Abstract
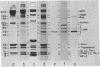
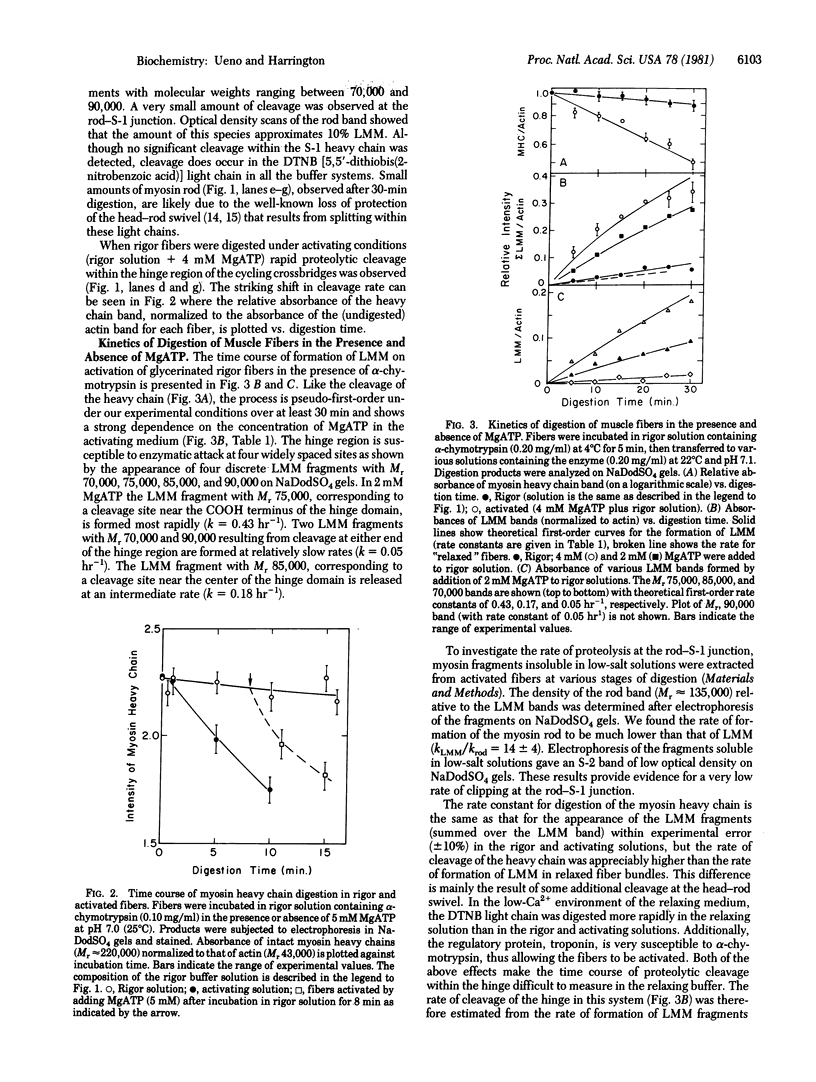
We have determined the rates of chymotryptic proteolysis of the myosin hinge region in glycerinated rabbit psoas fibers and myofibrils in in rigor-inducing, activating, and relaxing buffers. The time course of formation of light meromyosin (LMM) provides a specific probe for cleavage within the hinge domain. In rigor-inducing and relaxing buffers proteolysis within the hinge is depressed, but on activation LMM is formed at a markedly increased rate, which is dependent on the concentration of MgATP. Peptide bond cleavage occurs at four widely separated sites spanning the length of the hinge domain. Only a trivial amount of proteolysis occurs at the head--rod swivel or within the heavy chain of the head itself (S-1 subunit) in rigor-inducing and relaxing solvents, and we find no significant change on activation. The rate of formation of LMM in rigor-inducing buffer is unchanged by addition of MgADP, Pi, or magnesium adenosine 5'-[beta, gamma-imido]triphosphate or in activating solvent at zero overlap between thick and thin filaments. These results provide evidence for a conformational (helix--coil) transition within the myosin hinge upon activation of skeletal muscle.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagshaw C. R. On the location of the divalent metal binding sites and the light chain subunits of vertebrate myosin. Biochemistry. 1977 Jan 11;16(1):59–67. doi: 10.1021/bi00620a010. [DOI] [PubMed] [Google Scholar]
- Burke M., Himmelfarb S., Harrington W. F. Studies on the "hinge" region of myosin. Biochemistry. 1973 Feb;12(4):701–710. doi: 10.1021/bi00728a020. [DOI] [PubMed] [Google Scholar]
- Eisenberg E., Hill T. L. A cross-bridge model of muscle contraction. Prog Biophys Mol Biol. 1978;33(1):55–82. doi: 10.1016/0079-6107(79)90025-7. [DOI] [PubMed] [Google Scholar]
- Feldhau P., Fröhlich T., Goody R. S., Isakov M., Schirmer R. H. Synthetic inhibitors of adenylate kinases in the assays for ATPases and phosphokinases. Eur J Biochem. 1975 Sep 1;57(1):197–204. doi: 10.1111/j.1432-1033.1975.tb02291.x. [DOI] [PubMed] [Google Scholar]
- GERGELY J., GOUVEA M. A., KARIBIAN D. Fragmentation of myosin by chymotrypsin. J Biol Chem. 1955 Jan;212(1):165–177. [PubMed] [Google Scholar]
- Güth K. Polarization of tryptophan fluorescence measurements in muscle. A re-evaluation. Biophys Struct Mech. 1980;6(2):81–93. doi: 10.1007/BF00535746. [DOI] [PubMed] [Google Scholar]
- HUMMEL B. C. A modified spectrophotometric determination of chymotrypsin, trypsin, and thrombin. Can J Biochem Physiol. 1959 Dec;37:1393–1399. [PubMed] [Google Scholar]
