Abstract
Complex I (NADH:ubiquinone oxidoreductase) is the entry point for electrons into the respiratory electron transport chain; therefore, it plays a central role in cellular energy metabolism. Complex I from different organisms has a similar basic structure. However, an extra structural module, referred to as the γ-carbonic anhydrase (γCA) subcomplex, is found in the mitochondrial complex I of photoautotrophic eukaryotes, such as green alga and plants, but not in that of the heterotrophic eukaryotes, such as fungi and mammals. It has been proposed that the γCA subcomplex is required for the light-dependent life style of photoautotrophic eukaryotes, but this hypothesis has not been successfully tested. We report here a genetic study of the genes γCAL1 and γCAL2 that encode two subunits of the γCA subcomplex of mitochondrial complex I. We found that mutations of γCAL1 and γCAL2 in Arabidopsis (Arabidopsis thaliana) result in defective embryogenesis and nongerminating seeds, demonstrating the functional significance of the γCA subcomplex of mitochondrial complex I in plant development. Surprisingly, we also found that reduced expression of γCAL1 and γCAL2 genes altered photomorphogenic development. The γcal1 mutant plant expressing the RNA interference construct of the γCAL2 gene showed a partial constitutive photomorphogenic phenotype in young seedlings and a reduced photoperiodic sensitivity in adult plants. The involvement of the γCA subcomplex of mitochondrial complex I in plant photomorphogenesis and the possible evolutionary significance of this plant-specific mitochondrial protein complex are discussed.
The light-dependent development of plants, or photomorphogenesis, has been extensively studied in Arabidopsis (Arabidopsis thaliana). It is now clear that several nuclear photosensory receptors, such as red/far-red light receptor phytochromes, blue light receptor cryptochromes, and LOV domain F-box proteins ZEITLUPE (ZTL) and FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1) mediate the light-dependent modulation of gene expression to alter various physiological aspects to regulate photomorphogenesis (Franklin and Quail, 2010; Kami et al., 2010; Liu et al., 2011b; Li et al., 2012). Those photoreceptors interact with regulators of transcription or protein degradation to affect gene expression. For example, phytochromes interact with the basic helix-loop-helix (bHLH) transcription factor PHYTOCHROME-INTERACTING FACTORS (PIFs) in response to red/far-red light to trigger degradation of the PIF proteins and alter the transcription of the PIF target genes (Ni et al., 1998; Huq and Quail, 2002; Bauer et al., 2004; Leivar and Quail, 2011); cryptochromes interact with the bHLH transcription factor CRYPTOCHROME-INTERACTING BHLH1 and the SUPPRESSOR OF PHYTOCHROME A1/COP1 E3 ubiquitin ligase complex in response to blue light to regulate transcription and protein degradation, respectively (Yang et al., 2000; Wang et al., 2001; Liu et al., 2008, 2011a; Lian et al., 2011; Zuo et al., 2011); and ZTL and FKF1 interact with transcription regulators such as TIMING OF CAB EXPRESSION1, CYCLING DOF FACTOR1, GIGANTEA, and CONSTANS (CO) to modulate the transcription or protein degradation of the target genes and physiological responses to light (Más et al., 2003; Imaizumi et al., 2005; Kim et al., 2007; Sawa et al., 2007; Ito et al., 2012). Many photoreceptor target genes are regulators or enzymes of various metabolic processes, especially the energy metabolism in chloroplasts and mitochondria (Ma et al., 2001). For example, Arabidopsis cryptochromes regulate the transcription of the nucleus-encoded chloroplast σ factor, SIG5, to regulate plastid transcription and development to affect photomorphogenesis (Ruckle et al., 2007; Ruckle and Larkin, 2009). However, it is less clear how photoreceptors affect mitochondrial development or whether mitochondrial proteins affect photomorphogenesis.
The respiratory complex I (NADH:ubiquinone oxidoreductase or NADH dehydrogenase) is the entry point of electrons to the mitochondrial respiratory electron transport chain. Despite the similar function in energy metabolism, complex I of photoautotrophic organisms exhibits different subunits and structure features in comparison with its counterpart of heterotrophic organisms (Brauna and Zabaleta, 2007; Hunte et al., 2010; Klodmann et al., 2010). Notably, complex I of plants and algae contains a γ-carbonic anhydrase (γCA) subcomplex that is absent from complex I of fungi and animals (Dudkina et al., 2005; Sunderhaus et al., 2006; Brauna and Zabaleta, 2007; Bultema et al., 2009; Efremov et al., 2010; Hunte et al., 2010; Klodmann et al., 2010; Klodmann and Braun, 2011; Fig. 1A). In Arabidopsis, the γCA subcomplex contains three carbonic anhydrase subunits, γCA1 (At1g19580), γCA2 (At1g47260), and γCA3 (At5g66510), and two γCA-like subunits, γCAL1 (AT5G63510) and γCAL2 (AT3G48680). The γCA and γCAL genes are widely found in plant genomes but not in animal genomes examined thus far (Brauna and Zabaleta, 2007). Therefore, it was proposed that the γCA subcomplex may play a role in the light-dependent life style of plants (Brauna and Zabaleta, 2007). Several Arabidopsis mutants have been reported that affect proteins of complex I; these mutants showed growth retardation, abnormal leaf morphology, and poor germination (de Longevialle et al., 2007; Keren et al., 2012). A tobacco (Nicotiana tabacum) complex I-specific mutant, CMSII, exhibits decreased photosynthesis at atmospheric CO2 levels but not at higher CO2 levels (Sabar et al., 2000; Dutilleul et al., 2003), demonstrating a role of complex I in light-dependent energy metabolism. However, because those complex I mutants affect either multiple complex I proteins or proteins not specific to photoautotrophic organisms, how the plant-specific subunits of complex I, especially that of the γCA subcomplex, might affect plant development remains unknown. Indeed, no photomorphogenic phenotype has been reported for those tobacco or Arabidopsis mutants (Sabar et al., 2000; Dutilleul et al., 2003; de Longevialle et al., 2007; Keren et al., 2012). The T-DNA insertion mutations of Arabidopsis genes encoding the γCA subunits have been reported previously, but none of those mutants showed visible phenotypic alterations, making it difficult to directly test the physiological function of the γCA subcomplex (Perales et al., 2005). For example, mutants impaired in the γCA2 or γCA3 genes exhibited morphologic phenotypes indistinguishable from that of the wild-type plants (Perales et al., 2005). A suspension culture derived from the γca2 mutant showed clearly reduced growth rate and respiration, but it remains unclear whether such a defect is dependent on light and how the phenotype of the suspension culture is directly related to specific aspects of the development of whole plants.
