | ✓ |
| Septohippocampal Nucleus | ++ | ✓ | ✓+ | -- | ✓ | ✓ |
| Substantia Innominata | +++ | ✓ | ✓+ | + | ✓ | ✓ |
| Tania Tecta | ||||||
| Dorsal | ++ | ✓ | ✓+ | ++ | ✓ | ✓ |
| Ventral | + | ✓ | ✓ | -- | ✓ | -- |
| Ventral Pallidum | + | ✓ | ✓ | -- | ✓ | -- |
| Concordance to Vertes | =53/57 (0.93) | = 47/57 (0.825) | =44/57 (0.772) | =44/57 (0.772) | ||
| Diencephalon | ||||||
| Thalamus | ||||||
| Anterodorsal Nucleus | -- | -- | -- | -- | -- | -- |
| Anteromedial Nucleus | + | ✓ | ✓ | ++ | ✓ | ✓+ |
| Anteroventral Nucleus | -- | -- | ✓ | -- | -- | ✓ |
| Central Lateral Nucleus | -- | -- | -- | + | -- | -- |
| Central Medial Nucleus | ++ | -- | -- | ++ | -- | -- |
| Interanteromedial Nucleus | ++ | -- | -- | +++ | -- | -- |
| Intermediodorsal Nucleus | +++ | -- | ✓ | +++ | -- | ✓+ |
| Lateral Geniculate Nucleus | -- | ✓ | ✓ | -- | ✓ | ✓ |
| Lateral Habenula | + | ✓ | ✓ | + | ✓ | ✓ |
| Laterodorsal Nucleus | + | ✓ | ✓ | + | ✓ | ✓ |
| Lateroposterior Nucleus | + | - | ✓ | + | - | ✓ |
| Medial Geniculate Nucleus | -- | ✓ | -- | -- | ✓ | -- |
| Medial Habenula | -- | -- | ✓ | -- | -- | -- |
| Mediodorsal Nucleus | ++ | -- | ✓ | +++ | -- | -- |
| Paracentral Nucleus | -- | -- | -- | -- | -- | -- |
| Parafasicicular Nucleus | + | -- | -- | -+ | -- | -- |
| Paratential Nucleus | +++ | +++ | ||||
| Paraventricular Nucleus | +++ | -- | -- | +++ | -- | -- |
| Posterior Nucleus | -- | -- | -- | -- | -- | |
| Reticular Nucleus | -- | ✓ | ✓ | -- | ✓ | ✓ |
| Reuniens Nucleus | +++ | ✓ | ✓+ | +++ | ✓ | ✓ |
| Rhomboid Nucleus | ++ | -- | ✓ | + | ✓ | ✓+ |
| Submedial Nucleus | -- | -- | -- | -- | -- | -- |
| Ventral Anterior-lateral Nucleus | -- | -- | ✓ | -- | -- | ✓ |
| Ventral Basal Complex | -- | -- | ||||
| Concordance to Vertes | = 12/24 (0.50) | = 14/24 (0.583) | = 12/24 (0.50) | = 13/24 (0.54) | ||
| Hypothalamus | ||||||
| Anterior Nucleus | ++ | ✓ | ✓+ | -- | -- | ✓ |
| Dorsal Hypothalamic Area | ++ | + | ||||
| Dorsomedial Nucleus | +++ | ✓ | ✓+ | + | -- | ✓ |
| Lateral Nucleus | +++ | ✓ | ✓+ | ++ | ✓ | ✓ |
| Mammillary Bodies | + | -- | -- | -- | -- | -- |
| Paraventricular Nucleus | -- | -- | -- | -- | -- | -- |
| Perifornical Area | +++ | -- | -- | ++ | -- | -- |
| Posterior Nucleus | +++ | -- | -- | + | -- | -- |
| Premammillary Nucleus | + | -- | -- | -- | -- | -- |
| Supramammillary Nucleus | ++ | -- | -- | ++ | -- | -- |
| Ventromedial Nucleus | -- | -- | -- | -- | -- | |
| Concordance to Vertes | = 5/10 (0.50) | = 5/10 (0.50) | =5/9 (0.555) | = 5/9 (0.555) | ||
| Subthalamus | ||||||
| Fields of Forel | + | -- | -- | -- | -- | -- |
| Zona Incerta | + | -- | ✓ | -- | -- | -- |
| = 0/2 (0.00) | = ½ (0.5) | = 2/2 (1.00) | = 2/2 (1.00) | |||
| Brain Stem | ||||||
| Anterior Pretectal Nucleus | -- | -- | -- | -- | -- | -- |
| Barrington’s Nucleus | + | -- | -- | + | -- | -- |
| Cuneiform Nucleus | -- | -- | -- | + | -- | -- |
| Dorsal Motor Nucleus (Vagus) | + | -- | -- | -- | -- | -- |
| Dorsal Raphe Nucleus | + | ✓ | -- | +++ | ✓ | -- |
| Dorsal Tegmental Nucleus | -- | -- | -- | -- | -- | -- |
| Interpeduncular Nucleus | ++ | ✓ | -- | + | ✓ | -- |
| Laterodorsal Tegmental Nucleus | + | -- | -- | + | -- | -- |
| Locus Coeruleus | + | -- | -- | -- | -- | -- |
| Mesencephalic Reticular | + | -- | -- | + | -- | -- |
| Formation | ||||||
| Nucl+eus Ambiguus | -- | -- | -- | -- | -- | -- |
| Nucleus Incertus | + | -- | -- | ++ | -- | -- |
| Nucleus Gigantocellularis | -- | -- | -- | + | -- | -- |
| Nucleus Pons | -- | ✓ | -- | -- | ✓ | -- |
| Nucleus Pontis Caudalis | -- | -- | -- | -- | -- | -- |
| Nucleus Pontis Oralis | -- | -- | -- | -- | ✓ | -- |
| Nucleus Solitary Tract | + | -- | -- | -- | -- | -- |
| Parabrachial Nucleus | ++ | ✓ | -- | -- | ✓ | -- |
