Abstract
The phytohormone auxin (indole-3-acetic acid [IAA]) plays a fundamental role in vegetative and reproductive plant development. Here, we characterized a seed-specific viable maize (Zea mays) mutant, defective endosperm18 (de18) that is impaired in IAA biosynthesis. de18 endosperm showed large reductions of free IAA levels and is known to have approximately 40% less dry mass, compared with De18. Cellular analyses showed lower total cell number, smaller cell volume, and reduced level of endoreduplication in the mutant endosperm. Gene expression analyses of seed-specific tryptophan-dependent IAA pathway genes, maize Yucca1 (ZmYuc1), and two tryptophan-aminotransferase co-orthologs were performed to understand the molecular basis of the IAA deficiency in the mutant. Temporally, all three genes showed high expression coincident with high IAA levels; however, only ZmYuc1 correlated with the reduced IAA levels in the mutant throughout endosperm development. Furthermore, sequence analyses of ZmYuc1 complementary DNA and genomic clones revealed many changes specific to the mutant, including a 2-bp insertion that generated a premature stop codon and a truncated YUC1 protein of 212 amino acids, compared with the 400 amino acids in the De18. The putative, approximately 1.5-kb, Yuc1 promoter region also showed many rearrangements, including a 151-bp deletion in the mutant. Our concurrent high-density mapping and annotation studies of chromosome 10, contig 395, showed that the De18 locus was tightly linked to the gene ZmYuc1. Collectively, the data suggest that the molecular changes in the ZmYuc1 gene encoding the YUC1 protein are the causal basis of impairment in a critical step in IAA biosynthesis, essential for normal endosperm development in maize.
The phytohormone auxin, as a signaling molecule, controls and coordinates numerous aspects of plant growth and development. Indole-3-acetic acid (IAA) is the most predominant in planta auxin and regulates diverse processes, including cell division, cell elongation, formation and maintenance of meristems, vascular tissue differentiation, phototropism, flowering, and endosperm and embryo growth in developing seeds (Davies, 2010). Despite its critical roles, basic components of IAA biosynthesis are poorly understood, compared with transport and signaling aspects. However, the use of appropriate genetic screens in Arabidopsis (Arabidopsis thaliana) and the use of sensitive analytical tools in the identification of metabolic intermediates have led to significant advancements toward a better understanding of biosynthesis. Currently, there are four proposed Trp-dependent pathways of de novo IAA biosynthesis in plants (Woodward and Bartel, 2005; Pollmann et al., 2009; Normanly, 2010); of these, indole-3-pyruvic acid (IPA) was recently suggested to predominate in Arabidopsis (Mashiguchi et al., 2011; Won et al., 2011; Stepanova et al., 2011) and in pea (Pisum sativum) seeds (Tivendale et al., 2012).
The first step of the IPA pathway involves the conversion of Trp to IPA by Trp aminotransferases, first demonstrated in Arabidopsis by Stepanova et al. (2008) and Tao et al. (2008). The mutants of Arabidopsis Trp-aminotransferase (taa1) are defective in shade avoidance syndrome due to reduced levels of IAA. In maize (Zea mays), orthologs of the TAA1 gene include an endosperm-specific gene, ZmTar1 (for TA-Related1; Chourey et al., 2010) and Vanishing tassel2 (Vt2), which encode grass-specific Trp aminotransferases (Phillips et al., 2011). The vt2 mutant is marked by severe developmental abnormality, attributed to approximately 60% reduced IAA levels in the mutant seedlings. These results are significant in showing the functionality of the TAR enzyme and the IPA pathway in IAA biosynthesis in maize. Recently, it was suggested that the IPA pathway also involves the YUCCA (YUC) genes, which encode flavin monooxygenases that are now believed to catalyze the conversion of IPA to IAA (Phillips et al., 2011; Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011; Kriechbaumer et al., 2012). This is based in part on evidence that the Arabidopsis YUC2 protein, expressed in Escherichia coli, converted IPA to IAA in vitro (Mashiguchi et al., 2011). In Arabidopsis, three Yuc genes, Yuc-1, -4, and -10, are expressed in an overlapping fashion in developing seeds and are considered essential in embryogenesis (Cheng et al., 2007); however, single or double mutant yuc1 yuc4 do not show detectable defects in embryogenesis or seed phenotype.
Orthologs of the AtYuc genes are now described in several plant groups, including maize (Gallavotti et al., 2008; LeClere et al., 2010). The first Yuc-like gene in maize was isolated through positional cloning of the sparse inflorescence1 (spi1) locus; spi1 mutants showed auxin-deficient-related characteristics in the male inflorescence (Gallavotti et al., 2008). The second gene, ZmYuc1, is highly endosperm specific and its temporal expression pattern coincided with IAA biosynthesis at various stages of seed development (LeClere et al., 2010). In pea, two highly similar PsYuc-like genes, PsYuc1 and PsYuc2, showed seed- and root-specific expression, respectively (Tivendale et al., 2010). Metabolic studies in pea, however, showed that only the roots but not seeds can metabolize Trp to IAA through the proposed TAM pathway (Quittenden et al., 2009; Tivendale et al., 2010).
In contrast with many studies on auxin-related mutants that affect vegetative parts of the plant, very limited data are available on auxin mutants affecting seed development, even though seeds accumulate higher levels of IAA than any other tissue of the plant. In maize, endosperm synthesizes nearly 100- to 500-fold higher levels of IAA relative to vegetative tissues (Jensen and Bandurski, 1994; LeClere et al., 2008; Phillips et al., 2011). The significance of the large abundance of IAA in developing endosperm remains to be understood, except that it may be used during the very early stages of seed germination because >90% of the total IAA is in biologically inactive conjugated storage form (Jensen and Bandurski, 1994; LeClere et al., 2008). Such a role in germination is consistent with the fact that there are very few viable seed mutants reported in maize that are linked to IAA deficiency, although single-locus recessive mutants (defective kernels [dek]) with various abnormalities in either embryo or endosperm development and with low IAA levels (measured by ELISA) were reported by Lur and Setter (1993). It is significant in this regard that a viable defective endosperm-B18 (hereafter, de18) was identified as associated with IAA deficiency (Torti et al., 1986). Although not quantified by mass spectrometry, de18 endosperms contained total IAA levels (including conjugates) in the range of 6% to 0.3% of the wild type B37 (hereafter, De18) values, during 12 to 40 d after pollination (DAP). At the early stages, the mutant seed phenotype is <50% of the wild type in seed weight, and throughout seed development, mutant seeds are reduced in kernel size and accumulate less dry matter. Furthermore, application of the synthetic auxin, naphthalene acetic acid, to developing seeds largely rescued the de18 mutant phenotype, indicating impairment in IAA biosynthesis or metabolism as the cause of the phenotypic changes (Torti et al., 1986). Recent cellular-level studies also indicated the IAA deficiency of the de18 endosperm; high levels of immunosignal for IAA were detected in the basal endosperm transfer layer (BETL), aleurone, embryo surrounding region domains, and maternal chalazal tissue in De18 but not in the mutant (Forestan et al., 2010). Overall, the maize de18 and the pea tar2 (Tivendale et al., 2012) mutants are thus far the only seed-specific viable mutants linked to auxin deficiency. The objective of this study is to further extend our knowledge on IAA deficit in the de18 kernels, to specifically analyze temporal expression of two major IAA biosynthetic genes and to elucidate the possible molecular basis of the mutant. Our collective data, based on the cloning and sequencing of ZmYuc1 and on mapping studies, indicate that ZmYuc1 and De18 are tightly associated and that the aberrant YUC1 protein in de18 is the causal basis of IAA deficiency and the small seed phenotype in that mutant.