Figure 1.
γCAL1 and γCAL2 are mitochondrial proteins of the plant-specific γCA subcomplex of complex I. A, Diagram depicting the γCA subcomplex (black module indicated by arrows), the mitochondrial complex I of green algae (Polytomella spp.) and plants (Arabidopsis; Dudkina et al., 2005; Sunderhaus et al., 2006), but not in bacteria (Escherichia coli; Morgan and Sazanov, 2008), fungi (Neurospora spp.; Guénebaut et al., 1997), or mammals (bovine; Grigorieff, 1998). The structural outlines of complex I are redrawn from the published structure of the respective complex I. B to E, Confocal images showing the subcellular localization of the γCAL1-YFP and γCAL2-YFP (B–D) or γCAL1-GFP and γCAL2-GFP (E) fusion proteins in hypocotyl cells of 5-d-old seedlings (B and C) or protoplasts isolated from 3-week-old plants (E). CS16264 is a mitochondrial marker protein. Propidium iodide (PI) was used to stain the cell wall. BF, Bright field. Bars = 20 µm (B and C) and 25 µm (D). The boxed areas (B–D) are enlarged to show details. [See online article for color version of this figure.]
RESULTS
γCAL1 and γCAL2 Are Mitochondrial Proteins
We isolated clones corresponding to the Arabidopsis γCAL2 gene from a yeast two-hybrid screen in our search for proteins that interact with the blue light receptor CRY2 (Liu et al., 2008; Zuo et al., 2011; X. Yu, unpublished data). The Arabidopsis genome encodes three closely related γCAs, γCA1, γCA2, and γCA3, and two γCA-like proteins, γCAL1 and γCAL2, which together form the γCA subcomplex of the mitochondrial complex I (Perales et al., 2004; Klodmann et al., 2010; Klodmann and Braun, 2011; Supplemental Fig. S1). The amino acid sequences of the three γCA subunits contain all critical and evolutionarily conserved residues in the γCAs, whereas the two γCAL subunits lack two critical His residues that are conserved in the carbonic anhydrases and, therefore, are not likely to be catalytically active (Perales et al., 2004; Klodmann et al., 2010; Supplemental Fig. S1). γCA1, γCA2, and γCA3 are more closely related to each other (approximately 75% identity) than they are to γCAL1 and γCAL2 (approximately 30% identical; Supplemental Fig. S1). The γCAL1 and γCAL2 genes, which share approximately 90% amino acid sequence identity, appear to express constitutively without obvious responses to light or circadian rhythm (Supplemental Fig. S2). The γCAL1 and γCAL2 proteins have been previously identified in the Arabidopsis mitochondrial fraction by systematic mass spectrometry analyses (Sunderhaus et al., 2006; Klodmann et al., 2010). To verify the subcellular localization, we prepared and examined transgenic plants expressing γCAL1-YELLOW FLUORESCENT PROTEIN (YFP) and γCAL2-YFP proteins. We found that the γCAL1-YFP and γCAL2-YFP fusion proteins accumulate exclusively in mitochondria in the transgenic plants (Fig. 1, B–E). Given that Arabidopsis CRY2 performs all the known functions and undergoes light-dependent protein modifications in the nucleus (Yu et al., 2007), γCAL2 seems unlikely to be directly involved in the function or regulation of CRY2, and our initial yeast two-hybrid result is most likely an often-encountered artifact in such experiments. However, we cannot completely exclude the remote possibility that CRY2 and γCAL2 may interact in the cytosol before they are imported into their respective organelles. Because of a potential involvement of these mitochondrial proteins in light-dependent plant growth and development (Brauna and Zabaleta, 2007), we continued our study to investigate the physiological functions of γCAL1 and γCAL2.
γCAL1 and γCAL2 Are Essential Genes with Overlapping Functions
We first collected and analyzed a complete set of T-DNA insertion mutants of the genes encoding each of the five subunits of the Arabidopsis γCA subcomplex, including γca1, γca2, γca3, γcal1, and γcal2 (Supplemental Figs. S3 and S4). None of the monogenic mutants or the ca1ca3 double mutant showed easily discernible phenotypic alternation (Supplemental Fig. S4). These results are consistent with previous reports on phenotypes of ca2 and ca3 mutants (Perales et al., 2004). Transgenic plants overexpressing the γCAL1 and γCAL2 genes also showed no apparent phenotypic alterations (X. Yu, unpublished data).
