Abstract
The dual oxidase enzymes, DUOX, localized to the respiratory tract epithelium, are important components of innate host defense against bacteria and virus. However, little is known regarding the regulation of DUOX transcription. To better understand DUOX2-mediated mechanisms of antiviral host defense in the airway epithelium, we designed a bidirectional promoter luciferase reporter system to identify important cis-regulatory regions in the human DUOX2/DUOXA2 promoter. In this report, we demonstrate that the genomic region between the translation start sites of DUOX2 and DUOXA2 functions as a bidirectional promoter in human airway tissue. We also identified key regulatory regions on the DUOX2/DUOXA2 promoter that were necessary for both bidirectional and unidirectional transcriptional activity. Importantly, we discovered two functionally important single-nucleotide polymorphisms (SNPs) within the promoter that differentially regulated DUOX2/DUOXA2 transcription in response to exogenous double-stranded DNA. One of these SNPs, rs269855 (enriched in people of African descent), conferred the highest level of DUOX2 promoter activity. The clinical sequelae for individuals who carry this polymorphism remain to be determined.
Keywords: airway, bidirectional promoter, dual oxidase, single-nucleotide polymorphism, virus
Clinical Relevance
The research presents novel information on the regulation of dual oxidase 2 (DUOX2), an enzyme important in host defense against bacteria and viruses. The demonstration of single-nucleotide polymorphisms that play important roles in gene regulation may be relevant for a better understanding of the individual variability to virus infection.
The respiratory tract epithelium is the central component of airway host defense. It is elegantly equipped to protect us from invading organisms through its mucus layer, tight epithelial barrier, air–surface liquid rich in antimicrobial peptides, and Toll-like or nucleotide oligomerization domain–like receptors that can induce inflammatory cytokines and recruit inflammatory cells (1, 2). Mounting evidence suggests that the hydrogen peroxide (H2O2) specifically generated in airway epithelial cells by the nicotinamide adenine dinucleotide phosphate–reduced oxidase enzymes dual oxidase 1 or 2 (DUOX1 or DUOX2) is critical for many of these functions (3, 4).
The initial and best characterized role of DUOX in host defense involves the production of H2O2, which serves as a substrate for lactoperoxidase (LPO) to oxidize thiocyanate (SCN−) to hypothiocyanate (5, 6). When this oxidative antimicrobial model is tested in vitro, all three components (DUOX, LPO, and SCN−) are essential for the effective killing of both gram-positive and gram-negative bacteria (7–10). Moreover, the in vivo localization of these components is highly consistent with the proposed model (10). Recently, Fischer and colleagues demonstrated that this same oxidative system, by substituting I− for the pseudohalide SCN−, resulted in the production of virus-inactivating hypoiodous acid (11), which suggests that DUOX is critical for the direct inactivation of bacteria and virus (12, 13). In addition to its immediate antimicrobial effects, DUOX-generated H2O2 is implicated in critical signaling roles for many other host defense functions of the respiratory tract epithelium. These include mucus generation through epidermal growth factor receptor signaling (14, 15), the maintenance or repair of the epithelial barrier (16), Toll-like receptor signaling (17), and neutrophil recruitment (18).
Although DUOX has highly pleiotropic host defense activities in the airway epithelium, little is known regarding the relative importance of, or the regulatory factors required for, the transcription of each isoform. Here, we sought to identify the regulatory factors important for DUOX2 transcription. Importantly, the full functional expression of DUOX2 at the plasma membrane requires the coordinated regulation of its respective maturation factor DUOXA2 (19, 20). The head-to-head genomic organization of DUOX2 with DUOXA2 suggests that the two gene pairs are regulated through a shared bidirectional promoter (21–23). Consistent with this notion, a recent study demonstrated that a minimal promoter element of rat Duox2 exerted bidirectional activity in a rat thyroid cell line (24). Based on these observations, we designed a DUOX2/DUOXA2 bidirectional promoter reporter system that included cis-regulatory elements contained between the translation start sites of each gene. Using tracheobronchial epithelial (TBE) cells, we characterized DUOX2 and DUOXA2 coregulated transcription through a shared bidirectional promoter in human tissues, and identified key regulatory regions in this promoter.
Materials And Methods
Cell Culture
Human bronchial epithelium-1 (HBE1) cells, a papilloma virus–immortalized human TBE cell line (25), were used for all of our experiments. HBE1 cells were maintained in submerged culture conditions with Ham’s F12/Dulbecco’s Modified Eagle’s Medium (1:1) supplemented with transferrin (5 μg/ml), insulin (5 μg/ml), cholera toxin (10 ng/ml), epidermal growth factor (10 ng/ml), dexamethasone (0.1 μM), bovine hypothalamus extract (15 μg/ml), and plasmocin (2.5 mg/ml). As soon as HBE1 cells were confluent, all trans-retinoic acid (0.03 μM) was added for 10 days before harvesting.
DUOX2/DUOXA2 Promoter Reporter Constructs
DUOX2/DUOXA2 (D2DA2) promoter DNA sequences were isolated from NCI-H292 and HBE1 cells. These genomic DNA sequences varied from the human genome reference database at two sites (G/T or G/C haplotypes in HCI-H292 cells, or G/T haplotypes in HBE1 cells) that matched two single-nucleotide polymorphisms (SNPs) identified in the HapMap database (www.hapmap.org). The full-length bidirectional promoter reporter vector pTRE-RLuc-D2DA2-Luc (FL) was generated by two separate insertions. First, a pTRE-D2DA2-Luc vector expressing firefly luciferase in the DUOXA2 orientation was generated by replacing the cytomegalovirus minimal promoter from EcoR1/BamH1-cleaved pTRE-Tight-BI-Luc vector (Clontech Laboratories, Inc., Mountain View, CA) with DUOX2/DUOXA2 promoter region amplicons, using two primers to amplify genomic DNA from HBE1 cells: D2A2F19-EcoR1, CCGGAATTCTCCAGCCAACCCTGCAGCCTGCG; and D2A2R18-BamH1, CGCGGATCCCCGGCTGCACGCCGGCCGAGT. The Renilla luciferase reporter gene was isolated from an EcoRI/XbaI-digested pRL-null vector (Promega Corp., Madison, WI) and inserted into the second multiple cloning site of the EcoR1/XbaI-digested pTRE-D2DA2-Luc vector, which produced a bidirectional pTRE-RLuc-D2DA2-Luc vector (FL). Mutations were performed on the pTRE-RLuc-D2DA2-Luc vector, using the QuickChange Lightning Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA) to obtain four single-site mutations and four deletion constructs (Figures 5 and 6). The DNA sequencing of all PCR-amplified products and ligated plasmids was performed to ensure that no sequence errors were introduced during construction.