| Pedunculopontine Tegmental | ++ | -- | -- | + | -- | -- |
| Nucleus | ||||||
| Periaquaductal Gray Matter | + | -- | -- | +++ | -- | -- |
| Peripeduncular Nucleus | +++ | -- | -- | + | -- | -- |
| Reticular Tegmental Nucleus | + | ✓ | -- | -- | ✓ | -- |
| Retrorubral Areas | -- | -- | -- | + | -- | -- |
| Rostro-ventrolateral Medulla | -- | -- | -- | -- | -- | -- |
| Surpalemniscal Nucleus | -- | ++ | -- | -- | ||
| Superior Colliculus | -- | -- | -- | -- | -- | -- |
| Ventral Tegmental Area | ++ | ✓ | ✓ | ++ | ✓ | -- |
| Ventral Tegmental Nucleus | -- | -- | -- | -- | -- | -- |
| Concordance with Vertes | = 16/27 (0.592) | = 13/27 (0.481) | = 13/27 (0.481) | = 14/27 (0.519) | ||
| Total Concordance | = 86/120 (0.717) | = 80/120 (0.667) | = 76/119 (0.639) | = 78/119 (0.655) | ||
| True Detect | 55 | 56 | 40 | 39 | ||
| True No Detect | 31 | 24 | 36 | 39 | ||
| False Detect | 7 | 14 | 19 | 16 | ||
| False No Detect | 27 | 26 | 24 | 25 | ||
| TD/(TD+FD) | =55/62 (0.890) | =56/70 (0.80) | =40/59 (0.678) | = 39/55 (0.709) | ||
| TND/(TND+FND) | =31/58 (0.534) | =24/50 (0.480) | = 36/60 (0.60) | = 39/64 (0.609) |
Key To Table
Vertes: +++ robust
Vertes: ++ strong
Vertes: + Weak
Vertes: −None
Probtrack: ✓✓voxel of probtrack present in that area
Probtrack: ✓+ voxel of probtrack present in that area (but so is the other seed, but this area shows more voxels than other area)
Probtrack: −No voxels in that area
IL Connectivity via PT
Robust frontal projections included the frontal association cortex, the olfactory bulbs, and the dorsal tenia tecta. Furthermore, the tractography also showed connections to the medial and the ventral orbital cortices. Continuing caudally within the cortical regions, there were connections to the prelimbic cortex and the cingulate cortices, the agranular insular cortex, the ectorhinal and entorhinal cortex and the endopiriform cortex. Of note, there was less connectivity apparent in the medial agranular cortex corresponding to secondary motor area. Subcortically, tractography was consistent with connections to the nucleus accumbens, including both the core and the shell. There were robust connections throughout the medial aspects near the septum, including components of the basal forebrain (medial septal nucleus, the vertical and horizontal diagonal band nucleus), the lateral septum (dorsal, intermediate and ventral), the septohippocampal nucleus, the septofimbrial nucleus, the subfornical nucleus, and the ventral pallidum.
Focusing at the level of the mid-septum, there were cortical projections to the secondary motor cortex and the cingulate cortex, with subcortical projections to the bed nucleus of the stria terminalis (BNST, all major divisions), the substantia inominata, the horizontal limb of the diagonal band, the caudate-putamen, the lateral septal nuclei, the endoprirform cortex, and the magnocellular and lateral preoptic nucleus.
At the level of the hippocampus, the cortical projections were notable at the cingulate cortex and the adjoining retrosplenial agranular and granular cortices. In the subcortical regions, there were projections to the lateral geniculate nucleus, and also robustly to the basolateral, basomedial, lateral, and central amygdala, and both the retrosplenial agranular and granular cortex. There was a notable scarcity of connectivity to the thalamic, and hypothalamic regions, though low probability connectivity was noted in the anteromedial nucleus of the thalamus, the lateral geniculate nucleus, the lateral habenula, the laterodorsal nucleus, the reticular and the Reunions nucleus. There were also minor tracts in the hypothalamus include the anterior nucleus, the dorsomedial nucleus and the lateral nucleus.
