Abstract
We have created two neuron-specific mouse models of mitochondrial electron transport chain deficiencies involving defects in complex III (CIII) or complex IV (CIV). These conditional knockouts (cKOs) were created by ablation of the genes coding for the Rieske iron–sulfur protein (RISP) and COX10, respectively. RISP is one of the catalytic subunits of CIII and COX10 is an assembly factor indispensable for the maturation of Cox1, one of the catalytic subunits of CIV. Although the rates of gene deletion, protein loss and complex dysfunction were similar, the RISP cKO survived 3.5 months of age, whereas the COX10 cKO survived for 10–12 months. The RISP cKO had a sudden death, with minimal behavioral changes. In contrast, the COX10 cKO showed a distinctive behavioral phenotype with onset at 4 months of age followed by a slower but progressive neurodegeneration. Curiously, the piriform and somatosensory cortices were more vulnerable to the CIII defect whereas cingulate cortex and to a less extent piriform cortex were affected preferentially by the CIV defect. In addition, the CIII model showed severe and early reactive oxygen species damage, a feature not observed until very late in the pathology of the CIV model. These findings illustrate how specific respiratory chain defects have distinct molecular mechanisms, leading to distinct pathologies, akin to the clinical heterogeneity observed in patients with mitochondrial diseases.
INTRODUCTION
Genetic defects affecting the function of the electron transport chain and the oxidative phosphorylation (OXPHOS) system are known as mitochondrial disorders. This group of diseases involves defects in either the nuclear or the mitochondrial DNA (mtDNA) and is heterogeneous in nature. Mitochondrial diseases can affect single or multiple organs. Tissues with higher energetic demands, such as brain and muscle, are most commonly affected (1).
In the last few years, effort has been concentrated in understanding the molecular bases of the phenotypic variability of mitochondrial disorders. The heterogeneous nature of mitochondrial diseases poses a challenge for the development of effective treatments. Advances in this area have been hampered by the lack of appropriate animal models with a single respiratory defect. In the last few years, mouse models of mitochondrial diseases have started to emerge (2), allowing the testing of therapeutic approaches (3,4). Here we characterized two animal models of mitochondrial encephalopathy caused by complex III (CIII) or complex IV (CIV) deficiency in neurons. Surprisingly, we found significant differences in their phenotypes.
Mammalian CIII is composed of 11 subunits, with one of them, cytochrome b, being encoded by the mitochondrial genome. In the case of mammalian CIV, there are 13 subunits, of which three, which comprise the catalytic core, are encoded by the mtDNA. The rest of the subunits are encoded in the nucleus and imported into the mitochondria.
CIII deficiencies are relatively rare among the respiratory chain defects. They have been associated with mutations in catalytic and structural subunits of the complex and with assembly factors. CIII deficiencies comprise a broad spectrum of symptoms and exhibit tissue specificity (reviewed in 5). Among the clinical presentations that are associated with mutations in the catalytic subunit cytochrome b are: encephalopathy, Leber's hereditary optic neuropathy, cardiomyopathy and myopathy (6,7). Mutations in UQCRB and UQCRQ, structural subunits of CIII, cause hypoglycemia, lactic acidosis and psychomotor retardation, respectively (8). Mutations in the assembly factors (BCS1L and TTC19) also show various clinical presentations. BCS1L is a molecular chaperone that assists in the incorporation of the Rieske iron–sulfur protein (RISP, one of the catalytic subunits) and UQCR10 into the complex. Defects in BCS1L can cause Björnstad syndrome affecting multiple organs (muscle weakness, optic atrophy, encephalopathy, liver failure and tubolopathy) or GRACILE syndrome (growth restriction, aminoaciduria, cholestasis, iron overload, lactic acidosis and early death) affecting the liver. Defects in TTC19 cause the accumulation of CIII-assembly intermediates and lead to neurological abnormalities (reviewed in 9). The specific function of TTC19 remains unknown.
CIV deficiencies are more common defects of the electron transport chain. Mutations in COX subunits encoded by the mtDNA have been associated with encephalopathy, sideroblastic anemia, myopathy, myoglobinuria, Leigh-like syndrome, multi-systemic disease and metabolic acidosis among other pathologies. In the case of mutations in structural subunits, only two cases have been reported with defective COX6b1, supporting the idea that perhaps mutations in the structural components are not compatible with life. The majority of the cases of CIV deficiency correspond to defects in the auxiliary proteins. In yeast, over 40 assembly factors for CIV have been identified (10). CIV ancillary factors associated with disease are SURF1, SCO1, SCO2, LRPPRC, COX10, COX15, TACO1 and FASTKD2, and their clinical characteristics include Leigh syndrome, metabolic acidosis, hypertrophic cardiomyopathy, French-Canadian Leigh syndrome and encephalopathy (reviewed in 9).
In addition to specific mitochondrial disorders, impairment of mitochondrial function has been linked also to many neurodegenerative diseases and aging, possibly because impairment of the electron transport chain can produce excess free radicals leading to oxidative stress/damage (11). The role of oxidative damage in mitochondrial diseases has not been extensively documented in vivo and most of the studies refer to increased reactive oxygen species (ROS) production in cultured cells derived from patients with mitochondrial disorders.
To gain a better understanding on the pathophysiological mechanisms of mitochondrial diseases, we created two conditional knockout (cKO) models with either CIII or CIV defect in the same subgroup of neurons. The CIII deficiency was achieved by ablating the RISP, one of the catalytic subunits of the complex and the CIV deficiency by ablating the assembly factor COX10. COX10 encodes a heme a farnesyl transferase required for heme a biosynthesis. Heme a is an indispensable cofactor of Cox1, a catalytic subunit of CIV. Although the two mitochondrial defective mice recapitulate mitochondrial disease phenotypes, there were marked differences between them.
RESULTS
Neuron-specific CIII- and CIV-deficient mice
We created two mouse models of mitochondrial CIII and CIV deficiency in neurons using the Cre-loxP system. To produce the CIII-deficient mouse, a knock-in mouse homozygous for the floxed UQCRFS1 gene (12) was crossed with a transgenic mouse expressing the Cre recombinase under the control of the CaMKIIα (calcium-calmodulin kinase II alpha) promoter (13). The UQCRFS1 gene encodes the RISP, one of the catalytic subunits of CIII. To create the CIV-deficient mouse, we crossed a knock-in mouse homozygous for the floxed COX10 gene with the same Cre transgenic mouse mentioned above (14). Cox10 is indispensable for Cox1 maturation and stability.
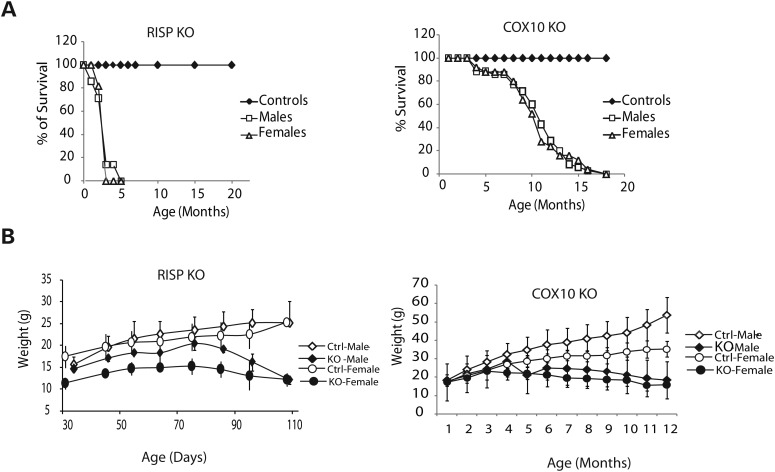
Both RISP and COX10 cKO mice had shortened life spans albeit the RISP cKO was markedly shorter than the COX10 cKO. RISP cKO died between 3 and 3.5 months of age whereas the COX10 cKO mice died between 8 and 12 months (Fig. 1A). RISP cKO weighted less than their control littermates, particularly RISP cKO females (Fig. 1B). In the case of COX10 cKO mice, a significance weight difference was observed only later in life (after 4 and 5 months of age for females and males, respectively, Fig. 1B).
Figure 1.
Survival and growth curves of RISP and COX10 cKO mice. (A) Comparison of the survival curve of the RISP and COX10 cKO mice. RISP cKO [closed diamonds, control male and female mice (n = 20 per group); open squares, cKO male mice (n = 7); open triangles, cKO female (n = 11)]. COX10 cKO [closed diamond, control male and female mice (n = 35 per group); open squares, cKO male mice (n = 35); open triangle, cKO female (n = 25)]. There is a significant difference in the survival of RISP compared with the COX10 cKO. The premature death of the RISP cKO occurs much earlier than the COX10 cKO. No difference between male and female survival was observed in both cKO models. (B) Growth curve of RISP and COX10 cKO. RISP cKO mice weight less than control counterparts from an early age, particularly female mice [open diamonds, control male (n = 7); closed diamonds, cKO males (n = 5); open circles, control females (n = 15); closed circles, cKO females (n = 7)]. COX10 cKO mice start to lose weight at ∼4–5 months of age [open diamonds, control male (n = 9); closed diamonds, cKO males (n = 8); open circles, control females (n = 13); closed circles, cKO females (n = 8)]. Values represent mean ± standard deviation.