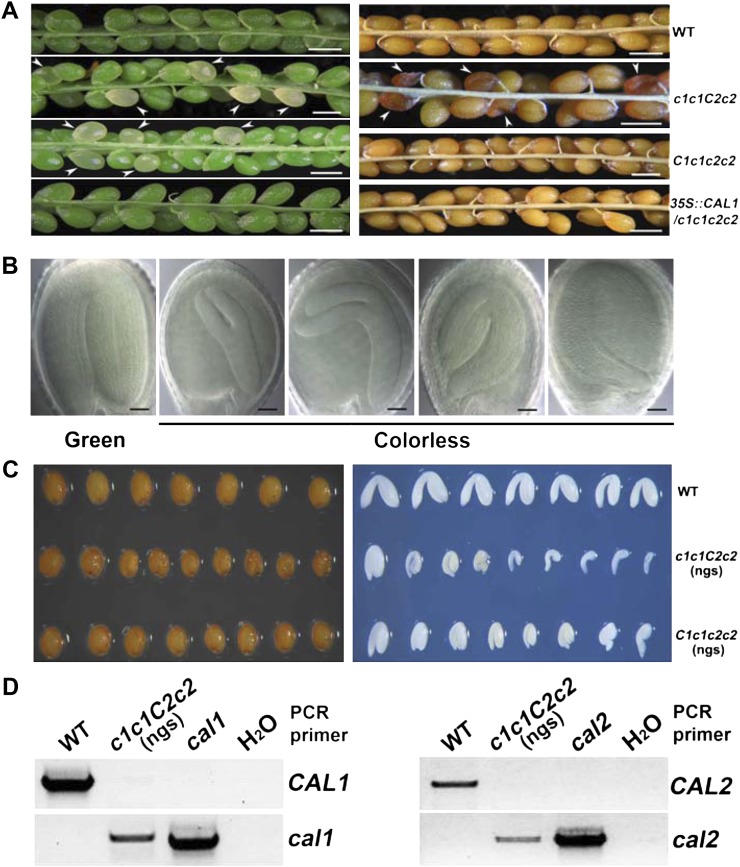
We reasoned that the γCAL1 and γCAL2 genes may have redundant functions, given that they share over 90% sequence identity (Supplemental Fig. S1), and prepared the double mutant by a conventional genetics method. However, we failed to identify the cal1cal2 double mutant after genotyping over 500 F2 progeny derived from reciprocal crosses of γcal1 and γcal2 mutants (X. Yu, unpublished data). Because plants homozygous for one γCAL gene but heterozygous for the other gene (c1c1C2c2 or C1c1c2c2) showed a normal phenotype, we suspect that the c1c1c2c2 double mutant may suffer from embryonic defects. To test this possibility, we examined young siliques derived from parents that are homozygous for the mutation of one γCAL gene but heterozygous for the mutation of the other gene (c1c1C2c2 or C1c1c2c2). We found that young siliques derived from the c1c1C2c2 or C1c1c2c2 parents contain approximately 20% to 25% colorless ovules (Fig. 2A, left), which appeared to turn deep brown at the later stage of silique development (Fig. 2A, right). Those colorless/deep-brown ovules were apparently delayed in embryogenesis to various extents, in comparison with that of the normal green ovules (Fig. 2, B and C). A statistical analysis demonstrated that the colorless/deep-brown phenotype of the c1c1C2c2 parents (23.91%) exhibits the 3:1 segregation ratio (Table I), indicating that those abnormal ovules are the double mutant progeny of the c1c1C2c2 parent. The colorless/deep-brown phenotype of the C1c1c2c2 parents (19.94%) appears to deviate from the 3:1 segregation (Table I). These results suggest that the cal2 mutation has a nearly normal penetrance for the colorless/deep-brown ovule phenotype, whereas the cal1 mutation may have a less than normal penetrance.
Figure 2.
The γCAL1 and γCAL2 subunits of the γCA subcomplex is essential for Arabidopsis development. A, The immature siliques of wild-type (WT) and rescued mutant (35S::CAL1/c1c1c2c2) parents that show uniform and green-colored ovules, and the siliques derived from C1c1c2c2 or c1c1C2c2 hemizygote parents that show segregating colorless ovules indicated by arrowheads (left). The mature siliques from the wild-type and 35S::CAL1/c1c1c2c2 parents show uniformly yellow seeds, but those from the c1c1C2c2 hemizygote parent show segregating deep-brown seeds indicated by arrowheads (right). No dark-brown seed was found in the mature siliques of the C1c1c2c2 hemizygote parents, but no double homozygous progeny were identified. Bars = 0.5 mm. B, Embryos of the segregating progeny of the c1c1C2c2 hemizygote. Ovules from a young silique of the c1c1C2c2 hemizygote parent were cleared with Hoyer’s solution and observed with a differential interference contrast microscope. The segregating white ovules arrested at various stages (c1c1c2c2) and the pale-green wild-type-looking ovule at the mature cotyledon stage from the same young silique are shown. Bars = 50 µm. C, Seeds (left) and embryos (right) of the nongerminating seeds (ngs) harvested from the C1c1c2c2 and c1c1C2c2 hemizygote parents. For the wild type, C1c1c2c2 and c1c1C2c2 seeds were placed on MS medium for 7 d. The nongerminating seeds were collected first for photographic record (left) and then dissected using a microscope. The dissected embryos corresponding to the seed (left) are shown on the right. D, Genotyping of the nongerminating seeds. Genomic DNA was isolated from the cal1 mutant, the cal2 mutant, and nongerminating seeds derived from the c1c1C2c2 parent. Genomic PCR was performed using the primers designed to amplify the wild-type γCAL1 (CAL1) and γCAL2 (CAL2) genes or the γcal1 (cal1) and γcal2 (cal2) mutant genes. [See online article for color version of this figure.]
Table I. Approximately 20% to 25% of ovules of the c1c2C2c2 and C1c1c2c2 hemizygote parents show defective embryogenesis.