The probability of connection fell off sharply in the more caudal regions of the brain, as is apparent both in Figure 3A and also Table 1 at the level of the thalamus, hypothalamus, and the brain stem. Of note, there were some isolated tracts that reached the substantia nigra reticularis and the dorsal raphe nucleus.
PL Connectivity via PT
Raw tractography results for the PL are largely inclusive of those presented for the IL seed; however, there were some notable differences. First, the PL connectivity results were quantitatively less robust (in any given voxel of overlap between IL and Pl connectivity, the IL usually had more streamlines passing through any given voxel). Qualitatively, however, there was nearly the same area of coverage. Most notably, the PL seed did not show connections to the agranular and dysgranular insular cortex (dorsal portion), the medial septal nucleus, and the anterior and dorsomedial hypothalamic nuclei. Conversely, the PL showed connectivity to the infralimbic cortex, the rhomboid nucleus and the nucleus pontis oralis.
Direct Contrast of IL and PL Connectivity
Preferential IL Connectivity
A contrast analysis to identify voxels that showed greater probability of connectivity from IL vs PL seeds was also conducted (Figure 6 and Table). Overall the IL seed showed stronger relative connectivity showing strong connectivity (moving rostral to caudal) in most of the frontal association cortex (excluding the more ventral areas), the orbital cortex (lateral, medial, ventral and ventrolateral), the secondary motor cortex, the piriform cortex, the insular cortex (agranular and dysgranular), the dorsal peduncular cortex, the cingulate cortex, minor tracts were seen in the somatosenory cortex (both primary and secondary), the retrosplenial cortex, the perirhinal cortex, the ectorhinal cortex, and the entorhinal cortex.
Progressing to the subcortical areas of the telencephalon, from rostral to caudal, there was stronger connectivity in the anterior olfactory nucleus, ventral tenia tecta, the dorsal endopiriform nucleus, the nucleus accumbens (shell and core), the dorsal tenia tecta, the caudate putamen, the septohippocampal nucleus, semilunar nucleus, the claustrum, the ventral pallidum, islands of Calleja, the lateral septum (including the dorsal, intermediate and ventral) nucleus, the medial septal nucleus, the septofimbrial nucleus the diagonal band nucleus (both horizontal and vertical), the BNST, the magnocellular, medial and lateral preoptic area, all portions of the amygdala (anterior, basolateral, basomedial, central, medial and lateral) (e.g., Figure 5), the globus pallidus, and the very most anterior portion of the hippocampus.
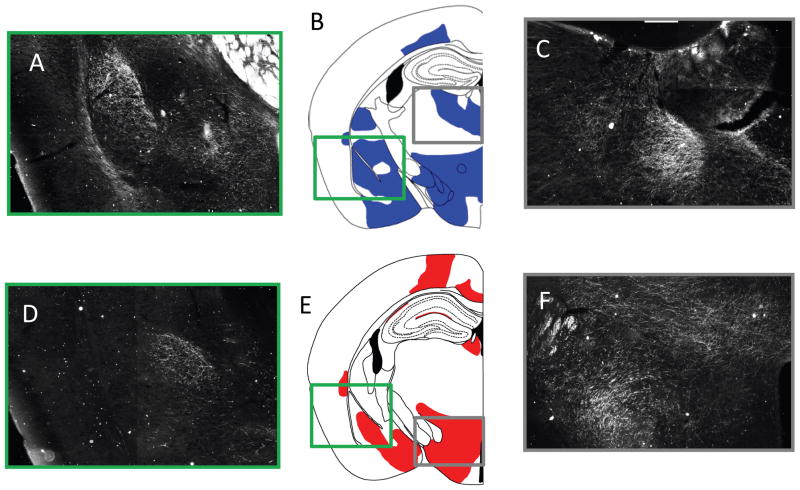
Figure 5. Comparison of the BDA projections of IL/PL.
A) Shows the BDA/ABC/DAB dark-field image composite of 20x images around the amygdala. B) Shows a representative image of the areas showing BDA/ABC/DAB projections from the PL, the green box surrounds the amygdala represented in Part A, while the grey box shows the tracts projecting to the mediodorsal thalamic nucleus highlighted in Part C. C) Shows the BDA/ABC/DAB dark field image composite of 20x images of the mediodorsal thalamic nucleus. D-F Shows the same approximately representative slices of the BDA/ABC/DAB staining from the IL injection site, with the exception that the grey box highlights the posterior hypothalamic regions and the dorsomedial hypothalamic nucleus. Comparable information from the DTI tractography conducted in mice can be found in Figure 3, particularly in rows 3 and 4.
Neither seed showed robust connectivity to the dienchephalon, though there were several regions common across the IL contrast. In brief, the following thalamic nuclei showed connectivity, the anteromedial, anterventeral, lateral geniculate, lateraldorsal, lateroposterior, mediodorsal, reticular, reunions, rhomboid, and ventral anterior lateral thalamic nuclei, and the medial and lateral habenula. There were also three nuclei of the hypothalamus that were showing tracts, the anterior nucleus, the dorsomedial nucleus, and the lateral nucleus. Finally, the only area of the brainstem that showed results was the ventral tegmental area.
