Abstract
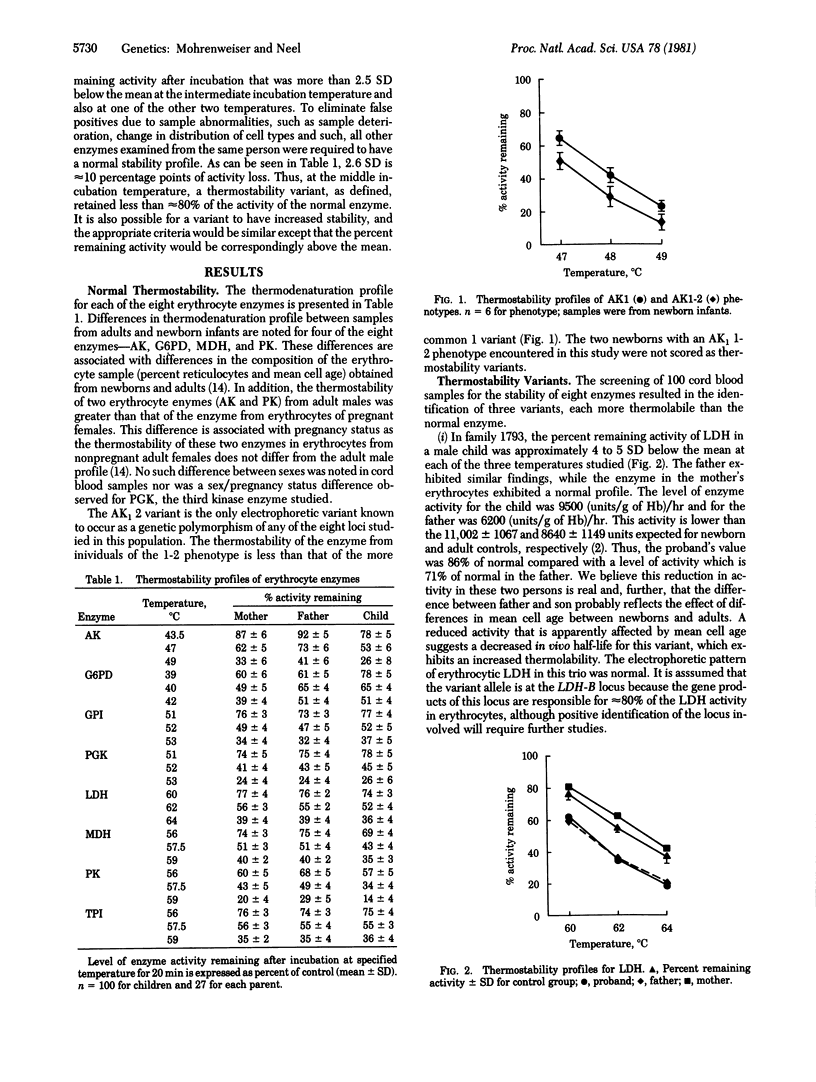
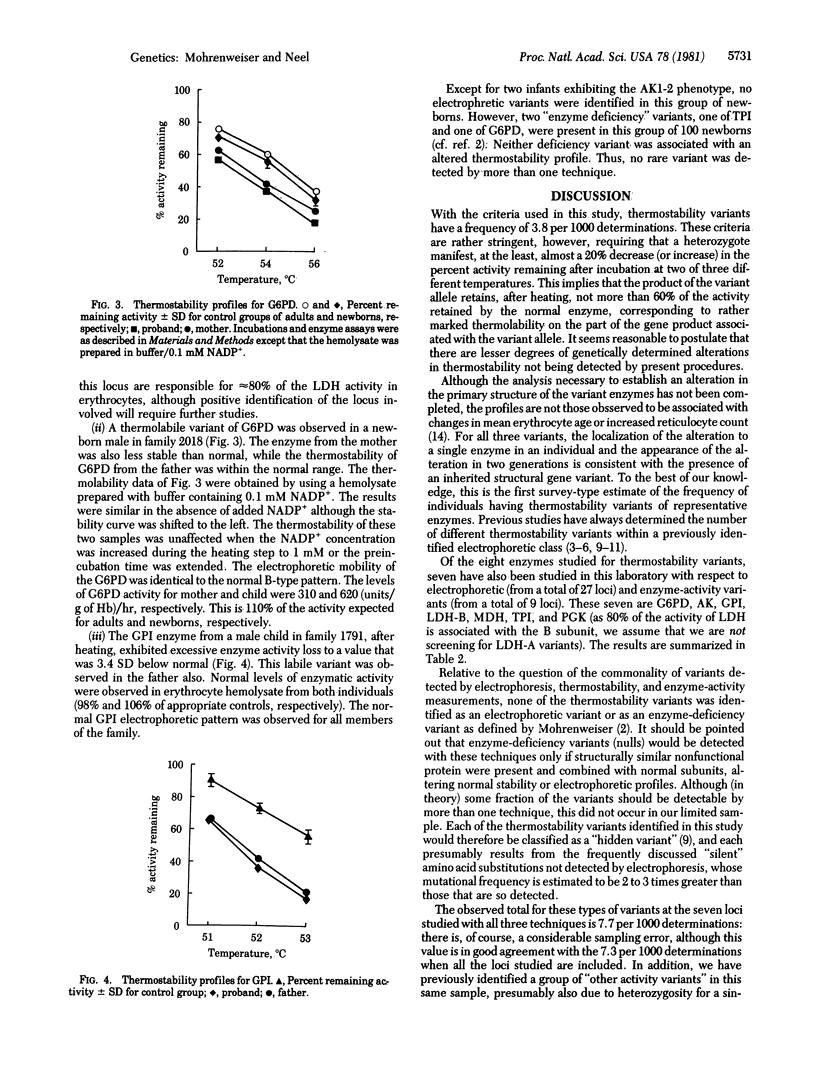
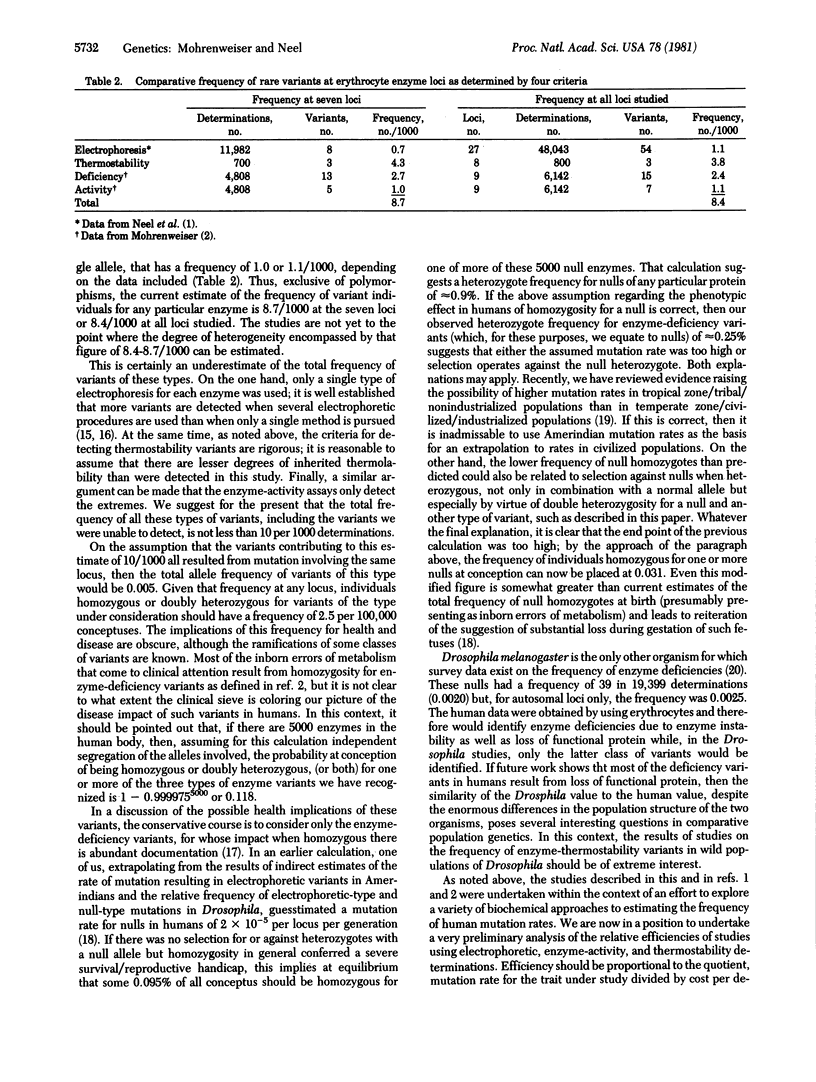
Eight erythrocyte enzymes were examined for thermostability in an unselected sample of 100 newborn infants. Three thermolabile variants, one each of lactate dehydrogenase, glucosephosphate isomerase, and glucose-6-phosphate dehydrogenase, were identified, none of which was detectable as a variant by standard electrophoretic techniques. All were inherited. This frequency of 3.8 heritable thermostabiliy variants per 1000 determinations is to be compared with a frequency of electrophoretically detectable variants of 1.1 per 1000 determinations, a frequency of 2.4 enzyme-deficiency variants per 1000 determinations, and a frequency of 1.1 hypo/hyperactive enzyme-activity variants per 1000 determinations in this human newborn population. The total measured frequency of individuals with rare enzyme deficiency or electrophoretic or thermostability (or both) variants at these loci is 8.3 per 1000 determinations. A similar distribution and frequency is seen when the comparison is limited to the seven loci studied by all techniques. It is clear that not all of the electrophoretic and thermostability variants present in the population are detected by the techniques used in this study. Accordingly, it is estimated that the true frequency of carriers of a rare variant for each of these enzyme-coding loci averages greater than 10/1000. Some implications of these frequencies for human disease are discussed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beckenbach A. T., Prakash S. Examination of Allelic Variation at the Hexokinase Loci of DROSOPHILA PSEUDOOBSCURA and D. PERSIMILIS by Different Methods. Genetics. 1977 Dec;87(4):743–761. doi: 10.1093/genetics/87.