Abstract
Glioblastoma (GBM) is a highly heterogeneous type of tumor characterized by genomic and signaling abnormalities affecting pathways involved in control of cell fate, including tumor-suppressor- and growth factor-regulated pathways. An aberrant miRNA expression has been observed in GBM, being associated with impaired cellular functions resulting in malignant transformation, proliferation and invasion. Here, we demonstrate for the first time that platelet-derived growth factor-B (PDGF-B), a potent angiogenic growth factor involved in GBM development and progression, promotes downregulation of pro-oncogenic (miR-21) and anti-oncogenic (miR-128) miRNAs, as well as upregulation/downregulation of several miRNAs involved in GBM pathology. Retrovirally mediated overexpression of PDGF-B in U87 human GBM cells or their prolonged exposure, as well as that of F98 rat glioma cells to this ligand, resulted in decreased miR-21 and miR-128 levels, which was associated with increased cell proliferation. Furthermore, siRNA-mediated PDGF-B silencing led to increased levels of miR-21 and miR-128, while miRNA modulation through overexpression of miR-21 did not alter the levels of PDGF-B. Finally, we demonstrate that modulation of tumor suppressors PTEN and p53 in U87 cells does not affect the decrease in miR-21 levels associated with PDGF-B overexpression. Overall, our findings suggest that, besides its role in inducing GBM tumorigenesis, PDGF-B may enhance tumor proliferation by modulating the expression of oncomiRs and tumor suppressor miRNAs in U87 human GBM cells.
INTRODUCTION
The human glioblastoma (GBM) represents the most common and lethal type of glioma (1). Despite recent improvement in imaging and surgical techniques, allowing more accurate diagnosis and treatment, current therapeutic options for GBM lack effective long-term impact on disease control and patient survival, and clinical recurrence is nearly universal (2,3). This clearly emphasizes the need for new and effective therapeutic strategies, as well as a better understanding of the molecular and cellular alterations that occur in GBM.
The discovery of miRNAs, a new class of small noncoding RNAs that regulate gene expression, has revealed an additional level of fine tuning of the genome. MiRNAs have been found to regulate post-transcriptionally the expression of over 30% of protein-coding genes (4) through imperfect pairing with the target mRNAs (5,6), and bioinformatic data indicate that each miRNA can control hundreds of gene targets, underscoring the potential influence of miRNAs in almost every genetic pathway (4,7). Accumulated evidence has shown that miRNA dysregulation is associated with cancer development and progression (8–10), including GBM pathogenesis (11–13). Studies by Holland and colleagues showed that miR-26a is amplified in high-grade gliomas and facilitates gliomagenesis in vivo (14). Godlewski et al. demonstrated that expression of miR-128, a highly downregulated miRNA in GBM, reduced significantly glioma cell proliferation in vitro and glioma xenograft growth in vivo (15). Nevertheless, the causes of the widespread disruption of miRNA expression in cancer cells are not completely understood and, most probably, various abnormalities in each tumor could contribute to the global miRNA-expression profile (16).
Relevant molecular alterations that govern GBM progression have already been identified, including mutation/deletion of p53 and PTEN and amplification/overexpression of the epidermal growth factor receptor (EGFR) (17,18). The platelet-derived growth factor (PDGF), a vast family of angiogenic growth factors, has also been proposed to play a role in GBM development and progression. Alterations in PDGF signaling, including overexpression of PDGF-A and B ligands or their receptors (PDGFR-α and -β), are commonly observed in high-grade gliomas (19–21); in vitro studies have also shown that PDGF directly stimulates the migration and proliferation of glial progenitors (22,23). Moreover, retrovirally mediated expression of PDGF-B in adult white matter, subventricular zone and brainstem progenitors induces tumors that closely resemble human GBM (24–27), thus emphasizing the importance of PDGF signaling in brain tumors.
Emerging evidence suggested that PDGF signaling modulates miRNAs in several biological processes. Davis et al. demonstrated that miR-221 is transcriptionally induced upon PDGF treatment in primary vascular smooth muscle cells (vSMC), leading to downregulation of the targets c-Kit and p27Kip1 and consequent induction of cell proliferation (28). Recent studies identified a group of miRNAs whose expression is altered shortly after PDGF stimulation and revealed that the EGFR expression and function are repressed by PDGF-induced miR-146b (29). Here, we demonstrate that PDGF-B overexpression modulates miRNA expression in human U87 GBM cells. Prolonged exposure of human and rat GBM cells to PDGF-B promotes miR-21 and miR-128 downregulation, this effect being specific for this ligand (PDGF-B), as concluded from the observation of increased levels of these miRNAs upon siRNA-mediated PDGF-B silencing. On the other hand, transient miR-21 overexpression does not significantly affect PDGF-B mRNA levels. Moreover, we demonstrate that PDGF-B-related miR-21 downregulation is not affected by the modulation of tumor suppressors PTEN and p53.
RESULTS
MiR-21 and miR-221 are significantly downregulated in U87 cells overexpressing PDGF-B
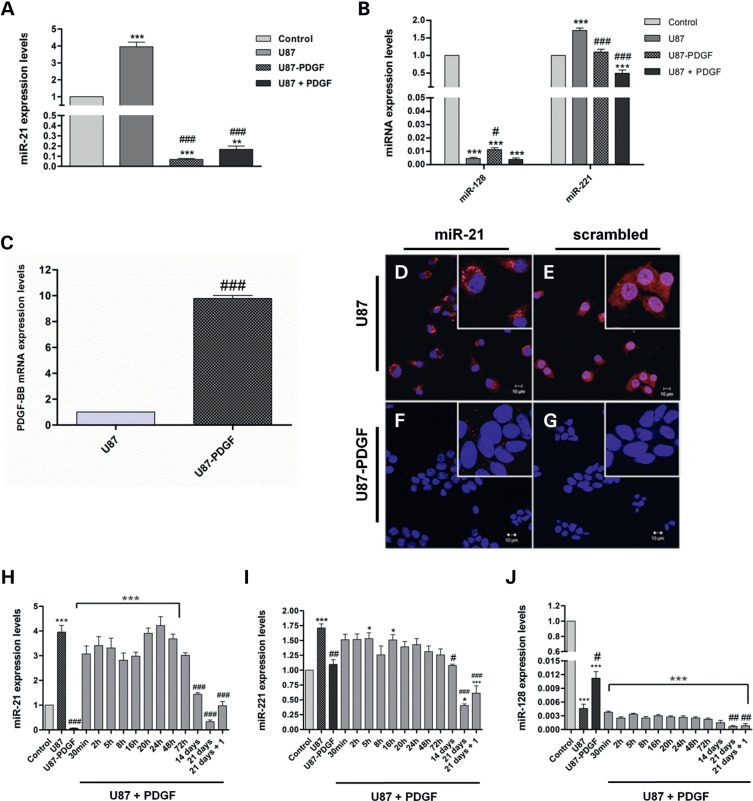
We have recently demonstrated that the pro-oncogenic miR-21 is overexpressed and the anti-oncogenic miR-128 is highly downregulated in several human GBM, which was corroborated by the analysis of miRNA-expression data in ∼200 human GBM samples from The Cancer Genome Atlas (30) (P. Costa, A. Cardoso, L. Pereira de Almeida, J. Bruce, P. Canoll, M. Pedroso de Lima, manuscript in preparation). Similarly, significant miR-21 overexpression and miR-128 downregulation were observed in the widely used U87 human GBM cell line (Figure 1A and B).
Figure 1.
PDGF-B-related modulation of miRNA-expression levels in U87 and U87-PDGF human GBM cells. (A) MiR-21 and (B) miR-128/miR-221 expression levels in parental U87 cells, PDGF-B-overexpressing U87 cells (U87-PDGF) and U87 cells cultured in the presence of 30 ng/ml PDGF-B for 60 days (U87 + PDGF), compared with control epileptic tissue. (C) PDGF-B mRNA levels in U87 and U87-PDGF. FISH staining in cultured (D and E) U87 and (F and G) U87-PDGF cells. Cells were stained with (D and F) miR-21 or (E and G) noncoding (scrambled) probes. Nuclear staining was accomplished using the cell-permeable DNA stain Hoescht 33342 (Blue). MiR-21 staining (red dots) is observed in U87 cells, predominantly in the cytoplasm, whereas only residual staining is detected in U87-PDGF cells. Control experiments targeting the endogenous U6snRNA (positive control) or without LNA probe (negative control) were performed in parallel (Supplementary Material, Figure S2D–G). Images were obtained by confocal microscopy with a 40× EC Plan-Neofluar. Scale corresponds to 10 µm. (H) MiR-21, (I) miR-221 and (J) miR-128 expression levels after culturing parental U87 cells in medium supplemented with PDGF-B (30 ng/ml) for different time periods; 21 days + 1: cells cultured for 21 days in PDGF-B-enriched medium followed by 7 days of culture in normal medium (PDGF-B-depleted medium). *P < 0.05, **P < 0.01, ***P < 0.001 compared with control epileptic tissue. #P < 0.05, ##P < 0.01, ###P < 0.001 compared with parental U87 cells.
