Abstract
Verticillium wilt disease is caused by fungi of the Verticillium genus that occur on a wide range of host plants, including Solanaceous species such as tomato and tobacco. Currently, the well characterized Ve1 gene of tomato is the only Verticillium wilt resistance gene cloned. During experiments to identify the Verticillium molecule that activates Ve1 resistance in tomato, RNA sequencing (RNA-Seq) of Verticillium-infected Nicotiana benthamiana was performed. In total, over 99% of the obtained reads were derived from N. benthamiana. Here, we report the assembly and annotation of the N. benthamiana transcriptome. In total, 142,738 transcripts >100 bp were obtained, amounting to a total transcriptome size of 38.7 Mbp, which is comparable to the Arabidopsis transcriptome. About 30,282 transcripts could be annotated based on homology to Arabidopsis genes. By assembly of the N. benthamiana transcriptome, we provide a catalogue of transcripts of a Solanaceous model plant under pathogen stress.
Keywords: RNA-Seq, Verticillium, assembly, genomics, tobacco, transcriptome
Verticillium spp are soil-borne pathogens that cause wilt disease on more than 200 plant species in temperate and sub-tropical regions. Verticillium wilts are notorious because they are difficult to control due to persistent resting structures that reside in the soil and that are difficult to eradicate with currently available control measures, absence of fungicides to cure infected plants, and the availability of relatively few sources of genetic resistance.1,2 Thus far, the only well characterized Verticillium resistance gene is the tomato Ve1 gene that provides resistance against race 1 isolates of V. dahliae and V. albo-atrum.3,4 Recently, a genome sequence has been determined for each of these two species.5 Until recently, the Verticillium molecule that is intercepted by the Ve1 immune receptor remained unknown. However, by population genome high-throughput sequencing this effector, designated Ave1 (for Avirulence on Ve1 tomato), was identified.6 Intriguingly, Ave1 was found to be homologous to a widespread family of plant proteins, suggesting that the fungus acquired Ave1 from plants through horizontal gene transfer.6
As part of the sequencing strategy to identify the Verticillium effector that is detected by the Ve1 immune receptor, RNA-Seq was performed on samples of V. dahliae-infected plants. The plant species used to generate the samples for RNA-Seq was Nicotiana benthamiana, which is able to accumulate more V. dahliae biomass upon infection than tomato or Arabidopsis, the other two model plants that have mostly been used to study this pathogen.3,5 N. benthamiana is an Australian endemic tobacco species which has been extensively used in research on plant-pathogen interactions.7 The popularity of N. benthamiana as a model to study such interactions can be attributed to the relative ease of genetic manipulation, such as transient overexpression and gene silencing, when compared with other Solanaceous species. Moreover, N. benthamiana has been used successfully to study interactions between various immune receptors and pathogen effectors as well as immune signaling. Here, we describe the transcriptome of N. benthamiana infected by V. dahliae as determined by RNA-Seq.6
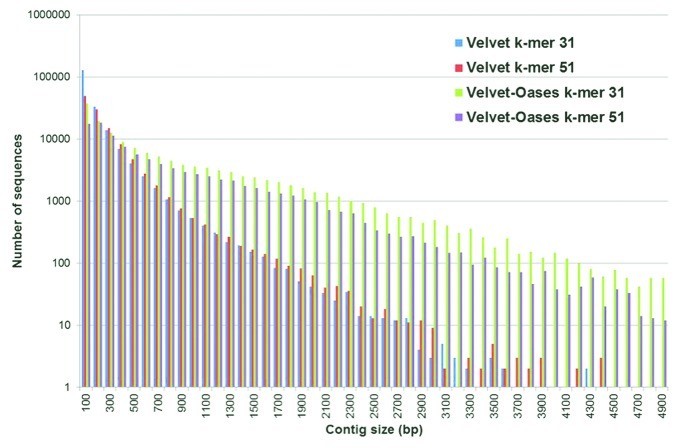
Deep RNA sequencing was performed on three-week-old N. benthamiana plants inoculated through root-dipping in a conidial suspension of V. dahliae strain JR2 as described previously, and harvested in a time course on 4, 8, 12 and 16 d post inoculation (DPI).5,6 Total RNA was extracted using the RNeasy Mini Kit (Qiagen), and cDNA synthesis, library preparation (200-bp inserts), and Illumina sequencing (90-bp paired-end reads) was performed at the Beijing Genomics Institute (BGI). For each sample, ~25 million paired-end 90 bp reads were obtained. A quality check was performed on the reads using the FastQC software (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/), resulting in a high quality score, and thus no reads were discarded. To reconstruct the N. benthamiana transcriptome, reads belonging to the N. benthamiana transcriptome were separated from those belonging to the V. dahliae transcriptome using Tophat.8 To this end, the complete RNA-Seq data set was mapped onto the draft genome sequence of Verticillium dahliae strain JR26 and the unmapped reads were isolated. Only 0.05% of the reads obtained at 4 DPI could be mapped on the V. dahliae genome, implying that the remaining 99.95% were plant derived. By 16 DPI, the amount of V. dahliae derived transcripts increased to 0.9%. By removing all reads mapping to V. dahliae, a total of 105,152,616 reads were retained which were used