Figure 5.
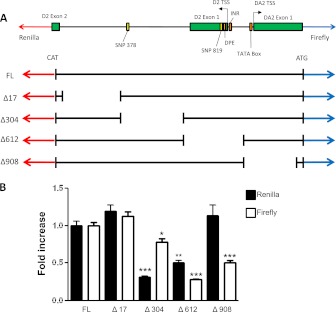
Effects of promoter deletions on basal DUOX2/DUOXA2 promoter activity. (A) Schematic representation of the DUOX2/DUOXA2 promoter/untranslated region (ATG) between the translation start sites (TSSs) of each gene, aligned with five different promoter–luciferase reporter constructs. The cloned region includes the first two exons of DUOX2 and the first exon of DUOXA2. Relative locations of the TSSs for DUOX2 and DUOXA2, the TATA box for DUOXA2, the initiator element (INR) and downstream promoter element (DPE) sites for DUOX2, and two single-nucleotide polymorphisms (SNPs) are shown. A small region (17 nucleotides) immediately proximal to the ATG of both the Renilla and firefly reporter was preserved in all four deletion constructs. (B) HBE1 cells were grown in submerged cell culture conditions and transfected in triplicate with one of five promoter constructs, followed by sequential measurements of Renilla and firefly luciferase activity at 12 hours. A β-galactosidase expression plasmid was cotransfected in all cells as an internal control. Normalized luciferase activities are shown relative to the full-length plasmid (set to 1), and represent means ± SEMs of three independent experiments. *P < 0.05, by ANOVA. **P < 0.01, by ANOVA. ***P < 0.001, by ANOVA.
Figure 6.
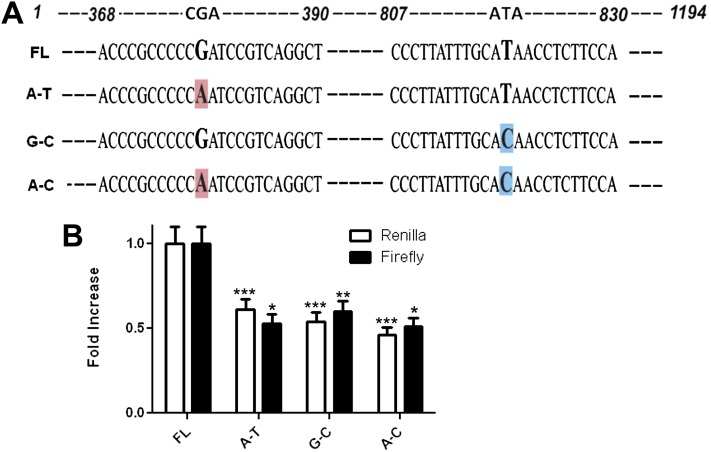
Effect of two SNPs on basal DUOX2/DUOXA2 promoter activity. (A) Schematic diagrams of the four possible DUOX2/DUOXA2 promoter SNP combinations. Site-directed mutagenesis was performed on the full-length (1,194 nucleotides) dual reporter construct to generate specific SNP combinations. Nucleotides are numbered from the beginning of the ATG coding for DUOX2 to the ATG coding for DUOXA2. Enlarged/shaded letters represent the specific nucleotides/SNPs that were mutated. (B) HBE1 cells were grown in submerged cell culture conditions and transfected with one of the four possible promoter constructs, followed by sequential measurements of Renilla and firefly luciferase activity at 12 hours. A β-galactosidase expression plasmid was cotransfected in all cells, and used to normalize luciferase activity between experiments. Normalized data are presented as fold increase compared with the FL construct (mean ± SEM; n = 3). *P < 0.05, by ANOVA. **P < 0.01, by ANOVA. ***P < 0.001, by ANOVA.
Transient Transfection and Luciferase Assay
HBE1 cells were transfected with various DUOX2/DUOXA2 promoter constructs, using the DharmaFECT Duo transfection reagent (ThermoFisher, Waltham, MA) for 8 hours before a media change. Firefly and Renilla luciferase activity was measured by luminometry at various time points after transfection, using the Dual-Glo Luciferase Assay System (Promega). The β-galactosidase expression plasmid was cotransfected in all cells to normalize transfection efficiency. β-galactosidase activity was measured at 405 nm (with an Emax Precision Microplate Reader; Molecular Devices, Sunnyvale, CA), using the β-Galactosidase Enzyme Assay System (Promega). All transfections were performed in triplicate for each experiment, and normalized to β-galactosidase activity for each individual well.
PCR Assays
Plasmid DNA was extracted from transfected HBE1 cells by resuspending the cell pellet, normalized to cell number before lysis, in 20 μl PCR lysis buffer (1% Triton X-100, 20 mM Tris-Cl, pH 8.5, and 2 mM EDTA, pH 8.0), incubating at 95°C for 10 minutes, placing on ice briefly, and collecting the supernatant after spinning down the cell debris. PCR was performed with plasmid-specific primers (Table 1), and amplicons were visualized by electrophoresis in a 1% agarose gel with ethidium bromide, or measured by quantitative real-time PCR (qRT-PCR), using SYBR Green qPCR Mastermix (Invitrogen, Carlsbad, CA) in an ABI 5700 Sequence Detection system (Applied Biosystems, Inc., Foster City, CA). For endogenous DUOX2 and DUOXA2 measurements, 3 μg of total RNA were reverse-transcribed with M-MLV-Reverse Transcriptase (Promega), using oligo-dT primers. Reactions were performed in 384-well optical reaction plates containing SYBR Green qPCR Mastermix in an ABI 5700 Sequence Detection system, as already described. The relative expression values for DUOX2 and DUOXA2 were normalized to glyceraldehyde 3–phosphate dehydrogenase, as previously described (26).
