Abstract
Background
Starch serves as a temporal storage of carbohydrates in plant leaves during day/night cycles. To study transcriptional regulatory modules of this dynamic metabolic process, we conducted gene regulation network analysis based on small-sample inference of graphical Gaussian model (GGM).
Results
Time-series significant analysis was applied for Arabidopsis leaf transcriptome data to obtain a set of genes that are highly regulated under a diurnal cycle. A total of 1,480 diurnally regulated genes included 21 starch metabolic enzymes, 6 clock-associated genes, and 106 transcription factors (TF). A starch-clock-TF gene regulation network comprising 117 nodes and 266 edges was constructed by GGM from these 133 significant genes that are potentially related to the diurnal control of starch metabolism. From this network, we found that β-amylase 3 (b-amy3: At4g17090), which participates in starch degradation in chloroplast, is the most frequently connected gene (a hub gene). The robustness of gene-to-gene regulatory network was further analyzed by TF binding site prediction and by evaluating global co-expression of TFs and target starch metabolic enzymes. As a result, two TFs, indeterminate domain 5 (AtIDD5: At2g02070) and constans-like (COL: At2g21320), were identified as positive regulators of starch synthase 4 (SS4: At4g18240). The inference model of AtIDD5-dependent positive regulation of SS4 gene expression was experimentally supported by decreased SS4 mRNA accumulation in Atidd5 mutant plants during the light period of both short and long day conditions. COL was also shown to positively control SS4 mRNA accumulation. Furthermore, the knockout of AtIDD5 and COL led to deformation of chloroplast and its contained starch granules. This deformity also affected the number of starch granules per chloroplast, which increased significantly in both knockout mutant lines.
Conclusions
In this study, we utilized a systematic approach of microarray analysis to discover the transcriptional regulatory network of starch metabolism in Arabidopsis leaves. With this inference method, the starch regulatory network of Arabidopsis was found to be strongly associated with clock genes and TFs, of which AtIDD5 and COL were evidenced to control SS4 gene expression and starch granule formation in chloroplasts.
Keywords: Arabidopsis thaliana, Constans-like, Indeterminate domain 5, Graphical Gaussian model, Starch synthase 4, Transcriptional regulation
Background
Starch is an insoluble glucose polymer stored in seeds and storage organs of plants. The starch molecule is composed of two types of glucose polymers—amylose and amylopectin—and is organized to form a distinct structure called a starch granule. It is commonly accepted that biosynthesis of starch takes place during the day using an excess sugar residue from photosynthesis as a substrate. At night, starch granules in leaves are decomposed to sugars to be transported to seeds or storage organs and stored as reserved carbohydrates or used as precursors in other metabolic pathways [1-3]. Besides starch synthase, various enzymes and proteins have been identified to play unexpected roles in starch biosynthesis, metabolism and granule formation [4-11].
With regard to regulation of starch biosynthesis and metabolism, post-translational protein modifications have major impacts on controlling the enzyme activities [12-14]. Allosteric regulation of ADP-glucose pyrophosphorylase (AGPase) [15-17] and redox modulation of pullulanase-type debranching enzymes [18,19], glucan-water-dikinase (GWD) [20] and β-amylase [21] indicate the significance of post-translational regulatory mechanisms. Protein phosphorylation and formation of multi-protein complexes of starch synthase (SS), branching enzyme (BE), debranching enzyme (DBE), and starch phosphorylase (SP) suggest tight linkages of metabolic pathways through modification and physical interactions of the enzymes (reviewed in [12]). In addition to post-translational mechanisms, genes encoding starch metabolic enzymes are also known to be regulated under transcriptional control. In barley, a sugar-inducible transcription factor (TF) in the WRKY family, SUSIBA 2, is reported to act as an activator in endosperm starch biosynthesis [22]. In rice, a complex of a MYC protein (OsBP-5) and an EREBP protein (OsEBP-89) is proposed to be a transcriptional regulator of the rice Wx gene, whose product, namely the granule-bound starch synthase (GBSS), is responsible for synthesis of amylose in mature seeds [23]. Additional finding in Arabidopsis indicates that expression of the GBSS-I gene is controlled by 2 main clock TFs, circadian clock associated 1 (CCA1: At2g46830) and late elongated hypocotyl (LHY: At1g01060) [24]. The roles of these TFs suggest the significance of transcriptional mechanisms, although gene regulatory networks of starch metabolism remain largely uncharacterized.