Preferential PL Connectivity
The PL tracts, although showing less robust preferential connectivity than IL throughout the brain, still had several notable areas of connection (Figure 4 and Table). Starting with the cortical areas and moving rostral to caudal, there were tracts in the ventral aspect of the frontal association cortex, the orbital cortex (lateral, medial, ventral and ventrolateral), the secondary motor cortex, the piriform cortex, the insular cortex (agranular and dysgranular), the dorsal peduncular cortex, the cingulate cortex, the retrosplenial cortex, the perirhinal cortex.
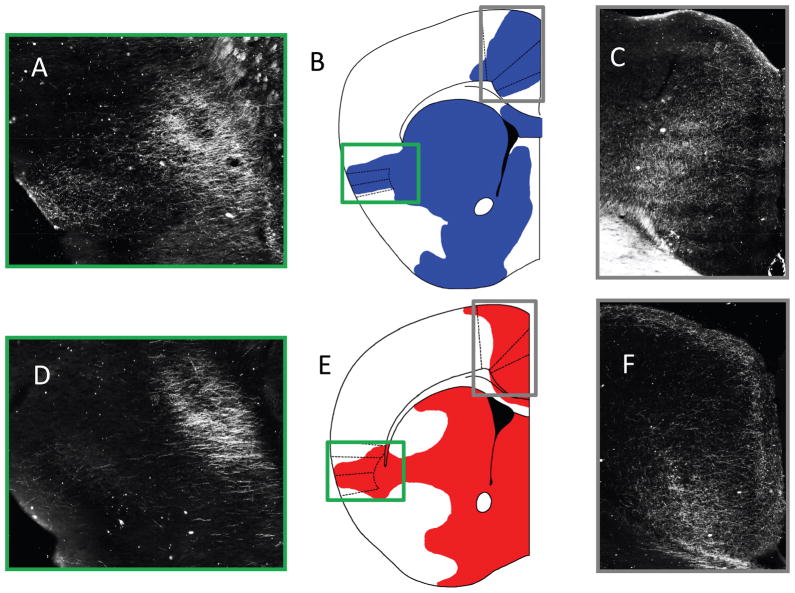
Figure 4. Comparison of the BDA projections of IL/PL.
A) Shows the BDA/ABC/DAB dark-field image composite of 20x images around the insula. B) Shows a representative image of the areas showing BDA/ABC/DAB projections from the PL, the green box surrounds the insula represented in Part A, while the grey box shows the cingulate projections highlighted in Part C. C) Shows the BDA/ABC/DAB dark field image composite of 20x images of the cingulate. D-F Shows the same approximate slices but with the IL injection site. Comparable information from the DTI tractography conducted in mice can be found in Figure 3, particularly in rows 3 and 4.
Progressing to the subcortical areas of the telencephalon, from rostral to caudal, there was connectivity in the anterior olfactory nucleus, the nucleus accumbens (shell and core), the dorsal tenia tecta, the caudate putamen (e.g., Figure 6), the septohipocampal nucleus, semilunar nucleus, the ventral pallidum, islands of Calleja, the lateral septum (including the dorsal, intermediate and ventral) nucleus, the septofimbrial nucleus, the horizontal diagonal band nucleus, the BNST, the magnocellular, medial and lateral preoptic area, most portions of the amygdala (basolateral, basomedial, central, medial and lateral), the globus pallidus, and the very most anterior portion of the hippocampus.
The diencephalon results for the PL seed mirrored those of the IL seed, with a significant drop off in the number of tracts passing through this area. The following thalamic nuclei were linked to the PL seed, the anteromedial, anteroventral, lateral geniculate, lateral dorsal, lateroposterior, reticular, reunions, rhomboid, and ventral anterior lateral thalamic nuclei, and the lateral habenula. Like the IL, there were also three nuclei of the hypothalamus that were showing tracts, the anterior nucleus, the dorsomedial nucleus, and the lateral nucleus. Finally, the PL showed no results in the region of the brainstem.
BDA Anterograde Tracing of Il/PL Connectivity
Of the four mice selected for PL injections (representative PL injection site shown in Figure 1), three mice had an injection site that was inclusive of the PL. The fourth animal did not yield staining either in the injection site or any of the tracts seen in the other three animals. With the more difficult IL injection site, only 3 of the 20 mice had injection (representative IL injection site shown in Figure 1) sites that were inclusive of the IL. Among the 17 animals where the injection site deviated from the intended IL target, three main problems were noted. First was injection into the ventricles, the second was a miss injection hitting more of corpus callosum and cingulate, the third was a null or poorly DAB staining injection. Overall the patterns of connectivity of the IL/PL of the mouse were largely analogous to those of the rat presented in Vertes (2004). More pertinent to the goal of this paper, the DTI probabilistic tractography results and BDA anterograde projections of the IL/PL, qualitatively had a high degree of concordance as shown in Figure 4 and 5 when compared to Figure 3. In particular, these figures are highlighting certain areas of interest including the insula, the amygdala, and the cingulate. Each of these areas is an important component of the circuit regulated by the IL and the PL. Importantly, and in accordance with the rat tracing results, the mouse IL/PL injection did show more medial projections that were missed, for the most part, by DTI, and highlighted in Figure 5. Furthermore, the IL injections did have less robust classical tracing results when compared to the PL.