There were no overt functional phenotypes in the first 3 months of age for either model. The RISP cKO displayed lower nocturnal ambulatory movement than their control littermates (Supplementary Material, Fig. S1). In the case of the COX10 cKO mice, a clear and reproducible behavioral difference was only detected at 4 months of age, including alternate cycles of hyperactivity. Analysis of nocturnal ambulatory movement at 2 months of age showed indistinguishable behavior between COX10 cKO and their control counterparts. At 3 months of age, the COX10 cKO started to decrease their nocturnal activity and by 4 months, cycles of hyperactivity were obvious (Supplementary Material, Fig. S1A). This behavioral abnormality continued for their life span and only when terminal the animals became markedly quiet.
Despite the absence of overt phenotypes at an early age, a motor coordination test in the rotarod revealed that the RISP cKO had decreased performance at 2 months of age with a rapid decline, being unable to perform the test 15 days later (mice were unable to stay in the rotating rod for more than few seconds without falling). In the case of the COX10 cKO, decrease in the rotarod performance had a later onset than the RISP cKO, with differences between COX10 cKO and control mice first observed at 3 and 4 months of age for males and females, respectively (Supplementary Material, Fig. S1B).
Effect of ablation of RISP and COX10 on OXPHOS complexes
We estimated the extent of gene ablation over time by amplification of the remaining floxed alleles in the two models and found them to be similar (Supplementary Material, Fig. S2). Assuming that both alleles were recombined, we calculated that the percentage of recombination in cortex was 50 ± 9.8 and 52 ± 3.2% (mean ± SD) for RISP and COX10, respectively, at 1 month of age (Supplementary Material, Fig. S2A). Similar values were obtained for hippocampus (54 ± 8 and 54 ± 3, respectively). The levels of floxed alleles remaining at 4 months were comparable to the levels observed at 1 month of age, indicating that most of the deletion of both RISP and COX10 had already occurred at an early age (Supplementary Material, Fig. S2B). However, lower percentage of deletion values were obtained for RISP cKO at 3 months, and there was a significant difference between RISP and COX10 deletion at 3–4 months of age in cortex but not in hippocampus (Supplementary Material, Fig. S2A). Since the determination of the deletion was performed in whole-tissue samples that include other cell types besides CamKIIα neurons, we assessed the specific effect of the gene ablation in neurons by immunohistochemistry using RISP and Cox1-specific antibodies. Supplementary Material, Fig. S3 shows confocal microscopy images of the immunostaining of brain sections of control and cKO mice with RISP (green) and Cox1 (red) antibodies. In merged images of control mice, neurons stained in yellow, indicating the presence of both RISP and Cox1 proteins; however, in the RISP cKO, neurons only stained in red or positive for Cox1 (arrowheads), indicating the absence of the RISP protein. Similarly, in the COX10 cKO, the merged image shows neurons stained in green or positive for RISP (arrowheads), indicating the absence of the Cox1 protein.
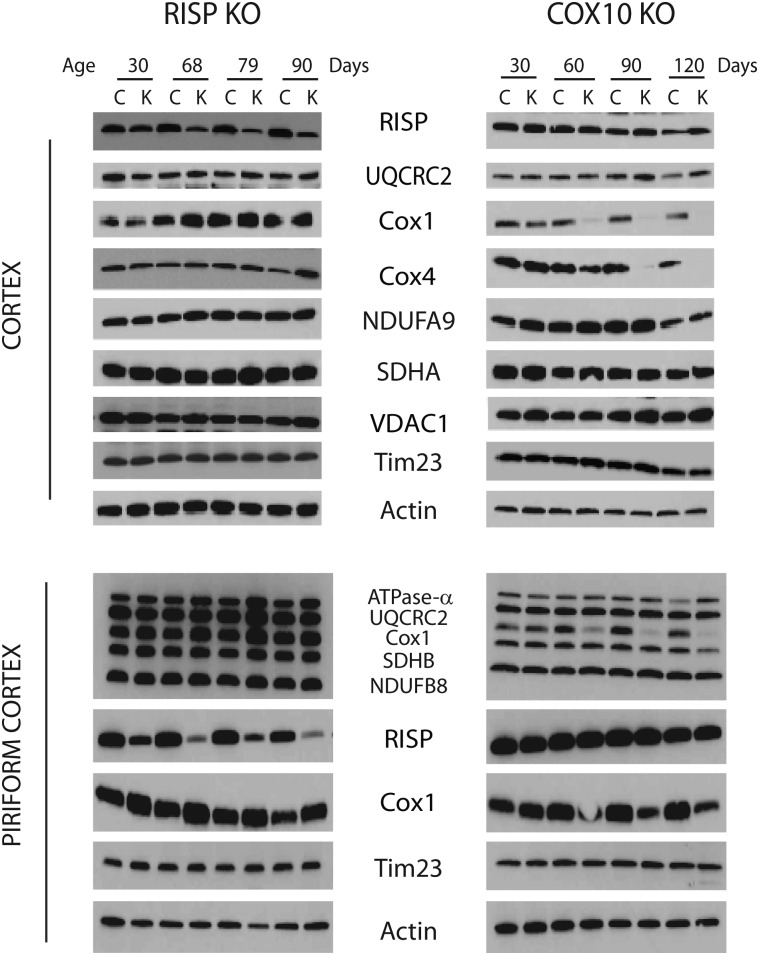
Western blot analysis of steady-state levels of respiratory complex subunits showed decreased levels of RISP at 1 month of age in hippocampus homogenates of RISP cKO, and the levels progressively decreased with age in both cortex and hippocampus, reaching ∼50 and 3% of control levels, respectively, at 3 months (Fig. 2 and Supplementary Material, Fig. S4). In cortex homogenates, the levels of UQCRC2 (another CIII subunit), NDUFA9 (CI subunits) and SDHA (CII subunit) normalized to actin remained comparable to control values whereas there was a slight increase in the levels of Cox1, Cox4 (CIV subunits), VDAC1 and Tim23 (1.4-, 3.6-, 1.7- and 1.3-fold, respectively) at 3 months of age (Fig. 2, top panel). In the case of hippocampus homogenates, an increase in all OXPHOS proteins ranging from 1.3- to 1.9-fold was observed with the exception of NDUFA9, the levels of which remained unchanged (Supplementary Material, Fig. S4). In the case of piriform cortex, increases of 2.3–3-fold in all mitochondrial proteins relative to actin were observed at 79 days (Fig. 2, bottom panel).
Figure 2.
Steady-state levels of mitochondrial oxidative phosphorylation complex subunits in RISP and COX10 cKO cortex. Immunoblots of cingulate and piriform cortex homogenates (20 μg protein) of RISP and COX10 cKO at different ages using antibodies against different subunits of mitochondrial respiratory complexes. RISP and UQCRC2 are subunits of CIII; Cox1 and Cox4 are subunits of CIV; NDUFA9 is a CI subunit; SDHA is a CII subunit. VDAC1 (voltage-dependent anion channel-1), Tim 23 (subunit of inner membrane import translocator) and actin were used as loading controls.
Ablation of COX10 affected the steady-state levels of Cox1 as well as Cox4 in a progressive manner, and these markers were barely detectable by 4 months of age in both cortex and hippocampus homogenates (Fig. 2 and Supplementary Material, Fig. S4). The levels of other OXPHOS subunits and mitochondrial membrane proteins were slightly increased in cortex homogenates (1.2- to 2.2-fold from control values when normalized to actin). In the case of hippocampus, there was a slight increase in the levels of OXPHOS complexes at 3 months of age (1.2- to 1.7-fold of control values), but there was a slight decrease at 4 months of age (0.84–0.98-fold of control values, Supplementary Material, Fig. S4).
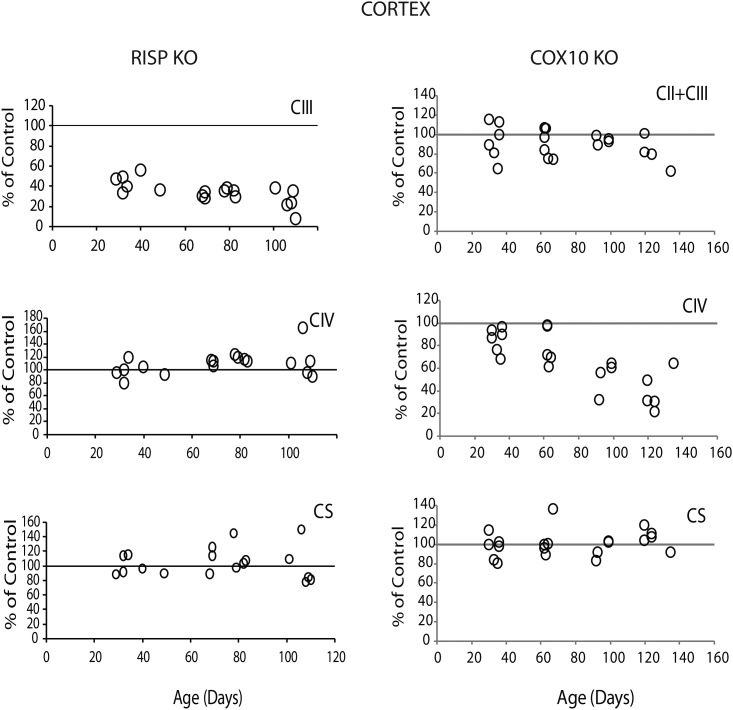
We also determined the effect of the deletion of RISP and COX10 on the enzymatic activity of different complexes of the electron transport chain. Figure 3 shows that, as expected, deletion of RISP had a severe impact on CIII activity. At 1 month of age, RISP cKO mice displayed ∼43 ± 8.8% (mean ± SD) of CIII activity of control levels in cortex homogenates. The enzymatic defect was progressive and reached ∼25 ± 12% of control values at 110 days of age. Interestingly, increases in the enzymatic activity of CIV and citrate synthase (CS) of ∼114 ± 6% and 115 ± 17%, respectively, were observed in the RISP cKO at ∼2.5 months of age (Fig. 3). Similar results were obtained in hippocampus homogenates where CIII was ∼40 ± 11% of control values at 1 month of age and progressed to 14 ± 6% of control values by 110 days, and CIV and CS were elevated after 70 days of age to 120 ± 17% and 121 ± 16% of control values, respectively (Supplementary Material, Fig. S5A).