RESULTS AND DISCUSSION
Free IAA Content in De18 and de18 Endosperms
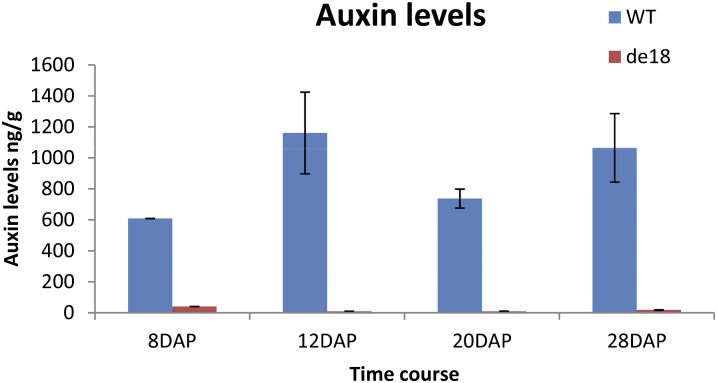
Figure 1 shows the level of free IAA in the De18 and de18 endosperm (B37 inbred background) at the 8, 12, 20, and 28 DAP stages. The mutant endosperm contained only 1% to 7% of the free IAA content of De18 at all four stages based on our ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analyses. The effect of genotype was significant at the 0.001 level, as shown by ANOVA. Interestingly, although Torti et al. (1986) reported dramatic reductions in total (free + conjugated) IAA across the entire developmental period of endosperm (12–40 DAP), they also reported that at 20 DAP, there was no reduction in free IAA; that result is not supported by our data. The de18 mutation also markedly reduced the content of total IAA (data not shown). Our data, obtained by mass spectrometry, show a consistent drop across all time points sampled.
Figure 1.
Concentrations of free IAA (ng g−1 dry matter) in De18 and de18 endosperms at 8, 12, 20, and 28 DAP, by UPLC-MS/MS. WT, Wild type. [See online article for color version of this figure.]
Spatial Pattern of Endopolyploid Cells and Cell Volume in De18 and de18 Endosperms
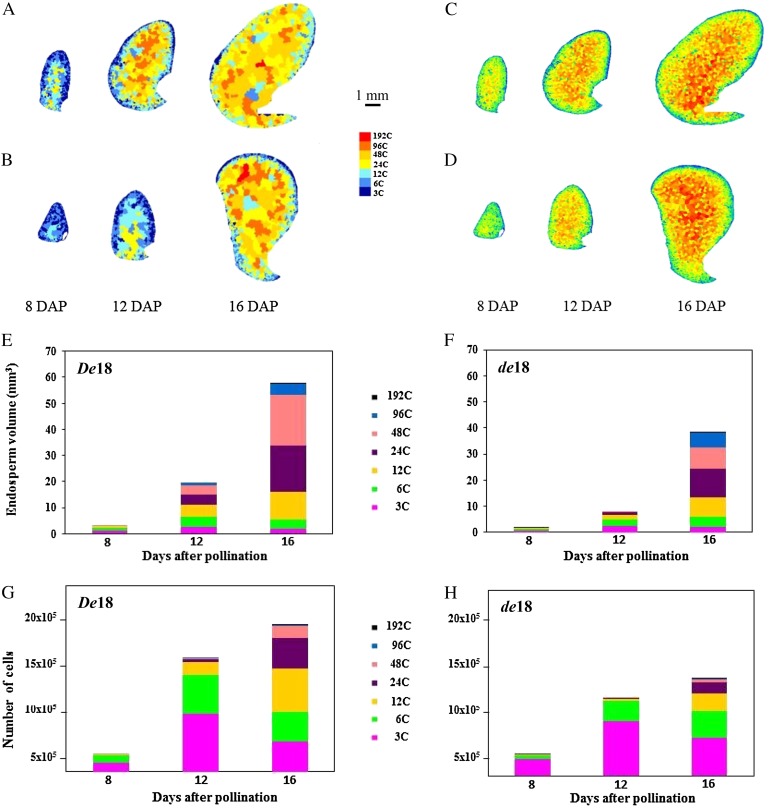
The spatial distribution of cells with different ploidy levels in the De18 and de18 endosperms at 8, 12, and 16 DAP is shown in Figure 2, A and B. Since endopolyploid cells are not known to undergo mitosis (Larkins et al., 2001; Sugimoto-Shirasu and Roberts, 2003), the endosperm was divided into a putative mitotic cell population (i.e. 3C and 6C cells) and a population that undergoes endoreduplication cycle (cells with higher than 6C-values). At 8 DAP, 24C nuclei were found only in De18 (Fig. 2A). The most significant differences between the two genotypes were detected at 12 DAP, where de18 showed a sharp deficiency of cells with 48C and higher C levels (Fig. 2B), whereas such cells in De18 were detected throughout the endosperm, except the outer layers, which were mainly in the 3C range. These results are consistent with previous data (Lur and Setter, 1993) that showed smaller nuclei in dek endosperm relative to the wild type. Nuclear diameter is known to increase in proportion to nuclear DNA content and the extent of endoreduplication during endosperm development (Kowles and Phillips, 1985). Cell volume (size) increased in both De18 and de18 genotypes at 8 and 12 DAP, (Fig. 2, C and D); it was, on average 0.1 and 1 µm3 in de18 compared with 0.5 and 2 µm3 in De18. At 16 DAP, the spatial distribution by cell volume in the two genotypes was similar: The largest cells were in the central region, whereas the smallest were at the outer periphery of the endosperm.
Figure 2.
A to D, Spatial distribution of cells at different ploidy classes (A and B) and volumes (C and D) on the longitudinal sections of De18 (A and C) and de18 (B and D) endosperms at 8 to 16 DAP. E to H, Number of cells by volume and total number of endosperm cells belonging to different C value classes in De18 (E and G) and de18 (F and H). Endopolyploid cells with C values >6 are represented by a color scale from azure (12C) to red (192C). The cell volume is presented with a color scale ranging from blue (1 × 10−3 µm3) to red (5,000 × 10−3 µm3).
The histograms in Figure 2, E to H, show a quantitative relationship between endopolyploidy level, endosperm volume, and number of cells. The de18 mutation reduced endosperm volume by 49%, 61%, and 33% at 8, 12, and 16 DAP, respectively, compared with De18 (Fig. 2, E and F). The histograms also depict relative differences in endoploidy levels with endosperm volume; the volume of the central endosperm with 12C and higher endopolyploid cells (24C and 48C) increased an average of 36 and 11 times in De18 and de18 endosperms, respectively, from 8 to 12 DAP (Fig. 2, E and F). At 16 DAP, 34% of the total wild-type endosperm volume consisted of 48C cells; the percentage diminished to 21% in the mutant.