for assembly of the N. benthamiana transcriptome using the Velvet-Oases package.9,10 The Velvet software uses a so-called de Bruijn graph for the construction of contigs. In general, de Bruijn graph based assembly software is not well suited for the assembly of transcripts because the algorithm does not account for alternative splicing and fluctuations in coverage depth within a contig, thus specific software for mRNA assembly have been developed. The Oases software uses the Velvet assembly as input and tries to connect contigs by using the information from the reads taking alternative splicing and dynamic expression levels into account.11 The assembly result largely depends on two parameters: the k-mer length and coverage cut-off. A k-mer is a fragment of a read of arbitrary length k, while the coverage cut-off is the minimal number of times that a nucleotide needs to be represented in a contig at a specific position in order to be included in the final assembly. The Perl script VelvetOptimizer.pl, included in the Velvet package, was used to optimize these two parameters by comparing the N50 (the size N at which 50% of the genome is assembled in contigs of size N or greater) and the total number of large contigs obtained upon assembly using different k-mer sizes. The k-mers tested ranged from 23 to 61 with 2 step intervals, while the coverage cut-off was automatically set by the software. The analysis showed that a k-mer setting of 51 resulted in a higher N50 and longer contigs when compared with the standard k-mer setting of 31 (Fig. 1). Although higher N50 and longer contigs were obtained with a k-mer setting of 51, assembly results obtained with both k-mer settings of 31 and 51 were used as input for the Oases software. Surprisingly, the Oases software produced better results using the Velvet k-mer 31 output rather than the 51 k-mer output (Fig. 1). In de Bruijn graph based software, longer k-mers provide a higher specificity for the placement of reads in a contig, while the sensitivity for the placement is lower due to a lower coverage as a result of the increased k-mer length. Oases, taking into consideration dynamic expression and multiple transcript isoforms, tries to position reads that can connect contigs that were generated by Velvet. Consequently, a more conservative assembly generated by Velvet using a k-mer of 31 leads to a more fragmented assembly into contigs which allows Oases more flexibility to connect these contigs into transcripts. The Oases assembly based on the Velvet k-mer 31 output resulted in 142,738 transcripts larger than 100 bp (N50 of 1,464 bp, N90 of 744 bp) (Fig. 1), the longest transcript being 15,046 bp, with an average GC content of 42.15%. Within the assembled sequences, 55,808 sequences represented only one transcript, while 18,830 sequences represented multiple (average ~4.5) transcript isoforms. In order to reduce transcript redundancy, the longest mRNA was selected in case multiple transcript isoforms were obtained. The selection resulted in a set of 74,638 non-redundant transcripts. Subsequently, the non-redundant transcripts were aligned to the V. dahliae genome5 and to the N. benthamiana genome version 0.3 (ftp://ftp.solgenomics.net/genomes/Nicotiana_benthamiana/assemblies/) in order to eliminate pathogen-derived transcripts that were not subtracted upon mapping of the short reads with Tophat and retain only transcripts matching to the N. benthamiana genome. This analysis showed that 1,597 out of the 74,638 non-redundant assembled transcripts had a significant hit to the V. dahliae genome and were therefore removed. All other transcripts matched to the N. benthamiana genome. Considering the 73,041 transcripts that matched to the N. benthamiana genome, the transcriptome size of N. benthamiana amounted to 38.7 Mbp, which equals ~1.3% of the total genome size and is close to the size of the Arabidopsis thaliana transcriptome.12 With this, we assembled ~4.5 times more non-redundant mRNAs than the number of ESTs that is already deposited in the Unigene database of N. benthamiana at the Solgenomics database (http://www.solgenomics.net/). However, most likely the number of transcripts obtained in this study is over-estimated due to partial assembly of mRNAs, leading to messengers that are split over multiple transcripts during the assembly process.
Figure 1. Size distribution of the assembled transcripts after Velvet and Oases analyses with different k-mer settings. The histogram represents the number of sequences of a particular contig size. The different colors show the output of Velvet or Velvet-Oases with different k-mer settings. The Velvet-Oasis combination with k-mer setting 31 results in the highest number of long contigs.