Although a large number of in vitro and in vivo studies have demonstrated that PDGF-B is an important mediator in GBM development and progression (27,31,32), its influence on miRNA expression in tumor cells remains to be clarified. In order to address the potential involvement of PDGF-B in miRNA expression in GBM, we measured the expression levels of miR-128, miR-21 and miR-221 in retrovirally modified U87 cells overexpressing PDGF-B (U87-PDGF), which were compared with those in control U87 cells transduced with a noncoding retroviral vector (no PDGF-B), or control epileptic tissue. Our results from quantitative real-time PCR (qPCR) experiments revealed that PDGF-B mRNA levels are significantly higher in U87-PDGF cells (∼10-fold) than in parental U87 cells (Figure 1C, P < 0.001). Moreover, U87-PDGF cells displayed an altered morphology and increased proliferation rate, compared with parental U87 cells (Supplementary Material, Figure S1). Surprisingly, miR-21 was significantly downregulated in U87-PDGF cells compared with parental U87 cells (∼58-fold decrease, P < 0.001) or control epileptic tissue, as shown in Figure 1A. Similarly, a considerable reduction in miR-21 staining, as assessed by FISH, was observed in cultured U87-PDGF cells (Figure 1F) when compared with control U87 cells (Figure 1D), thus supporting our findings of miR-21 downregulation in U87 cells overexpressing PDGF-B. As shown in Supplementary Material, Figure S2A, no miR-21 staining was detected in control P19 cells, which express low levels of this miRNA. Our observation of miR-221 downregulation in U87-PDGF cells compared with parental U87 cells suggests that PDGF-B-related miRNA downregulation is not miR-21 specific (Figure 1B). On the other hand, the robust downregulation of miR-128 in parental U87 cells was slightly reduced in U87-PDGF cells (Figure 1B).
Culturing U87 human and F98 rat glioma cells in PDGF-B-enriched medium promotes downregulation of miR-21 and miR-128 expression levels
In order to mimic the autocrine production of PDGF-B by U87-PDGF cells, parental U87 cells were grown in medium supplemented with PDGF-B (30 ng/ml) for 60 days and miR-128, miR-21 and miR-221 levels were subsequently assessed. As shown in Figure 1, miR-21 (Figure 1A) and miR-221 (Figure 1B) levels were significantly reduced in U87 cells under these conditions, when compared with those observed when the cells were maintained in PDGF-B-depleted medium or control epileptic tissue (P < 0.001), whereas miR-128 levels were not considerably changed (Figure 1B). Furthermore, the decrease in miR-21 and miR-221 expression levels was clearly time-dependent, a significant reduction in the miR-21 and miR-221 levels being observed when culturing cells in PDGF-B for long periods of time (Figure 1H and I).
Interestingly, when PDGF-B-enriched medium was replaced by normal culture medium and cells were grown for 1 week, a slight recovery in miR-21 and miR-221 levels was observed (Figure 1H and I, P < 0.05); this effect was further enhanced by culturing U87 cells in normal medium for 30 days (not shown), suggesting that PDGF-B-related miRNA modulation may constitute a reversible and time-dependent mechanism affected by the presence of this ligand in the extracellular environment.
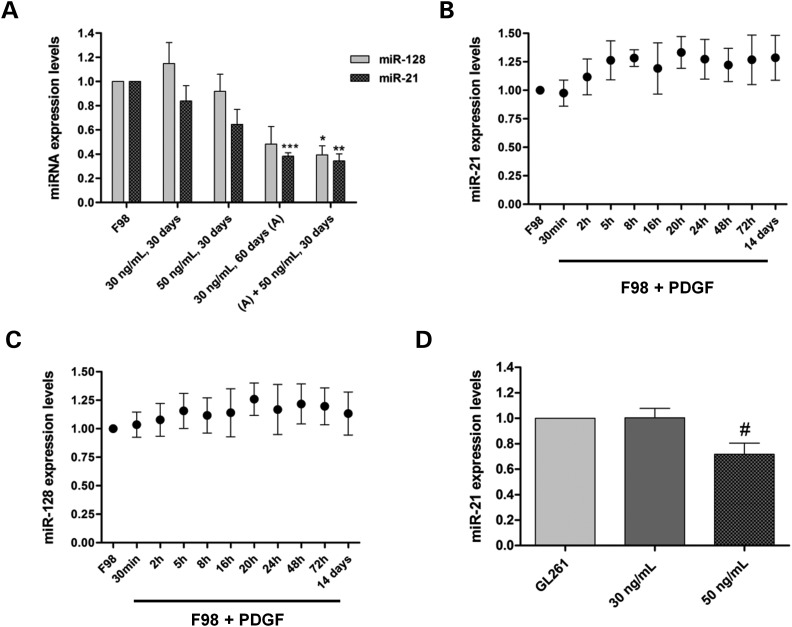
Based on our results on miRNA downregulation promoted by PDGF-B in human GBM cells, we tried to clarify whether this effect would be cell-specific or could also be observed in glioma cells from a different origin. For this purpose, F98 rat glioma cells were cultured in medium supplemented with PDGF-B (30 or 50 ng/ml) for different periods of time and subsequently assessed for the effect on miR-128 and miR-21 expression levels, when compared with untreated cells. As shown in Figure 2A, a considerable decrease in miR-21 (P < 0.001) and miR-128 levels (P > 0.05) was observed when the cells were cultured for 60 days in the medium containing 30 ng/ml PDGF-B, which was enhanced when the cells were further cultured for 30 days in 50 ng/ml PDGF-B. Similarly to what was observed for U87 cells, miR-21 and miR-128 expression levels did not change significantly upon culturing F98 cells in PDGF-B-enriched medium for short periods of time (Figure 2B and C). A significant decrease in miR-21 levels (P < 0.05) was also observed in GL261 mouse glioma cells cultured for 30 days in medium containing 50 ng/ml PDGF-B (Figure 2D), when compared with untreated cells.
Figure 2.
PDGF-B-related modulation of miRNA-expression levels in F98 rat and GL261 mouse glioma cells. (A) MiR-21 and miR-128 expression levels in F98 cells cultured in PDGF-B-depleted medium (control) or cultured in medium supplemented with PDGF-B (30 or 50 ng/ml) for different time periods; (A) +50 ng/ml, 30 days: cells cultured for 60 days in PDGF-B-enriched medium (30 ng/ml), followed by a further incubation for 30 days in medium containing a higher concentration of PDGF-B (50 ng/ml). (B) MiR-21 and (C) miR-128-expression levels in F98 cells cultured in medium supplemented with PDGF-B (30 ng/ml) for different time periods, compared with those obtained in F98 cells cultured in PDGF-B-depleted medium. 14 days: cells cultured for 14 days in PDGF-B-enriched medium. (D) MiR-21-expression levels in GL261 glioma cells cultured in PDGF-B-depleted medium (control) or cultured in medium supplemented with PDGF-B (30 or 50 ng/ml) for 30 days. *P < 0.05, **P < 0.01, ***P < 0.001 compared with control F98 cells; #P < 0.05 compared with control GL261 cells.
Overall, our results suggest that PDGF-B promotes a time-dependent downregulation of miRNAs in glioma cells.