The number of cells in both genotypes showed a large increase from 8 to 12 DAP (Fig. 2, G and H), consistent with previous data that show a peak phase of cell division in maize endosperm during this stage (Sabelli and Larkins, 2009), which also coincides with the highest IAA levels (Fig. 1; Torti et al., 1986). At 12 DAP, the de18 mutant showed a lower number of total cells than De18 (1.3 × 105 versus 0.8 × 105). Endopolyploidy at 8 DAP in both genotypes was restricted to 3C and 6C cells and did not appear to be markedly different (Fig. 2, G and H). At 12 and 16 DAP, there was a major quantitative change: De18 endosperm showed a sharp increase relative to de18 in the number of cells with nuclei of 12C and higher values. Each ploidy group was represented with a higher number of cells in De18 than in the mutant. Moreover, endosperm volume in both genotypes was well correlated with number of cells (r2 + 0.80 and 0.88 for De18 and de18, respectively) throughout development. The reduction in endosperm volume in the mutant at 16 DAP (33%) probably leads to the reduction in kernel dry weight at maturity at 40 DAP (Torti et al., 1986).
Overall, these data show that IAA may also affect endoreduplication, in addition to the well-established role in cell proliferation and elongation. Larger cells were associated with a higher C value (Sugimoto-Shirasu and Roberts, 2003). Nuclear-cytoplasmic ratio is believed to be critical and may be a driving force that leads to increased cell size. The role of IAA in these processes is not as well understood. However, an EBP1-like protein (the ErbB3 epidermal growth factor receptor binding protein) that controls cell size is known to be stabilized by auxin in Arabidopsis and potato (Solanum tuberosum; Horváth et al., 2006). Although our data here show reduced and delayed endoreduplication with the IAA deficiency, the reverse is seen in Arabidopsis where yuc mutants display increased 64C and 128C ploidy peaks in cotyledons of 14-d-old plants. The low level of auxin promoted the transition from mitotic cycles to endoreduplication (Ishida et al., 2010). Clearly, the role of IAA on endoploidy in developing seeds remains to be clarified.
Quantitative PCR Analysis of Select Putative IAA Biosynthesis Genes in Developing Endosperm
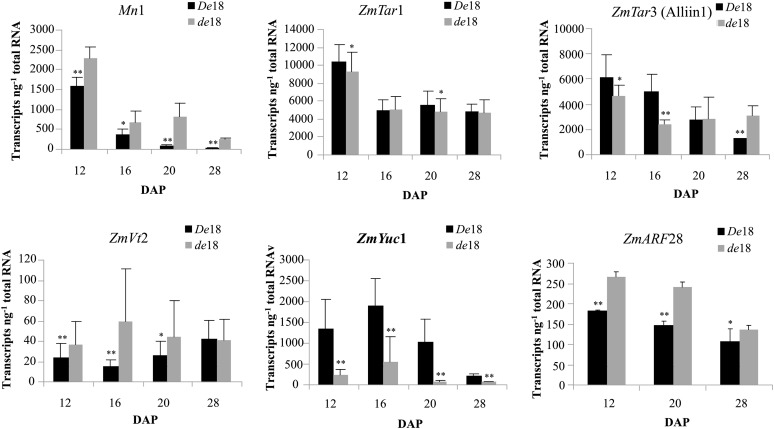
Gene expression analyses by absolute quantitative PCR (qPCR) approach are shown in Figure 3, over a time course of 12 to 28 DAP. The Mn1 expression is included here as an internal control to monitor the overall quality of RNA as well as the qPCR reactions due to its routine use in our studies that provide a known range of temporal expression in kernel development. Additionally, our previous data (LeClere et al., 2010) showed that the Mn1 gene is also involved in sugar-auxin cross talk due to the reduced IAA levels in the mn1 mutant. The Mn1 expression here showed the expected temporal pattern as well as the transcript abundance in the De18 endosperm: highest at 12 DAP, gradual reductions at 16 and 20 DAP, and lowest levels at 28 DAP. The mutant, de18, showed a similar pattern except for a higher Mn1 expression for all four stages (Fig. 3). The significance of this increase in the context of IAA deficiency in the mutant is currently not known.
Figure 3.
qPCR determinations of transcript abundance of various genes in De18 and de18 endosperm at four developmental stages (DAP). Expression levels are shown as number of copies of transcripts per nanogram of total RNA. Each histogram bar is a mean of three biological replicates and each with three technical replications.
Trp Aminotransferases
Trp aminotransferase of Arabidopsis (TAA1) is a major enzyme of IAA biosynthesis for rapid changes in IAA levels due to the shade avoidance syndrome (Stepanova et al., 2008; Tao et al., 2008). Both in Arabidopsis (Tao et al., 2008) and maize (Phillips et al., 2011), taa1 and vt2 mutants, respectively, are associated with reduced IAA levels. Homology searches against maize translated sequences (tBLASTn, E-value cutoff: 1e−20) led to five maize orthologs (Table I). Two that have been described previously, Tar1 and Vt2, encoded TAR1 and VT2 proteins, respectively (Chourey et al., 2010; Phillips et al., 2011). The third gene, ZmTar3 (ZmAlliin1, GRMZM2G141810) is reported here, to our knowledge for the first time. Phylogenetic analysis of the TARs, based on protein alignments of available sequences, indicated two distinct clusters, TAA-related and Alliin-lyase precursor-like genes (Chourey et al., 2010; Phillips et al., 2011). Table I shows DNA and protein homologies among various Trp-aminotransferases, including TAA1, which shares high homology with all three ZmTAR genes and proteins. The gene expression analyses (Fig. 3) show high level expressions of both Tar1 and Tar3 relative to other genes throughout the four stages, and there was no major difference in the levels of Tar1 expression in the two genotypes. Tar3 showed significant reduction at 16 DAP and slight reduction at 12 DAP in the mutant compared with De18, and the pattern in the two genotypes was reversed at 28 DAP; significance of these temporal changes is unclear. Simultaneous expressions of these two orthologs that share high level of protein similarity/identity is indicative of redundancy in developing endosperm and/or that the product IPA may be used in additional pathways. Combined expressions of the two Tar genes at 12 DAP was coincident with the highest IAA levels seen at this stage of endosperm development (Torti et al., 1986; LeClere et al., 2008). The qPCR values for Vt2 were exceedingly low compared to the other two orthologs, indicating it may be specific to other parts of the plant. A similar tissue dichotomy is also seen with ZmYuc1 and its ortholog, Spi, which are endosperm and vegetative tissue specific, respectively (Gallavotti et al., 2008; LeClere et al., 2010).
Table I. DNA-protein sequence homologies.
Protein sequence homologies are indicated in boldface (identity/similarity); DNA sequence homologies are indicated in roman.