Figure 3.
Mitochondria respiratory complex activities in RISP and COX10 cKO cortex. Enzymatic activity of CIII and CIV and citrate synthase (CS) were measured spectrophotometrically in cortex homogenates of RISP and COX10 cKO mice at different ages. Mean values of specific activity of the KO mice were compared with mean values of age-matched control mice and represented as percentage of control. Each symbol corresponds to one cKO mouse (some symbols overlap, n = 6 per group). A progressive decline of CIII and CIV activity is observed in RISP and COX10 cKO, respectively.
Likewise, the ablation of COX10 produced a progressive CIV deficiency. At 1 month of age, cortex homogenates of COX10 cKO showed ∼80 ± 10% of control levels of CIV activity, and by 4 months of age, the values decreased to ∼33 ± 11% of control levels (Fig. 3). Similar results were observed in hippocampus homogenates, although the differences with control values were more pronounced (Supplementary Material, Fig. S5A). The CIV deficiency in single neurons was also confirmed immunohistochemically using the double activity stain of CII/CIV (SDH/COX activity, Fig. 4D). Regarding the enzymatic activity of other respiratory complexes, the COX10 cKO did not show a significant increase in CII + CIII or CS activities in contrast to the results observed for the RISP cKO (Fig. 3).
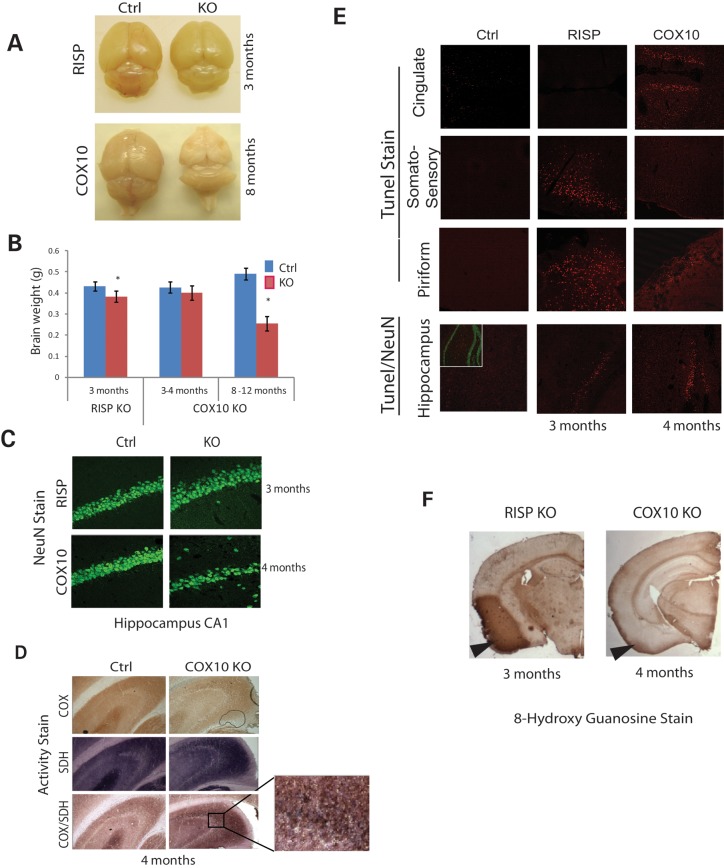
Figure 4.
Pathological features of RISP and COX10 cKO brains. (A) Gross anatomy of RISP and COX10 cKO brain. No overt differences were observed in RISP or COX10 cKO brain at 3 and 4 months, respectively. However, at 8 months, severe cortical atrophy occurred in the COX10 cKO. (B) Weights of RISP and COX10 cKO brains were compared with control brains at different ages. The RISP cKO brain weighted significantly less than that of control at 3 months of age, although no differences were apparent in gross anatomy. No changes were observed up to 4 months of age in the COX10 cKO brain. RISP control (n = 5) and cKO (n = 5); 3–4 months COX10 control (n = 6) and cKO (n = 5); 8–12 months (n = 3). (C) Frozen brain sections were immunohistochemically stained with the NeuN antibody. Images showed the hippocampal CA1 region of RISP and COX10 cKO at 3 and 4 months of age. (D) Brain sections of control and COX10 cKO mice (hippocampus) were stained for CIV (COX), CII (SDH) or combined activity stain. Insert shows magnification of the CA3 region with COX-negative neurons stained in blue/purple by the SDH stain. (E) TUNEL staining of control and cKO mice in different brain regions (cingulate, somatosensory and piriform cortex and hippocampus/dentate gyrus). Insert in the bottom panel shows NeuN stain (green) in control mice. No signs of TUNEL-positive cells were detected in earlier ages in both RISP and COX10 cKO mice (data not shown). (F) Immunohistochemical stain of the 8-hydroxy guanosine (nucleic acid oxidative damage marker) in RISP and COX10 cKO mice at 3 and 4 months of age, respectively.
Increases in the levels of other respiratory complexes and CS in RISP cKO are suggestive of mitochondrial proliferation. Therefore, we examined the levels of mtDNA/nDNA by quantitative PCR. Supplementary Material, Fig. S5B shows that indeed, the ratio of ND1 to actin was significantly higher in the RISP cKO than in control mice at 3 months of age, whereas this was not the case for the COX10 cKO mice. As mentioned above, this was reflected by a slight increase in mitochondrial proteins (Fig. 2). There was no clear evidence of mitochondrial proliferation (mtDNA/nDNA) at earlier ages (1–2 months) in either cKO mice (data not shown).
Pathological features of ablation of RISP and COX10 in brain
Gross examination of the RISP and COX10 cKO brains at 3 and 4 months of age, respectively, did not show obvious alterations in structure (Fig. 4A); however, the brain of the RISP cKO weighted less than that of the control counterpart (Fig. 4B). COX10 cKO brain weight did not show any changes at 1–4 months of age. However, by 8 months of age, severe cortical atrophy was observed and brains weighted about half of controls (Fig. 4A and B). To assess the extent of neuronal cell death, we performed TUNEL staining in brain sections at different ages and found that the RISP cKO mice consistently had TUNEL-positive cells in the somatosensory cortex, piriform cortex and hippocampus at 3 months (Fig. 4E), right before their death but not at earlier ages (data not shown). In the case of the COX10 cKO, the TUNEL-positive cells were consistently found in the cingulate cortex, piriform cortex and hippocampus/dentate gyrus at 4 months (Fig. 4E), long before their death but not at earlier time points (data not shown). Immunohistochemical staining with the neuronal marker, NeuN, revealed no obvious differences between control and RISP cKO mice at 3 months in the hippocampus although TUNEL-positive cells were present, indicating that cell death was a recent event (Fig. 4C). In contrast, the hippocampus CA1 region of the COX10 cKO showed evident neuronal loss at 4 months (Fig. 4C).