SiRNA-mediated PDGF-B silencing increases miR-21 and miR-128 expression levels in U87-PDGF cells
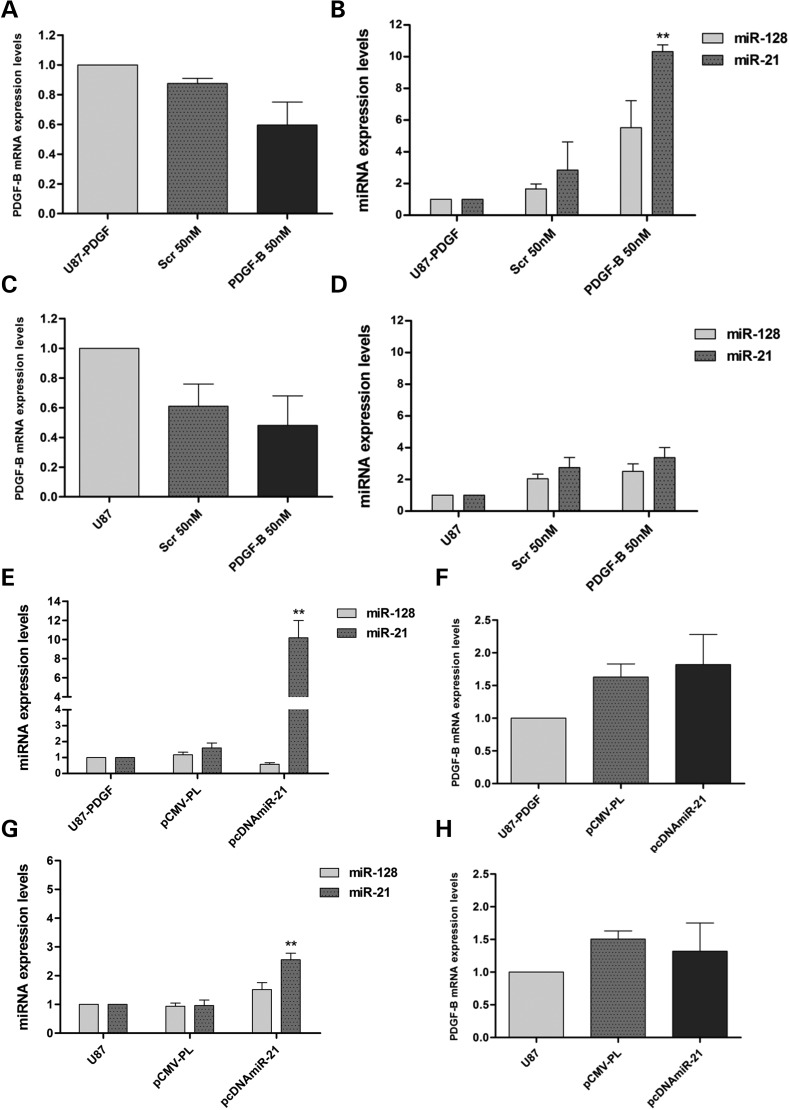
As our results demonstrated that PDGF-B promotes the downregulation of miRNAs in human and rat glioma cells, we evaluated the effect of modulating the autocrine production of PDGF-B in U87-PDGF cells, via siRNA-mediated PDGF-B mRNA silencing, on miR-128 and miR-21 expression levels. As shown in Figure 3A, cell transfection with a PDGF-B-specific siRNA sequence (50 nm) resulted in a pronounced, although not significant, decrease in PDGF-B mRNA levels, when compared with those obtained upon transfection with a noncoding siRNA sequence (∼28%, P > 0.05); no further decrease was observed when U87-PDGF cells were transfected with 100 nm siRNAs (data not shown). Nonetheless, miR-21 levels were significantly upregulated in U87-PDGF cells transfected with anti-PDGF-B siRNAs (10.33 ± 0.73), compared with those transfected with a noncoding sequence (1.606 ± 0.537, P < 0.01); although not significant, miR-128 upregulation was also observed under these conditions (Figure 3B). These results suggesting that PDGF-B mRNA knockdown in U87-PDGF cells induced the upregulation of miR-21 and miR-128 levels prompted us to test whether PDGF-B mRNA silencing in cells expressing lower levels of this growth factor would also promote such considerable miR-21 upregulation. As observed in Figure 3C, transfection of parental U87 cells with anti-PDGF-B siRNAs also resulted in PDGF-B mRNA downregulation, although no significant increase in miR-21 and miR-128 levels was observed, when compared with those obtained in cells transfected with a noncoding siRNA (Figure 3D).
Figure 3.
MiR-21- and miR-128 expression levels following siRNA-mediated PDGF-B mRNA silencing and plasmid-mediated miR-21 overexpression. For siRNA transfection, 24 h before any experiment U87 and U87-PDGF cells were plated on 12-well plates at a density of 7 × 104 and 6 × 104 cells/well, respectively and total RNA was extracted 24 h after transfection. PDGF-B mRNA-expression levels in (A) U87-PDGF and (C) parental U87 cells after transfection with 50 nm anti-PDGF-B siRNA (PDGF-B 50 nm) or a noncoding sequence (Scr 50 nm). MiR-21 and miR-128 expression levels in (B) U87-PDGF and (D) U87 cells after siRNA transfection. For plasmid transfection of U87 and U87-PDGF cells, 24 h before any experiment cells were plated onto 6-well plates at a density of 1 × 105 and 8 × 104 cells/well (respectively) and total RNA was extracted 48 h after transfection. MiR-21- and miR-128-expression levels in (E) U87-PDGF and (G) U87 cells after transfection with the human miR-21-coding plasmid pcDNA3.1-miR-21 (pcDNAmiR-21) or noncoding plasmid (pCMV-PL). PDGF-B mRNA levels in (F) U87-PDGF and (H) U87 cells after transfection. **P < 0.01, ***P < 0.001 compared with untreated U87-PDGF cells.
Plasmid-induced miR-21 upregulation in U87-PDGF cells does not significantly alter PDGF-B mRNA levels
Based on our findings of the modulating effect of PDGF-B on miRNA expression in GBM cells and since miRNAs regulate gene expression post-transcriptionally through imperfect pairing with the 3′ UTR of their target mRNAs (4,33), we sought to identify possible interactions between miR-21/miR-128 and the PDGF-B mRNA. Although bioinformatic analysis using mirWalk (34) and other tools that predict miRNA targets has not revealed any conserved sites at 3′ UTR of the human PDGF-B mRNA for either of these miRNAs (Supplementary Material, Table S1), we examined whether a transient upregulation of miR-21 in human GBM cells would have any effect on PDGF-B mRNA expression and cell proliferation. As shown in Figure 3E, transfection of U87-PDGF cells with the plasmid pcDNA3.1-miR-21 resulted in a significant increase in miR-21 levels (10.18 ± 3.12), when compared with those observed with the noncoding plasmid pCMV-PL (1.6 ± 0.54, P < 0.01). An increase in miR-21 levels was also obtained upon transfection of U87 cells (Figure 3G), although such increase was not so pronounced, as these cells express higher basal levels of miR-21. As observed, in both the cell lines, miR-21 upregulation did not affect significantly the levels of PDGF-B mRNA (Figure 3F and H). Moreover, no significant changes in viability were observed in U87-PDGF cells transfected with miR-21-coding pcDNA3.1-miR-21, when compared with that observed in cells transfected with control pCMV-PL, whereas a small, although significant, increase in viability was observed in U87 cells transfected with the same formulations (∼9%, P < 0.05) (Supplementary Material, Figure S3A).
PDGF-B overexpression promotes the upregulation and downregulation of several miRNAs in U87 GBM cells
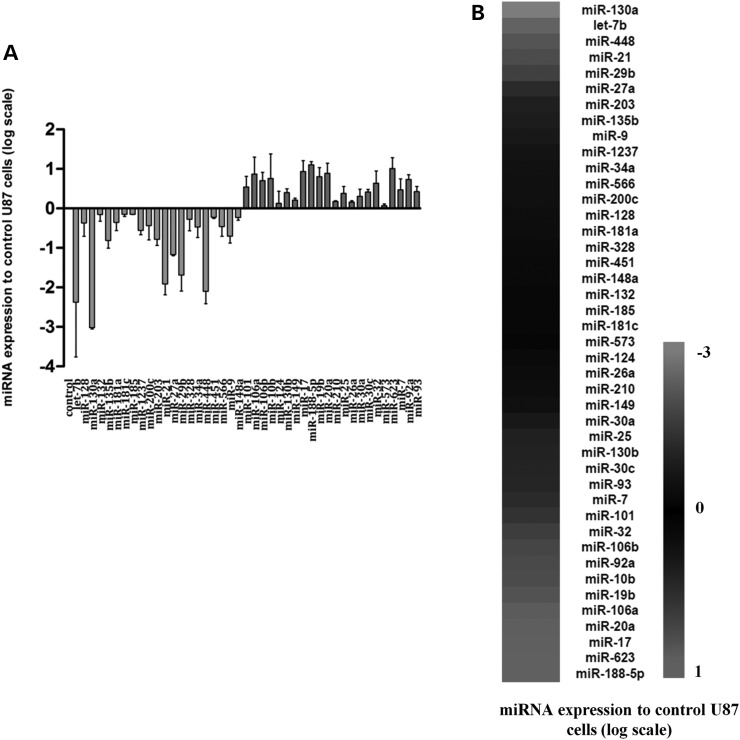
As our results demonstrated that PDGF-B-overexpression promotes ligand-specific downregulation of the pro-oncogenic miR-21 and the anti-oncogenic miR-128 in glioma cells, which was associated with increased cell proliferation (Supplementary Material, Fig. S1), we evaluated whether PDGF-B would modulate the expression of other miRNAs involved in GBM pathology that could contribute to a more tumorigenic phenotype. As shown in Figure 4 and Supplementary Material, Table S2, several miRNAs, including the anti-oncogenic let-7b, miR-130a and miR-29b, were found to be downregulated in U87-PDGF cells, when compared with parental U87 cells, whereas the pro-oncogenic miR-17, miR-20a, miR-10b and the anti-oncogenic miR-101 were upregulated under the same conditions. Furthermore, miR-188-5p and miR-623, were considerably overexpressed in U87-PDGF cells, compared with control U87 cells. Although no role in GBM has been reported for these miRNAs, increased expression of miR-188 has been associated with enhanced cellular proliferation in ovarian cancer (35).