The young siliques of the indicated genotypes were dissected using a microscope to examine the ovules. The numbers of normal (green) and defective (colorless) ovules were calculated and are shown. The frequency of the colorless ovules of c1c1C2c2 (23.91%) is consistent with the 3:1 segregation ratio. The frequency of colorless ovules observed in young siliques of the C1c1c2c2 hemizygote parent (19.94%) deviates from the 3:1 segregation, indicating a less than perfect penetrance of the ycal mutation. The frequency of colorless ovules observed in young siliques derived from the 35S::CAL1/c1c1c2c2 transgenic plant (1.53%) is similar to that of the wild-type control (0.35%), indicating rescue of the colorless ovules of the c1c1c2c2 double mutant by the 35S::CAL1 transgene.
| Parameter | Wild Type | cal1 | cal2 | c1c1C2c2 | C1c1c2c2 | 35S::CAL1/c1c1c2c2 |
|---|---|---|---|---|---|---|
| Total ovules | 862 | 834 | 853 | 1,150 | 1,329 | 720 |
| Green ovules | 859 | 831 | 847 | 875 | 1,064 | 709 |
| Colorless ovules | 3 | 3 | 6 | 275 | 265 | 11 |
| Colorless (%) | 0.35 | 0.36 | 0.70 | 23.91 | 19.64 | 1.53 |
| χ2 (3:1) | – | – | – | 0.72 | 18.15 | – |
Surprisingly, in contrast to a common embryonic defective mutant, we did not observe an abnormal number of aborted embryos in the mature siliques of the c1c1C2c2 and C1c1c2c2 plants (Fig. 2A, right). We reasoned that the colorless/deep-brown ovules might complete their development to set seeds, but the double mutant seeds may fail to germinate. This explains why we were unable to find the double mutant in the F2 plants. To test this interpretation, we examined the germination rate of the seeds harvested from the selfed c1c1C2c2 and C1c1c2c2 parents (Table II). In contrast to the wild-type seeds that showed the germination rate of higher than 99%, about 20% to 25% of seeds derived from the selfed c1c1C2c2 and C1c1c2c2 parents failed to germinate (Table II). As a control, the seeds resulting from the reciprocal crosses between the wild-type and the c1c1C2c2 and C1c1c2c2 plants showed normal germination rates (Supplemental Table S1), suggesting that the nongerminating seeds derived from selfed c1c1C2c2 and C1c1c2c2 parents are the double mutants. Similar to the colorless/deep-brown ovule phenotype, the cal2 mutation had a nearly normal penetrance for the nongermination phenotype, whereas the cal1 mutation had a less than normal penetrance for the germination phenotype (Supplemental Table S1).
Table II. Approximately 20% to 25% of seeds of the c1c2C2c2 and C1c1c2c2 hemizygote parents failed to germinate.
Seeds harvested from individual siliques of the indicated genotype were germinated on MS plates for 7 d, and the frequency of nongerminating seeds was calculated on the basis of a single silique. The frequency of nongerminating seeds harvested from the c1c1C2c2 hemizygote parents (24.58) is consistent with the 3:1 segregation ratio. The frequency of nongerminating seeds harvested from the C1c1c2c2 hemizygote parent (20.31%) deviates from the 3:1 segregation, indicating a less than perfect penetrance of the γcall mutation. The frequency of nongerminating seeds harvested from the 35S::CAL1/c1c1c2c2 transgenic plant (0.32%) is similar to that of the wild-type control (0.39), indicating rescue of the colorless ovules of the c1c1c2c2 double mutant by the 35S::CAL1 transgene.
| Parameter | Wild Type | cal1 | cal2 | c1c1C2c2 | C1c1c2c2 | 35S::CAL1/c1c1c2c2 |
|---|---|---|---|---|---|---|
| Germinating seeds | 514 | 746 | 593 | 454 | 467 | 627 |
| Nongerminating seeds | 2 | 2 | 1 | 148 | 119 | 2 |
| Nongerminating seeds frequency (%) | 0.39 | 0.27 | 0.17 | 24.58 | 20.31 | 0.32 |
| χ2 (3:1) | – | – | – | 0.05 | 6.88 | – |
We confirmed that the colorless ovule and nongerminating seed phenotypes both result from the c1c1c2c2 double mutation by genotyping and complementation tests (Figs. 2D and 3; Tables I and II). We first collected the nongerminating seeds from the progeny derived from selfed c1c1C2c2 parents and genotyped those seeds. Indeed, the nongerminating seeds derived from the c1c1c2C2 hemizygote parents contained T-DNA inserts in both the γcal1 and γcal2 mutant loci, and they lacked the intact γCAL1 and γCAL2 genes (Fig. 2D). For the complementation assay, we transformed the C1c1c2c2 plants with the 35S::CAL1 transgene and genotyped the transgenic individuals (Basta resistant) in the T2 generation (Fig. 3). All T2 Basta-resistant individuals grew normally and showed normal ovules and seed germination (Tables I and II). We randomly selected some T2 Basta-resistant individuals to identify those that showed homozygous γcal1 and γcal2 mutations (c1c1c2c2; Fig. 3A). In contrast to our previous failure in identifying any c1c1c2c2 individuals homozygous for both the γcal1 and γcal2 mutations in over several hundred progeny of the C1c1c2c2 parent, we readily identified individuals homozygous for both mutations (c1c1c2c2) in the Basta-resistant transgenic progeny of the C1c1c2c2 parent (Fig. 3A). We then examined the existence of the 35S::CAL1 transgene in plants of the c1c1c2c2 genotype and found that all c1c1c2c2 double mutant plants also contained the 35S::CAL1 transgene (Fig. 3B). This result confirmed that the c1c1c2c2 double mutant was rescued by the 35S::CAL1 transgene. We conclude that the cal1cal2 double mutation caused the defective embryogenesis and nongerminating seed phenotypes and that the γCAL1 and γCAL2 genes are essential for plant development.
Figure 3.
Genotyping showing the complementation of the γcal1γcal2 (c1c2) double mutant. A, The c1c1C2c2 plants were transformed with the 35S::CAL1 transgene. The genomic DNA was isolated from arbitrarily selected T2 Basta-resistant plants and PCR amplified using the primers specific to the CAL2 gene or the cal2 mutant locus. B, The individuals identified as the cal2 mutant in A were further genotyped for the cal1 mutant locus and the 35S::CAL1 transgene. Primers specific for the CAL2, cal2, CAL1, and cal1 loci as well as the 35S::CAL1 transgene are indicated on the right. WT, Wild type.