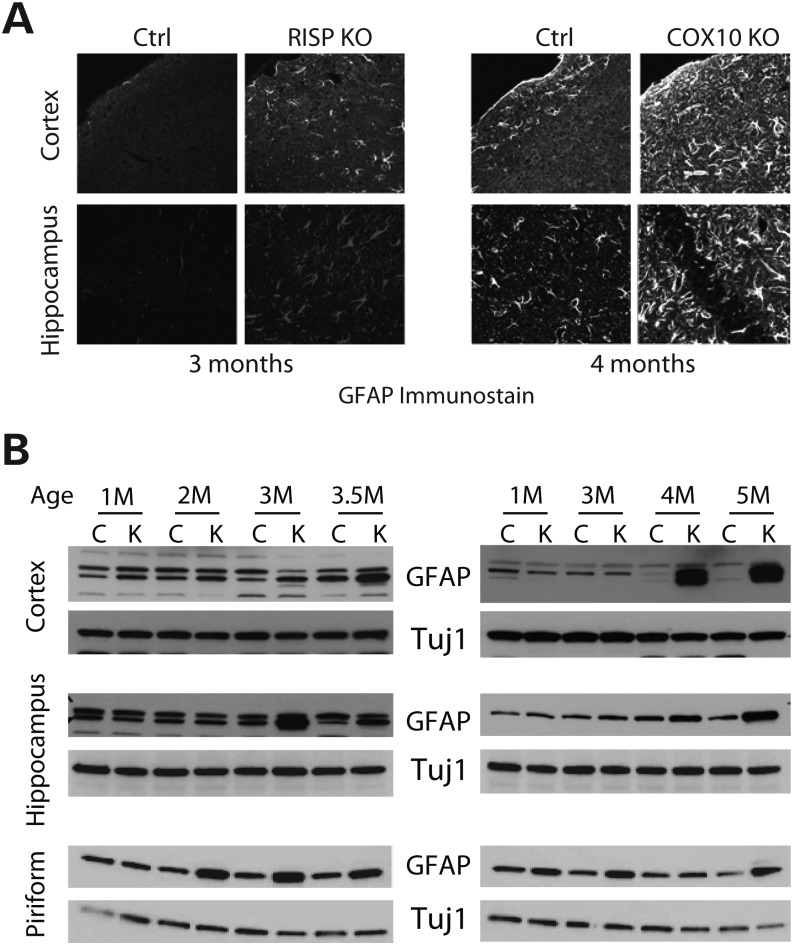
The contribution of glia to the neurodegenerative process was examined by both immunohistochemistry and western blot (Fig. 5). Both RISP and COX10 cKO mice showed increased immunoreactivity to GFAP in both cortex and hippocampus when compared with control mice (Fig. 5A). Analysis of the steady-state levels of GFAP showed that RISP cKO mice had a slight increase in GFAP levels (normalized to Tuj1) in cortex, hippocampus and piriform cortex ranging from 1.6- to 2.7-fold of controls at 3–3.5 months of age (Fig. 5B). In the case of the COX10 cKO, a dramatic increase in GFAP levels (16.9–18.4-fold) was observed in 4–5-month-old mice cortex homogenates. Although increased GFAP was observed in hippocampus and piriform cortex (4.9–6.8-fold), the levels were not as high as the ones observed in cortex (Fig. 5B).
Figure 5.
Reactive glia in RISP and COX10 cKO mice. (A) Immunohistochemical stain of frozen brain sections of control, RISP and COX10 cKO mice at 3 and 4 months, respectively, with the GFAP antibody showing reactive glia. (B) Steady-state levels of GFAP and Tuj1 (astrocytic and neuronal markers, respectively) in RISP and COX10 cKO mice at different ages in homogenates of different brain regions.
Progressive neurodegenerative process in RISP and COX10 cKO mice
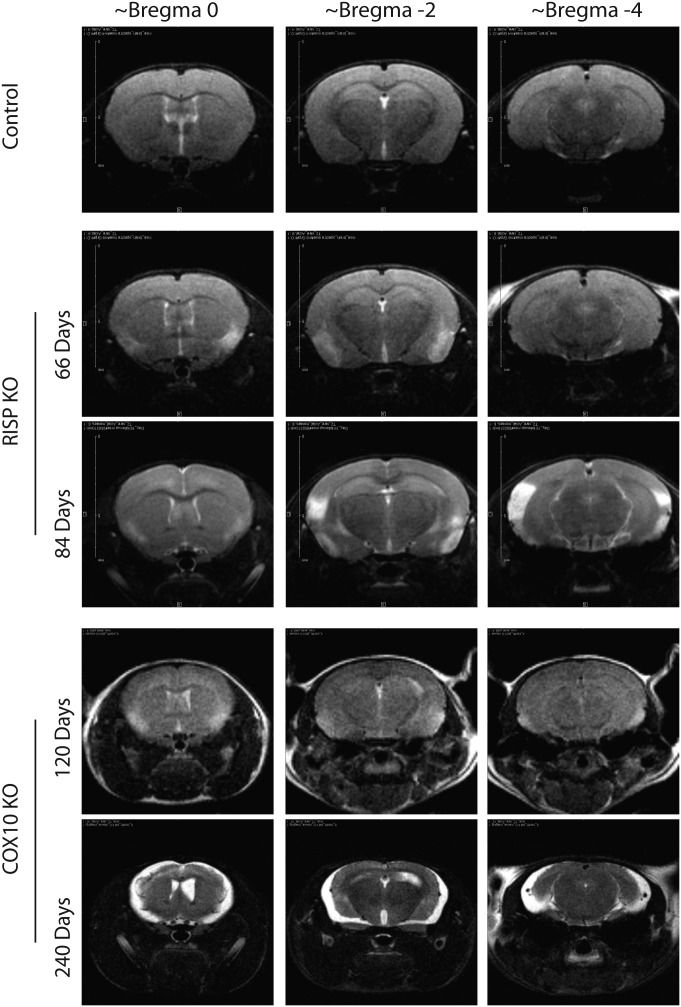
To follow the progression of the neurodegenerative process, we performed magnetic resonance imaging (MRI) in the same animals at different ages. Figure 6 shows different brain regions of control, RISP cKO (66 and 84 days old) and COX10 cKO (120 and 240 days old). The RISP cKO mice showed lesions in the piriform area at 66 days of age. Eighteen days later, these lesions worsened and extended to posterior parts of the brain. In the case of the COX10 cKO, small lesions were observed in the piriform cortex at 4 months of age (Fig. 6). By 6 months, lesions in the striatum, outer cortical layers and hippocampus were evident (data not shown) and by 8 months, there was massive degeneration of the cortex (Fig. 6). These results are consistent with cell death observed at 3 and 4 months in the RISP and COX10 cKOs, respectively (Fig. 4E).
Figure 6.
In vivo nuclear MRI of RISP and COX10 cKO brain. Progression of encephalopathy was followed in the same animals at different ages using T2-weighted MRI datasets. Representative images of different brain regions are shown. RISP cKO at 66 days of age shows hyper-intense areas in the piriform cortex that extend to different areas and posterior regions of the brain 18 days later. COX10 cKO mice also demonstrate lesions in piriform cortex at 120 days of age, and by 240 days, severe striatum and cortical damage was observed.
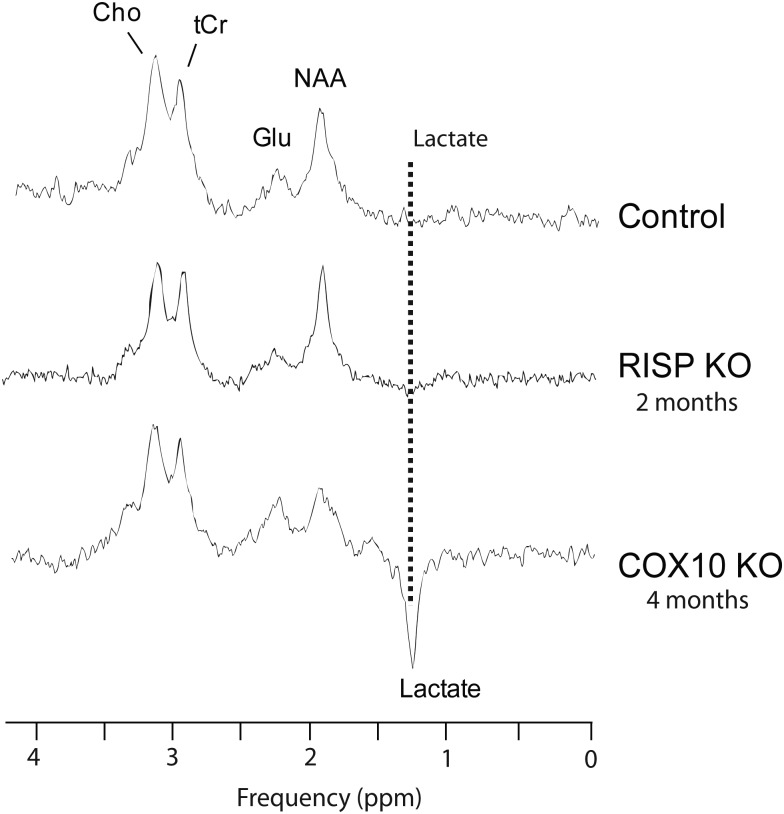
In vivo proton magnetic resonance spectroscopy (MRS) of the mice brain permitted the detection of choline (Cho), total creatine (tCr), glutamate (Glu), n-acetyl aspartate (NAA) and lactate (Fig. 7). The COX10 KO showed metabolic changes with elevated levels of lactate (plotted in the inverse phase to distinguish it from the lipids resonance). However, we were unable to detect brain lactate in control or in RISP cKO mice by this method (Fig. 7). MRS also revealed a decrease in NAA in the COX10 cKO. NAA is synthesized in the mitochondria, and it is considered to be only present in neurons and dendrites and thus a marker for the neuronal population. NAA reduction has been associated with energetic impairments (reviewed in 15).
Figure 7.