Figure 4.
QPCR quantification of 44 miRNAs in U87-PDGF cells, using pre-designed miRNA PCR plates. (A) Column graphic and (B) heatmap representation of the relative miRNA-expression levels in PDGF-B-overexpressing U87 cells (U87-PDGF), compared with parental U87 cells (n = 3). The Ct values were obtained for each sample (threshold = 40 cycles) and normalized to references U6snRNA, snord44 and snord49A; the GeNorm module was used to determine the most stable reference gene (snord44). The relative miRNA-expression values were calculated using the qBasePlus software, and the statistical significance was assessed using one-way ANOVA combined with Benferroni's post-hoc test (Supplementary Material, Table S2). MiR-367 was not detected in both U87 and U87-PDGF cells and, therefore, is not represented in the chart.
PDGF-driven mouse tumor models overexpress miR-21
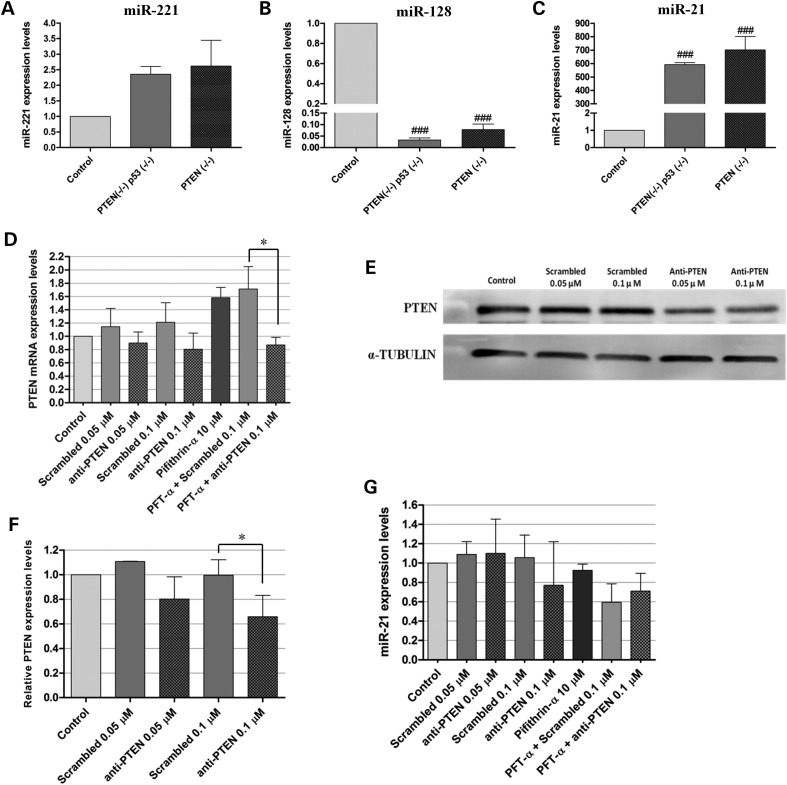
The observation of a reduction in the levels of the oncogenic miR-21 in U87-PDGF cells (compared with control epileptic tissue), as opposed to what has been published regarding the expression of this miRNA inGBM, prompted us to evaluate if such reduction would also be observed in a PDGF-driven animal model. In this regard, we have recently developed a retrovirally induced mouse GBM model characterized by the deletion of the tumor suppressors PTEN and p53 and overexpression of PDGF-B (PDGF+PTEN−/−p53−/−), which displays molecular and histopathological features that closely resemble human GBM (36). A similar GBM model with deleted PTEN and functional p53 (PDGF+PTEN−/−) was also established and characterized. The expression levels of miR-128, miR-21 and miR-221 were measured by qPCR in primary cultures prepared from tumors of both the models and compared with those of control samples, obtained from double-floxed (PTEN−/−p53−/−) mouse brains following animal injection with a control vector (no PDGF). As shown in Figure 5, miR-221 was overexpressed (Figure 5A) and miR-128 was highly downregulated (Figure 5B) in both primary cultures, without significant differences between double-floxed and PTEN-floxed samples. As opposed to what was observed in U87-PDGF cells, miR-21 was found to be highly and differentially overexpressed in both PDGF-driven primary cultures when compared with control, with an average-fold change of 592.3 ± 35.14 in double-floxed samples (P < 0.001) and 703.3 ± 243.8 in PTEN-floxed samples (P < 0.001) (Figure 5C). As U87-PDGF cells express wild-type PTEN and p53, we ascertained whether the status of these tumor suppressors could explain such considerable differences in miR-21 levels.
Figure 5.
MiRNA-expression levels in primary cultures established from PDGF-B-driven mouse GBM tumor models and modulation of PTEN and p53 in U87-PDGF cells. (A) miR-221 (B) miR-128 and (C) miR-21-expression levels in PDGF-B-overexpressing double-floxed (PTEN−/−p53−/−) or PTEN-floxed (PTEN−/−) primary cultures (n = 3) prepared from mouse tumors (as described in Materials and Methods), compared with double-floxed (PTEN−/−p53−/−) mouse brains following animal injection with a control vector (no PDGF) (n = 4). For siRNA transfection and/or pifithrin-α incubation, 24 h before any experiment U87-PDGF cells were plated onto 12-well plates at a density of 6 × 104 cells/well (RNA quantification) or 6-well plates at 1.1 × 105 cells/well (protein quantification). Complexes, prepared as described in Materials and Methods, were added to the cells at a final concentration of 0.05 or 0.1 μm and the medium was replaced by fresh DMEM after 4 h. Twenty-four hours after transfection, cells were exposed to 10 μm pifithrin-α for a period of 24 h, which was followed by the extraction of RNA and protein. (D) PTEN mRNA levels after transfection with anti-PTEN siRNAs (0.05/0.1 μm) or a noncoding sequence (Scrambled), either per se or in combination with exposure to 10 μm pifithrin-α. (E) Representative gel showing PTEN protein levels 48 h after transfection with anti-PTEN or scrambled siRNAs (n = 3). (F) Quantification of PTEN silencing observed in (E), corrected for individual α-tubulin signal intensity. The results are presented as PTEN-expression levels relative to control. (G) MiR-21 expression levels after transfection with anti-PTEN siRNAs (0.05/0.1 μm) or a noncoding sequence (Scrambled), either per se or in combination with exposure to 10 μm pifithrin-α. *P < 0.05 compared with the indicated condition. ###P < 0.001 compared with control mouse brain.
PTEN and p53 modulation does not significantly affect miR-21 levels in PDGF-B-overexpressing U87 cells
The modulation of tumor suppressors PTEN and p53 in U87-PDGF cells was achieved via siRNA-mediated PTEN silencing and/or pifithrin-α-mediated inhibition of p53.
As shown in Figure 5, cell transfection with a PTEN-specific siRNA sequence (50 nm) resulted in a small decrease in PTEN mRNA (0.90 ± 0.164, Figure 5D) and protein levels (0.802 ± 0.181, Figure 5E and F), when compared with those obtained upon transfection with a noncoding siRNA sequence. A more pronounced decrease in both the mRNA (0.805 ± 0.243, Figure 5D) and protein levels (0.657 ± 0.175, Figure 5E and F) (P < 0.05) was observed when U87-PDGF cells were transfected with 100 nm PTEN-specific siRNAs, resulting in a small, although not significant, decrease in miR-21-expression levels (0.77 ± 0.45, Figure 5G); such effect was, nevertheless, proportional to the degree of siRNA-mediated PTEN inhibition obtained. Modulation of p53 was achieved by using pifithrin-α, a specific DNA binding inhibitor of p53 transcriptional activity. Several studies performed in different cell types using pifithrin-α have not reported toxicity when the drug was applied at a range of concentrations up to 30 μm (37–39). Similarly, no signs of toxicity were observed when U87-PDGF cells were treated with 10 μm of pifithrin-α, while a marked decrease in cell viability could be observed in cells treated with 30 μm or higher concentrations of the drug (Supplementary Material, Figure S3B). In agreement with the results obtained in the above-mentioned PTEN-silencing experiments, miR-21 expression levels did not significantly change in U87-PDGF cells exposed to 10 μm of pifithrin-α either per se (0.925 ± 0.064) or in combination with siRNA-mediated PTEN silencing (0.71 ± 0.184), when compared with those obtained upon transfection with a PTEN-specific siRNAs without drug (0.77 ± 0.45) (Figure 5G). These results suggest that PTEN and p53 modulation do not significantly affect the decrease in miR-21 levels associated with PDGF-B overexpression in U87 cells. However, the limited efficiency of siRNA-mediated PTEN silencing should be considered when analyzing these findings.