DISCUSSION
In this work we have developed a high-throughput DTI imaging methodology in the mouse and were able to delineate connectivity of the infralimbic and prelimbic cortex in the mouse, regions separated by less than one mm. These regions have been shown to be critically involved in regulation of emotion regulation (1–11), and new approaches which can examine connectivity of these mPFC regions to areas like the cingulate, insula and the amygdala in a hypothesis-neutral fashion are critical to furthering understanding their function. Our methodology provided a good balance of tissue contrast, SNR, resolution and throughput allowing us to perform high resolution DTI on 6 mouse brains in about 55 hours. Our results are largely consistent with both previously published gold standard HRP-staining techniques (Vertes, 2004) and also our own mouse IL/PL tracing using DAB for anterograde tracing. The contrasting method employed in this paper, similar to those used in our previous work (Gutman et al., 2009) was a critical first step in highlighting the unique connectivity of these two regions that lie in close proximity. Despite the potential confounds including the adjacency of these regions working within a standardized space and potential confounds related to partial voluming, this contrast-based approach was able to differentiate regions more connected to the IL vs. PL seeds.
A readily apparent feature of the raw data is the large degree of overlap in terms of the projections from the IL and PL (as expected from previous anatomical tracing work in rats and also seen in the mouse work presented here), though there are known differential projections. In order to parse out differences in connectivity it was important to understand how differences may be lost. In the probabilistic tractography method used in this paper, each seed voxel generates tracts, which in this case will pass through the adjacent seed in some instances and replicate the most likely tracts generated through that area (IL tracts passing through the PL and taking on tracts similar to those of the PL, and vice versa). Therefore, one potential solution was to compare the number of tracts passing through a voxel for the IL and PL and assigning the voxel to either the IL or PL depending on which seed generated the highest number of tracts in that area. This notion of a voxel containing more tracts from one seed versus the other seed has been referred to as “stronger connectivity”, however, it is important to note that it has not yet been established that this corresponds to stronger connectivity in terms of classical tracing studies (a higher density of axons terminating on a particular brain region). This was done for all 10 brains used in the analysis, and the tracts were overlaid on one another with a scale of 0–10 brains showing that assignment in that voxel [See Figure 6]. We have also presented preliminary BDA tracing work in both the mouse Il and PL showing both that there was agreement between mouse and rat IL/PL projections, and also that the DTI was able to recapitulate these results, particularly with connectivity to areas including the insula, amygdala, and cingulate known to have a strong role in the circuit postulated to regulate emotional activity. With regards to the IL and PL classical connectivity strength it should be noted that the results from the IL appear less robust in Figure 4 and 5, however given the smaller size of IL we reduced the injection volume of BDA from 0.25 to 0.15 microliters potentially reducing the amount of tracer that was available for uptake. Furthermore, the IL BDA injection results shows some hippocampal projections, which is inconsistent with known anatomical connections, we ascribed these connections to a group of neurons in the lateral septum that also showed tracer uptake in the mouse selected for analysis (for the rat see Kohler et al., 1984).
One of the key developments that made this study feasible was the method for obtaining high resolution DTI data from multiple brains during a single scan. High spatial resolution and many diffusion weighted directions are imperative for DTI studies in small animals. The high spatial resolution reduces partial volume effects that reduce the signal from small white matter tracts, and the greater number of diffusion weight directions combined with the probabilistic tractography algorithm facilitates the identification of twisting white matter pathways. However, to obtain the desired resolution, scan times become prohibitively long (~55 hours). We designed our protocol to take advantage of the fact that acquiring more data points in the readout direction is nearly free in terms of enhancing acquisition time, so that an entire tube of six brains (as opposed to two) could be scanned over a single weekend. It should be noted in the particular setup utilized in this study the limit to the resolution was a slice thickness of 161 micron, which limited the in plane resolution to 161 micron by 161 microns to ensure the voxels were isotropic. Therefore, scanning one brain will reduce the time for imaging (by half), but it would not have increased the isotropic resolution or signal to noise of the individually imaged brain. Furthermore, techniques like a two coil system utilizing both a receiver coil and transmitter coil that were available at the imaging center, while increasing the signal to noise, also limit the depth of imaging before signal dropout. As a result the two coil system could not provide the whole brain coverage available in this study.