In vivo proton nuclear MRS of control, RISP and COX10 cKO mice. Representative proton spectra of control, RISP and COX10 cKO mice where the detection of some brain metabolites by MRS is demonstrated (n = 3–4 for each group). Peaks corresponding to choline (Cho), total creatine (tCr), glutamate (Glu), n-acetyl aspartate (NAA) and lactate (represented as the inverted peak to differentiate it from lipid signal resonance) are shown. COX10 cKO mice have increased lactate levels. No peak corresponding to lactate was detected in either control or the RISP cKO mice.
Oxidative Damage in RISP and COX10 cKO mice
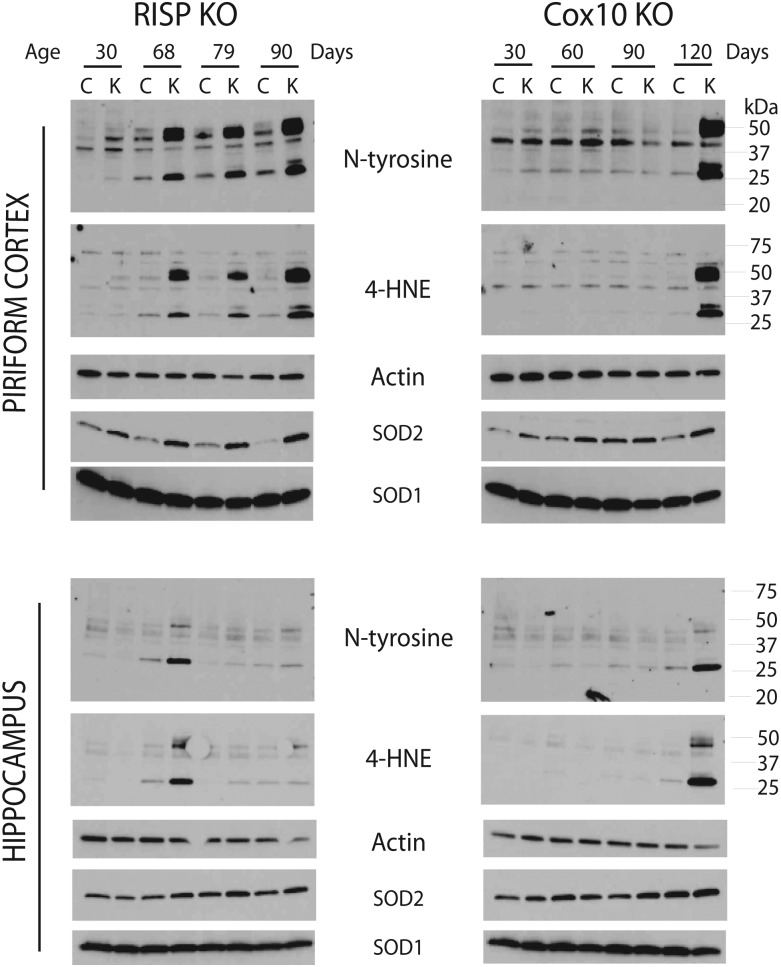
To gain a better understanding of the pathological differences of the CIII- and CIV-deficient mice, we examined the extent of oxidative damage in both mice models. Immunohistochemistry of brain sections with 8-hydroxyguanosine, a marker of nucleic acid oxidation, showed strong staining in the piriform cortex of the 3-month-old RISP cKO mice. COX10 cKO brain was only weakly stained in this region (Fig. 4F, arrowheads). Examination of other oxidative stress markers such as protein adducts of nitrosylation (N-tyrosine) and lipid peroxidation (4-hydroxynonenal, 4-HNE) by western blot showed that at 1 month of age, the RISP cKO mice already have higher levels of nitrosylated proteins and 4-HNE protein adducts than control mice in the piriform cortex. The levels of nitrosylated proteins normalized to actin ranged from 2.8- to 5.0-fold higher and 4-HNE adducts were 1.7–14.8 times higher than control values in the RISP cKO at different ages (Fig. 8). In the case of the COX10 mice, an increase in protein oxidation (8.1- and 12.6-fold increase in N-tyrosine and 4-HNE, respectively) was only observed later, at 4 months, and not at earlier ages. In addition, the RISP cKO mice also had a dramatic increase in the levels of mitochondrial superoxide dismutase 2 (SOD2) ranging from 3- to 31-fold increase at different ages in the piriform cortex when compared with control mice. In contrast, the levels of SOD1 were only slightly increased (1.7-fold) in RISP cKO mice (Fig. 8). Likewise, the levels of SOD2 were also increased in the COX10 cKO, although the changes were modest (2.6–3.6-fold higher than controls) when compared with the levels observed in the RISP cKO. SOD1 levels were slightly lower in COX10 cKO compared with control mice (Fig. 8).
Figure 8.
Oxidative stress markers in RISP and COX10 cKO mice. Steady-state levels of different oxidative markers were analyzed by western blot in piriform cortex and hippocampus homogenates (20 μg of protein) of control, RISP and COX10 cKO at different ages. Increased levels of N-tyrosine and 4-hydroxy nonenal (4-HNE) protein adducts indicate RNS species and lipid peroxidation in RISP cKO mice from an early age. SOD1 and SOD2, superoxide dismutase 1 and 2. Actin was used as loading control.
We compared the steady-state level of mitochondrial proteins of control and cKO mice in the piriform cortex, cingulate cortex and hippocampus in both RISP and COX10 cKO mice. From the molecular markers analyzed, there were no obvious differences that could explain the increased susceptibility of the piriform cortex in the cKO mice (Supplementary Material, Fig. S6).
DISCUSSION
In the present study, we compared two animal models of mitochondrial encephalopathy caused by defects in mitochondrial respiratory complexes III or IV produced by the ablation of UQCRFS1 or COX10, respectively. To date, there are no reports of any patient with mutations in the UQCRFS1 gene (encoding for RISP), perhaps accounting for its incompatibility with life. Mutations in the COX10 gene in humans result in a fatal outcome early in life (2–24 months of age) and have been associated with leukodystrophy, tubolopathy, hypertrophic cardiomyopathy and Leigh-like syndrome (16–18).
To gain insight into the heterogeneous nature of mitochondrial diseases and pathophysiological mechanisms underlying the phenotype of a single mitochondrial respiratory complex defect, we crossed our knock-in floxed UQCRFS1 (12) and COX10 (19) mice with the same CamKIIα-Cre expressing transgenic line to be able to establish a direct comparison between the two mitochondrial deficiencies (13). We used the CamKIIα-Cre to drive the postnatal deletion of the floxed alleles in forebrain neurons affecting mainly hippocampus and cortex; however, in humans, mitochondrial disorders of the nervous system affect other structures like striatum, basal ganglia, brain stem and cerebellum. For this reason, our mouse models might display limited recapitulation of the human disorders; however, they prove useful to study the molecular mechanisms leading to the heterogeneous nature of mitochondrial encephalopathies. In addition, although our studies compare the deletion of a structural subunit (RISP) with the deletion of an assembly factor (COX10), we do not think that the differences in phenotypes are due to this fact. We have confirmed previous observations that COX10 is indispensable for stability of the structural subunit Cox1. Therefore, both models have defects in a structural/catalytic subunit, and their corresponding steady-state levels (Fig. 2 and Supplementary Material, Fig. S4) were affected in a similar way.
Our results showed that the pathologic mechanisms in mitochondrial diseases are not solely caused by the depletion of ATP due to a dysfunctional OXPHOS, but also by differential regional susceptibility to oxidative stress. Our results are supported by several in vitro studies involving deficiencies in other respiratory complexes or electron carriers (CI, CV or CoQ deficiencies) where the partial defect in the different cell lines generated an increase in free radicals leading to oxidative stress and cell death (20–22). Mitochondria are regarded as the main generators of ROS and reactive nitrogen species (RNS) in the cell. In particular, the established site for ROS production is respiratory complexes I, II and III, although other mitochondrial enzymes such as α-ketoglutarate dehydrogenase, electron transfer flavoprotein and dihydro-orotate dehydrogenase are able to produce superoxide when donating electrons to CoQ (23,24). CI and CII produce superoxide radicals in the mitochondrial matrix and CIII produces superoxide in both the matrix and the intermembrane space. In addition, CIV also contributes to the mitochondrial free radical species pool by catalyzing the production of nitric oxide (NO). Superoxide radicals can react with NO and produce peroxynitrate (24,25).
Free radicals participate in intracellular signaling in a variety of metabolic processes but during pathological conditions when they are produced in excess, ROS and RNS are the cause of oxidative stress. Since free radicals can cause extensive damage in the structure/function of proteins, nucleic acids and membrane lipids, they are considered the culprit of degenerative conditions, although more concrete evidence of their effects in vivo and their role in mitochondrial diseases is needed.
Our RISP cKO mice showed extensive oxidative stress already detectable from early age preceding neuronal death. In contrast, in the COX10 cKO mice, detection of oxidative damage coincided with neuronal death. In particular, the piriform cortex was the main region affected in the RISP cKO and to a less extent in the COX10 cKO. The oxidative stress detected included damage to nucleic acids, lipids and proteins as shown by 8-hydroxyguanosine staining and nitro-tyrosine and HNE adducts western blots (Figs 4F and 8).