TABLE 1.
SEQUENCES FOR THE PRIMERS USED TO AMPLIFY PLASMID DNA, DUOX2, DUOXA2, OR GAPDH BY PCR OR QUANTITATIVE RT-PCR
| Gene | Forward | Reverse |
| Plasmid | CCAGCTTGCTGGCTTGC | TTTTGGCGTCTTCCATCG |
| DUOX2 | ACGCAGCTCTGTGTCAAAGGT | TGATGAACGAGACTCGACAGC |
| DUOXA2 | CACCTCTTATCCTCGGCGAC | GAAGGAGTCCAGATTGGGTCC |
| GAPDH | TCCTCCACCTTTGACGCTG | ACCACCCTGTTGCTGTAGCC |
Definition of abbreviations: DUOX, dual oxidase enzyme; GAPDH, glyceraldehyde 3–phosphate dehydrogenase; Plasmid, plasmid DNA.
All primers are oriented 5′–3′.
Statistical Analyses
All statistical analyses were performed using Prism 5 software (GraphPad, San Diego, CA). Data are presented as original values or as means with standard errors of the mean (SEMs); n refers to the number of replicates. Effects of multiple treatments were tested using ANOVA, followed by the Tukey multiple comparison test. The resultant P values are provided, and P < 0.05 was considered significant.
Results
Promoter Activity Measurements
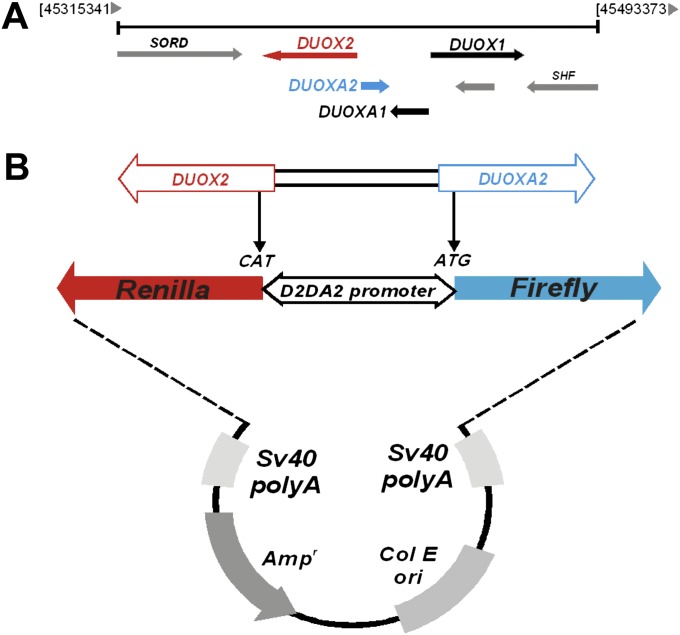
To determine whether the intergenic region between the human DUOX2 and DUOXA2 genes functioned as a bidirectional promoter, we cloned the chromosomal fragment between the start codons of DUOX2 and DUOXA2 into the pTRE-Tight-BI-Luc expression vector (Clontech), followed by the sequential insertion of the Renilla luciferase reporter gene (Figure 1; see Materials and Methods) to generate a bidirectional promoter reporter vector pTRE-RLuc-D2DA2-Luc (FL). Using this construct, we were able to determine changes in directional transcription through the DUOX2/DUOXA2 promoter, based on relative levels of Renilla (DUOX2) or firefly (DUOXA2) luciferase activity.
Figure 1.
Cloning strategy for the dual oxidase enzyme–2 (DUOX2)/DUOXA2 (D2DA2) promoter–reporter system. (A) Genomic organization of DUOX1 and DUOX2 on chromosome 15 highlights the structural relationship between the two genes and their corresponding maturation factors. (B) The region between the translation start sites in exon 2 of DUOX2 and exon 1 of DUOXA2 was cloned from human tracheobronchial epithelial cells and inserted into the pTRE-tight-BI-Luc expression vector (Clontech) with modifications (see Materials and Methods) to assess bidirectional promoter activity.
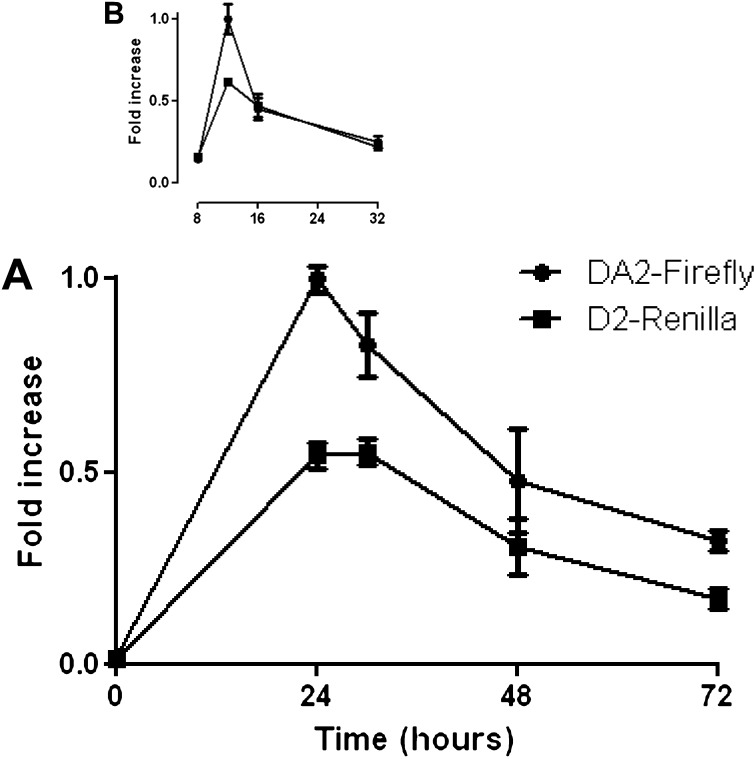
After the transfection of HBE1 cells with the FL vector, we observed bidirectional transcriptional activity at 24 hours, with a decrease in firefly or Renilla luciferase expression at 48 and 72 hours (Figure 2A). To better characterize the time of peak promoter activity, we repeated the transfection experiments and harvested cells between 8 and 32 hours after transfection (Figure 2B). These data suggested that promoter activity was present as early as 8 hours after transfection, and that it reached its peak at or before 12 hours. The subsequent decrease in luciferase activity suggested that either transfection itself stimulated DUOX2/DUOXA2 promoter activity with subsequent return to baseline levels at 72 hours, or else a loss of plasmid material occurred, either by degradation or dilution, over time. DNA was harvested from transfected HBE1 cells at various time points corresponding to the times we measured promoter activity. The plasmid content was qualitatively similar, based on the gel electrophoresis of amplicons after 30 cycles of PCR, using plasmid-specific primers (Figure 3A). Similarly, qRT-PCR determined that plasmid concentrations remained relatively unchanged throughout the transfection experiments (Figures 3B and E1 in the online supplement). These data suggest that the decrement in luciferase activity at 48 and 72 hours was not attributable to the subsequent loss of plasmid material, but rather to a return to baseline activity after a transient increase in promoter activity that peaked at 12 hours.