While in vivo experiments obviously have some inherent advantages, in particular the ability to look at longitudinal changes, the associated logistical challenges including keeping the animals warm, use of anesthesia, movement artifacts, as well as difficulty in moving live animals from different animal facilities for scanning even within a given academic environment are some clear advantages of ex vivo approaches. Also the technique as described is readily amenable to post-imaging analysis as the agarose can be easily removed allowing the brains to be sectioned and stained for further higher-resolution analysis. In fact a key motivation for the development and implementation of our approach was to allow the addition of MRI imaging to the traditional workflow used in our lab in as rapid a fashion as possible; thus we avoided the use of additional contrast agents that could potentially have affected downstream assays and also took the brains out of the skull. While this step induces cortical shape change, the resulting MRI images will share similar morphology/dimensions to histological sections which would be used for correlative analysis.
A number of high-throughput setups have been employed in the community to facilitate this type of research, and our work is heavily influenced by these sorts of schemes. Schneider et al (Schneider and Bhattacharya, 2004) has demonstrated that up to 32 ex-vivo embryos can be scanned in a single overnight session. Special setups that allow multiple animals (e.g. 16 mice), under generalized anesthesia to be scanned in vivo have also been developed (Henkelman, 2010; Lerch et al., 2011).
In silico tractography results have been validated in a number of species. Using a porcine-brain as a model, comprehensive analysis was performed directly comparing histological tract tracing results using standard methods with post-mortem DWI imaging, as well as MEMRI techniques (22). The results of both biotinylated dextran amine (BDA) and MEMRI techniques produced results that are consistent with those obtained ex vivo via tractography, although there were some significant differences in some of the circuits evaluated (Dyrby et al., 2007). Dauguet et al have also shown good visual concordance between DTI and 3D histological tract tracing in the macaque (Dauguet et al., 2006, 2007), and similar studies have also been done in the rat (Leergaard et al., 2010). Recent work from Calamante and colleagues (2011) have also recently validated their novel track density imaging methodology in the mouse showing high concordance between mouse histology and computed tract structure. Our work largely builds on this past study, suggesting valid results can still be obtained in the much smaller mouse brain, allowing whole brain structure and connectivity analysis in this widely used genetic and behavioral model. Future approaches examining differential mouse IL/PL connectivity as a function of genetic background and stress should further our understanding emotion regulation and improve translational approaches to human mPFC functioning.
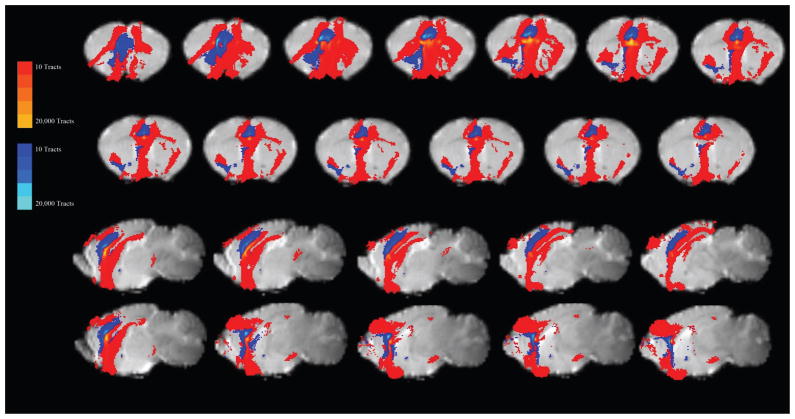
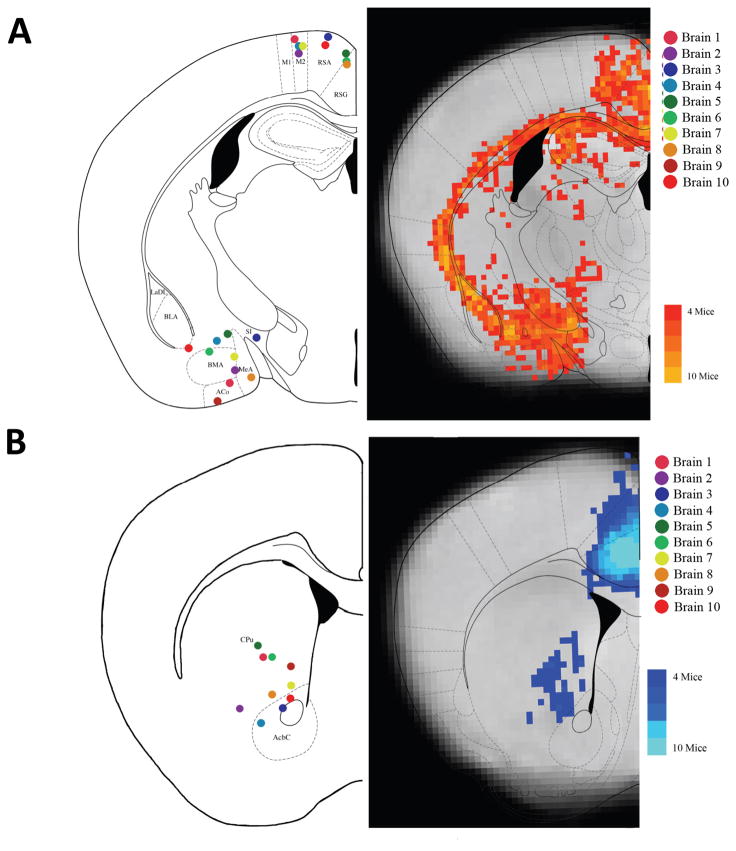
Figure 7. Combined results for IL to Amygdala and PL to Striatum.