RISP is thought to be required for the production of ROS as RISP knockdown experiments showed the inability of cells to produce the ROS required for hypoxic signaling (26,27). However, RISP KO mouse lung fibroblasts showed increased levels of superoxide, presumably originating from sources other than CIII (28).
In the COX10 cKO mice, we observed a delayed onset of oxidative stress correlating with the first signs of neurodegeneration when compared with the RISP cKO. However, our previous studies showed that the neuronal ablation of COX10 reduced the oxidative stress and concomitantly the number of plaques caused by the expression of mutant β-amyliod precursor protein and presenilin-1 in an Alzheimer's mouse model (14). A similar phenotype was observed in the neuron-specific Tfam (mitochondrial transcription factor) cKO mouse. Neuronal Tfam cKO developed a late-onset encephalopathy; however, there was no evidence of oxidative stress even at the latest stages of disease (29). Other mitochondrial encephalopathy mouse model involving a defect in CI by disruption of the NDUFS4 gene in both neurons and glia (nestin promoter-driven deletion) was associated with increased protein oxidation in the olfactory bulb of the KO mice (30).
We also observed a specific vulnerability of different brain regions to specific defects in the respiratory chain. The piriform cortex was the main region affected in the CIII defect whereas retrosplineal/cingulate cortex and, to a less extent, piriform cortex were more affected by the CIV deficiency. This also has been observed in other models of OXPHOS deficiency. For example, in the Tfam cKO (deletion also driven by the CamKIIα promoter), a massive cell loss was found in cingulated cortex and hippocampus (29). In the NDUFS4 KO, spongiform vacuolization was observed in the brainstem (vestibular nuclei), inferior olive, cerebellar vermis and olfactory bulb (30). In an mtDNA depletion model caused by expression of mitochondria-targeted endonucleases in CamKIIα neurons, the striatum was severely affected when compared with cortex and hippocampus (31).
Such differences in regional vulnerability are common in neurodegenerative diseases associated with defects in the mitochondrial electron transport chain. Interestingly, as observed in the COX10 cKO mice, Alzheimer's disease (AD) patients and AD mice models show metabolic abnormalities in the posterior cingulated cortex. These metabolic abnormalities in AD patients as well as in young adults at high risk of developing late-onset AD (apolipoprotein E ɛ4 carriers) are related to decreased glucose metabolism and decreased CIV activity in this brain region (32,33). In the case of Huntington's disease (HD), striatal GABAergic neurons are particularly affected as disease progresses. The vulnerability of this neuronal population in HD has been attributed to alterations in the mitochondrial calcium homeostasis and more recently to impairments on mitochondrial trafficking (34). Preferential vulnerability of the piriform cortex, as observed in the RISP cKO mouse, has been described in animal models of temporal lobe epilepsy (35,36). Interestingly, the structural damage to piriform cortex and other limbic structures (hippocampus, amigdaloid nuclei and entorhinal cortex) during drug-induced status epilepticus is preceded by metabolic activation (37). Moreover, the excitotoxicity of kainic acid (an epileptogenic drug) is associated with increased oxidative damage and does not correlate with decreased antioxidant defenses in affected brain regions (38,39). It is thought that the increased oxidative damage is caused by a marked increase in the levels of intracellular calcium during overstimulation of receptors and perhaps reduced mitochondrial calcium buffering capacity (40).
Another interesting aspect concerning the mitochondrial diseases is a phenomenon observed in many mitochondrial myopathies, where increased mitochondrial biogenesis takes place to compensate for the respiratory defect. No clear documentation of this phenomenon has been reported in mitochondrial encephalopathies. However, brain tissue appears to have the capability to increase mitochondrial biogenesis as it has been recently shown in a Huntington's mouse model by inducing the expression of PGC-1α, the master regulator of mitochondrial biogenesis, with bezafibrate treatment (41). Our previous studies showed mitochondrial proliferation as a compensatory mechanism in our muscle and liver COX10 cKO models; however this did not occur in the neuron-specific COX10 cKO, at least at the ages tested (1–4 months of age). In contrast, increase in the mtDNA/nDNA ratio, CS and CIV activities and a slight increase in steady-state levels of mitochondrial proteins were observed in the RISP cKO mouse, suggesting that increased mitochondrial biogenesis can take place in neurons, and could be a potential compensatory mechanism.
The RISP and the COX10 cKO mice characterized in this study illustrate how different OXPHOS defects can have different pathogenic mechanisms. These well-characterized models will be instrumental in the development of new therapeutic approaches.
MATERIALS AND METHODS
Generation of RISP and COX10 cKO mice
Conditional RISP KO mice were generated by crossing heterozygous floxed UQCRFS1 (12) with a transgenic mouse expressing Cre recombinase under the CamKIIα promoter kindly supplied by Dr Zeitling (University of Virginia) (13). The COX10 conditional KO was generated by crossing the homozygous floxed mice (19) with the CamKIIα transgenic mouse.
All experiments were conducted according to protocols approved by the University of Miami Institutional Animal Care and Use Committee. The mice colony was maintained in a virus antigen-free facility at the Division of Veterinary Resources at the University of Miami in controlled 12 h light/dark cycle at room temperature. Food (standard rodent diet) and water were administered ad libitum.
RISP cKO mice had to be maintained in a mixed genetic background (129svj and C57Bl6) since mice backcrossed to pure C57Bl6 genetic background were sterile. Homozygous floxed UQCRFS1 were incapable of producing offspring. In the case of COX10 cKO, the colony is maintained in C57Bl6 background. We did not observe obvious phenotypic differences in COX10 animals of mixed compared with pure background. Crosses to obtain cKO mice were performed using female animals expressing Cre and not males, since there is a CamKIIα-driven Cre expression in testis/sperm tissue. Both cKO mice were born in the expected Mendelian ratios. As control animals for RISP and COX10 cKO, we used littermate mice (same genetic background as the cKO) with the RISPflox/flox or COX10flox/flox genotype. When the previous genotypes were not obtained, we also used RISPflox/wt;Cre+ or COX10flox/wt;Cre+, respectively, as controls since there was no difference in the values of the enzymatic activity of either CIII or CIV when compared with the values obtained for the respective homozygous floxed mice.
Determination of enzyme activities
Homogenates of different brain regions (cortex and hippocampus) were prepared in PBS containing complete protease inhibitors cocktail (Roche Diagnostics) in a volume of ∼10× the weight. Homogenates were prepared in microcentrifuge tubes using a motor-driven pestle and tissue was disrupted by 10–15 strokes. Homogenates were centrifuged at 100× g for 5 min and supernatants were used for enzymatic assays. CIII or ubiquinol cytochrome c reductase, CIV or cytochrome c oxidase and CS activities were measured spectrophotometrically as described previously (42). Protein concentrations were determined using the Bio-Rad Bradford Assay Kit and BSA as standard. Specific activity was determined and values represented as percentage of control values performed simultaneously.
Western blot analysis
Proteins were separated by SDS–PAGE in 4–20% acrylamide gels and transferred to PVDF membranes. Membranes were blocked with 5% non-fat milk in 0.1% Tween 20 in PBS (PBST) and subsequently incubated with specific antibodies. Horseradish peroxidase-conjugated secondary antibodies were used and the signal was developed by chemiluminescence using the Superwest signal reagent (Pierce) or RapidStep (EDM). Antibodies against RISP, UQCRC2, Cox1, Cox4, NDUFA9, SDHA and VDAC1 were obtained from Mitosciences; Actin from Sigma; Tim23 from BD Biosciences; GFAP from Cell Signaling; Tuj1, N-tyrosine and 4-HNE from Abcam; SOD1 from Research Diagnostics and SOD2 from Upstate. All antibodies were used in a 1/1000 dilution (in 0.15% milk in PBST) except for N-tyrosine used at 1/400 and 4-HNE at 1/80 in blocking buffer (5% milk in PBST).
Rotarod analysis
Mice motor coordination was tested at different ages using a Rotarod (IITC 755, IITC Inc., CA, USA) set at a ramp speed of 6–20 rpm over 180 s. The test consisted of three trials performed for each animal at the corresponding age and the latency to fall was recorded. Animals were trained in the rotarod two times of three trials each about 2 weeks prior to the first test.
Activity cage analysis
Spontaneous ambulatory movement of mice was recorded using the Opto-M3 activity meter (Columbus Instruments) equipped with infrared beams along the cage. Movement was counted as the number of times the infrared beams were disrupted. Mice were housed individually in a new cage 30 min prior to their daily dark cycle and ambulatory counts were recorded for a period of 12 h (7 pm to 7 am).
Quantitative PCR of genomic DNA
Genomic DNA was extracted from cortex and hippocampus using standard proteinase K, phenol, chloroform extraction and isopropyl alcohol precipitation. The ratio of mitochondrial to nuclear DNA was determined by quantitative real-time PCR using 10 ng of genomic DNA in a 20 µl reaction mixture using SsoFast EvaGreen Supermix (Biorad) following PCR conditions stipulated by the manufacturer in a CFX96 Real Time PCR system (Biorad). Primers for the mtDNA were ND1-F: 5′-CAG CCT GAC CCA TAG CCA TA-3′ and ND1-B: 5′-ATT CTC CTT CTG TCA GGT CGA A-3′ and for the genomic DNA β-actin-F: 5′-GCG CAA GTA CTC TGT GTG GA-3′ and β-actin-B: 5′-CAT CGT ACT CCT GCT TGC TG-3′. DNA amounts were quantified using the delta–delta Ct method and expressed as a ratio of ND1/Actin.