DISCUSSION
Several lines of evidence have shown that PDGF signaling plays an important role in the development and progression of high-grade gliomas (20,25–27). The results obtained in the present study show that PDGF modulates the expression of several oncomiRs and tumor-suppressor miRNAs in human GBM cells, which may constitute a mechanism for the enhancement of PDGF-driven GBM tumorigenesis.
MiRNA dysregulation has shown to be an important hallmark in the oncogenic process. Different miRNA signatures distinguish normal and neoplastic tissues, as well as tumors of various differentiation states (40); alterations in miRNA signaling have also been associated with several aspects of carcinogenesis, including tumor invasion and metastasis and patient outcome (41–44). Nevertheless, the underlying mechanisms of miRNA dysregulation in human cancer are still partially unknown.
PDGF, a widely studied pro-oncogenic growth factor, has been shown to modulate cellular responses in different biological processes, including proliferation and migration of SMC and osteogenesis, through miRNA induction/repression (28,45,46). Interestingly, PDGF has been recently shown to modulate EGFR expression and function in GBM and ovarian cancer cells by enhancing miR-146b expression (29). Conversely, here we demonstrate that prolonged exposure of GBM cells to PDGF-B promotes ligand-specific downregulation of oncogenic miR-21 and anti-oncogenic miR-128; this was associated with increased cell proliferation as retrovirally modified U87 cells overexpressing PDGF-B (U87-PDGF) display increased proliferation rate, compared with parental U87 cells. Furthermore, our results provide evidence that miR-21 and miR-128 dysregulation in GBM cells is specifically modulated by PDGF-B, while miRNA modulation does not have any effect on PDGF-B mRNA levels, thus suggesting that PDGF-B-mediated miRNA downregulation is a one-way mechanism of gene regulation.
Several studies, including our recent data (P. Costa, A. Cardoso, L. Pereira de Almeida, J. Bruce, P. Canoll, M. Pedroso de Lima, manuscript in preparation), have demonstrated that miR-21 is highly overexpressed in a number of human malignancies, including GBM (12,47,48), which is associated with tumor development and proliferation. Surprisingly, here we observed a significant reduction in miR-21 levels which may be associated with the increased cell proliferation and tumorigenic potential in PDGF-overexpressing U87 cells. In this regard, we tried to clarify two main questions, the first one being related to the ability of PDGF-B to modulate the expression of other miRNAs that, by balancing the effect of decreased miR-21 expression, would contribute to a more tumorigenic phenotype.
Interestingly, several members of the miR-106b family, a pro-oncogenic microRNA family with seed region homology that includes the oncogenic polycistrons miR-17–92 and miR-106-363 (49), were found to be modulated following PDGF-B exposure. Mir-17, miR-19b, miR-20a and miR-92a, encoded by the polycistronic miR-17-92 cluster, miR-106a and miR-106b, as well as miR-10b, were upregulated in U87-PDGF cells, compared with control U87 cells. The miR-17-92 cluster is frequently overexpressed in human cancers and functions pleiotropically during both normal development and malignant transformation to promote proliferation, increase angiogenesis and sustain cell survival (50–53); increased levels of miR-17 and miR-106b also were observed in GBM stem cell-containing CD133+ cell populations and miR-17 inhibition reduced neurosphere formation and stimulated cell differentiation (54). MiR-106a, erratically expressed in human GBM (55,56), is part of the miR-106-363 cluster, whose oncogenic potential has been demonstrated in leukemias and other cancer types (57). Nevertheless, high levels of miR-17, miR-106a and miR-20a have been correlated with increased survival in glioma patients (55). Studies by Gabriely et al. revealed that survival of GBM patients expressing high levels of miR-10 family members (miR-10a/b) is significantly reduced in comparison to patients with low miR-10 levels.
On the other hand, let-7b, miR-29b and miR-130a were downregulated following PDGF-B overexpression. Let-7b was shown to have an anti-tumorigenic effect on GBM cells as its expression reduced not only the in vitro proliferation and migration of GBM cells, but also the size of the tumors produced after transplantation into nude mice (58). Similarly, by targeting podoplanin, a protein related to cellular invasion in astrocytic tumors, miR-29b was shown to inhibit invasion, apoptosis and proliferation of GBM. MiR-130a, expressed at low levels in lung cancer cell lines, was able to reduce TRAIL resistance in NSCLC cells through the c-Jun-mediated downregulation of miR-221 and miR-222 (59).
Based on the aforementioned evidence, it is reasonable to assume that PDGF-B drives U87-PDGF cells towards a more tumorigenic phenotype not by reducing miR-21 expression levels but by promoting the upregulation of several oncomiRs and downregulation of tumor-suppressor miRNAs. Nevertheless, the mechanisms by which PDGF-B regulates the expression of each miRNA remain unclear and their clarification will be crucial to understand why miR-21 needs to be downregulated in this tumorigenic phenotype.
It would be equally important to quantify the global miRNA levels in both U87 and U87-PDGF cells, as relevant studies have associated a reduction in global miRNA levels with increased tumorigenic potential (40,60,61).
The second main question we addressed focused on the mechanisms by which PDGF-B promoted miR-21 downregulation in U87 cells. As opposed to human U87-PDGF cells, miR-21 was highly overexpressed in PDGF-driven, PTEN/p53-null and PTEN-null mouse GBM cells. Considering that U87-PDGF cells express wild-type PTEN and p53, we transiently modulated the expression of both tumor suppressors in U87-PDGF cells, although no significant changes in miR-21-expression levels have been detected. These observations suggest that the effect of PDGF-B on miRNA expression varies according to the type of target cell and genetic background. However, since we were not able to completely silence PTEN and p53 in U87-PDGF cells, at least partially due to the reduced number of cells that internalizes the fluorescently labeled siRNAs, future studies should involve the development of PTEN/p53-null U87-PDGF cells to fully elucidate the role of these tumor suppressors in PDGF-related miRNA modulation.
Studies from Hata's group may provide a clue to clarify the mechanisms by which PDGF-B promotes miR-21 downregulation in U87-PDGF cells. Transforming growth factor beta (TGF-β) and BMP were shown to trigger vSMC differentiation to a more contractile phenotype by SMAD-mediated increase in miR-21 expression (62). Moreover, by inducing miR-24, PDGF-B was shown to reduce the expression of SMAD proteins and decrease bone morphogenic protein (BMP) and TGF-β signaling, thus promoting a synthetic phenotype in vSMCs (63). Therefore, by antagonizing TGF-β, PDGF-B was able to modulate the levels of miR-21 in vSMCs. It is possible that such a mechanism can also be occurring in U87-PDGF cells. Another explanation for the observed reduction in miR-21 levels relies on the existence of miRNA ‘sponge’ modulators (64), which include both mRNAs and noncoding RNAs. Depending on their expression levels and on the total number of functional miRNA-binding sites, ‘sponge’ modulators can decrease the number of free miRNA molecules available to repress other functional targets (64). Therefore, it is reasonable to assume that by activating several signaling pathways, including Ras and ERK1/2 (65), PDGF-B could be modulating a network of genes that would ultimately result in the overexpression of a miR-21 ‘sponge’ and consequent decrease in miR-21 levels.
In summary, our findings provide new insights into the PDGF signaling in brain tumors and implicate ncRNAs in the PDGF-mediated regulation of tumorigenesis, which can be of great importance for the development of targeted anticancer therapies towards the highly heterogeneous GBM multiforme.
MATERIALS AND METHODS
Materials
Pifithrin-α was acquired from Sigma (Munich, Germany) and stock solutions were prepared in DMSO (Sigma) and used within a 3-day period. The anti-PDGF-B siRNA and a noncoding sequence were acquired from Ambion (Austin, TX, USA). The anti-PTEN siRNA and a noncoding sequence were kindly donated by GenePharma (Shangai, China). The plasmids pcDNA3.1-miR-21 and pCMV-PL were obtained from Addgene (Cambridge, MA, USA). The locked nucleic acid (LNA)-modified anti-miR-21 oligonucleotides and a noncoding (scrambled) sequence were acquired from Exiqon (Vedbaek, Denmark). Digoxigenin-labeled LNA detection probes for miR-21, scrambled control and U6snRNA were also acquired from Exiqon. All sequences are displayed in Supplementary Material, Table S3. Custom-designed miRNA PCR plates (Pick&Mix miRNA PCR panels) were acquired from Exiqon. Total RNA from adult human brain was acquired from Clontech (Mountain View, CA, USA). All other reagents were obtained from Sigma unless stated otherwise.