Figure 2.
DUOX2/DUOXA2 bidirectional promoter activity. Human bronchial epithelium-1 cells were grown in submerged cell culture conditions and transfected with the bidirectional reporter plasmid pTRE-RLuc-D2DA2-Luc (FL). (A) Simultaneous measurements of firefly and Renilla luciferase activity were performed at four different time points between 24 and 72 hours. (B) Simultaneous measurements of firefly and Renilla luciferase activity were performed at four different time points between 8 and 32 hours. Normalized luciferase activities are shown relative to the maximum luciferase activity measured (set to 1). Means ± SEMs from three separate samples per time point are shown. Data from one experiment are shown, and are representative of five (A) or three (B) independent experiments.
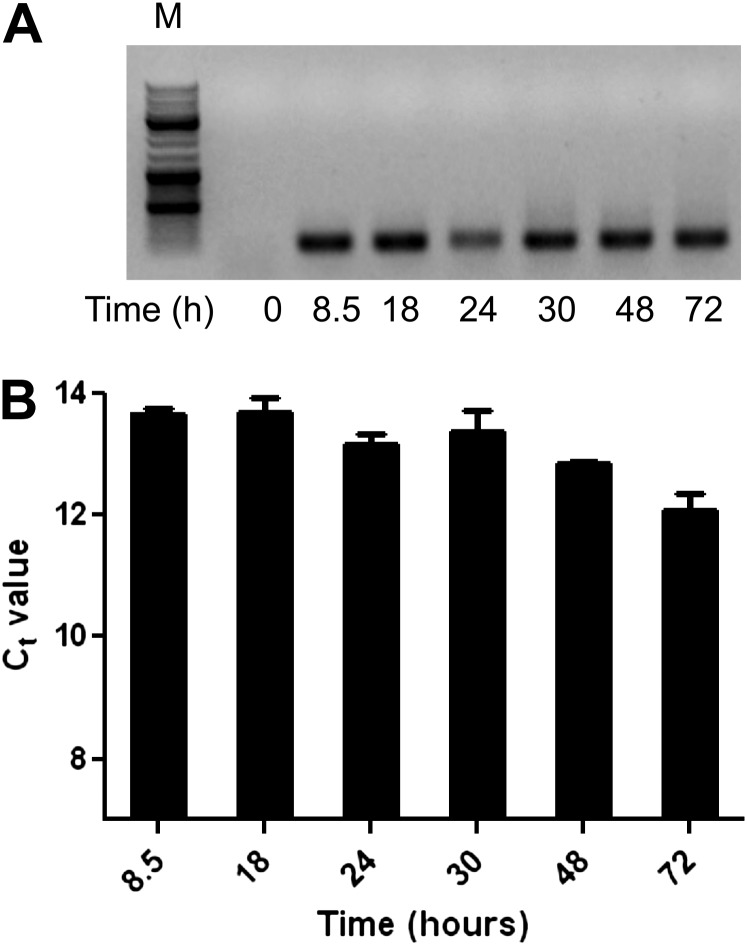
Figure 3.
Plasmid concentrations in HBE1 cells. HBE1 cells in submerged cell culture conditions were transfected at 60–70% confluence with the bidirectional reporter plasmid, and total cellular DNA was prepared at various time points after transfection. Cells were counted and underwent lysis with equal buffer volumes for each sample, to ensure similar inputs of total DNA before PCR amplification. (A) Thirty cycles of PCR using plasmid-specific primers were performed, followed by gel electrophoresis in 1% agarose. M, molecular weight marker (three heavy bands represent molecular weights of 3 kB, 1 kB, and 0.5 kB, respectively). (B) Means ± SEMs of threshold cycles (Ct; inversely proportional to the log2 of plasmid DNA) were measured by quantitative RT-PCR of three separate samples per time point. Data from one experiment are shown, and are representative of three independent experiments.