Decreased Expression of γCAL1 and γCAL2 Altered Plant Light Responses
Since the monogenic γcal1-1 and γcal2-1 mutants showed no phenotype alteration whereas the c1c1c2c2 double mutant failed to germinate, we sought to generate intermediate phenotypes to facilitate functional analysis using a “knockdown” approach. We prepared γCAL2 RNA interference (RNAi) transgenic lines in the wild type or the cal1 mutant backgrounds, which are referred to as c2i or c1c2i knockdown lines, respectively. Both knockdown lines showed similar phenotypes (Figs. 4 and 5; Supplemental Fig. S5; data not shown), but we focused on the c1c2i knockdown lines for the following analyses (Figs. 4–7). The c1c2i knockdown lines showed reduced expression of the γCAL1 and γCAL2 genes without affecting the mRNA expression of the three γCA genes of the γCA subcomplex (Fig. 5A; Supplemental Fig. S6). The phenotypic alterations of the c1c2i knockdown plants are observed in multiple independent transgenic lines (Figs. 4–7), arguing that the phenotypic alterations described are due to the reduced expression of the γCAL1 and γCAL2 genes. Consistent with the observation that the c1c1c2c2 double mutant failed to germinate, the c1c2i knockdown lines showed delayed germination (Fig. 4). After germination, the c1c2i plants appeared to grow normally, except that they exhibited smaller stature (Fig. 5; Supplemental Fig. S5). By the time the wild-type plants developed four true leaves, the c1c2i plants had only two true leaves, and the leaves of the c1c2i plants remained markedly smaller than those of the wild type throughout the life cycle (Fig. 5; Supplemental Fig. S5). It remains unclear whether the smaller stature of the c1c2i plants results from delayed germination or retarded growth or both. In addition to the effect on the organ and plant size, decreased expression of the γCAL1 and γCAL2 genes also affects leaf morphology and leaf senescence (Supplemental Fig. S7).
Figure 4.
The delayed germination phenotype of the c1c2i knockdown seeds. Seeds of the indicated genotypes (wild type [WT], cal1, and c1c2i) were placed on MS medium, and the photographs were taken at the times indicated. Seeds or seedlings of independent c1c2i mutant transgenic lines show similar phenotypes of delayed germination or retarded growth, respectively. Bars = 1 mm. [See online article for color version of this figure.]
Figure 5.
The growth retardation phenotypes of transgenic lines expressing the CAL2-RNAi construct in the cal1 mutant background (c1c2i). A, Quantitative PCR showing decreased mRNA expression of the CAL1 (left) and CAL2 (right) genes in independent transgenic lines expressing the CAL2-RNAi construct in the cal1 mutant background (c1c2i). B, Phenotypes of the c1c2i mutant transgenic seedlings grown for 7 d in a long-day (16 h of light/8 h of dark) photoperiod (left) or a short-day (8 h of light/16 h of dark) photoperiod (right). C, Phenotypes of the c1c2i mutant transgenic seedlings grown for 11 d in a long-day photoperiod. WT, Wild type. [See online article for color version of this figure.]
Figure 7.
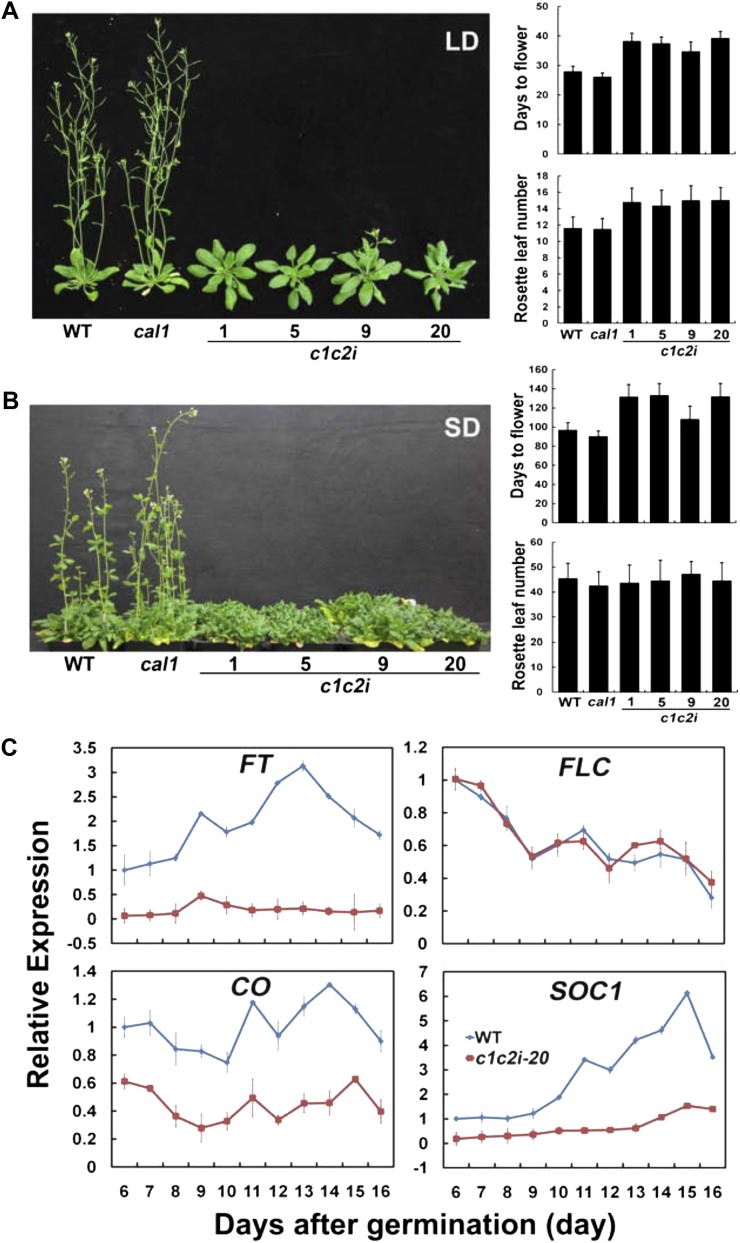
The photoperiod-dependent delay of floral initiation of the c1c2i knockdown plants. A, Plants of the indicated genotypes were grown in a long-day (LD) photoperiod (16 h of light/8 h of dark; left). The flowering time was measured as the days to flowering and the numbers of rosette leaves at the time of flowering (right). B, Plants of the indicated genotypes grown in a short-day (SD) photoperiod (8 h of light/16 h of dark; left). The flowering times were measured as days to flowering and the number of rosette leaves, and sd values are shown (n ≥ 20). C, Quantitative PCR results showing the mRNA expression of the indicated flowering-time genes in wild-type (WT) and c1c2i plants. Plants were grown in continuous white light, and tissues were collected at the indicated days after germination for RNA isolation and quantitative PCR analysis. The relative expression of the indicated genes is normalized to the level of the UBQ5 control. [See online article for color version of this figure.]