Cell lines and culturing conditions
Double-floxed (PTEN−/−p53−/−) (MGPP1) and PTEN-floxed (PTEN−/−) mouse GBM cell lines overexpressing PDGF-B (PDGF-B+/+) were derived from established mouse GBM models and maintained in culture, as described previously (36). The F98 rat glioma cell line was kindly donated by Dr Hélène Elleaume (European Synchrotron Radiation Facility, Grenoble, France). The GL261 mouse glioma cell line was kindly donated by Dr Perez-Castillo (Universidad Autónoma de Madrid, Madrid, Spain). The U87 human GBM cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). Glioma cell lines were maintained in DMEM (Invitrogen, Carlsbad, CA, USA), supplemented with 10% heat-inactivated FBS (Gibco, Paisley, Scotland), 100 U/ml penicillin (Sigma), 100 µg/ml streptomycin (Sigma) and cultured at 37°C under a humidified atmosphere containing 5% CO2. An undifferentiated P19 embryonal carcinoma cell line, kindly donated by Dr Richard Cerione (Cornell University, NY, USA), was maintained in alpha minimum essential medium (Gibco); supplements and growth conditions were similar to those used for glioma cells. Primary rat cortical astrocyte cultures were prepared from the cerebral cortices of newborn pups according to the established protocols (66). Cell plating densities for all our experiments are indicated in Supplementary Material, Methods.
Cell line production and PDGF concentration measurement
A modified U87 cell line overexpressing PDGF-B (U87-PDGF) was derived from U87 cells after infection with a previously described PDGF-B retrovirus (25). Briefly, a 0.8 kb fragment encoding PDGF-B-hemagglutinin (HA) was ligated into the MCS1 region of the retroviral vector PQ-MCS1-IRES-eGFP. U87 cells infected with the same retroviral vector but without the PDGF-B-HA construct were used as a control. Cell lines were maintained in culture as described above for glioma cells. Cells plated onto 10 cm culture dishes at 90% confluency were incubated for 48h in serum-free basal media. PDGF-B determination was performed in the conditioned media via ELISA (R&D Systems, Minneapolis, MN, USA). Parental U87 conditioned media had undetectable levels of PDGF-B, while U87-PDGF conditioned media had a mean concentration of 353 ng/ml. We have previously shown that the U87 cell line does not secrete significant levels of PDGF-A (25).
Cell transfection
For siRNA and plasmid transfection, complexes with siRNAs and Lipofectamine RNAiMax or DNA with Lipofectamine 2000 (respectively) (Invitrogen) were prepared following the manufacturer's instructions and added to cells, maintained in OptiMEM medium (Gibco), at a final concentration of 50/100 nM siRNA or 1 µg plasmid/100.000 cells. After 4 h incubation, cells were washed with phosphate buffered saline and further cultured in fresh DMEM medium for 24 (anti-PDGF-B siRNA transfection) or 48 h (anti-PTEN siRNA or plasmid transfection), before RNA extraction. Anti-miR-21 oligonucleotide transfection followed a similar procedure but lipoplexes were prepared with DLS liposomes, as described by Trabulo et al. (67). This formulation has already shown a great efficacy in delivering single-stranded oligonucleotides into cultured cells.
Cell viability
Cell viability was evaluated by a modified Alamar blue assay, as described previously (68). Briefly, 1 ml of 10% (v/v) Alamar blue dye in complete DMEM medium was added to each well and cells were incubated at 37°C until the development of a pink coloration. Two hundred microliters of supernatant were collected from each well, transferred to 96-well plates and the absorbance at 570 and 600 nm was measured in a microplate reader (SpectraMaz Plus 384, Molecular devices). Cell viability was calculated as a percentage of control cells using the formula: (A570 − A600) of treated cells × 100/(A570 − A600) of control cells.
RNA extraction and cDNA synthesis in cultured cells
Total RNA from cultured cells was extracted using the miRCURY RNA extraction kit (Exiqon). After RNA quantification, different transcription protocols were performed depending on the type of RNA to be determined by qPCR.
For miRNA quantification, cDNA conversion was performed using the Universal cDNA synthesis kit (Exiqon). For each sample, cDNA was produced from 25 ng of total RNA (10 ng for the miRNA PCR panel assay) in an iQ5 thermocycler, by applying the following protocol: 60 min at 42°C and 5 min at 95°C. The cDNA was further diluted 1:60 (1:100 for the miRNA PCR panel assay) with RNase-free water prior to quantification by qPCR.
For mRNA quantification, cDNA conversion was performed using the iScript™ cDNA synthesis kit (Bio-Rad). For each sample, cDNA was produced from 1 µg of total RNA in an iQ5 thermocycler, by applying the following protocol: 5 min at 25°C, 30 min at 42°C and 5 min at 85°C. The cDNA was further diluted 1:3 with RNase-free water prior to quantification by qPCR.
QPCR quantification of miRNA expression
MiRNA quantification in cultured cells was performed in an iQ5 thermocycler using 96-well microtitre plates and the SYBR® Green master mix (Exiqon). The primers for the target miRNAs (miR-128, miR-21 and miR-221) and the references U6snRNA, snord44 and snord110 were also acquired from Exiqon. A master mix was prepared for each primer set, containing a fixed volume of SYBR Green master mix and the appropriate amount of each primer to yield a final concentration of 150 nm. For each reaction, performed in duplicate, 12 μl of master mix were added to 8 μl of template cDNA. Reaction conditions consisted of enzyme activation and well-factor determination at 95°C for 10 min followed by 40 cycles at 95°C for 10 s (denaturation) and 60 s at 60°C (annealing and elongation). The melting curve protocol started immediately after and consisted of 1 min heating at 55°C followed by eighty 10 s steps, with 0.5°C increase in temperature at each step. The threshold values for threshold cycle determination (Ct) were generated automatically by the iQ5 Optical System Software. Relative miRNA levels were determined following the Pfaffl method for the relative miRNA quantification in the presence of target and reference genes with different amplification efficiencies (69). The amplification efficiency for each target or reference gene was determined according to the formula: E = 10(−1/S), where S is the slope of the standard curve obtained for each gene.
QPCR quantification of mRNA expression
MRNA quantification in cultured cells was performed in an iQ5 thermocycler using 96-well microtitre plates and the iQ™ SYBR® Green supermix kit (Bio-Rad). The primers for the target genes (PDGF-BB, PTEN) and the reference gene (HPRT1) were pre-designed by Qiagen. A master mix was prepared for each primer set, containing a fixed volume of SYBR Green master mix, water and the appropriate amount of each primer to yield a final concentration of 150 nm. For each reaction, performed in duplicate, 20 μl of master mix were added to 5 μl of template cDNA. The reaction conditions consisted of enzyme activation and well-factor determination at 95°C for 1 and 30 s followed by 40 cycles at 95°C for 10 s (denaturation), 30 s at 55°C (annealing) and 30 s at 72°C (elongation). The melting curve protocol consisted of 1 min heating at 55°C followed by eighty 10 s steps, with 0.5°C increase in temperature at each step. The percentage of PDGF-BB knockdown was determined following the Pfaffl method for relative mRNA quantification in the presence of target and reference genes with different amplification efficiencies (69).
MiRNA PCR panels
MiRNA quantification using the 96-well miRNA PCR plates (Exiqon) was performed in an iQ5 thermocycler using the SYBR® Green master mix (Exiqon). The primers for the target miRNAs and the references are displayed in Supplementary Material, Table S2. A master mix was prepared for each sample, containing equal volumes (1:1) of SYBR Green master mix and diluted cDNA. For each reaction, performed in duplicate, 10 μl of master mix were added per well. Reaction conditions and melting curve protocol were similar to those described for qPCR quantification of miRNA expression. The threshold values for threshold cycle determination (Ct) were generated automatically by the iQ5 Optical System Software. The GeNorm module (70) was used to determine the most stable reference gene. Relative miRNA levels calculation and statistical analysis were performed using the software qBasePlus software (Biogazelle, Gent, Belgium).