To quantify the deletion of the floxed RISP and COX10 alleles in knockout animals, 20 ng of genomic DNA was amplified using corresponding primers and PCR conditions mentioned above. Standard curves were obtained using genomic DNA from RISPflox/flox, RISPflox/wt, RISPflox/wt;Cre+, RISPwt/wt and COX10flox/flox, COX10flox/wt, COX10flox/wt;Cre+ and COX10wt/wt, respectively. Values were normalized to an unrelated gene (EGF) as described previously (43). The forward primer used to amplify the RISP and COX10 floxed alleles was common to both genes since its target sequence comprises part of the loxP site. The common forward primer sequence is 5′-CGG GGA TCA ATT CGA GCT CGG CC-3′. The reverse primers were specific for each gene. The sequence for the reverse primers is RISP-B: 5′-AAC TTC CTA CAT GGT TGT TTC AAT T-3′ and COX10-B: 5′-CAC TGA CGC AGC GCC AGC ATC TT-3′, respectively. Primer sets produced an amplicon of 191 and 167 bp for RISP and COX10, respectively. The percentage of deletion was calculated by assuming that recombination occurs in both floxed alleles in cre-expressing neurons (43).
Immunohistochemistry
Frozen brains sections were obtained in a cryostat and processed for immunohistochemistry using NeuN (Chemicon), 8-hydroxy guanosine (QED Bioscience), RISP (Mitosciences) and Cox1 (Alexa-conjugated, Molecular Probes) antibodies. Either fluorescent-labeled or HRP-labeled secondary antibodies were used for immunodetection using standard immunohistochemical techniques. To detect cell death, the ‘in situ cell death detection kit—TMR red’ from Roche was used following manufacturer's instructions.
Cytochrome c oxidase or COX activity stain was performed in cryostat sections (∼20 µm) as described in Fukui et al. (14). Briefly, sections were incubated in 100 mm sodium phosphate buffer pH 7.4 containing 0.5 mg/ml diaminobenzidine, 0.2 mg/ml cytochrome c and 40 mg/ml sucrose at 37°C for 50 min. Succinate dehydrogenase or SDH activity stain was performed as described in Diaz et al. (19) by incubating sections in 10 mm sodium phosphate buffer pH 7.5 containing 1.6 mg/ml EDTA, 0.65 mg/ml KCN, 0.06 mg/ml phenazine methosulfate, 1.3 mg/ml succinic acid and 1.2 mg/ml nitroblue tetrazolium at 37°C for 20 min. For double COX/SDH stain, sections were first stained with COX reagents, washed three times in PBS and then incubated with SDH reagents.
In vivo MRI and proton MRS
MRI and MRS studies were conducted in the University of Miami High Field MRI Biomedical Research Core Facility equipped with a 4.7 T horizontal bore Bruker magnet designed for small animal experimentation. Mice were anesthetized by spontaneous breathing of 1.3–1.6% isofluorane and oxygen mixture through a nose cone using an Ohmeda veterinary anesthesia unit. The mice were placed in an MRI-compatible cradle, their body temperature was maintained at 37°C by a water bath circulation system and their respiratory rate was monitored by a pneumatic pillow and an accompanying pressure transducer. Both temperature and breathing were recorded by a small animal monitoring system (SA Instruments Inc.). Anesthesia was adjusted to obtain a respiratory rate of about 90 breaths per minute. Following localizer scans, high-resolution axial and coronal T2-weighted datasets were acquired in order to visualize any cerebral or structural atrophy and to investigate for potential focal pathologies (44). High-resolution T2 axial datasets were manually segmented for whole-brain volume for comparisons between groups and time points. Water-suppressed point-resolved spectroscopy was acquired in order to glean the metabolic status from the mouse brain (45). The MRI dataset was collected with the following settings: a voxel size of 4 × 4 × 2 mm, a bandwidth of 3 kHz, 4096 acquisition points, a TR period of 2.5 s, a TE period of 144 ms and 256 averages. To optimize the quality of the MRS data, the magnetic field was shimmed to a line width of <20 Hz measured at full-width at half-maximum on the unsuppressed water signal. The voxel was centered in the cortex and hippocampus regions. Spectral data were processed using software developed by JMRUI (Java MR User Interface: http://www.mrui.uab.es/mrui/mrui_Overview.shtml). To obtain estimates of the lactate levels in vivo, a long TE time of 144 ms was employed to differentiate the lactate from lipid signal contamination and is shown as an inverted peak in the spectra (46).
SUPPLEMENTARY MATERIAL
FUNDING
This research was supported by the following grants: The James and Esther King Research Program, Florida Department of Health (grant number 08KN-01 to F.D.); National Institute of Health (grant numbers GM101225 to F.D., PHS NS041777, NS079965, EY010804 and AG036871 to C.T.M.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Milena Pinto for critical discussion of the manuscript and Aline Hida for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.DiMauro S., Schon E.A. Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. doi:10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- 2.Torraco A., Diaz F., Vempati U.D., Moraes C.T. Mouse models of oxidative phosphorylation defects: powerful tools to study the pathobiology of mitochondrial diseases. Biochim. Biophys. Acta. 2009;1793:171–180. doi: 10.1016/j.bbamcr.2008.06.003. doi:10.1016/j.bbamcr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerr D.S. Treatment of mitochondrial electron transport chain disorders: a review of clinical trials over the past decade. Mol. Genet. Metab. 2010;99:246–255. doi: 10.1016/j.ymgme.2009.11.005. doi:10.1016/j.ymgme.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Wenz T., Williams S.L., Bacman S.R., Moraes C.T. Emerging therapeutic approaches to mitochondrial diseases. Dev. Disabil. Res. Rev. 2010;16:219–229. doi: 10.1002/ddrr.109. doi:10.1002/ddrr.109. [DOI] [PubMed] [Google Scholar]
- 5.Benit P., Lebon S., Rustin P. Respiratory-chain diseases related to complex III deficiency. Biochim. Biophys. Acta. 2009;1793:181–185. doi: 10.1016/j.bbamcr.2008.06.004. doi:10.1016/j.bbamcr.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Johns D.R., Neufeld M.J. Cytochrome b mutations in Leber hereditary optic neuropathy. Biochem. Biophys. Res. Commun. 1991;181:1358–1364. doi: 10.1016/0006-291x(91)92088-2. doi:10.1016/0006-291X(91)92088-2. [DOI] [PubMed] [Google Scholar]
- 7.Keightley J.A., Anitori R., Burton M.D., Quan F., Buist N.R., Kennaway N.G. Mitochondrial encephalomyopathy and complex III deficiency associated with a stop-codon mutation in the cytochrome b gene. Am. J. Hum. Genet. 2000;67:1400–1410. doi: 10.1086/316900. doi:10.1086/316900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haut S., Brivet M., Touati G., Rustin P., Lebon S., Garcia-Cazorla A., Saudubray J.M., Boutron A., Legrand A., Slama A. A deletion in the human QP-C gene causes a complex III deficiency resulting in hypoglycaemia and lactic acidosis. Hum. Genet. 2003;113: 118–122. doi: 10.1007/s00439-003-0946-0. [DOI] [PubMed] [Google Scholar]
- 9.Diaz F., Kotarsky H., Fellman V., Moraes C.T. Mitochondrial disorders caused by mutations in respiratory chain assembly factors. Semin. Fetal Neonatal Med. 2011;16:197–204. doi: 10.1016/j.siny.2011.05.004. doi:10.1016/j.siny.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrientos A., Gouget K., Horn D., Soto I.C., Fontanesi F. Suppression mechanisms of COX assembly defects in yeast and human: insights into the COX assembly process. Biochim. Biophys. Acta. 2009;1793:97–107. doi: 10.1016/j.bbamcr.2008.05.003. doi:10.1016/j.bbamcr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 12.Garcia S., Diaz F., Moraes C.T. A 3′ UTR modification of the mitochondrial Rieske iron sulfur protein in mice produces a specific skin pigmentation phenotype. J. Invest. Dermatol. 2008;128:2343–2345. doi: 10.1038/jid.2008.64. doi:10.1038/jid.2008.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dragatsis I., Zeitlin S. CaMKIIalpha-Cre transgene expression and recombination patterns in the mouse brain. Genesis. 2000;26:133–135. doi: 10.1002/(sici)1526-968x(200002)26:2<133::aid-gene10>3.0.co;2-v. doi:10.1002/(SICI)1526-968X(200002)26:2<133::AID-GENE10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 14.Fukui H., Diaz F., Garcia S., Moraes C.T. Cytochrome c oxidase deficiency in neurons decreases both oxidative stress and amyloid formation in a mouse model of Alzheimer's disease. Proc. Natl Acad. Sci. USA. 2007;104:14163–14168. doi: 10.1073/pnas.0705738104. doi:10.1073/pnas.0705738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffett J.R., Ross B., Arun P., Madhavarao C.N., Namboodiri A.M. N-Acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog. Neurobiol. 2007;81:89–131. doi: 10.1016/j.pneurobio.2006.12.003. doi:10.1016/j.pneurobio.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonicka H., Leary S.C., Guercin G.H., Agar J.N., Horvath R., Kennaway N.G., Harding C.O., Jaksch M., Shoubridge E.A. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 2003;12:2693–2702. doi: 10.1093/hmg/ddg284. doi:10.1093/hmg/ddg284. [DOI] [PubMed] [Google Scholar]