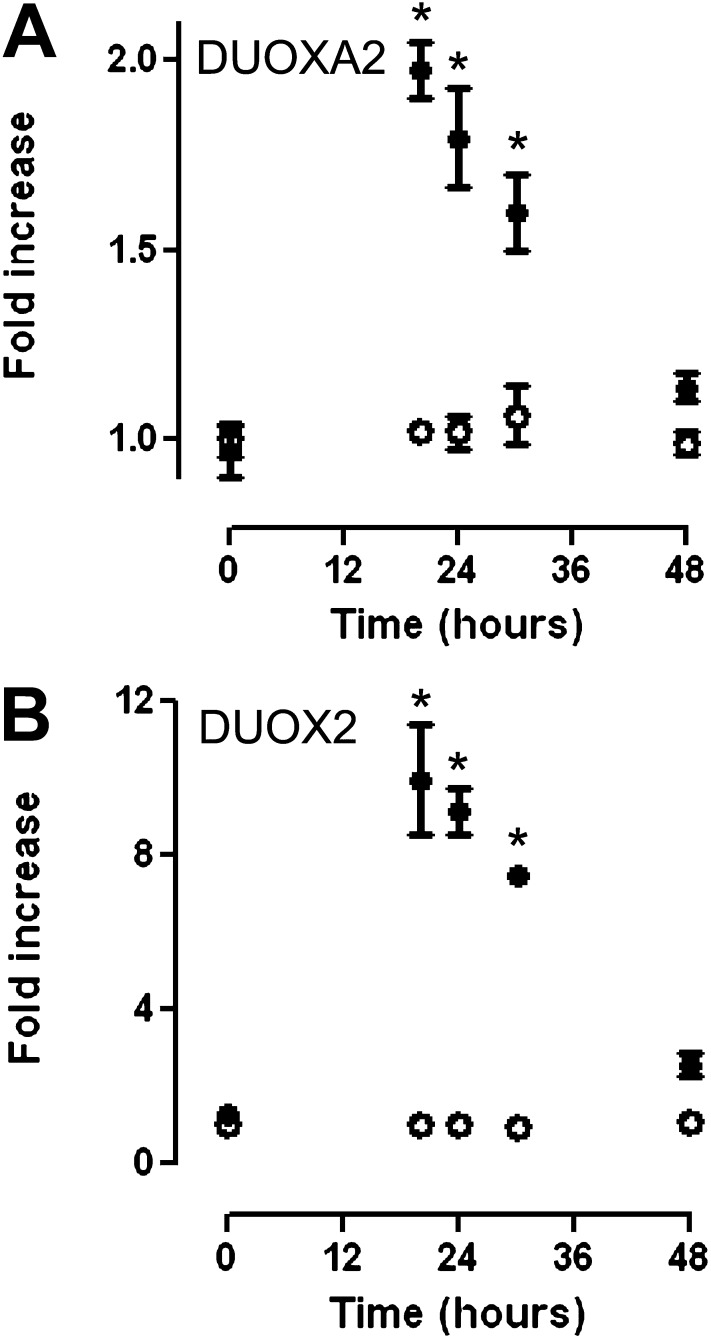
The observed peak luciferase activity at 12 hours suggested that the transfection procedure itself may temporarily activate the promoter. Because DUOX2/DUOXA2 transcription is increased after viral exposure in primary TBE cells and HBE1 cells (26) (data not shown), we postulated that exogenous double-stranded (ds)DNA provided a stimulus for DUOX2/DUOXA2 transcription. To assess this possibility, we measured the expression of endogenous DUOX2/DUOXA2 mRNA at multiple time points after transfection with dsDNA (the FL vector). The presence of plasmid DNA (FL vector) during transfection resulted in an approximately tenfold increase in DUOX2 and a twofold increase in DUOXA2 at 20 hours, and a return to near-baseline gene expression by 48 hours. The addition of transfection reagents without plasmid DNA did not modulate DUOX2 or DUOXA2 mRNA expression (Figure 4). These results were highly similar to our promoter data (Figure 2A), which suggested that exogenous dsDNA is a stimulus for DUOX2/DUOXA2 transcription.
Figure 4.
Effect of double-stranded (ds)DNA transfection on DUOXA2 and DUOX2 mRNA transcription. HBE1 cells in submerged cell culture conditions were transfected with FL vector (solid circles) or transfection reagents without vector (open circles), and RNA was extracted at various time points (20–48 hours) after transfection. DUOX2 and DUOXA2 mRNA concentrations were measured by quantitative RT-PCR and normalized to glyceraldehyde 3–phosphate dehydrogenase (GAPDH) expression. Fold increase is based on DUOX mRNA concentrations in untransfected cells at each time point. Fold increase for untransfected cells is based on DUOX mRNA concentrations at 0 hours. Means ± SEMs for triplicate samples of two independent experiments are shown. *P < 0.05, by ANOVA.
DUOX2/DUOXA2 Promoter Deletions
To determine which regions were required for DUOX2/DUOXA2 bidirectional promoter activity in response to exogenous dsDNA, we created four different promoter deletion constructs (Figure 5A). Each construct, missing approximately 300 base pairs (bp) from the full-length promoter, was cotransfected into HBE1 cells with a plasmid expressing β-galactosidase. Renilla and firefly luciferase concentrations were measured 12 hours after transfection, to identify which promoter regions were essential for the observed change in promoter activity. As shown in Figure 5B, the position of the deletion exerted significant effects on which transcriptional direction was attenuated. The deletion FLΔ304 exerted the largest effect on attenuating normalized Renilla luciferase activity (DUOX2 direction), whereas deletions FLΔ612 and FLΔ908 were more important for firefly luciferase expression (DUOXA2 direction). That deletion FLΔ612 significantly hampered transcription in both directions suggests that this region contains cis-elements that are critical for the initiation of transcription in either direction.
Effects of SNPs on Basal DUOX2/DUOXA2 Promoter Activity
During our initial cloning of the DUOX2/DUOXA2 promoter, sequence analyses of genomic DNA from two different cell lines identified two single-nucleotide changes in the DUOX2/DUOXA2 promoter region (see Materials and Methods). These SNPs, SNP 378 and SNP 819, corresponded to rs269855 (A/G) and rs2576089 (C/T) in the HapMap database (www.hapmap.org). We performed site-directed mutagenesis to generate four different promoter–reporter constructs to determine the contribution of each SNP to exogenous dsDNA-stimulated DUOX2/DUOXA2 promoter activity (Figure 6). The FL vector (comprising the 378G-819T haplotype) provided the highest peak luciferase activity in either direction. Mutagenesis of the FL vector with any other SNP combination (378A-819T, 378G-819C, or 378A-819C) resulted in an approximately 50% reduction in normalized Renilla and firefly luciferase activity, compared with the 378G-819T haplotype (Figure 6B).