To assess the robustness of the assembly, the transcripts were blasted to the N. benthamiana unigenes in the Solgenomics database (http://www.solgenomics.net) and an overall high identity (99.3%) was found. Moreover, the longest 200 N. benthamiana unigenes were compared with their homologous transcripts, showing that 72 out of 200 transcripts were covering > 90% of the unigene length. Furthermore, the length of the assembled N. benthamiana transcripts were compared with their Arabidopsis homologs. The core eukaryotic gene set of Arabidopsis is composed of 459 genes (http://korflab.ucdavis.edu/) which were aligned to the N. benthamiana transcriptome using the tBLASTn algorithm.13,14 Contigs mapping to all of the 459 genes were identified, with an overall sequence length coverage of about 74%, while for about 58% of the genes a sequence length coverage > 90% was obtained. Thus, our assembly covered homologs for all core Arabidopsis genes and about half of the transcripts had a length that is comparable to their Arabidopsis homologs. The longest transcript of 15,046 bp was a homolog of the gene encoding the auxin transport protein BIG-like in Glycine max (soybean), which is one of the longest transcripts in plants. The identity between the two homologs was about 61% at the protein level, and the assembled transcript corresponded to about 95% of the Glycine max BIG-like protein.
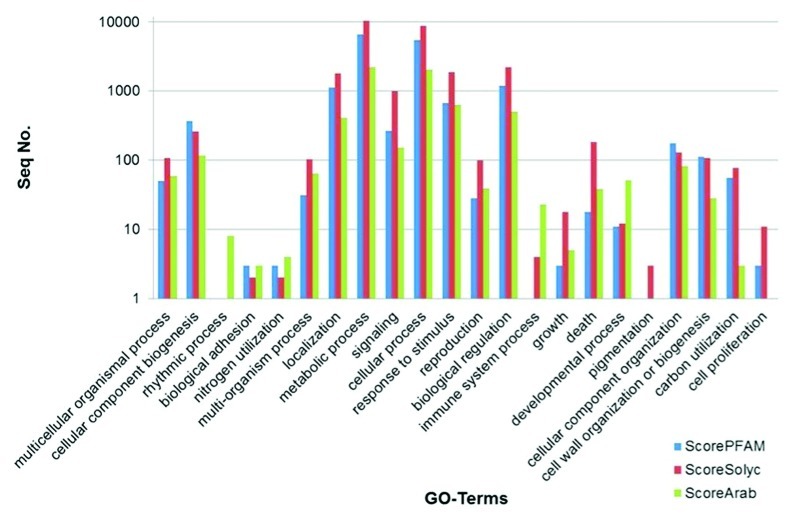
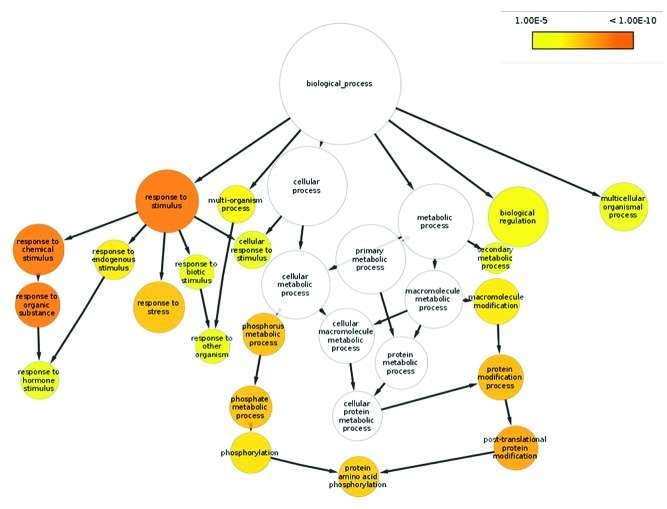
To annotate the N. benthamiana transcriptome, the assembled transcripts were translated into proteins by making a 6-frame translation and selecting the longest open reading frame. The proteins were subsequently aligned by BLASTp software to the Arabidopsis protein database, the tomato (Solanum lycopersicum) protein database and the PFAM database using the HMMER software (Fig. 2). Out of 73,041 N. benthamiana transcripts, 30,282 matched to an Arabidopsis protein, 31,976 matched to a tomato protein and 23,614 matched to a PFAM model. Subsequently, because of optimal curation and integration into software programmes, the annotation based on Arabidopsis proteins was used for GO-term enrichment analysis. The assembled transcriptome was used as a reference for digital expression quantification. The reads, derived from the different time points, were remapped to the assembled N. benthamiana transcriptome using the Bowtie2 software with the –M option set to 1 in order to map reads to a single transcript16 (Table 1). The Cufflinks package was used to determine the digital expression of each of the transcripts. The software Cuffdiff, which is part of Cufflinks, was run with the –time-series option activated in order to compare the samples in sequential time, and not the default option all vs. all. The expression analysis and the annotation were combined to produce a data set in which only annotated differentially expressed transcripts were present. This resulted in 3,923 (12.9%) transcripts that were induced between 4 and 16 DPI and 1,673 (5.5%) transcripts that were repressed. Next, gene ontology term enrichment was analyzed. A threshold of 10-fold induction or repression was chosen to select the transcripts to be used in the gene ontology term enrichment analysis using the BiNGO software.17 Out of the 5,596 differentially expressed transcripts, 1,267 genes were selected as induced and 163 as repressed at 16 DPI. This analysis identified an enrichment (p-value 10−11) of GO terms associated to biotic stress (Fig. 3) in the group of induced genes, which was expected based on earlier transcriptome analysis on V. dahliae-infected tomato.18 No specific enrichment was found for the repressed genes. Collectively, our data provide a robust catalog of transcripts of the Solanaceous model plant N. benthamiana undergoing pathogen stress that can be used for annotation of the N. benthamiana genome and for mining of candidate genes in pathogen defense.