The c1c2i seedlings grown in the dark exhibited typical etiolated phenotypes similar to the wild-type seedlings, including elongated hypocotyls, unexpanded cotyledons, and normal apical hook, although the c1c2i seedlings had slightly shorter hypocotyls than the wild-type control seedlings (Fig. 6A). However, when grown under light, the c1c2i seedlings developed a more pronounced shorter hypocotyl phenotype, and they also showed shorter petioles (Fig. 6, A and B). A fluence-rate analysis using different monochromatic lights demonstrates that c1c2i seedlings are hypersensitive to light in a wavelength-independent manner (Fig. 6B). This result demonstrates a functional association between the mitochondrial γCA subcomplex and photomorphogenesis. Light-grown c1c2i seedlings also showed increased anthocyanin accumulation (Fig. 6A). The gene encoding the key enzyme of anthocyanin synthesis, chalcone synthase, was expressed higher in c1c2i than in the wild-type seedlings (Fig. 6C). Similar to the hypocotyl phenotype, CHS expression was markedly exaggerated in light-grown c1c2i seedlings relative to the etiolated c1c2i seedlings.
Figure 6.
The wavelength-independent hypersensitive light responses of the c1c2i knockdown seedlings. A, Representative seedling phenotypes of the indicated genotypes. Independent lines of the c1c2i seedlings and the controls were grown in the dark or in continuous blue light (25 µmol m−2 s−1), far-red light (2.5 µmol m−2 s−1), or red light (15 µmol m−2 s−1) for 5 d. Seedlings of four independent c1c2i lines (lines 1, 5, 9, and 20) are shown. B, Hypocotyl lengths of seedlings of the indicated genotypes. Seedlings were grown for 5 d under the different fluence rates indicated, and sd values are shown (n ≥ 20). C, Quantitative PCR results showing the increased expression of the light-induced CHS gene in c1c2i seedlings. Wild-type (WT) and c1c2i seedlings were grown in the dark (D7–D9) or under continuous blue light (B7–B9) for 7, 8, or 9 d before sample harvest for RNA isolation. The relative expression of CHS is normalized by UBQ5. [See online article for color version of this figure.]
In addition to the altered photomorphogenic development of young seedlings, decreased expression of γCAL1 and γCAL2 also affects photoperiod-dependent reproductive development of the knockdown plants (Fig. 7). When grown in long-day photoperiods, the c1c2i plants clearly exhibited a late-flowering phenotype measured by both “days to flower” and “rosette leaf number” (Fig. 7A). Although the c1c2i plants grown in short-day photoperiods also took longer to flower than did the wild-type control as measured by days to flower, the c1c2i and wild-type plants developed the same number of rosette leaves at the time of flowering in short days (Fig. 7B). This result argues that the apparent late-flowering phenotype of the c1c2i plants grown in the short-day photoperiod is due to a retardation of growth rather than a delay in floral initiation per se (Koornneef et al., 1991). Consistent with the notion that γCAL1 and γCAL2 expression affects photoperiodic flowering, genes closely associated with the photoperiodic control of floral initiation, such as CO, SOC1, and FT, showed markedly reduced mRNA expression in the c1c2i plants, whereas another flowering-time gene, FLC, which is an autonomous pathway gene not directly involved in the photoperiodic control of flowering time, showed normal expression in the c1c2i plants (Searle and Coupland, 2004; Sung and Amasino, 2005; Bäurle and Dean, 2006; Fig. 7C). Importantly, the development-dependent change of mRNA expression for the photoperiodic pathway genes FT and SOC1, but not the autonomous pathway gene FLC, is almost abolished in the c1c2i plants (Fig. 7C). Given the essential role of γCAL1 and γCAL2 in plant development (Figs. 1–5), this result argues that the developmental impacts of these two mitochondrial proteins affect the photoperiodic pathway more than the autonomous pathway that controls floral initiation. Taken together, we conclude that γCAL1 and γCAL2 play important roles in light-dependent plant growth and development.
DISCUSSION
We have demonstrated in this report the physiological role of a plant-specific γCA subcomplex of mitochondrial complex I in Arabidopsis. We found that γCAL1 and γCAL2 are not only essential for plant development, they are also involved in the light regulation of seedling development and photoperiodic control of flowering. The association of γCAL1 and γCAL2 with light-dependent plant development appears to provide a functional and evolutionary explanation for why the γCA subcomplex is found only in the mitochondrial complex I of photoautotrophic organisms. On the other hand, our study also raised several questions that remain to be further investigated.