|
Protein |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | ZmTAR1 | AtTAA | ZmVT2 | ZmTAR3 | ZmYUC1 | ZmYUC2 | ZmYUC3 | ZmSpi1 | SbYUC1 | OsYUC1 | AtYUC10 | AtYUC11 | PsYUC1 |
| TAR | |||||||||||||
| ZmTAR1 | 40/54 | 49/62 | 52/66 | ||||||||||
| AtTAA | 46 | 35/46 | 38/50 | ||||||||||
| ZmVT2 | 52 | 41 | 46/57 | ||||||||||
| ZmTAR3 | 58 | 48 | 58 | ||||||||||
| YUCCA | |||||||||||||
| ZmYUC1 | 46/56 | 55/67 | 43/58 | 71/76 | 63/75 | 47/64 | 50/66 | 38/51 | |||||
| ZmYUC2 | 53 | 40/56 | 52/64 | 38/50 | 43/57 | 36/54 | 41/56 | 44/52 | |||||
| ZmYUC3 | 61 | 48 | 40/56 | 53/63 | 60/72 | 51/65 | 49/62 | 37/51 | |||||
| ZmSpi1 | 50 | 61 | 49 | 36/51 | 42/58 | 37/55 | 41/59 | 45/57 | |||||
| SbYUC1 | 69 | 42 | 55 | 40 | 61/70 | 44/59 | 44/60 | 40/54 | |||||
| OsYUC1 | 60 | 47 | 56 | 44 | 59 | 49/66 | 49/68 | 37/51 | |||||
| AtYUC10 | 51 | 44 | 56 | 45 | 48 | 50 | 49/65 | 35/49 | |||||
| AtYUC11 | 54 | 46 | 53 | 46 | 48 | 51 | 56 | 36/49 | |||||
| PsYUC1 | 41 | 39 | 43 | 42 | 48 | 40 | 44 | 42 | |||||
ZmYuc1
A phylogenetic analysis of the YUCs, based on protein alignments of available sequences, has been reported previously (Gallavotti et al., 2008; LeClere et al., 2010). DNA and protein sequence homologies of various Yuc genes is shown in Table I. We identified up to eight orthologs of Yuc in maize, including Spi. Three Yuc genes (ZmYuc2 and ZmYuc3, GRMZM2G159393 and GRMZM2G107761, respectively; and ZmSPI1) were studied here as they share the highest homologies with the endosperm-specific ZmYuc1 (Table I). However, only the Yuc1 profile is shown in Figure 3 because Yuc2 and Yuc3 showed exceedingly low values (<10 transcripts) compared with Yuc1 even though they share significant sequence homologies (Table I). The developmental profile of Yuc1 in De18 is similar to the our previous data for the inbred W22, Mn1 genotype (LeClere et al., 2010; Chourey et al., 2010): High transcript abundance was detected during 12 to 16 DAP, coincident with cell division and cell elongation phase in the developing endosperm, which also shows the highest IAA levels (Fig. 1). Most importantly, the de18 mutant showed low transcript abundance of the Yuc1 gene at all four stages relative to De18. The qPCR values of both genotypes were also similarly reduced for the 8 DAP caryopses. Clearly, Yuc1 expression in the mutant was uniformly reduced throughout endosperm development, consistent with the free and total IAA levels that showed large reductions in the de18 mutant at all stages of development (Fig. 1).
We also examined the Arf28 gene that codes for AUXIN RESPONSE FACTOR28, a transcription factor linked to ZmYuc1 on chromosome 10 (Fig. 4A, SNP #1306). The de18 mutant showed statistically significant higher levels of Arf28 transcripts than the De18 genotype at all four stages. The possible significance of such a response is unclear as it contrasts with another transcription factor of Trp-dependent auxin biosynthesis, TERMINAL FLOWER2 (TFL2), where the mutant gene (tfl2) is correlated with lower rate of auxin biosynthesis and a down-regulation of the AtYuc genes (Rizzardi et al., 2011).
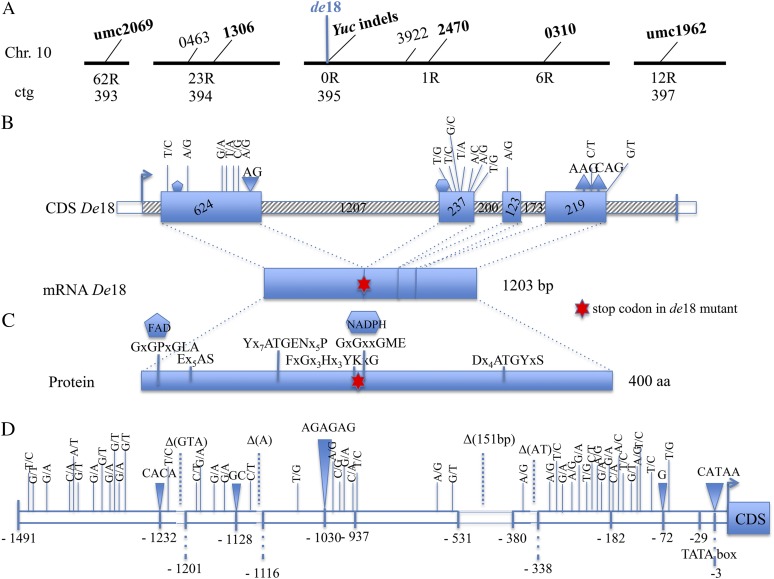
Figure 4.
Fine mapping of de18 on chromosome 10 (A), sequence analysis of the ZmYuc1 coding region (B), protein structure of ZmYUC1 (C), and noncoding putative promoter (D). A, Schematic representation of the region containing the Yuc gene on chromosome 10 (not to scale). Contig 393 maps to bin 10.2, while contigs 394, 395, and 397 map to bin 10.3 (each black bar represents a contig). The simple sequence repeat and the SNP markers used for genotyping are in bold; #1306 represents the gene Arf28. The number of recombinants (R) is shown below each marker. B, Yuc1 gene structure in De18; region between arrow and left vertical bar represents mRNA. Exons and untranslated regions/introns are represented as solid and shaded rectangles respectively. Numbers within the boxes denote exon and intron length (derived from B73 MaizeGDB) in base pairs. Downward and upward triangles represent insertion and deletion, respectively, in the de18 mutant. SNPs present in mutant are shown with thin bar. CDS, Coding sequence. C, Locations of conserved motifs present in YUCs protein. Among all motifs, N-terminal GxGPxGLA and middle GxGxxGME motifs are postulated as nucleotide binding motifs FAD and NADPH, respectively. In de18, NADPH binding motif is absent because of a 2-bp insertion in the mRNA at position 532. aa, Amino acids. D, Promoter region of 1491 bp of the Yuc1 gene. Polymorphisms in de18 respect to De18 are shown. Downward triangles indicate insertions, dashed lines deletions, and continuous lines SNPs. [See online article for color version of this figure.]
Mapping of the de18 Locus
Mapping data were obtained using the de18 allele backcrossed into the B37 background six times. The de18 mutant was mapped to chromosome 10, bin 10.03, by linkage with the simple sequence repeat marker umc1962 (Pasini et al., 2008). By screening a mapping population of 291 individuals from the cross (A69YDe18 × de18), the de18 locus was mapped between marker umc1962 (12 recombinants, 1.54 centimorgans [cM]) and umc2069 (62 recombinants, 9.79 cM; Fig. 4A). Using single nucleotide polymorphism (SNP) markers identified in neighboring genes in the region, the number of Rs was narrowed down on the north side of de18 with the SNP marker 1306 that showed 23 R (3.95 cM, contig 394). More closely to de18 on the contig 395, south side, were the SNP markers 0310 (6 recombinants, 1.03 cM) and 2470 (1 recombinant, 0.17 cM; Fig. 4A). A candidate gene search in the contig 395 revealed the location of the ZmYuc1 gene. To seek a possible unique SNP within the de18 allele for fine mapping, full-length Yuc1 complementary DNA (cDNA) and genomic clones were obtained by sequencing from both De18 and de18. All clones were identical within each genotype, but there were major differences between the normal and the mutant genotypes, as shown in Figure 4B and Supplemental Figure S1. Compared with the normal, the mutant sequences showed many SNPs in each of the four exons (16 in total; Supplemental Fig. S1). The main polymorphisms were an A↔G transition at nucleotide 82 (nonsynonymous) and an AG insertion at position 532. Moreover, a large deletion of 151 bp was found in the YUC promoter of de18 between −380 and −531 bp (Fig. 4D; Supplemental Fig. S2). The AG insertion and the 151-bp deletion were used as markers for mapping. The (A69Y × de18) F3 population was genotyped using primers surrounding the insertion/deletions (indels). No recombinants were detected, indicating that the Yuc1 indels were fully linked to the mutant (0 recombinants, 0 cM). To further confirm these data, a second (B37 × de18) F3 population was screened with the two previously mentioned microsatellites, the SNP marker 0310 and the Yuc1 indels. Four recombinants were detected using umc2069 (5.71 cM), two recombinants with both SNP 0310 and umc1962 (2.86 cM). Again, no recombinants were found testing the individuals with the Yuc1 indel markers, confirming that de18 is tightly in linkage with the Yuc1 gene.