Figure 2. Functional annotation of the predicted Nicotiana benthamiana proteome by Blast analysis to the Arabidopsis protein database (scoreArab), the tomato protein database (scoreSolyc) and the PFAM database (scorePFAM). Sequences are organized by gene ontology.
Table 1. Number of N. benthamiana reads used for the digital expression analysis.
| Sample time point (days post infection) | Total number of reads | Reads remapped to the transcriptome (% of total) |
|---|---|---|
| 4 |
26,193,846 |
2,111,671 (8%) |
| 8 |
26,167,768 |
10,126,370 (38.7%) |
| 12 |
26,146,456 |
9,985,223 (38.2%) |
| 16 | 26,644,546 | 9,708,919 (36.4%) |
Figure 3. Gene ontology enrichment identified in the Verticillium-infected Nicotiana benthamiana transcriptome by the BiNGO software. The gene ontology term enrichment analysis was performed on the 1,267 genes that displayed 10-fold induction at 16 d post inoculation when compared with the standard gene ontology term representation of Arabidopsis genes. Circle sizes indicate the relative number of genes that represent a gene ontology term, and the color gradient indicates the statistical robustness of the over-representation.
Acknowledgments
We thank Dr. T.J.A. Borm and Dr. R.G.F. Visser, Department of Plant Breeding of Wageningen UR, for access to the Selma computer.
Data deposition
The RNA-Seq data are available at ftp://ftp.solgenomics.net/transcript_sequences/by_species/Nicotiana_benthamiana/illumina/Verticillium_dahliae.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/21014
References
- 1.Fradin EF, Thomma BPHJ. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol. 2006;7:71–86. doi: 10.1111/j.1364-3703.2006.00323.x. [DOI] [PubMed] [Google Scholar]
- 2.Stout MJ, Thaler JS, Thomma BPHJ. Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu Rev Entomol. 2006;51:663–89. doi: 10.1146/annurev.ento.51.110104.151117. [DOI] [PubMed] [Google Scholar]
- 3.Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GCM, Joosten MH, Thomma BP. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 2011;156:2255–65. doi: 10.1104/pp.111.180067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CDM, Nazar RN, Robb J, et al. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009;150:320–32. doi: 10.1104/pp.109.136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, Thomma BPHJ, et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011;7:e1002137. doi: 10.1371/journal.ppat.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, et al. Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci U S A. 2012;109:5110–5. doi: 10.1073/pnas.1119623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. Nicotiana benthamiana: its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact. 2008;21:1015–26. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- 8.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–92. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haridas S, Breuill C, Bohlmann J, Hsiang T. A biologist’s guide to de novo genome assembly using next-generation sequence data: A test with fungal genomes. J Microbiol Methods. 2011;86:368–75. doi: 10.1016/j.mimet.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 12.Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–7. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 14.Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol. 2012;30:83–9. doi: 10.1038/nbt.2022. [DOI] [PubMed] [Google Scholar]
- 16.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–9. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 18.van Esse HP, Fradin EF, de Groot PJ, de Wit PJGM, Thomma BPHJ. Tomato transcriptional responses to a foliar and a vascular fungal pathogen are distinct. Mol Plant Microbe Interact. 2009;22:245–58. doi: 10.1094/MPMI-22-3-0245. [DOI] [PubMed] [Google Scholar]