It remains unclear how mitochondrial proteins γCAL1 and γCAL2 affect the light regulation of plant growth. The mRNA abundances of the nuclear genes γCAL1 and γCAL2 are not apparently regulated by light (Supplemental Fig. S2). It is possible that light may affect the protein modification or abundance of γCAL1 and γCAL2. Our attempts to raise antibodies against the γCAL1 and γCAL2 proteins were unsuccessful, so these hypotheses cannot be directly tested at present. Alternatively, the γCA subcomplex may affect the composition and activity of complex I, which in turn affects energy metabolism in response to light, such as photosynthesis. Indeed, the c1c2i plants exhibited a modest but statistically significant (P < 0.05) reduction in the photochemical efficiency of PSII, especially at a relatively high fluence rate of light (Supplemental Fig. S8). This result is consistent with the previous finding that the mitochondrial complex I is important for photosynthesis (Sabar et al., 2000; Dutilleul et al., 2003). The trivial explanation of a change in the composition of light harvesting and photosystem stoichiometry is unlikely, because the chlorophyll a/b ratios are unchanged and blue native gel electrophoresis of thylakoid membranes does not reveal any differences in the formation of supercomplexes of photosystems and light-harvesting complexes (Supplemental Fig. S9). There remains the possibility that the biogenesis or maintenance (repair cycle) of PSII is impacted, which is consistent with the exacerbated phenotype with a higher light fluence (Supplemental Fig. S8). The reduced photochemical efficiency of PSII of the c1c2i plants may result from an effect on photorespiration that occurs partly in the mitochondrion. It is known that complex I affects photorespiration (Sabar et al., 2000; Dutilleul et al., 2003), and it has also been proposed that the γCA subcomplex may play a role in photorespiration (Brauna and Zabaleta, 2007). The γCA subunits (γCA1–3) of the γCA subcomplex may, by converting CO2 to HCO3−, facilitate the recycling of CO2 released at the decarboxylation step of photorespiration to the chloroplast. Therefore, we investigated whether the γCA subcomplex may affect photorespiration by testing whether the c1c2i phenotype might be rescued by a high concentration of CO2. It is well known that a high CO2 concentration can suppress photorespiration and photorespiration-dependent phenotypes of photorespiration-defective mutants (Somerville and Ogren, 1980; Foyer et al., 2009; Peterhansel et al., 2010). However, increased CO2 concentration failed to rescue the reduced PSII efficiency of the c1c2i plants (Supplemental Fig. S9). Consistent with this result, the c1c2i plants grown at a high level of CO2 (5%) also showed a similar morphological phenotype as that grown in normal air (approximately 0.03% CO2; Supplemental Fig. S10). Therefore, the reduced PSII efficiency and growth defects of the c1c2i plants may not be simply explained by the defective photorespiration. It should be noted that our result does not necessarily imply a lack of function of the γCA subcomplex in photorespiration, because all three γCA genes are apparently expressed normally in the c1c2i mutant (Supplemental Fig. S5).
The biochemical function that γCAL may play in complex I is not clear at present, nor is it clear why the reduced level of γCAL1 and γCAL2 proteins affect photomorphogenesis. As expected, γCAL2 showed no carbonic anhydrase activity (Supplemental Fig. S11A). On the other hand, it is known that the γCAs contain the hexapeptide repeats ([LIV]-X2-[STAV]-X) that form a left-handed β-helix structure (Gaedeke et al., 2001; Perales et al., 2004; Ferry, 2010). Sequence analysis and structural modeling indicate that both γCAL proteins contain the hexapeptide repeats and the left-handed β-helix folds (Supplemental Fig. S11B). The left-handed β-helix structure is widely found in members of the acyltransferase superfamily (Raetz and Roderick, 1995). This superfamily contains enzymatically unrelated proteins, including acetyltransferases, such as Ser acetyltransferases of bacteria and mitochondrial Ser acetyltransferases of Arabidopsis (Parisi et al., 2004; Pye et al., 2004; Haas et al., 2008). The left-handed β-helix structure constitutes the active site of the acetyltransferases (Pye et al., 2004). Interestingly, γCAL2 appears to exhibit a protein acetyltransferase activity in vitro (Supplemental Fig. S11C). The in vivo substrate of γCAL2 remains unclear at present. Our attempts to test a possible acetyltransferase activity with other subunits of complex I was unsuccessful, which may be due to the difficulties in the purification of native subunits of this membrane complex. Given that fatty acids of seed lipids are converted to acetyl-CoA during germination for the synthesis of compounds essential for germination and early seedling growth, a potential acetyltransferase activity of γCAL2, which utilizes acetyl-CoA as a substrate in vitro, may partially explain the germination defect of the γacl1γcal2 double mutant. It is conceivable that the energy metabolism is important for photomorphogenesis in general, and certain malfunctions in mitochondria may adversely affect photomorphogenesis. Specifically, it is tempting to speculate that γCAL proteins may acetylate other mitochondrial proteins in response to the changing light environment and acetyl-CoA concentration to indirectly affect plant photomorphogenesis. This proposition, however, remains to be further investigated.
MATERIALS AND METHODS
Most Arabidopsis (Arabidopsis thaliana) γca and γcal mutants and all transgenic lines described in this study are from the Columbia accessions, except the γcal2 mutant, which is from the Wassilewskija accession. All the mutants were obtained from the Arabidopsis Biological Resource Center. Plant transformation and genetic, photomorphogenic, DNA, and RNA analyses were as described previously (Yu et al., 2007; Liu et al., 2008; Li et al., 2011). For the embryonic phenotype analyses, young (green) and mature (brown) siliques (15–20 per sample) were collected from 3- to 4-week-old plants, opened with a needle using a dissecting microscope, and examined with the microscope. To measure germination rates, seeds from individually collected siliques were grown in Murashige and Skoog (MS) medium for 7 d under continuous light before counting the germinating seeds. Additional methods and primers (Supplemental Table S2) used in the genomic and quantitative PCR can be found in the Supplemental Materials and Methods S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence analysis of the γCAL and γCA proteins.
Supplemental Figure S2. Light-independent mRNA expression of γCAL1 and γCAL2.
Supplemental Figure S3. Genotyping of the γcal1 and γcal2 mutants.
Supplemental Figure S4. Phenotypes of adult plants of the indicated genotypes.
Supplemental Figure S5. mRNA expression of the γCA genes are not affected in the c1c2i knockdown plants.
Supplemental Figure S6. Analyses of transgenic lines expressing the CAL2-RNAi construct in the wild-type background (c2i).
Supplemental Figure S7. Morphological defects of leaves of the c1c2i knockdown plants.
Supplemental Figure S8. The growth retardation phenotype of the c1c2i knockdown plants is not rescued by the high-CO2 growth condition.