- Harrington W. F. A mechanochemical mechanism for muscle contraction. Proc Natl Acad Sci U S A. 1971 Mar;68(3):685–689. doi: 10.1073/pnas.68.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington W. F. On the origin of the contractile force in skeletal muscle. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5066–5070. doi: 10.1073/pnas.76.10.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselgrove J. C., Huxley H. E. X-ray evidence for radial cross-bridge movement and for the sliding filament model in actively contracting skeletal muscle. J Mol Biol. 1973 Jul 15;77(4):549–568. doi: 10.1016/0022-2836(73)90222-2. [DOI] [PubMed] [Google Scholar]
- Huxley A. F., Simmons R. M. Proposed mechanism of force generation in striated muscle. Nature. 1971 Oct 22;233(5321):533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- Huxley H. E. The mechanism of muscular contraction. Science. 1969 Jun 20;164(3886):1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MIHALYI E., SZENT-GYORGYI A. G. Trypsin digestion of muscle proteins. I. Ultracentrifugal analysis of the process. J Biol Chem. 1953 Mar;201(1):189–196. [PubMed] [Google Scholar]
- Potekhin S. A., Privalov P. L. Teplovaia denaturatsiia alpha-tropomiozina i ego fragmentov. Biofizika. 1978 Mar-Apr;23(2):219–223. [PubMed] [Google Scholar]
- Potekhin S. A., Trapkov V. A., Privalov P. L. Stadiinost' teplovoi denaturatsii spiral'nykh fragmentov miozina. Biofizika. 1979 Jan-Feb;24(1):46–50. [PubMed] [Google Scholar]
- Rome E. Light and X-ray diffraction studies of the filament lattice of glycerol-extracted rabbit psoas muscle. J Mol Biol. 1967 Aug 14;27(3):591–602. doi: 10.1016/0022-2836(67)90061-7. [DOI] [PubMed] [Google Scholar]
- Sutoh K., Sutoh K., Karr T., Harrington W. F. Isolation and physico-chemical properties of a high molecular weight subfragment-2 of myosin. J Mol Biol. 1978 Nov 25;126(1):1–22. doi: 10.1016/0022-2836(78)90276-0. [DOI] [PubMed] [Google Scholar]
- Swenson C. A., Ritchie P. A. Conformational transitions in the subfragment-2 region of myosin. Biochemistry. 1980 Nov 11;19(23):5371–5375. doi: 10.1021/bi00564a035. [DOI] [PubMed] [Google Scholar]
- Tsong T. Y., Karr T., Harrington W. F. Rapid helix--coil transitions in the S-2 region of myosin. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1109–1113. doi: 10.1073/pnas.76.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H., Harrington W. F. Cross-bridge movement and the conformational state of the myosin hinge in skeletal muscle. J Mol Biol. 1981 Jul 15;149(4):619–640. doi: 10.1016/0022-2836(81)90350-8. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Weeds A. G., Pope B. Studies on the chymotryptic digestion of myosin. Effects of divalent cations on proteolytic susceptibility. J Mol Biol. 1977 Apr;111(2):129–157. doi: 10.1016/s0022-2836(77)80119-8. [DOI] [PubMed] [Google Scholar]
- Yanagida T. Angles of nucleotides bound to cross-bridges in glycerinated muscle fiber at various concentrations of epsilon-ATP, epsilon-ADP and epsilon-AMPPNP detected by polarized fluorescence. J Mol Biol. 1981 Mar 15;146(4):539–560. doi: 10.1016/0022-2836(81)90046-2. [DOI] [PubMed] [Google Scholar]