A) The IL results in terms of the number of mice showing voxels with at least 10 tracts passing through. The probtrack results from the IL seed were thresholded at 10 or more tracts and where binarized (voxels showing connectivity of greater than ten tracts were set to a value of 1, those with no tracts or less than 10 tracts were set to a value of 0), the binarized results from each mouse were then summed creating a density map of the number of mice showing tracts in that voxel. This schematic was selected to show the connectivity to the BLA, BMA, and BLV. Additionally, for each brain individually, areas of interest, (like the RSA/RSG and BLA/BMA), were examined individually, and the voxel with the highest number of tracts passing through that area was plotted on the corresponding anatomical atlas image (on the left). B) The PL results in terms of the number of mice showing voxels with at least 10 tracts passing through. This schematic was selected to show the connectivity to the Caudate-Putamen and the Cingulate.
Acknowledgments
Funding for this project was provided by two Venture Grants from the Science and Technology Center (The Center for Behavioral Neuroscience of the National Science Foundation under Agreement No. IBN-9876754). Support was also provided by the National Institutes of Health (MH071537, DA019624), the Burroughs Wellcome Fund and the National Primate Research Center base grant #RR-00165.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, Matthews PM, Brady JM, Smith SM. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med. 2003;50:1077–1088. doi: 10.1002/mrm.10609. [DOI] [PubMed] [Google Scholar]
- Choi DC, Maguschak KA, Ye K, Jang SW, Myers KM, Ressler KJ. Prelimbic cortical BDNF is required for memory of learned fear but not extinction or innate fear. Proc Natl Acad Sci U S A. 107:2675–2680. doi: 10.1073/pnas.0909359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran KA, Quirk GJ. Activity in prelimbic cortex is necessary for the expression of learned, but not innate, fears. J Neurosci. 2007;27:840–844. doi: 10.1523/JNEUROSCI.5327-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauguet J, Peled S, Berezovskii V, Delzescaux T, Warfield SK, Born R, Westin CF. 3D histological reconstruction of fiber tracts and direct comparison with diffusion tensor MRI tractography. Med Image Comput Comput Assist Interv. 2006;9:109–116. doi: 10.1007/11866565_14. [DOI] [PubMed] [Google Scholar]
- Dauguet J, Peled S, Berezovskii V, Delzescaux T, Warfield SK, Born R, Westin CF. Comparison of fiber tracts derived from in-vivo DTI tractography with 3D histological neural tract tracer reconstruction on a macaque brain. Neuroimage. 2007;37:530–538. doi: 10.1016/j.neuroimage.2007.04.067. [DOI] [PubMed] [Google Scholar]
- Douaud G, Jbabdi S, Behrens TE, Menke RA, Gass A, Monsch AU, Rao A, Whitcher B, Kindlmann G, Matthews PM, Smith S. DTI measures in crossing-fibre areas: increased diffusion anisotropy reveals early white matter alteration in MCI and mild Alzheimer's disease. Neuroimage. 2011;55:880–890. doi: 10.1016/j.neuroimage.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrby TB, Baare WF, Alexander DC, Jelsing J, Garde E, Sogaard LV. An ex vivo imaging pipeline for producing high-quality and high-resolution diffusion-weighted imaging datasets. Hum Brain Mapp. 2011;32:544–563. doi: 10.1002/hbm.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrby TB, Sogaard LV, Parker GJ, Alexander DC, Lind NM, Baare WF, Hay-Schmidt A, Eriksen N, Pakkenberg B, Paulson OB, Jelsing J. Validation of in vitro probabilistic tractography. Neuroimage. 2007;37:1267–1277. doi: 10.1016/j.neuroimage.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Gutman DA, Holtzheimer PE, Behrens TE, Johansen-Berg H, Mayberg HS. A tractography analysis of two deep brain stimulation white matter targets for depression. Biol Psychiatry. 2009;65:276–282. doi: 10.1016/j.biopsych.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkelman RM. Systems biology through mouse imaging centers: experience and new directions. Annu Rev Biomed Eng. 2010;12:143–166. doi: 10.1146/annurev-bioeng-070909-105343. [DOI] [PubMed] [Google Scholar]