Western blot analysis
Total protein extracts were prepared from cultured U87-PDGF cells homogenized at 4°C in lysis buffer (50 mm Tris pH 8.0, 150 mm NaCl, 50 mm EDTA, 0.5% sodium deoxycholate and 1% Triton X-100) containing a protease inhibitor cocktail (Sigma), 2 mm DTT and 0.1 mm PMSF. The protein content was determined using the Bio-Rad Dc protein assay (Bio-Rad) and 30 μg of total protein were resuspended in a loading buffer (20% glycerol, 10% SDS, 0.1% bromophenol blue), incubated for 5 min at 95°C and loaded onto a 10% polyacrylamide gel. After electrophoresis, the proteins were blotted onto a PVDF membrane according to the standard protocols. After blocking in 5% nonfat milk, the membrane was incubated with an anti-PTEN antibody (1:1000) (Cell Signaling, Beverly, MA, USA) overnight at 4°C, and with the appropriate alkaline phosphatase-labeled secondary antibody (1:20 000) (Amersham, Uppsala, Sweden) for 2 h at room temperature. Equal protein loading was shown by reprobing the membrane with an anti-α-tubulin antibody (1:10 000) (Sigma) and with the same secondary antibody. The blots were washed several times with saline buffer (TBS/T, 25 mm Tris-HCl, 150 mm NaCl, 0.1% Tween-20), incubated with ECF (alkaline phosphatase substrate; 20 μl of ECF/cm2 of membrane) for 5 min at room temperature and then subjected to fluorescence detection at 570 nm using a VersaDoc imaging system model 3000 (Bio-Rad). For each membrane, the analysis of band intensity was performed using the Quantity One software (Bio-Rad).
Fluorescence in situ hybridization (FISH) in cultured cells
FISH analysis was performed in cultured adherent cells as described by Pena et al. (71), with some modifications incorporated from the protocol of Lu et al. (72). The detailed protocol is described in Supplementary Materials, Methods.
Statistical analysis
All data are presented as means ± standard deviation of at least three independent experiments, each performed in triplicate, unless stated otherwise. One-way analysis of variance (ANOVA) combined with the Tukey post-hoc test was used for multiple comparisons in cell culture experiments and considered significant when P < 0.05. Statistical differences are presented at probability levels of P < 0.05, P < 0.01 and P < 0.001. Calculations were performed with standard statistical software (Prism 5, GraphPad, San Diego, CA, USA).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a grant from the Portuguese Foundation for Science and Technology (PTDC/BIO/65627/2006). P.M.C. is a recipient of a fellowship from the Portuguese Foundation for Science and Technology (SFRH/BD/45902/2008). The work performed at Columbia University (P.C.) was supported by a grant from the National Institute of Health/National Institute of Neurological Disorders and Stroke (1R01NS066955-01).
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Orlando Gil (Columbia University, NY, USA) for technical support and Dr Luisa Cortes (Center for Neuroscience and Cell Biology) for the assistance with the confocal microscopy imaging.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Furnari F.B., Fenton T., Bachoo R.M., Mukasa A., Stommel J.M., Stegh A., Hahn W.C., Ligon K.L., Louis D.N., Brennan C., et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. doi:10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Khasraw M., Lassman A.B. Advances in the treatment of malignant gliomas. Curr. Oncol. Rep. 2010;12:26–33. doi: 10.1007/s11912-009-0077-4. doi:10.1007/s11912-009-0077-4. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. doi:10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. doi:10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Bartel D.P., Chen C.Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. doi:10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 6.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. doi:10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A., Slack F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. doi:10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. doi:10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 9.Iorio M.V., Ferracin M., Liu C.G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. doi:10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 10.Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. doi:10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novakova J., Slaby O., Vyzula R., Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochem. Biophys. Res. Commun. 2009;386:1–5. doi: 10.1016/j.bbrc.2009.06.034. doi:10.1016/j.bbrc.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 12.Ciafre S.A., Galardi S., Mangiola A., Ferracin M., Liu C.G., Sabatino G., Negrini M., Maira G., Croce C.M., Farace M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. doi:10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 13.Silber J., Lim D.A., Petritsch C., Persson A.I., Maunakea A.K., Yu M., Vandenberg S.R., Ginzinger D.G., James C.D., Costello J.F., et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. doi:10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huse J.T., Brennan C., Hambardzumyan D., Wee B., Pena J., Rouhanifard S.H., Sohn-Lee C., le Sage C., Agami R., Tuschl T., et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. doi:10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godlewski J., Nowicki M.O., Bronisz A., Williams S., Otsuki A., Nuovo G., Raychaudhury A., Newton H.B., Chiocca E.A., Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. doi:10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 16.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. doi:10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 17.Ohgaki H., Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. doi:10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagarajan R.P., Costello J.F. Epigenetic mechanisms in glioblastoma multiforme. Semin. Cancer Biol. 2009;19:188–197. doi: 10.1016/j.semcancer.2009.02.005. doi:10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Hermanson M., Funa K., Hartman M., Claesson-Welsh L., Heldin C.H., Westermark B., Nister M. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52:3213–3219. [PubMed] [Google Scholar]
- 20.Brennan C., Momota H., Hambardzumyan D., Ozawa T., Tandon A., Pedraza A., Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. doi:10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. doi:10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armstrong R.C., Harvath L., Dubois-Dalcq M.E. Type 1 astrocytes and oligodendrocyte-type 2 astrocyte glial progenitors migrate toward distinct molecules. J. Neurosci. Res. 1990;27:400–407. doi: 10.1002/jnr.490270319. doi:10.1002/jnr.490270319. [DOI] [PubMed] [Google Scholar]
- 23.Frost E.E., Zhou Z., Krasnesky K., Armstrong R.C. Initiation of oligodendrocyte progenitor cell migration by a PDGF-A activated extracellular regulated kinase (ERK) signaling pathway. Neurochem. Res. 2009;34:169–181. doi: 10.1007/s11064-008-9748-z. doi:10.1007/s11064-008-9748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhrbom L., Hesselager G., Nister M., Westermark B. Induction of brain tumors in mice using a recombinant platelet-derived growth factor B-chain retrovirus. Cancer Res. 1998;58:5275–5279. [PubMed] [Google Scholar]
- 25.Assanah M., Lochhead R., Ogden A., Bruce J., Goldman J., Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J. Neurosci. 2006;26:6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. doi:10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Assanah M.C., Bruce J.N., Suzuki S.O., Chen A., Goldman J.E., Canoll P. PDGF stimulates the massive expansion of glial progenitors in the neonatal forebrain. Glia. 2009;57:1835–1847. doi: 10.1002/glia.20895. doi:10.1002/glia.20895. [DOI] [PubMed] [Google Scholar]
- 27.Masui K., Suzuki S.O., Torisu R., Goldman J.E., Canoll P., Iwaki T. Glial progenitors in the brainstem give rise to malignant gliomas by platelet-derived growth factor stimulation. Glia. 2010;58:1050–1065. doi: 10.1002/glia.20986. doi:10.1002/glia.20986. [DOI] [PubMed] [Google Scholar]
- 28.Davis B.N., Hilyard A.C., Nguyen P.H., Lagna G., Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J. Biol. Chem. 2009;284:3728–3738. doi: 10.1074/jbc.M808788200. doi:10.1074/jbc.M808788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao M., Rossi S., Chelladurai B., Shimizu M., Ntukogu O., Ivan M., Calin G.A., Matei D. PDGF induced microRNA alterations in cancer cells. Nucleic Acids Res. 2011;39:4035–4047. doi: 10.1093/nar/gkq1305. doi:10.1093/nar/gkq1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network, C.G.A.R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calzolari F., Malatesta P. Recent insights into PDGF-induced gliomagenesis. Brain Pathol. 2010;20:527–538. doi: 10.1111/j.1750-3639.2009.00335.x. doi:10.1111/j.1750-3639.2009.00335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torisu R., Suzuki S.O., Masui K., Yoshimoto K., Mizoguchi M., Hashizume M., Canoll P., Goldman J.E., Sasaki T., Iwaki T. Persistent roles of signal transduction of platelet-derived growth factor B in genesis, growth, and anaplastic transformation of gliomas in an in-vivo serial transplantation model. Brain Tumor Pathol. 2011;28:33–42. doi: 10.1007/s10014-010-0006-0. doi:10.1007/s10014-010-0006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cannell I.G., Kong Y.W., Bushell M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 34.Dweep H., Sticht C., Pandey P., Gretz N. miRWalk—database: prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J. Biomed. Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. doi:10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Sirotkin A.V., Laukova M., Ovcharenko D., Brenaut P., Mlyncek M. Identification of microRNAs controlling human ovarian cell proliferation and apoptosis. J. Cell. Physiol. 2010;223:49–56. doi: 10.1002/jcp.21999. [DOI] [PubMed] [Google Scholar]