Of particular interest, SNP 378 and SNP 819 are present in the two regions that produced the most profound decrement in promoter activity when deleted, as demonstrated by constructs FLΔ304 and FLΔ918, respectively (Figure 5). Together, these data confirm that the region between nucleotides 304 and 912 of the bidirectional promoter is essential for the observed bidirectional promoter activity, and suggest that these two SNP mutations target critical regulatory regions in the promoter in response to foreign nucleic acid.
Discussion
Based on current data, the coordinated expression of DUOX2 and its maturation factor, DUOXA2, are clearly required for fully functional enzymatic activity at the plasma membrane (19, 20). This essential biological feature and the genomic organization of DUOX2/DUOXA2 strongly suggest that these proteins are regulated through a bidirectional promoter. We have verified that the chromosomal region between the start codons of DUOX2 and DUOXA2 exhibits bidirectional transcription in human airway cells. We identified key regions of this promoter that are essential for unidirectional or bidirectional transcriptional activity. Importantly, we identified two SNPs that significantly affect DUOX2/DUOXA2 promoter activity in response to exogenous dsDNA.
Key features of the DUOX2/DUOXA2 promoter region are highly consistent with structural features that are common to other well-characterized bidirectional promoters, including divergent genomic organization, minimal genomic distance between the head-to-head genes, and CpG island enrichment (27–30). The currently available complete mRNA sequences for DUOX2 and DUOXA2 (National Center for Biotechnology Information [NCBI] numbers NM_014080 and NM_207581, respectively) place the transcriptional start sites (TSSs) for the two divergently arranged genes 163 bp apart, which is less than the 0.3–1 kb that is typical for bidirectional genes (Figure E1). Moreover, we identified a distinct CpG island region using two different CpG island prediction programs (31, 32) and highly restrictive criteria (GC content > 60%, observed CpG/expected CpG ratio > 0.7, and CpG island length > 200 bp; Figure E2). Both programs predicted highly similar CpG island regions. The methylation status of this region was inversely correlated with DUOX2 expression levels in cancer tissues (33), suggesting this CpG-rich region is functionally significant. In addition, several alternative TSSs have been proposed or experimentally verified for DUOX2 (19, 34), suggesting that transcriptional initiation is broadly distributed in this region (Figure E2), a feature common to CpG island promoters and bidirectional promoters in particular (35).
Despite evidence that CpG island regions are present in the DUOX2/DUOXA2 promoter, it remains unclear whether CpG island regions are primarily important for DUOX2/DUOXA2 transcriptional activity. Firstly, the beginning of exon 1 for the canonical DUOX2 mRNA sequence (NM_014080) is outside the predicted GC-rich region (Figure E2). CpG islands will typically cover exon 1 of both bidirectionally regulated genes (29, 30), making it less likely that CpG motifs are relevant for both DUOX2 and DUOXA2. Secondly, most bidirectional promoters with enriched CpG islands are “TATA-less” promoters. In contrast, the DUOX2/DUOXA2 promoter has a highly conserved TATA box in the DUOXA2 direction, making this the more likely mechanism of transcriptional initiation. Finally, deletion of the TATA-containing promoter region (the FL Δ908 vector) in our study resulted in a significant reduction in DUOXA2 promoter activity (Figure 5). Together, these data suggest that the DUOX2/DUOXA2 promoter is part of a minority group of bidirectional promoters that are primarily regulated through the TATA box element (29).
Using a similar strategy, Christophe-Hobertus and Christophe demonstrated bidirectional promoter activity of the rat Duox2/Duoxa2 promoter, using a thyroid cell line (24). They observed that a minimal promoter element between the DUOXA2 TATA box and a putative initiator element (Inr) for DUOX2 transcription was sufficient for bidirectional promoter activity. These sequences, in addition to a downstream promoter element (DPE) that is typically closely linked with the Inr element (36), were also present in our human samples and the mouse genome, indicating that a similar regulatory strategy is used in multiple species and multiple tissue types (Figure E3). Indeed, loss of either the Inr/DPE region (FLΔ612) or the TATA box region (FLΔ908), resulted in reduced DUOX2 or DUOXA2 transcription, respectively (Figure 5). Together, these observations support the notion that the DUOX2/DUOXA2 promoter uses the Inr/DPE site coupled with a TATA box motif primarily to initiate bidirectional promoter activity. However, regulatory regions outside this minimal promoter convey important regulatory activity in human tissue.
As shown by deletion FLΔ304, the loss of a promoter segment well outside the minimal promoter exerted significant effects on both DUOX2 and DUOXA2 transcriptional activity (Figure 5). In particular, loss of the 378G SNP within this deleted region appeared to be responsible for most of this enhanced activity (Figure 6). The combination of the 378G allele with the 819T allele resulted in the highest level of bidirectional promoter activity. All other SNP combinations resulted in comparable levels of promoter activity, suggesting that the 378G-819T fragment cloned from HBE1 cells was unusually active. Therefore, the 378Α-819Τ haplotype, containing the major alleles at both SNP sites, likely represents the “normal” response to foreign DNA, and SNP819 provides important regulatory information only in the context of a 378G allele. Interestingly, the region surrounding SNP378 is unique to the human sequence in comparison with the rodent sequence, whereas the region containing SNP819 is shared with human, mouse, and rat species (Figure E4). Although the specific changes in transcription factor binding activity at these SNP sites remain unknown, a few potential candidates are suggested by DNA sequence analyses.
Using a web-based DNA binding element prediction program (AliBaba2.1; http://www.gene-regulation.com/index2.html), we compared how the SNP mutations altered putative transcription factor binding sites. For SNP378, the A to G transition resulted in a gain of the activating protein-2 alpha (AP-2α) and early growth response protein 2 (EGR-2) putative binding sites. For SNP819, the T to C transition resulted in the loss of transcription factor Sry-related-high mobility group box-9 (SOX-9) and an isotype switch from CCAAT-enhancer-binding protein (C/EBP)α to C/EBPβ. The importance of the AP-2 and C/EBP transcription factor families for host–virus interactions makes these regions specifically intriguing.