- Johansen-Berg H, Behrens TE. Just pretty pictures? What diffusion tractography can add in clinical neuroscience. Curr Opin Neurol. 2006;19:379–385. doi: 10.1097/01.wco.0000236618.82086.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLumiere RT, Smith KC, Kalivas PW. Neural circuit competition in cocaine-seeking: roles of the infralimbic cortex and nucleus accumbens shell. Eur J Neurosci. 2012;35(4):614–22. doi: 10.1111/j.1460-9568.2012.07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaLumiere RT, Niehoff KE, Kalivas PW. The infralimbic cortex regulates the consolidation of extinction after cocaine self-administration. Learn Mem. 2010;17:168–175. doi: 10.1101/lm.1576810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leergaard TB, White NS, de Crespigny A, Bolstad I, D'Arceuil H, Bjaalie JG, Dale AM. Quantitative histological validation of diffusion MRI fiber orientation distributions in the rat brain. PLoS One. 2010;5:e8595. doi: 10.1371/journal.pone.0008595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Sled JG, Henkelman RM. MRI phenotyping of genetically altered mice. Methods Mol Biol. 2011;711:349–361. doi: 10.1007/978-1-61737-992-5_17. [DOI] [PubMed] [Google Scholar]
- Li L, Preuss TM, Rilling JK, Hopkins WD, Glasser MF, Kumar B, Nana R, Zhang X, Hu X. Chimpanzee (Pan troglodytes) precentral corticospinal system asymmetry and handedness: a diffusion magnetic resonance imaging study. PLoS One. 2010;5:e12886. doi: 10.1371/journal.pone.0012886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddux JM, Holland PC. Effects of dorsal or ventral medial prefrontal cortical lesions on five-choice serial reaction time performance in rats. Behav Brain Res. 2011;221:63–74. doi: 10.1016/j.bbr.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J, Skaggs H, Churchwell J, Powell DA. Medial prefrontal cortex and pavlovian conditioning: trace versus delay conditioning. Behav Neurosci. 2002;116:37–47. [PubMed] [Google Scholar]
- Milad MR, Quirk GJ. Neurons in medial prefrontal cortex signal memory for fear extinction. Nature. 2002;420:70–74. doi: 10.1038/nature01138. [DOI] [PubMed] [Google Scholar]
- Milad MR, Vidal-Gonzalez I, Quirk GJ. Electrical stimulation of medial prefrontal cortex reduces conditioned fear in a temporally specific manner. Behav Neurosci. 2004;118:389–394. doi: 10.1037/0735-7044.118.2.389. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis IL, Takashima A. The role of the ventromedial prefrontal cortex in memory consolidation. Behav Brain Res. 2011;218:325–334. doi: 10.1016/j.bbr.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin G. The Mouse Brain in Stereotaxic Coordinates. Elsevier; 2004. [Google Scholar]
- Peters J, LaLumiere RT, Kalivas PW. Infralimbic prefrontal cortex is responsible for inhibiting cocaine seeking in extinguished rats. J Neurosci. 2008;28:6046–6053. doi: 10.1523/JNEUROSCI.1045-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DA, Skaggs H, Churchwell J, McLaughlin J. Posttraining lesions of the medial prefrontal cortex impair performance of Pavlovian eyeblink conditioning but have no effect on concomitant heart rate changes in rabbits (Oryctolagus cuniculus) Behav Neurosci. 2001;115:1029–1038. doi: 10.1037//0735-7044.115.5.1029. [DOI] [PubMed] [Google Scholar]
- Quirk GJ, Russo GK, Barron JL, Lebron K. The role of ventromedial prefrontal cortex in the recovery of extinguished fear. J Neurosci. 2000;20:6225–6231. doi: 10.1523/JNEUROSCI.20-16-06225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilling JK, Glasser MF, Preuss TM, Ma X, Zhao T, Hu X, Behrens TE. The evolution of the arcuate fasciculus revealed with comparative DTI. Nat Neurosci. 2008;11:426–428. doi: 10.1038/nn2072. [DOI] [PubMed] [Google Scholar]
- Rocha A, Kalivas PW. Role of the prefrontal cortex and nucleus accumbens in reinstating methamphetamine seeking. Eur J Neurosci. 2010;31(5):903–9. doi: 10.1111/j.1460-9568.2010.07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JE, Bhattacharya S. Making the mouse embryo transparent: identifying developmental malformations using magnetic resonance imaging. Birth Defects Res C Embryo Today. 2004;72:241–249. doi: 10.1002/bdrc.20017. [DOI] [PubMed] [Google Scholar]
- Vertes RP. Differential projections of the infralimbic and prelimbic cortex in the rat. Synapse. 2004;51:32–58. doi: 10.1002/syn.10279. [DOI] [PubMed] [Google Scholar]
- Vidal-Gonzalez I, Vidal-Gonzalez B, Rauch SL, Quirk GJ. Microstimulation reveals opposing influences of prelimbic and infralimbic cortex on the expression of conditioned fear. Learn Mem. 2006;13:728–733. doi: 10.1101/lm.306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Benner T, Sorensen AG, Wedeen VJ. Diffusion Tookit: A Software Package for Diffusion Imaging Data Processing and Tractography. ISMRM; 2007. [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, Pandya DN, Hagmann P, D'Arceuil H, de Crespigny AJ. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41:1267–1277. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Zhang J, Richards LJ, Yarowsky P, Huang H, van Zijl PC, Mori S. Three-dimensional anatomical characterization of the developing mouse brain by diffusion tensor microimaging. Neuroimage. 2003;20:1639–1648. doi: 10.1016/s1053-8119(03)00410-5. [DOI] [PubMed] [Google Scholar]