Inference methods for construction of gene regulatory networks have been extensively developed after genome-wide microarray repositories became publicly available [25-31]. Reverse-engineering approaches of Arabidopsis gene regulatory network reconstruction utilizing large-scale microarray experiments have been previously proposed and revealed the Arabidopsis regulatory network models from different viewpoints [28-32]. Carrera and colleagues [30] applied a qualitative network model based on a probabilistic model and linear regression to 1,436 Arabidopsis microarrays, and analyzed topological parameters of the network. Their result showed that genes having cellular functions involved in responses and adaptation to environmental changes tended to have higher connectivity than genes not related to stress responses. Another approach of the gene network reconstruction from large-scaled Arabidopsis microarrays is proposed by Mao et al.[32]. They constructed a genome-wide co-expression network of Arabidopsis from 1,094 microarrays based on Pearson correlation and analyzed modular structures of the networks that are functionally related. Significantly enriched pathway terms were then analyzed for the predicted modules. One module was defined to be enriched in starch metabolism; 9 out of 10 genes contained in this module were related to starch metabolism. Since the genes in the same module were predicted from co-expression across various conditions, it is suggested that these starch metabolic genes are potentially co-regulated. The other method successfully utilized to construct a regulatory network is gene expression analysis using a modified graphical Gaussian model (GGM) [33,34]. The modified GGM is considered appropriate for analysis of microarray data that usually has a high-dimensionality problem (i.e. the number of genes is much higher than the number of measurements). This technique has been applied to 2,045 Arabidopsis microarrays to construct a gene network which can be subdivided to sub-network structures [29]. A number of sub-networks identified through this approach are suggested to be related to metabolism and stress responses. One of them is considered as a starch catabolism sub-network, where 7 out of 15 genes present in the network are apparently relevant to starch degradation pathways [29].
Given these backgrounds, systematic analysis of microarray data appears to provide insights into gene regulatory networks of starch metabolism in Arabidopsis. In this work, an inference model was constructed from a diurnal cycle microarray dataset to identify candidate transcriptional regulators of plant starch metabolism. This approach is based on evidence that biosynthesis and degradation of leaf starch is completed within 24 hours [3,35-37], and hypothesizes that regulators of our interests are co-expressed and oscillated with genes involved in starch metabolic processes under a day-night cycle [24,36-38]. Firstly, we identified genes whose expression profiles changed over an observed 24-hr time period. Among these temporally regulated ‘significant’ genes, a set of starch metabolic genes, TFs, and clock genes was utilized for the construction of gene association networks using the small sample inference framework of GGM [33,34]. Subsequently, a few pairs of TFs and starch metabolic genes were selected based on their correlation coefficients from global co-expression profiles. Finally, validation of the relationships between the selected TFs and starch metabolic genes were carried out using TF loss-of-function mutant lines. The results obtained from this study have led us to identify the involvement of TFs, indeterminate domain5 (AtIDD5: At2g02070) and constans-like (COL: At2g21320), in transcriptional regulation of an Arabidopsis starch metabolic gene. The work presented here provides a model for systematic understanding of regulatory networks of starch metabolic pathway applicable for modification of starch synthesis and accumulation.
Results and Discussion
Initial screening of significant genes in a diurnal cycle
A time-series significant analysis was performed using the Extraction of Differential Gene Expression (EDGE) software package. This software can identify differentially expressed genes from both typical and time-course microarray experiments [39,40]. In this research, the software was applied to detect changes in Arabidopsis gene expression occurring within a 24 hour period [36]. This data set is the time-course measurement of transcripts extracted from fully-expanded leaves of Arabidopsis grown under a 12-hour-light and 12-hour-dark (12 L/12D) condition. The samples are taken after 1, 2, 4, 8 and 12 hours in darkness or light, starting from the end or beginning of the light period, respectively. The concept of EDGE is based on the hypothesis testing of differential gene expression patterns fitted by natural cubic spline interpolation. Genes whose expression patterns deviate from a standard line within a 24-hour period would be detected as differentially expressed genes under a diurnal cycle. From approximately 22,000 Arabidopsis genes on the ATH-1 Affymetrix genome arrays, 1,480 genes were detected as significant genes (Q < 0.01). These significant genes were clustered by k-means clustering, then functionally classified in MapMan, a visualization tool for functional classification of Arabidopsis[41]. Genes encoding TFs and those related to starch metabolism were used as inputs for GGM network construction.