That transfection of exogenous DNA-induced DUOX2 and DUOX2 mRNA expression (Figure 4) suggests that transfection itself may induce antiviral pathways that maximally up-regulate the DUOX2/DUOXA2 promoter in response to “virus.” This suggestion is consistent with the observation that cytoplasmic dsDNA will induce Type I interferons similar to viral RNA (37–39). Moreover, the time course of DUOX2/DUOXA2 mRNA induction after transfection matched our previous studies with virus-induced DUOX2 mRNA expression (26) (and unpublished data).
Simultaneous plasmid transfection with rhinovirus infection, a known stimulus for DUOX2 mRNA transcription (26), did not further induce bidirectional promoter activity (Figure E5), which suggests that transfection maximally induced “virus-mediated” promoter activity. Similarly, cotreatment with IFN-γ, another stimulus for DUOX2 expression, did not further enhance promoter activity (Figure E5). This may also reflect a promoter that is maximally stimulated, but more likely, the IFN-γ regulatory region is outside this bidirectional promoter region (40). Consistent with that notion, we were unable to locate IFN-responsive sites contained within the promoter region that we isolated for these studies. Future studies that use stable cell lines expressing our bidirectional promoter construct, infect or treat transformed cells at later time points or incorporate a longer promoter sequence will be needed to understand rhinovirus-mediated or cytokine-mediated mechanisms to up-regulate DUOX2/DUOXA2 expression.
Because the two SNPs, rs269855 (378A/G) and rs2576089 (819C/T), exerted profound effects on transcriptional activity in response to exogenous dsDNA, we speculate that these SNPs are partly responsible for the variable response to virus infection. In particular, the 378G-819T haplotype demonstrated a twofold increase in promoter activity compared with the other SNP combinations (Figure 6). We speculate that the GT haplotype confers an overly exuberant response to foreign DNA. This will potentially result in a pathologic response to viral infection through unnecessarily high levels of oxidative stress or excessive H2O2-mediated signaling (17, 18, 41). The distribution of haplotypes in the human population suggests that homozygosity for the GT haplotype is rare (42) (Table E1), which supports our hypothesis. Future experiments combining cell culture and human studies will help clarify these possibilities.
Here, we demonstrated that DUOX2 and DUOXA2 are coregulated through a shared bidirectional promoter in human respiratory epithelium. Although a regulatory unit of this promoter is shared with rodents, suggesting shared mechanisms of DUOX2 regulation in multiple species and tissues, we clearly demonstrated the presence of important regulatory regions outside the confines of this minimal promoter. These regulatory regions can direct the bidirectional or unidirectional transcription of each gene. Although the bidirectional promoter ensures the coordinated expression of DUOX2 and DUOXA2, the presence of unidirectional transcription suggests a more complicated relationship between the two genes than previously appreciated. In addition, we identified two critical SNPs in the DUOX2/DUOXA2 promoter that may provide novel insights toward a better understanding of the individual variability to viral infection.
Supplementary Material
Footnotes
This work was supported by grant HL085311 (R.W.H.) from the National Heart, Lung and Blood Institute of the National Institutes of Health.
Originally Published in Press as DOI: 10.1165/rcmb.2012-0370OC on May 16, 2012
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Parker D, Prince A. Innate immunity in the respiratory epithelium. Am J Respir Cell Mol Biol 2011;45:189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans SE, Xu Y, Tuvim MJ, Dickey BF. Inducible innate resistance of lung epithelium to infection. Annu Rev Physiol 2010;72:413–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischer H. Mechanisms and function of DUOX in epithelia of the lung. Antioxid Redox Signal 2009;11:2453–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Vliet A. NADPH oxidases in lung biology and pathology: host defense enzymes, and more. Free Radic Biol Med 2008;44:938–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salathe M, Holderby M, Forteza R, Abraham WM, Wanner A, Conner GE. Isolation and characterization of a peroxidase from the airway. Am J Respir Cell Mol Biol 1997;17:97–105 [DOI] [PubMed] [Google Scholar]
- 6.Conner GE, Salathe M, Forteza R. Lactoperoxidase and hydrogen peroxide metabolism in the airway. Am J Respir Crit Care Med 2002;166:S57–S61 [DOI] [PubMed] [Google Scholar]
- 7.Wijkstrom-Frei C, El-Chemaly S, Ali-Rachedi R, Gerson C, Cobas MA, Forteza R, Salathe M, Conner GE. Lactoperoxidase and human airway host defense. Am J Respir Cell Mol Biol 2003;29:206–212 [DOI] [PubMed] [Google Scholar]
- 8.Conner GE, Wijkstrom-Frei C, Randell SH, Fernandez VE, Salathe M. The lactoperoxidase system links anion transport to host defense in cystic fibrosis. FEBS Lett 2007;581:271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moskwa P, Lorentzen D, Excoffon KJ, Zabner J, McCray PB, Jr, Nauseef WM, Dupuy C, Banfi B. A novel host defense system of airways is defective in cystic fibrosis. Am J Respir Crit Care Med 2007;175:174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL. Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J 2003;17:1502–1504 [DOI] [PubMed] [Google Scholar]
- 11.Fischer AJ, Lennemann NJ, Krishnamurthy S, Pocza P, Durairaj L, Launspach JL, Rhein BA, Wohlford-Lenane C, Lorentzen D, Banfi B, et al. Enhancement of respiratory mucosal antiviral defenses by the oxidation of iodide. Am J Respir Cell Mol Biol 2011;45:874–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Botteaux A, Hoste C, Dumont JE, Van Sande J, Allaoui A. Potential role of Noxes in the protection of mucosae: H(2)O(2) as a bacterial repellent. Microbes Infect 2009;11:537–544 [DOI] [PubMed] [Google Scholar]