- 36.Lei L., Sonabend A.M., Guarnieri P., Soderquist C., Ludwig T., Rosenfeld S., Bruce J.N., Canoll P. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One. 2011;6:e20041. doi: 10.1371/journal.pone.0020041. doi:10.1371/journal.pone.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King J.C., Lu Q.Y., Li G., Moro A., Takahashi H., Chen M., Go V.L., Reber H.A., Eibl G., Hines O.J. Evidence for activation of mutated p53 by apigenin in human pancreatic cancer. Biochim. Biophys. Acta. 2012;1823:593–604. doi: 10.1016/j.bbamcr.2011.12.008. doi:10.1016/j.bbamcr.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy P.J., Galigniana M.D., Morishima Y., Harrell J.M., Kwok R.P., Ljungman M., Pratt W.B. Pifithrin-alpha inhibits p53 signaling after interaction of the tumor suppressor protein with hsp90 and its nuclear translocation. J. Biol. Chem. 2004;279:30195–30201. doi: 10.1074/jbc.M403539200. doi:10.1074/jbc.M403539200. [DOI] [PubMed] [Google Scholar]
- 39.Waters F.J., Shavlakadze T., McIldowie M.J., Piggott M.J., Grounds M.D. Use of pifithrin to inhibit p53-mediated signalling of TNF in dystrophic muscles of mdx mice. Mol. Cell Biochem. 2010;337:119–131. doi: 10.1007/s11010-009-0291-2. doi:10.1007/s11010-009-0291-2. [DOI] [PubMed] [Google Scholar]
- 40.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. doi:10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 41.Chang K.W., Liu C.J., Chu T.H., Cheng H.W., Hung P.S., Hu W.Y., Lin S.C. Association between high miR-211 microRNA expression and the poor prognosis of oral carcinoma. J. Dent. Res. 2008;87:1063–1068. doi: 10.1177/154405910808701116. doi:10.1177/154405910808701116. [DOI] [PubMed] [Google Scholar]
- 42.Heinzelmann J., Henning B., Sanjmyatav J., Posorski N., Steiner T., Wunderlich H., Gajda M.R., Junker K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J. Urol. 2011;29:367–373. doi: 10.1007/s00345-010-0633-4. doi:10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 43.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. doi:10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 44.Zhang H., Li Y., Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937–948. doi: 10.1038/onc.2009.406. doi:10.1038/onc.2009.406. [DOI] [PubMed] [Google Scholar]
- 45.Quintavalle M., Elia L., Condorelli G., Courtneidge S.A. MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J. Cell Biol. 2010;189:13–22. doi: 10.1083/jcb.200912096. doi:10.1083/jcb.200912096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goff L.A., Boucher S., Ricupero C.L., Fenstermacher S., Swerdel M., Chase L.G., Adams C.C., Chesnut J., Lakshmipathy U., Hart R.P. Differentiating human multipotent mesenchymal stromal cells regulate microRNAs: prediction of microRNA regulation by PDGF during osteogenesis. Exp. Hematol. 2008;36:1354–1369. doi: 10.1016/j.exphem.2008.05.004. doi:10.1016/j.exphem.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Folini M., Gandellini P., Longoni N., Profumo V., Callari M., Pennati M., Colecchia M., Supino R., Veneroni S., Salvioni R., et al. miR-21: an oncomir on strike in prostate cancer. Mol. Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. doi:10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rask L., Balslev E., Jorgensen S., Eriksen J., Flyger H., Moller S., Hogdall E., Litman T., Nielsen B.S. High expression of miR-21 in tumor stroma correlates with increased cancer cell proliferation in human breast cancer. APMIS. 2011;119:663–673. doi: 10.1111/j.1600-0463.2011.02782.x. doi:10.1111/j.1600-0463.2011.02782.x. [DOI] [PubMed] [Google Scholar]
- 49.Ivanovska I., Ball A.S., Diaz R.L., Magnus J.F., Kibukawa M., Schelter J.M., Kobayashi S.V., Lim L., Burchard J., Jackson A.L., et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. doi:10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. doi:10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong L., Lai M., Chen M., Xie C., Liao R., Kang Y.J., Xiao C., Hu W.Y., Han J., Sun P. The miR-17–92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–8557. doi: 10.1158/0008-5472.CAN-10-1938. doi:10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L., Li C., Zhang R., Gao X., Qu X., Zhao M., Qiao C., Xu J., Li J. miR-17–92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309:62–70. doi: 10.1016/j.canlet.2011.05.017. doi:10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 53.Olive V., Jiang I., He L. mir-17–92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. doi:10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schraivogel D., Weinmann L., Beier D., Tabatabai G., Eichner A., Zhu J.Y., Anton M., Sixt M., Weller M., Beier C.P., et al. CAMTA1 is a novel tumour suppressor regulated by miR-9/9* in glioblastoma stem cells. EMBO J. 2011;30:4309–4322. doi: 10.1038/emboj.2011.301. doi:10.1038/emboj.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srinivasan S., Patric I.R., Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. doi:10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang G., Zhang R., Chen X., Mu Y., Ai J., Shi C., Liu Y., Sun L., Rainov N.G., Li H., et al. MiR-106a inhibits glioma cell growth by targeting E2F1 independent of p53 status. J. Mol. Med. 2011;89:1037–1050. doi: 10.1007/s00109-011-0775-x. doi:10.1007/s00109-011-0775-x. [DOI] [PubMed] [Google Scholar]
- 57.Landais S., Landry S., Legault P., Rassart E. Oncogenic potential of the miR-106–363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. doi:10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 58.Lee S.T., Chu K., Oh H.J., Im W.S., Lim J.Y., Kim S.K., Park C.K., Jung K.H., Lee S.K., Kim M., et al. Let-7 microRNA inhibits the proliferation of human glioblastoma cells. J. Neurooncol. 2011;102:19–24. doi: 10.1007/s11060-010-0286-6. doi:10.1007/s11060-010-0286-6. [DOI] [PubMed] [Google Scholar]
- 59.Acunzo M., Visone R., Romano G., Veronese A., Lovat F., Palmieri D., Bottoni A., Garofalo M., Gasparini P., Condorelli G., et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–642. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L., Volinia S., Bonome T., Calin G.A., Greshock J., Yang N., Liu C.G., Giannakakis A., Alexiou P., Hasegawa K., et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl Acad. Sci. USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. doi:10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. doi:10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 62.Davis B.N., Hilyard A.C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. doi:10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan M.C., Hilyard A.C., Wu C., Davis B.N., Hill N.S., Lal A., Lieberman J., Lagna G., Hata A. Molecular basis for antagonism between PDGF and the TGFbeta family of signalling pathways by control of miR-24 expression. EMBO J. 2010;29:559–573. doi: 10.1038/emboj.2009.370. doi:10.1038/emboj.2009.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sumazin P., Yang X., Chiu H.S., Chung W.J., Iyer A., Llobet-Navas D., Rajbhandari P., Bansal M., Guarnieri P., Silva J., et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. doi:10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White N.M., Fatoohi E., Metias M., Jung K., Stephan C., Yousef G.M. Metastamirs: a stepping stone towards improved cancer management. Nat. Rev. Clin. Oncol. 2011;8:75–84. doi: 10.1038/nrclinonc.2010.173. doi:10.1038/nrclinonc.2010.173. [DOI] [PubMed] [Google Scholar]
- 66.Banker G., Goslin K. Culturing nerve cells. 2nd edn. Cambridge, MA: The MIT Press; 1998. [Google Scholar]
- 67.Trabulo S., Resina S., Simoes S., Lebleu B., Pedroso de Lima M.C. A non-covalent strategy combining cationic lipids and CPPs to enhance the delivery of splice correcting oligonucleotides. J. Control. Release. 2010;145:149–158. doi: 10.1016/j.jconrel.2010.03.021. doi:10.1016/j.jconrel.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 68.Cardoso A.L., Simoes S., de Almeida L.P., Pelisek J., Culmsee C., Wagner E., Pedroso de Lima M.C. siRNA delivery by a transferrin-associated lipid-based vector: a non-viral strategy to mediate gene silencing. J. Gene Med. 2007;9:170–183. doi: 10.1002/jgm.1006. doi:10.1002/jgm.1006. [DOI] [PubMed] [Google Scholar]
- 69.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. doi:10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. 1–11 doi:10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pena J.T., Sohn-Lee C., Rouhanifard S.H., Ludwig J., Hafner M., Mihailovic A., Lim C., Holoch D., Berninger P., Zavolan M., et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. doi:10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu J., Tsourkas A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 2009;37:e100. doi: 10.1093/nar/gkp482. doi:10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.