Cloning and Sequence Polymorphism of YUC1 in De18 and de18 Mutant
Figure 4C depicts several changes in the predicted YUC1 protein in the mutant (Supplemental Fig. S1; cDNA and predicted protein data). The most critical change was the two-base (AG) insertion at position 532 in the first exon causing a frameshift in protein coding right before the putative NADPH binding motif and a premature stop codon at position 637. As a consequence, the predicted YUC protein in the de18 was of 212 amino acids compared with the normal-sized 400 amino acids in the De18. Of the known five critical motifs for YUC protein to be functional, the predicted YUC1 in de18 lacked the FAD or NADPH binding motif GXGXXG and C-terminal ATG motif. The NADPH binding motif is highly conserved and critical for normal functioning, and changes in these motifs are known to cause loss of activity. In Arabidopsis, mutation in the NADPH motif completely destroyed the YUC1 function (Hou et al., 2011). Similarly, in maize, the Spi mutant encoding a Yuc ortholog is due to a point mutation in the FAD binding domain and is associated with morphological changes that are consistent with auxin deficiency (Gallavotti et al., 2008). However, developing leaves of spi1 plants still contain 82% of the normal free IAA level (Phillips et al., 2011). The C-terminal ATG motif is believed to link the NADPH to the active site. Clearly, the predicted structural changes in the YUC1 protein in de18 are expected to yield an inactive enzyme. To the best of our knowledge, all previous data showing YUC as a rate-limiting step in IAA biosynthesis in Arabidopsis (Zhao et al., 2001), petunia (Petunia hybrida; Tobeña-Santamaria et al., 2002), and maize (Gallavotti et al., 2008) are based on either slight reductions in the IAA levels or gain-of-function experiments wherein ectopic overexpression of the Yuc gene lead to higher levels of free IAA and/or display high auxin phenotypes. The data with the de18 mutant show that the predicted loss of YUC1 protein is the causal basis of severe IAA deficiency of the seed.
The noncoding promoter region of the Yuc1 gene in the B37 genotype was 1473 bp in length (Fig. 4D; Supplemental Fig. S2). The Yuc promoter of de18 was 139 bp shorter (1334 bp) because of a large deletion of 151 bp, eight indels, and 45 SNPs. Density of SNPs and indels was higher both in the proximal (from −381 to −1 bp) and in the distal regions (from −1,491 to −1,021 bp) of the promoter (Fig. 4D). No nucleotide variations were detected in the central region of 406 bp. A 5-bp insertion at position −3 bp caused a shift of the putative TATA box site (TATAAA) from −29 bp in De18 to −34 bp in the mutant de18 Yuc1 promoter. The number of all sequence variations (SNPs and indels) was 54 with a frequency of 1 per 27 bp, which is higher compared with the frequency of polymorphic sites detected in noncoding regions of maize (Ching et al., 2002).
Further analyses of the Yuc1 promoter for putative binding sites showed several AAAG cis-elements that are recognized by the transcription factor Dof (DNA binding with one finger) gene family: three opaque-2 binding sites and an enhancer Q-element (Supplemental Table S1). The Dof recognition site also formed the core sequence of the prolamin box sequence or PBF (TGTAAAG) that was found in many genes coding for seed storage proteins (Marzábal et al., 2008). Members of the Dof family of transcription factors are reported to play a crucial role in regulating the expression of genes involved in grain filling and ripening (Gaur et al., 2011). Nineteen putative Dof core sites are found in De18, while in de18, Dof core sites decreased to 10. In the de18 mutant, the large rearrangements of the promoter and the decrement in the number of putative transcription factors binding sites could affect the regulation of the Yuc1 gene, leading to a decrease in transcript accumulation.
The study of the Yuc1 promoter sequence was extended to the other inbred lines B73 and W22. Sequence results showed no differences among the wild types tested, and the above-described polymorphisms (SNPs and indels) were found only between De18 and de18 genotypes.
Protein Level Expression of ZmYuc1 as Recombinant Proteins in E. coli
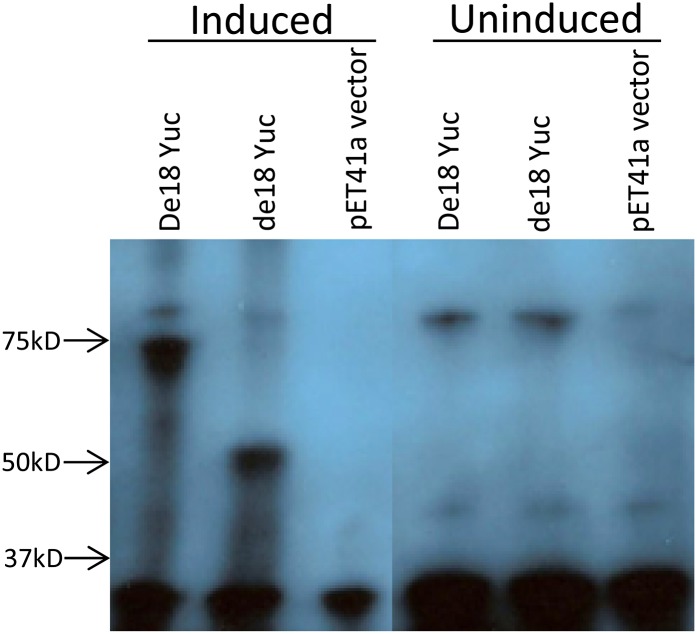
To test whether the predicted sizes of the YUC1 protein in De18 and de18 are consistent with the expressed proteins, we expressed full-length cDNA clones of ZmYuc1 from these two genotypes, fused to a glutathione S-transferase (GST-ZmYUC1) tag, in the pET system (Novagen). An immunoprofile on SDS-PAGE (western blot) from induced and uninduced E. coli extracts is shown in Figure 5. A normal-sized fusion protein of approximately 70 kD representing a GST tag of 26.0 kD plus the predicted size of 43.7 kD for YUC1 was readily detectable as a major band in cultures with the Yuc1 clone from the De18 endosperm. By contrast, the same protein band was missing in extracts from the de18 cultures; instead, we observed a major band of approximately 50.0 kD, as expected from the predicted truncated YUC1 protein of 24.14 kD plus the GST tag. Both of these two major bands were missing in the uninduced cultures as well as those with vectors lacking the cDNA inserts. Clearly, the truncated YUC1 protein was encoded by the mutant yuc1 gene of the de18 endosperm. Two additional bands, a major band of <35 kD and a minor band of >75 kD, were seen in all lanes, presumably due to bacterial proteins reacting with the antibody but independent of the ZmYuc1 gene.
Figure 5.
Western-blot analysis of E. coli-expressed proteins from full-length cDNA clones of the ZmYuc1 gene from De18 and de18 endosperm and the corresponding empty vector, as shown; each lane represents crude extract of 1µg of total protein. [See online article for color version of this figure.]