Supplemental Figure S9. Reduced expression of the γCAL1 and γCAL2 genes modestly affects the maximum quantum efficiency of PSII.
Supplemental Figure S10. Normal photosynthetic apparatus of the c1c2i mutant plants.
Supplemental Figure S11. Possible biochemical functions of the γCAL proteins.
Supplemental Table S1. Normal gametogenesis of the c1c2C2c2 and C1c1c2c2 hemizygotes.
Supplemental Table S2. Oligonucleotide primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Fulu Chen for his contribution to this work and Figure 2A.
Glossary
- MS
Murashige and Skoog
- YFP
yellow fluorescent protein
References
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäurle I, Dean C. (2006) The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Brauna H-P, Zabaleta E. (2007) Carbonic anhydrase subunits of the mitochondrial NADH dehydrogenase complex (complex I) in plants. Physiol Plant 129: 114–122 [Google Scholar]
- Bultema JB, Braun HP, Boekema EJ, Kouril R. (2009) Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim Biophys Acta 1787: 60–67 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small ID. (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. (2005) Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc Natl Acad Sci USA 102: 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G. (2003) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Baradaran R, Sazanov LA. (2010) The architecture of respiratory complex I. Nature 465: 441–445 [DOI] [PubMed] [Google Scholar]
- Ferry JG. (2010) The gamma class of carbonic anhydrases. Biochim Biophys Acta 1804: 374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Bloom AJ, Queval G, Noctor G. (2009) Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu Rev Plant Biol 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedeke N, Klein M, Kolukisaoglu U, Forestier C, Müller A, Ansorge M, Becker D, Mamnun Y, Kuchler K, Schulz B, et al. (2001) The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J 20: 1875–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N. (1998) Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J Mol Biol 277: 1033–1046 [DOI] [PubMed] [Google Scholar]
- Guénebaut V, Vincentelli R, Mills D, Weiss H, Leonard KR. (1997) Three-dimensional structure of NADH-dehydrogenase from Neurospora crassa by electron microscopy and conical tilt reconstruction. J Mol Biol 265: 409–418 [DOI] [PubMed] [Google Scholar]
- Haas FH, Heeg C, Queiroz R, Bauer A, Wirtz M, Hell R. (2008) Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells. Plant Physiol 148: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Zickermann V, Brandt U. (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329: 448–451 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH. (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297 [DOI] [PubMed] [Google Scholar]
- Ito S, Song YH, Imaizumi T. (2012) LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Mol Plant 5: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C. (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araujo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O. (2012) nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J 71: 413–426 [DOI] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Klodmann J, Braun HP. (2011) Proteomic approach to characterize mitochondrial complex I from plants. Phytochemistry 72: 1071–1080 [DOI] [PubMed] [Google Scholar]
- Klodmann J, Sunderhaus S, Nimtz M, Jänsch L, Braun HP. (2010) Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell 22: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang Q, Yu X, Liu H, Yang H, Zhao C, Liu X, Tan C, Klejnot J, Zhong D, et al. (2011) Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (Trp) triad-dependent photoreduction. Proc Natl Acad Sci USA 108: 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C. (2011a) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M, Lin C. (2011b) The action mechanisms of plant cryptochromes. Trends Plant Sci 16: 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW. (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Kim WY, Somers DE, Kay SA. (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Morgan DJ, Sazanov LA. (2008) Three-dimensional structure of respiratory complex I from Escherichia coli in ice in the presence of nucleotides. Biochim Biophys Acta 1777: 711–718 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Parisi G, Perales M, Fornasari MS, Colaneri A, González-Schain N, Gómez-Casati D, Zimmermann S, Brennicke A, Araya A, Ferry JG, et al. (2004) Gamma carbonic anhydrases in plant mitochondria. Plant Mol Biol 55: 193–207 [DOI] [PubMed] [Google Scholar]
- Perales M, Eubel H, Heinemeyer J, Colaneri A, Zabaleta E, Braun HP. (2005) Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J Mol Biol 350: 263–277 [DOI] [PubMed] [Google Scholar]
- Perales M, Parisi G, Fornasari MS, Colaneri A, Villarreal F, González-Schain N, Echave J, Gómez-Casati D, Braun HP, Araya A, et al. (2004) Gamma carbonic anhydrase like complex interact with plant mitochondrial complex I. Plant Mol Biol 56: 947–957 [DOI] [PubMed] [Google Scholar]
- Peterhansel C, Horst I, Niessen M, Blume C, Kebeish R, Kurkcuoglu S, Kreuzaler F. (2010) Photorespiration. The Arabidopsis Book 8: e0130, doi/10.1199/tab.0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye VE, Tingey AP, Robson RL, Moody PC. (2004) The structure and mechanism of serine acetyltransferase from Escherichia coli. J Biol Chem 279: 40729–40736 [DOI] [PubMed] [Google Scholar]
- Raetz CR, Roderick SL. (1995) A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science 270: 997–1000 [DOI] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM. (2007) Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, Larkin RM. (2009) Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol 182: 367–379 [DOI] [PubMed] [Google Scholar]
- Sabar M, De Paepe R, de Kouchkovsky Y. (2000) Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol 124: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, Coupland G. (2004) Induction of flowering by seasonal changes in photoperiod. EMBO J 23: 1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. (1980) Photorespiration mutants of Arabidopsis thaliana deficient in serine-glyoxylate aminotransferase activity. Proc Natl Acad Sci USA 77: 2684–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderhaus S, Dudkina NV, Jänsch L, Klodmann J, Heinemeyer J, Perales M, Zabaleta E, Boekema EJ, Braun HP. (2006) Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J Biol Chem 281: 6482–6488 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM. (2005) Remembering winter: toward a molecular understanding of vernalization. Annu Rev Plant Biol 56: 491–508 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW. (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Yang H-Q, Wu Y-J, Tang R-H, Liu D, Liu Y, Cashmore AR. (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C. (2007) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C. (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.