- 13.Bae YS, Choi MK, Lee WJ. Dual oxidase in mucosal immunity and host–microbe homeostasis. Trends Immunol 2010;31:278–287 [DOI] [PubMed] [Google Scholar]
- 14.Shao MX, Nadel JA. Dual oxidase 1–dependent MUC5AC mucin expression in cultured human airway epithelial cells. Proc Natl Acad Sci USA 2005;102:767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boots AW, Hristova M, Kasahara DI, Haenen GR, Bast A, van der Vliet A. ATP-mediated activation of the NADPH oxidase DUOX1 mediates airway epithelial responses to bacterial stimuli. J Biol Chem 2009;284:17858–17867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wesley UV, Bove PF, Hristova M, McCarthy S, van der Vliet A. Airway epithelial cell migration and wound repair by ATP-mediated activation of dual oxidase 1. J Biol Chem 2007;282:3213–3220 [DOI] [PubMed] [Google Scholar]
- 17.Yu M, Lam J, Rada B, Leto TL, Levine SJ. Double-stranded RNA induces shedding of the 34-kDa soluble TNFR1 from human airway epithelial cells via TLR3–TRIF–RIP1–dependent signaling: roles for dual oxidase 2– and caspase-dependent pathways. J Immunol 2011;186:1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joo JH, Ryu JH, Kim CH, Kim HJ, Suh MS, Kim JO, Chung SY, Lee SN, Kim HM, Bae YS, et al. Dual oxidase 2 is essential for the Toll-like receptor 5–mediated inflammatory response in airway mucosa. Antioxidants Redox Signal 2012;16:57–70 [DOI] [PubMed] [Google Scholar]
- 19.Grasberger H, Refetoff S. Identification of the maturation factor for dual oxidase: evolution of an eukaryotic operon equivalent. J Biol Chem 2006;281:18269–18272 [DOI] [PubMed] [Google Scholar]
- 20.Morand S, Ueyama T, Tsujibe S, Saito N, Korzeniowska A, Leto TL. DUOX maturation factors form cell surface complexes with DUOX affecting the specificity of reactive oxygen species generation. FASEB J 2009;23:1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adachi N, Lieber MR. Bidirectional gene organization: a common architectural feature of the human genome. Cell 2002;109:807–809 [DOI] [PubMed] [Google Scholar]
- 22.Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. An abundance of bidirectional promoters in the human genome. Genome Res 2004;14:62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang MQ, Elnitski LL. Prediction-based approaches to characterize bidirectional promoters in the mammalian genome. BMC Genomics 2008;9:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christophe-Hobertus C, Christophe D. Delimitation and functional characterization of the bidirectional THOX-DUOXA promoter regions in thyrocytes. Mol Cell Endocrinol 2010;317:161–167 [DOI] [PubMed] [Google Scholar]
- 25.Yankaskas JR, Haizlip JE, Conrad M, Koval D, Lazarowski E, Paradiso AM, Rinehart CA, Jr, Sarkadi B, Schlegel R, Boucher RC. Papilloma virus immortalized tracheal epithelial cells retain a well-differentiated phenotype. Am J Physiol 1993;264:C1219–C1230 [DOI] [PubMed] [Google Scholar]
- 26.Harper RW, Xu C, Eiserich JP, Chen Y, Kao CY, Thai P, Setiadi H, Wu R. Differential regulation of dual NADPH oxidases/peroxidases, DUOX1 and DUOX2, by Th1 and Th2 cytokines in respiratory tract epithelium. FEBS Lett 2005;579:4911–4917 [DOI] [PubMed] [Google Scholar]
- 27.Takai D, Jones PA. Origins of bidirectional promoters: computational analyses of intergenic distance in the human genome. Mol Biol Evol 2004;21:463–467 [DOI] [PubMed] [Google Scholar]
- 28.Gardiner K. Human genome organization. Curr Opin Genet Dev 1995;5:315–322 [DOI] [PubMed] [Google Scholar]
- 29.Yang MQ, Taylor J, Elnitski L. Comparative analyses of bidirectional promoters in vertebrates. BMC Bioinformatics 2008;9:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang MQ, Elnitski LL. Diversity of core promoter elements comprising human bidirectional promoters. BMC Genomics 2008;9:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet 2000;16:276–277 [DOI] [PubMed] [Google Scholar]
- 32.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA 2002;99:3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luxen S, Belinsky SA, Knaus UG. Silencing of DUOX NADPH oxidases by promoter hypermethylation in lung cancer. Cancer Res 2008;68:1037–1045 [DOI] [PubMed] [Google Scholar]
- 34.Pachucki J, Wang D, Christophe D, Miot F. Structural and functional characterization of the two human THOX/DUOX genes and their 5′-flanking regions. Mol Cell Endocrinol 2004;214:53–62 [DOI] [PubMed] [Google Scholar]
- 35.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 2006;38:626–635 [DOI] [PubMed] [Google Scholar]
- 36.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem 2003;72:449–479 [DOI] [PubMed] [Google Scholar]
- 37.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006;441:101–105 [DOI] [PubMed] [Google Scholar]
- 38.Takeshita F, Ishii KJ. Intracellular DNA sensors in immunity. Curr Opin Immunol 2008;20:383–388 [DOI] [PubMed] [Google Scholar]
- 39.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 2006;24:93–103 [DOI] [PubMed] [Google Scholar]
- 40.Wu Y, Antony S, Juhasz A, Lu J, Ge Y, Jiang G, Roy K, Doroshow JH. Up-regulation and sustained activation of STAT1 are essential for interferon-gamma (IFN-gamma)–induced dual oxidase 2 (DUOX2) and dual oxidase A2 (DUOXA2) expression in human pancreatic cancer cell lines. J Biol Chem 2011;286:12245–12256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipinski S, Till A, Sina C, Arlt A, Grasberger H, Schreiber S, Rosenstiel P. DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J Cell Sci 2009;122:3522–3530 [DOI] [PubMed] [Google Scholar]
- 42.Gaunt TR, Rodriguez S, Day IN. Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool “cubex”. BMC Bioinformatics 2007;8:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.