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein S. C., Throckmorton L. H., Hubby J. L. Still more genetic variability in natural populations. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3928–3931. doi: 10.1073/pnas.70.12.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme F., Selander R. K. Estimating total genic diversity in the house mouse. Biochem Genet. 1978 Apr;16(3-4):287–297. doi: 10.1007/BF00484085. [DOI] [PubMed] [Google Scholar]
- Cobbs G., Prakash S. An Experimental Investigation of the Unit Charge Model of Protein Polymorphism and Its Relation to the Esterase-5 Locus of DROSOPHILA PSEUDOOBSCURA, DROSOPHILA PERSIMILIS, and DROSOPHILA MIRANDA. Genetics. 1977 Dec;87(4):717–742. doi: 10.1093/genetics/87.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielek S., Mohrenweiser H. W. Erythrocyte enzyme deficiencies assessed with a miniature centrifugal analyzer. Clin Chem. 1979 Mar;25(3):384–388. [PubMed] [Google Scholar]
- Johnson G. B. Hidden alleles at the alpha-glycerophosphate dehydrogenase locus in Colias butterflies. Genetics. 1976 May;83(1):149–167. doi: 10.1093/genetics/83.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G. B. Structural flexibility of isozyme variants: genetic variants in Drosophila disguised by cofactor and subunit binding. Proc Natl Acad Sci U S A. 1978 Jan;75(1):395–399. doi: 10.1073/pnas.75.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrenweiser H. W. Frequency of enzyme deficiency variants in erythrocytes of newborn infants. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5046–5050. doi: 10.1073/pnas.78.8.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel J. V., Mohrenweiser H. W., Meisler M. H. Rate of spontaneous mutation at human loci encoding protein structure. Proc Natl Acad Sci U S A. 1980 Oct;77(10):6037–6041. doi: 10.1073/pnas.77.10.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel J. V. Mutation and disease in man. Can J Genet Cytol. 1978 Sep;20(3):295–306. doi: 10.1139/g78-033. [DOI] [PubMed] [Google Scholar]