Co-expression analysis of 1,480 significant genes using k-means clustering
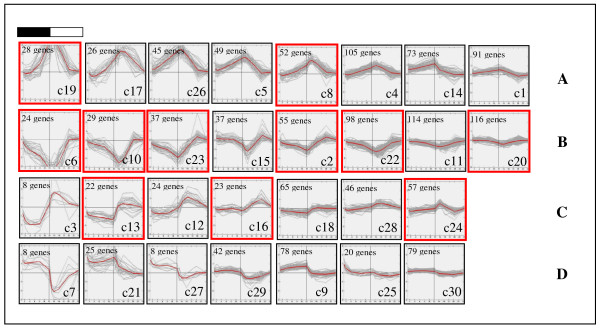
Since the leaf starch content of Arabidopsis increases in the light period and decreases in the dark period [36], we hypothesized that the high expression levels of genes in the starch biosynthetic pathway would be observed during the day, while those in the degradation pathway would occur at night. To test this hypothesis, the expression profile of all the significant genes were examined by k-means clustering (k = 30). All clusters were subjectively divided into 4 groups (Figure 1). Group A includes the genes whose mRNA levels are increased in the dark and decreased in the light. In contrast, the group B members showed their mRNAs increased in the light and decreased in the dark. Group C consists of the gene members whose expression remains relatively stable except their mRNA levels were observed to increase at the dark-to-light transition phase. Group D is similar to group C but the expression goes down at the dark-to-light transition phase. According to our hypothesis, the starch biosynthetic genes are expected to be clustered in group B and C since the gene expression increases when the light period starts. On the other hand, the genes in starch degradation whose expression are expected to increase in the dark period should be the members of group A and D.
Figure 1.
Thirty clusters of 1,480 significant genes using k-means clustering (k = 30). The number of gene members is shown at the top left of each cluster. The clusters where starch genes are present are indicated by red boxes. Four groups of 30 clusters were categorized by their expression patterns affected on light. (A) Tightly dark-positively and light-negatively regulated genes: the clusters of genes induced in the dark and repressed in the light periods. (B) Tightly dark-negatively and light-positively regulated genes: the clusters of genes repressed in the dark and induced in the light periods. (C) Light-positively regulated genes: the cluster of genes whose expressions are constant but the levels are elevated at dark to light phase transition. (D) Light-negatively regulated genes: the cluster of genes whose expressions are constant but the levels decline at dark to light phase transition. The black and white bar on the top of the figure corresponds to the dark and light period.
According to Smith et al.[36], there are 48 genes related to starch metabolism. In the group of 1,480 significant genes identified in this study, 21 out of 48 starch metabolic genes were present, 9 of which are related to starch synthesis and 12 of which are known to function in starch degradation pathway (Table 1). By using k-means clustering, these starch metabolic genes were classified into 11 different clusters (Figure 1). Most of the starch synthesis genes (7 out of 9 genes) are observed in the clusters of groups B and C. This result supports the hypothesis that starch biosynthetic genes should be up-regulated during the day time. The two starch biosynthetic genes that do not follow this rule are those coding for granule-bound starch synthase (GBSS: At1g32900) and starch synthase 2 (SS2: At3g01180). The expressions of both genes show distinct diurnal patterns distinguishable from other starch synthase genes [36]. Since their products are reported to be embedded in the starch granules that are daily destroyed at the night period [36], their high expression levels after the onset of the light are considered necessary to regenerate GBSS and SS2 proteins for starch biosynthesis during the light period. In contrast, only 2 of 12 total genes encoding for enzymes in starch degradation pathways, α-amylase 2 (a-AMY2: At1g76130) and β-amylase 9 (b-AMY9: At5g18670), showed expression patterns correlated with the starch content profile. One explanation is that starch degrading enzymes might need a lag time in post-transcriptional or translational processes to become catalytically functional.
Table 1.