CONCLUSION
The de18 mutant has been associated with auxin deficiency for some time (Torti et al., 1986), although in previous studies the hormone was not quantified by mass spectrometry. The reduction in free IAA content in de18 endosperm in the analyses here, based on UPLC-MS/MS, is the largest recorded for any auxin mutant, and large reductions were also observed in total IAA content. The dry weight of de18 endosperms is typically only about 60% of the wild-type weight (Torti et al., 1986). This effect may well be a consequence of reduced IAA content, since the weight of de18 endosperms was increased by application of the synthetic auxins naphthalene acetic acid (Torti et al., 1986) or 2,4-dichlorophenoxyacetic acid (Lur and Setter, 1993). Certainly it would be expected that the reduction in IAA content in the mutant is sufficient to affect the phenotype, since much smaller changes are thought to be responsible for marked effects on vegetative development in maize (Phillips et al., 2011).
Developing seeds and grains in particular undergo at least four highly specialized functions: endoreduplication, massive synthesis of starch and storage protein during grain filling, programmed cell death, and maturation, in addition to cell proliferation and cell elongation. The cellular data (Fig. 2) suggest that many of these functions are affected at the individual cell level in all storage cells of the de18 mutant. As indicated previously, IAA is also detected in BETL, aleurone layer, and the embryo-surrounding region domain of developing endosperm in De18 but not in the mutant, de18. Furthermore, Forestan et al. (2009) described a complete lack of ZmPIN1 expression in the mutant BETL compared to the De18 transfer cells. Given that the PIN protein is essential in polar auxin transport, it is not surprising that the de18 BETL is also deficient in the expression of a BETL marker gene, BETL1. Although nothing is known on the physiological role of the BETL1 protein in endosperm development, an aberrant BETL, as in the de18 mutant, is always associated with defective, shriveled, or miniature endosperm due to the possible impairment of critical transport functions associated with these cells in seed development (Kang et al., 2009, and refs. therein).
The associated changes in the coding sequence and Dof sites in the promoter of ZmYuc1 and its sugar responsiveness (LeClere et al., 2010) suggest that IAA may also orchestrate many storage functions and regulate genes related to endosperm development. Consistent with these molecular observations are the IAA profiles that show increases during storage phase, 20 to 28 DAP, in developing endosperm (LeClere et al., 2008). Overall, these data suggest a plausible role for the observed high abundance of IAA throughout endosperm development. Another noteworthy observation is that the high transcript abundance of the two ZmTar genes relative to ZmYuc1 (Fig. 3) did not compensate for the loss of Yuc1 function. Our results are consistent with recent suggestions (Mashiguchi et al., 2011; Phillips et al., 2011; Stepanova et al., 2011; Won et al., 2011) that YUC proteins operate in the IPA pathway. Clearly, the DE18/ZmYUC1 gene plays an essential role in IAA biosynthesis in maize endosperm, and the reduction in IAA content in de18 endosperm is the largest recorded for a nonlethal seed mutant in plants.
MATERIALS AND METHODS
Plant Material
The de18 allele was obtained from a collection of de mutants (Pasini et al., 2008). The de18 allele was back crossed six times to the inbred B37 to obtain De18 and de18 homozygous kernels in a similar genetic background. F3 populations were produced by crossing the mutant with A69Y and B37 inbred lines. All plants were grown in the field and self-pollinated, and immature kernels were harvested from 8 to 28 DAP.
IAA Extraction and Analysis
Fifty to two hundred milligrams of lyophilized material was ground and extracted with 80% methanol. Internal standard (13C6 IAA) was added, and samples were purified and analyzed by UPLC-MS/MS as described previously (Tivendale et al., 2012). Total IAA levels (free plus conjugates) were determined by adding 13C6 IAA internal standard to aliquots and then hydrolyzing with 7 n NaOH at 100°C for 3 h. Extracts were then purified using Alltech SAX cartridges, prior to UPLC-MS/MS.
Cytometric Analysis
Developing caryopses were harvested from 8 to 16 DAP and immediately fixed in cold FAA fixative (3.7% formaldehyde, 5% acetic acid, and 50% ethanol) for 24 h at 4°C, dehydrated in a series of ethanol and tertiary butyl alcohol (Sigma-Aldrich), and embedded in Paraplast (Sherwood Medical). Paraffin-embedded caryopses were sectioned to 12- to 20-μm-thick sections on a rotary microtome (Microm 325; Carl Zeiss). Longitudinal sections of seeds were dewaxed in xylene, rehydrated through an ethanol series to water, hydrolyzed in 5 m HCl for 75 min at 20°C, stained with Feulgen reagent for 120 min at 20°C, washed for 45 min in six changes of SO2 water, dehydrated in an ethanol series, and then mounted in DPX (Fisons, Scientific Equipment). The image analysis system consisted of the Axioskop 2 MOT microscope (Carl Zeiss) equipped with a CCD camera (DXC-950P; Sony) and the KS400 software package (Carl Zeiss Vision). Macro programs were developed for the different image analysis applications and described below. The cell walls were visualized using epifluorescence (UV excitation) and interactively outlined. The recorded cytometric parameters were cell area, minimum and maximum cell diameter, and cell centroid coordinates. The total number of cells in the endosperm was estimated essentially as previously described (Bengough et al., 2001) by rotation of the median longitudinal section for 180°. The volume of individual cells was estimated by multiplying the cell area either by the minimum cell diameter (cell area larger than 180 μm2) or by the maximum cell diameter (cell area smaller than 180 μm2). Cell size and endopolyploidy levels were measured in the same sections. Nuclear DNA amount was measured with image densitometry using the interphase-peak method adapted for use with tissue sections (Vilhar et al., 2001, 2002). Integrated optical density (IOD) was measured on a series of grayscale images of the sections recorded with a 40× objective. IOD was used to estimate the relative amount of DNA in individual nuclei. The coordinates of the nuclei were also recorded. IOD values for nuclei in endosperm sections were plotted as frequency polygons with the logarithmic scale for IOD, revealing peaks of increasing endopolyploidy levels at theoretically equal distances. For comparison of different sections, the nuclear DNA amount was expressed in C value units, where 1C represented the nuclear DNA content of a nonreplicated haploid genome. The 3C value was assigned to the mode of the first peak in the IOD frequency histogram as an internal standard for each section (Barlow, 1977). The nuclei were separated into C value classes at midpoints between peak modes into 3C, 6C, 12C, 24C, 48C, 96C, and 192C classes. The endopolyploidy data were linked with cell size using the nearest neighbor method (Vilhar et al., 2002).