- 17.Coenen M.J., van den Heuvel L.P., Ugalde C., Ten Brinke M., Nijtmans L.G., Trijbels F.J., Beblo S., Maier E.M., Muntau A.C., Smeitink J.A. Cytochrome c oxidase biogenesis in a patient with a mutation in COX10 gene. Ann. Neurol. 2004;56:560–564. doi: 10.1002/ana.20229. doi:10.1002/ana.20229. [DOI] [PubMed] [Google Scholar]
- 18.Valnot I., von Kleist-Retzow J.C., Barrientos A., Gorbatyuk M., Taanman J.W., Mehaye B., Rustin P., Tzagoloff A., Munnich A., Rotig A. A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:1245–1249. doi: 10.1093/hmg/9.8.1245. doi:10.1093/hmg/9.8.1245. [DOI] [PubMed] [Google Scholar]
- 19.Diaz F., Thomas C.K., Garcia S., Hernandez D., Moraes C.T. Mice lacking COX10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum. Mol. Genet. 2005;14:2737–2748. doi: 10.1093/hmg/ddi307. doi:10.1093/hmg/ddi307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baracca A., Sgarbi G., Mattiazzi M., Casalena G., Pagnotta E., Valentino M.L., Moggio M., Lenaz G., Carelli V., Solaini G. Biochemical phenotypes associated with the mitochondrial ATP6 gene mutations at nt8993. Biochim. Biophys. Acta. 2007;1767:913–919. doi: 10.1016/j.bbabio.2007.05.005. doi:10.1016/j.bbabio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Barrientos A., Moraes C.T. Titrating the effects of mitochondrial complex I impairment in the cell physiology. J. Biol. Chem. 1999;274:16188–16197. doi: 10.1074/jbc.274.23.16188. doi:10.1074/jbc.274.23.16188. [DOI] [PubMed] [Google Scholar]
- 22.Quinzii C.M., Lopez L.C., Gilkerson R.W., Dorado B., Coku J., Naini A.B., Lagier-Tourenne C., Schuelke M., Salviati L., Carrozzo R., et al. Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. FASEB J. 2010;24:3733–3743. doi: 10.1096/fj.09-152728. doi:10.1096/fj.09-152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand M.D. The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. doi:10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. doi:10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poyton R.O., Ball K.A., Castello P.R. Mitochondrial generation of free radicals and hypoxic signaling. Trends Endocrinol. Metab. 2009;20:332–340. doi: 10.1016/j.tem.2009.04.001. doi:10.1016/j.tem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Bell E.L., Klimova T.A., Eisenbart J., Moraes C.T., Murphy M.P., Budinger G.R., Chandel N.S. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J. Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. doi:10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandel N.S. Mitochondrial complex III: An essential component of universal oxygen sensing machinery? Respir. Physiol. Neurobiol. 2010 doi: 10.1016/j.resp.2010.08.004. 174, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz F., Enriquez J.A., Moraes C.T. Cells lacking Rieske iron–sulfur protein have a reactive oxygen species-associated decrease in respiratory complexes I and IV. Mol. Cell. Biol. 2012;32:415–429. doi: 10.1128/MCB.06051-11. doi:10.1128/MCB.06051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorensen L., Ekstrand M., Silva J.P., Lindqvist E., Xu B., Rustin P., Olson L., Larsson N.G. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J. Neurosci. 2001;21:8082–8090. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana A., Kruse S.E., Kapur R.P., Sanz E., Palmiter R.D. Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc. Natl Acad. Sci. USA. 2010;107:10996–11001. doi: 10.1073/pnas.1006214107. doi:10.1073/pnas.1006214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickrell A.M., Fukui H., Wang X., Pinto M., Moraes C.T. The striatum is highly susceptible to mitochondrial oxidative phosphorylation dysfunctions. J. Neurosci. 2011;31:9895–9904. doi: 10.1523/JNEUROSCI.6223-10.2011. doi:10.1523/JNEUROSCI.6223-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valla J., Berndt J.D., Gonzalez-Lima F. Energy hypometabolism in posterior cingulate cortex of Alzheimer's patients: superficial laminar cytochrome oxidase associated with disease duration. J. Neurosci. 2001;21:4923–4930. doi: 10.1523/JNEUROSCI.21-13-04923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valla J., Yaari R., Wolf A.B., Kusne Y., Beach T.G., Roher A.E., Corneveaux J.J., Huentelman M.J., Caselli R.J., Reiman E.M. Reduced posterior cingulate mitochondrial activity in expired young adult carriers of the APOE epsilon4 allele, the major late-onset Alzheimer's susceptibility gene. J. Alzheimer's Dis. JAD. 2010;22:307–313. doi: 10.3233/JAD-2010-100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira J.M. Nature and cause of mitochondrial dysfunction in Huntington's disease: focusing on huntingtin and the striatum. J. Neurochem. 2010;114:1–12. doi: 10.1111/j.1471-4159.2010.06741.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen S., Kobayashi M., Honda Y., Kakuta S., Sato F., Kishi K. Preferential neuron loss in the rat piriform cortex following pilocarpine-induced status epilepticus. Epilepsy Res. 2007;74:1–18. doi: 10.1016/j.eplepsyres.2006.11.008. doi:10.1016/j.eplepsyres.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Druga R., Kubova H., Suchomelova L., Haugvicova R. Lithium/pilocarpine status epilepticus-induced neuropathology of piriform cortex and adjoining structures in rats is age-dependent. Physiol. Res. 2003;52:251–264. [PubMed] [Google Scholar]
- 37.Dube C., Boyet S., Marescaux C., Nehlig A. Progressive metabolic changes underlying the chronic reorganization of brain circuits during the silent phase of the lithium-pilocarpine model of epilepsy in the immature and adult rat. Exp. Neurol. 2000;162:146–157. doi: 10.1006/exnr.2000.7324. doi:10.1006/exnr.2000.7324. [DOI] [PubMed] [Google Scholar]
- 38.Candelario-Jalil E., Al-Dalain S.M., Castillo R., Martinez G., Fernandez O.S. Selective vulnerability to kainate-induced oxidative damage in different rat brain regions. J. Appl. Toxicol. 2001;21:403–407. doi: 10.1002/jat.768. doi:10.1002/jat.768. [DOI] [PubMed] [Google Scholar]
- 39.Cheng Y., Sun A.Y. Oxidative mechanisms involved in kainate-induced cytotoxicity in cortical neurons. Neurochem. Res. 1994;19:1557–1564. doi: 10.1007/BF00969006. doi:10.1007/BF00969006. [DOI] [PubMed] [Google Scholar]
- 40.Carriedo S.G., Yin H.Z., Sensi S.L., Weiss J.H. Rapid Ca2+ entry through Ca2+-permeable AMPA/Kainate channels triggers marked intracellular Ca2+ rises and consequent oxygen radical production. J. Neurosci. 1998;18:7727–7738. doi: 10.1523/JNEUROSCI.18-19-07727.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johri A., Calingasan N.Y., Hennessey T.M., Sharma A., Yang L., Wille E., Chandra A., Beal M.F. Pharmacologic activation of mitochondrial biogenesis exerts widespread beneficial effects in a transgenic mouse model of Huntington's disease. Hum. Mol. Genet. 2012;21:1124–1137. doi: 10.1093/hmg/ddr541. doi:10.1093/hmg/ddr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrientos A., Fontanesi F., Diaz F. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr. Protoc. Hum. Genet. 2009;19 doi: 10.1002/0471142905.hg1903s63. Unit19 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Funfschilling U., Supplie L.M., Mahad D., Boretius S., Saab A.S., Edgar J., Brinkmann B.G., Kassmann C.M., Tzvetanova I.D., Mobius W., et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature. 2012;485:517–521. doi: 10.1038/nature11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colgan N.C., Cronin M.M., Gobbo O.L., O'Mara S.M., O'Connor W.T., Gilchrist M.D. Quantitative MRI analysis of brain volume changes due to controlled cortical impact. J. Neurotrauma. 2010;27:1265–1274. doi: 10.1089/neu.2009.1267. doi:10.1089/neu.2009.1267. [DOI] [PubMed] [Google Scholar]
- 45.Bottomley P.A. Spatial localization in NMR spectroscopy in vivo. Ann. NY Acad. Sci. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. doi:10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 46.Star-Lack J., Spielman D., Adalsteinsson E., Kurhanewicz J., Terris D.J., Vigneron D.B. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5 T using PRESS excitation with applications to the study of brain and head and neck tumors. J. Magn. Reson. 1998;133:243–254. doi: 10.1006/jmre.1998.1458. doi:10.1006/jmre.1998.1458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.