- Neel J. V., Rothman E. Is there a difference among human populations in the rate with which mutation produces electrophoretic variants? Proc Natl Acad Sci U S A. 1981 May;78(5):3108–3112. doi: 10.1073/pnas.78.5.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. S., Hubby J. L., Lewontin R. C. Molecular heterosis for heat-sensitive enzyme alleles. Proc Natl Acad Sci U S A. 1974 May;71(5):1808–1810. doi: 10.1073/pnas.71.5.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. S., Hubby J. L., Throckmorton L. H. The study of genic variation by electrophoretic and heat denaturation techniques at the octanol dehydrogenase locus in members of the Drosophila virilis group. Genetics. 1975 Jul;(3):637–650. doi: 10.1093/genetics/80.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thörig E. W., Schoone A. A., Scharloo W. Variation between electrophoretically identical alleles at the alcohol dehydrogenase locus in Drosophila melanogaster. Biochem Genet. 1975 Oct;13(9-10):721–731. doi: 10.1007/BF00484929. [DOI] [PubMed] [Google Scholar]
- Trippa G., Loverre A., Catamo A. Thermostability studies for investigating non-electrophoretic polymorphic alleles in Drosophila melanogaster. Nature. 1976 Mar 4;260(5546):42–44. doi: 10.1038/260042a0. [DOI] [PubMed] [Google Scholar]
- Voelker R. A., Langley C. H., Brown A. J., Ohnishi S., Dickson B., Montgomery E., Smith S. C. Enzyme null alleles in natural populations of Drosophila melanogaster: Frequencies in a North Carolina population. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1091–1095. doi: 10.1073/pnas.77.2.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]