RNA Extraction, Reverse Transcription-PCR, and qPCR
Total RNA was extracted using the lithium chloride method as described (Ausubel et al., 1993). Five micrograms of total RNA was digested with RNase free DNase I (Invitrogen) and reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen). We used the absolute qPCR method because it has served well as a reliable estimate of gene expression based on Mn1 (LeClere et al., 2010) and ZmYuc1 and ZmTar1 expression (Chourey et al., 2010) in developing endosperm of different stages, which showed excellent concordance between the low-resolution northern-blot hybridization and absolute qPCR values. By contrast, we have not been able to find a reliable housekeeping gene, including GRP2 (Sekhon et al., 2011), with stable qPCR values throughout endosperm development. The reverse transcription was performed at 50°C for 50 min by using oligo(dT) primers for all genes in 20-μL total volume. qPCR was performed using the DyNAmo HS SYBR Green qPCR kit (Finnzymes) and Chromo 4 CFD supported by Opticon Monitor Software version 2.03 (MJ Research). The PCR reactions contained 500 nm of gene-specific primers and 3 µL of the 10-fold diluted reverse transcription reaction in a final volume of 20 μL. The qPCR was achieved through initial denaturation and enzyme activation for 10 min at 95°C, followed by 35 cycles of 15 s of denaturation at 94°C, 20 s annealing at 56°C, and 25 s extension at 72°C. Following the extension step, optical data were acquired by Opticon Monitor software (version 2.03; MJ Research), and the PCR reactions were subject to melting curve analysis beginning at 65°C through 95°C, at 0.1°C s−1. For calculation of transcript number in subsequent ZmTAR1 qPCR, the full-length PCR product was cloned into TOPO vector (Invitrogen) and quantified based on absorbance. A dilution series was made, qPCR was performed, and a standard curve was plotted. The slope of this line was used to calculate absolute transcript number for PCR reactions performed under identical conditions and expressed as transcripts per micrograms of total RNA. Gene-specific primers for qPCR are reported in Supplemental Table S2.
Cloning and Sequencing of Yuc Alleles
Total RNA was extracted from the frozen maize (Zea mays) kernel as described (Chourey et al., 2010). Five micrograms of total RNA was reverse transcribed using qScript cDNA SuperMix (Quanta Biosciences) according to the manufacturer’s protocol. A 1235-bp cDNA fragment was PCR amplified using the following primer pairs: 5′-GATGACGGAGAAGGTGCTAGTTCT-3′ (sense) and 5′-CGACGATCGAGCTAGCAGATAGAT-3′ (antisense). The 1235 bp included the full-length Yuc1 gene of 1197 bp. The thermal cycling protocol entailed activation of AccuPrime Pfx DNA polymerase (proofreading enzyme from Invitrogen) at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 15 s, primer annealing at 56°C for 20 s, and extension at 68°C for 1 min 20 s each cycle. The amplification reactions were finally extended for 10 min at 68°C and held at 4°C. The Taq polymerase amplification product was ligated with zero Blunt II-TOPO vector from a cloning kit (Invitrogen) and grown overnight in one shot TOP 10 chemically competent cells on Luria-Bertani (LB) kanamycin plates. Single separated clones were picked up from culture overnight in LB kanamycin liquid media. Plasmid isolated with the Qiagen miniprep kit was digested with EcoRI at 37°C for 1 h, and four clones with right full-length insert (1197 bp) were chosen to sequence. Sequencing reactions were performed using M13F or M13R primer with Applied Biosystems BigDye terminator (v1.1 cycle). Sixteen clones were sent to the Interdisciplinary Center for Biotechnology Research read sequences and analyzed using Chromas and SDSC Workbench software. Protein sequences were aligned by ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) using default parameters and analyzed using bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
In addition, seven overlapping Yuc1-specific PCR primers (Supplemental Table S3) were designed to amplify and sequence the Yuc gene from genomic DNA in B37 and de18. The resulting nucleotide sequences were aligned by Muscle algorithm (Edgar, 2004) using Seaview graphical interface (Gouy et al., 2010). Polymorphisms and indels were detected with the aid of DnaSP software (Librado and Rozas, 2009).
The promoter region of 1471 bp of the Yuc1 gene was sequenced in both mutant and wild-type (B37) genotypes. Four different couples of primers were used to amplify the whole promoter region of De18, while six primers were necessary to amplify the same region in the de18 (Supplemental Table S4). A BLAST search in the PlantGDB (www.plantgdb.org) and Gramene (www.gramene.org) databases was performed to confirm the position of De18 and de18 promoter sequences on the physical maize map of chromosome 10. Sequence alignment and DNA polymorphism were performed as reported in gene cloning and sequencing section. The promoter sequences of both De18 and de18 were analyzed using PlantPAN (http://plantpan.mbc.nctu.edu.tw/) to find putative transcription factor binding sites. PlantPAN searches for regulatory elements among 59 maize transcription factors were collected from PLACE, TRANSFAC Public Version 7.0, JASPER, and AGRIS databases (Chang et al., 2008).
Protein Expression SDS-PAGE and Western Blots
A polyclonal YUC1 antibody against Escherichia coli-expressed full-length ZmYuc1 cDNA clone was commercially synthesized (Rockland Immunochemicals) and validated by western-blot analyses. Full-length cDNA clones of ZmYuc1 from De18 and de18 endosperm were expressed as GST fusion, GST-ZmYuc1, in E. coli (BL21) using the pET41b system following the manufacturer’s protocol (Novagen). Crude extracts were made from induced and uninduced cultures; induction included 1 mm isopropylthio-β-galactoside treatment for 4 h at 37°C. Samples were prepared for SDS-PAGE as described previously, blotted to polyvinylidene difluoride membranes (Chourey et al., 2010), and reacted with primary polyclonal antibody ZmYUC1 at a dilution of 1:2,000. Anti-rabbit horseradish peroxidase-labeled secondary antiserum was used for YUC1 detection at a 1:5,000 dilution from Sigma-Aldrich using the Pico kit (Pierce).
Mapping of de18
The de18 mutant, introgressed with six back crosses into the inbred B37, was crossed with the inbred A69Y. The F1 generated was selfed to obtain F2 and F3 progenies. This F3 population included 617 individuals divided into 326 De18 and 291 de18. A second F3 population was obtained from the cross (de18 × B37) and included 18 De18 and 35 de18. Seedlings from each F3 family were grown in the greenhouse and harvested for DNA extraction using the DNeasy 96 plant kit (Qiagen). All PCRs were carried out in a Mastercycler (Eppendorf). Two microsatellites (umc1962 and umc2069) were used for mapping, based on previous genetic data (Pasini et al., 2008). SNP markers were used for fine mapping. All genotyping was performed by KBiosciences using the KASPar technology, and data were visualized by SNPviewer v1.99 (http://www.kbioscience.co.uk). The Panzea database was exploited to identify SNPs (www.panzea.org), and primers flanking the SNP were designed to confirm the position and the polymorphism between wild-type lines (B37 and A69Y) and mutant de18. After this screening, the following five SNPs were found polymorphic: SNP 2470 at position 18,670,208 (located in intron 5 of GRMZM2G068382, coding for a hypothetical protein), SNP 0310 at position 20,409,355 (in exon 9 of GRMZM2G040968, coding for a phosphoglucan water dikinase), SNP 0463 at position 13,493,417 (in exon 2 of GRMZM2G110381, coding for a putative yippee-like protein), SNP 1306 at position 13,784,369 (in intron 12 of GRMZM2G006042, coding for ARF28), and SNP 3922 at position 17,678,943 (in 3′ untranslated region of GRMZM2G127386, protein homologous to the nitrilase-associated protein of rice [Oryza sativa] and sorghum [Sorghum bicolor]). Three of them (1306, 2470, and 0310) were used for genotyping the two F3 populations because of their location in different contigs. The remaining two SNPs were not tested in the populations because they are located at less than 1 Mb from the nearest SNP marker. Most importantly, the 2-bp insertion in exon 1 of the Yuc1 gene and the deletion in the promoter sequence (GRMZM2G091819, positions 16,518,075 and 16,517,095) were used as markers.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. cDNA and protein sequence alignments of Zmyuc1 alleles.
Supplemental Figure S2. Promoter sequence alignment of Zmyuc1 alleles.
Supplemental Table S1. Transcriptional factors binding sites of Zmyuc1 promoter.
Supplemental Table S2. Gene specific primers for qPCR.
Supplemental Table S3. Zmyuc1 gene specific PCR primers.
Supplemental Table S4. Zmyuc1 promoter specific PCR primers.
Supplementary Material
Acknowledgments
We thank Prof. Francesco Salamini (Edmund Mach foundation, Trento, Italy) for sharing the genetic material, Drs. Daryl R. Pring and Jinping Wang (University of Florida) for critical review of the article, and Lindsey Myers (University of Florida) and Associate Professor Noel Davies (Central Science Laboratory, University of Tasmania) for technical assistance. P.S.C. and A.M. designed the research. J.B., A.L., Q.-B.L., and S.D.C. performed research. A.K. contributed a new analytical tool. D.K., Q.-B.L., P.S.C., and J.B. analyzed sequence data. P.S.C., A.M., and. J.R. wrote the article.
Glossary
- IAA
indole-3-acetic acid
- IPA
indole-3-pyruvic acid
- DAP
d after pollination
- BETL
basal endosperm transfer layer
- UPLC-MS/MS
ultraperformance liquid chromatography-tandem mass spectrometry
- qPCR
quantitative PCR
- cM
centimorgans
- GST
glutathione S-transferase
- IOD
Integrated optical density
- indel
insertion/deletion
- SNP
single nucleotide polymorphism
- cDNA
complementary DNA
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (1993) Current Protocols in Molecular Biology. Wiley, New York
- Barlow PW. (1977) The time-course of endoreduplication of nuclear DNA in the root cap of Zea mays. Eur J Cell Biol 16: 98–105 [Google Scholar]
- Bengough AG, Iijima M, Barlow PW. (2001) Image analysis of maize root caps: estimating cell numbers from 2-D longitudinal sections. Ann Bot (Lond) 87: 693–698 [Google Scholar]
- Chang WC, Lee TY, Huang HD, Huang HY, Pan RL. (2008) PlantPAN: plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics 9: 561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YF, Dai XH, Zhao YD. (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching A, Caldwell KS, Jung M, Dolan M, Smith OS, Tingey S, Morgante M, Rafalski AJ. (2002) SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genet 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey PS, Li QB, Kumar D. (2010) Sugar-hormone cross-talk in seed development: two redundant pathways of IAA biosynthesis are regulated differentially in the invertase-deficient miniature1 (mn1) seed mutant in maize. Mol Plant 3: 1026–1036 [DOI] [PubMed] [Google Scholar]
- Davies PJ. (2010) Plant Hormones: Biosynthesis, Signal Transduction, Action! Springer, Dordrecht, The Netherlands
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Meda S, Varotto S. (2010) ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol 152: 1373–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. (2008) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur VS, Singh US, Kumar A. (2011) Transcriptional profiling and in silico analysis of Dof transcription factor gene family for understanding their regulation during seed development of rice Oryza sativa L. Mol Biol Rep 38: 2827–2848 [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 [DOI] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang YX, Hamburger AW, Bakó L, Visser RG, Bachem CW, Bögre L. (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XH, Liu SN, Pierri F, Dai XH, Qu LJ, Zhao YD. (2011) Allelic analyses of the Arabidopsis YUC1 locus reveal residues and domains essential for the functions of YUC family of flavin monooxygenases. J Integr Plant Biol 53: 54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Adachi S, Yoshimura M, Shimizu K, Umeda M, Sugimoto K. (2010) Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71 [DOI] [PubMed] [Google Scholar]
- Jensen PJ, Bandurski RS. (1994) Metabolism and synthesis of indole-3-acetic acid (IAA) in Zea mays. Plant Physiol 106: 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B-H, Xiong Y, Williams DS, Pozueta-Romero D, Chourey PS. (2009) Miniature1-encoded cell wall invertase is essential for assembly and function of wall-in-growth in the maize endosperm transfer cell. Plant Physiol 151: 1366–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowles RV, Phillips RL. (1985) DNA amplification patterns in maize endosperm nuclei during kernel development. Proc Natl Acad Sci USA 82: 7010–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Wang P, Hawes C, Abell BM. (2012) Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J 70: 292–302 [DOI] [PubMed] [Google Scholar]
- Larkins BA, Dilkes BP, Dante RA, Coelho CM, Woo YM, Liu Y. (2001) Investigating the hows and whys of DNA endoreduplication. J Exp Bot 52: 183–192 [PubMed] [Google Scholar]
- LeClere S, Schmelz EA, Chourey PS. (2008) Cell wall invertase-deficient miniature1 kernels have altered phytohormone levels. Phytochemistry 69: 692–699 [DOI] [PubMed] [Google Scholar]
- LeClere S, Schmelz EA, Chourey PS. (2010) Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol 153: 306–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 [DOI] [PubMed] [Google Scholar]
- Lur HS, Setter TL. (1993) Endosperm development of maize defective-kernel (Dek) mutants: auxin and cytokinin levels. Ann Bot (Lond) 72: 1–6 [Google Scholar]
- Marzábal P, Gas E, Fontanet P, Vicente-Carbajosa J, Torrent M, Ludevid MD. (2008) The maize Dof protein PBF activates transcription of gamma-zein during maize seed development. Plant Mol Biol 67: 441–454 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini L, Stile MR, Puja E, Valsecchi R, Francia P, Carletti G, Salamini F, Marocco A. (2008) The integration of mutant loci affecting maize endosperm development in a dense genetic map using an AFLP-based procedure. Mol Breed 22: 527–541 [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P. (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S, Düchting P, Weiler EW. (2009) Tryptophan-dependent indole-3-acetic acid biosynthesis by ‘IAA-synthase’ proceeds via indole-3-acetamide. Phytochemistry 70: 523–531 [DOI] [PubMed] [Google Scholar]
- Quittenden LJ, Davies NW, Smith JA, Molesworth PP, Tivendale ND, Ross JJ. (2009) Auxin biosynthesis in pea: characterization of the tryptamine pathway. Plant Physiol 151: 1130–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzardi K, Landberg K, Nilsson L, Ljung K, Sundås-Larsson A. (2011) TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J 65: 897–906 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. (2009) The development of endosperm in grasses. Plant Physiol 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin HN, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM. (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM. (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong FX, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale ND, Davidson SE, Davies NW, Smith JA, Dalmais M, Bendahmane AI, Quittenden LJ, Sutton L, Bala RK, Le Signor C, et al. (2012) Biosynthesis of the halogenated auxin, 4-chloroindole-3-acetic acid. Plant Physiol 159: 1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivendale ND, Davies NW, Molesworth PP, Davidson SE, Smith JA, Lowe EK, Reid JB, Ross JJ. (2010) Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol 154: 1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobeña-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JN, Souer E, Koes R. (2002) FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev 16: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti G, Manzocchi L, Salamini F. (1986) Free and bound indole-acetic acid is low in the endosperm of the maize mutant defective endosperm-B18. Theor Appl Genet 72: 602–605 [DOI] [PubMed] [Google Scholar]
- Vilhar B, Greilhuber J, Koce JD, Temsch EM, Dermastia M. (2001) Plant genome size measurement with DNA image cytometry. Ann Bot (Lond) 87: 719–728 [Google Scholar]
- Vilhar B, Kladnik A, Blejec A, Chourey PS, Dermastia M. (2002) Cytometrical evidence that the loss of seed weight in the miniature1 seed mutant of maize is associated with reduced mitotic activity in the developing endosperm. Plant Physiol 129: 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YD, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.