Twenty-one differentially expressed starch genes (Q < 0.01) under the diurnal condition
| AGI | Name | Description | Cluster | Group |
|---|---|---|---|---|
| At1g32900 |
GBSS |
Granule-bound starch synthase |
19 |
A |
| At1g76130 |
a-AMY2 |
α-amylase, putative |
8 |
A |
| At3g01180 |
SS2 |
Starch synthase 2 |
8 |
A |
| At5g18670 |
b-AMY9 |
β-amylase, putative (BMY3) |
19 |
A |
| At1g03310 |
ISA2/DBE1 |
Isoamylase, putative |
2 |
B |
| At1g10760 |
GWD1 |
Glucan water dikinase |
10 |
B |
| At1g69830 |
a-AMY3 |
α-amylase, putative |
6 |
B |
| At2g36390 |
SBE2-1/BE3 |
1,4-α-glucan branching enzyme |
22 |
B |
| At2g40840 |
DPE2 |
Glycoside hydrolase family 77 protein |
23 |
B |
| At3g29320 |
StP, Plast |
Glucan phosphorylase, putative |
6 |
B |
| At3g46970 |
Stp, Cyt |
Starch phosphorylase, putative |
6 |
B |
| At4g09020 |
ISO3 |
Isoamylase, putative |
6 |
B |
| At4g18240 |
SS4 |
Starch synthase 4 |
22 |
B |
| At5g11720 |
AGLU like-4 |
α-glucosidase1(AGLU1) |
20 |
B |
| At5g24300 |
SS1 |
Starch synthase 1 |
2 |
B |
| At5g26570 |
GWD/ PWD |
Glycoside hydrolase starch-binding domain-containing protein |
10 |
B |
| At5g51820 |
PGM |
Phosphoglucomutase |
22 |
B |
| At5g64860 |
DPE1 |
disproportionating enzyme, putative |
10 |
B |
| At2g32290 |
b-AMY6 |
β-amylase, putative |
13 |
C |
| At2g39930 |
ISA1 |
Isoamylase, putative |
16 |
C |
| At4g17090 | b-AMY3 | β-amylase (CT-BMY) | 24 | C |
Cluster indicates the cluster number analyzed by k-means clustering (k = 30). All clusters were subjectively divided into 4 groups—A, B, C, and D—according to the expression pattern in each cluster (Figure 1).
Functional categories of 1,480 significant genes
The 1,480 significant genes were categorized according to the functions defined in MapMan (Table 2). The largest functional group containing 471 genes, which accounted for ~32% of total significant genes, was classified as “Not assigned”. Out of the 471 genes in this group, 276 genes (~59%) were identified as unknown expressed proteins. The second largest group was the “RNA” group. This group contains 177 genes whose functions are related to RNA processing, transcription process, and regulation of transcription. As a result, 106 TF genes were assigned in the RNA group. Among 24 significant genes in the “Major carbohydrate metabolism” group (Table 2), 21 genes are starch-related, while the other 3 are related to sucrose metabolism. Additionally, our significant gene set also includes both clock and clock-regulated genes such as circadian clock associated 1 (CCA1: At2g46830), late elongated hypocotyl (LHY: At1g01060), early-flowering 3 (ELF3: At2g25930), phytochrome B (PHYB: At2g18790), timing of cab expression 1 (TOC1: At5g61380), and casein kinase II beta-chain (CKB3: At3g60250). These TF, clock, and starch metabolic genes whose mRNA levels are significantly modulated during a diurnal cycle were used for reconstruction of transcriptional regulatory network of starch metabolism.
Table 2.
Functional categories of 1,480 significant genes defined by the MapMan tool
| Functional category | Number of genes |
Group |
|||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Photosynthesis |
33 |
9 |
8 |
12 |
4 |
| Major carbohydrate metabolism |
24 |
6 |
15 |
3 |
0 |
| Minor carbohydrate metabolism |
17 |
6 |
3 |
4 |
4 |
| Glycolysis |
10 |
0 |
9 |
1 |
0 |
| Fermentation |
2 |
0 |
2 |
0 |
0 |
| Gluconeogenesis/glyoxylate cycle |
3 |
1 |
0 |
0 |
2 |
| Oxidative pentose phosphate pathway |
4 |
0 |
3 |
0 |
1 |
| TCA / organism Transformation |
11 |
3 |
4 |
3 |
1 |
| Mitochondrial electron transport /ATP synthesis |
7 |
3 |
2 |
0 |
2 |
| Cell wall |
19 |
6 |
6 |
2 |
5 |
| Lipid metabolism |
41 |
13 |
12 |
9 |
7 |
| N-metabolism |
2 |
1 |
0 |
0 |
1 |
| Amino acid metabolism |
38 |
14 |
13 |
5 |
6 |
| S-assimilation |
2 |
2 |
0 |
0 |
0 |
| Metal handling |
7 |
3 |
3 |
1 |
0 |
| Secondary metabolism |
32 |
21 |
5 |
5 |
1 |
| Hormone metabolism |
36 |
11 |
13 |
8 |
4 |
| Co-factor and vitamin metabolism |
8 |
2 |
4 |
0 |
2 |
| Tetrapyrrole synthesis |
6 |
3 |
2 |
1 |
0 |
| Stress |
61 |
11 |
24 |
16 |
10 |
| Redox regulation |
26 |
9 |
11 |
3 |
3 |
| Polyamine metabolism |
6 |
4 |
1 |
1 |
0 |
| Nucleotide metabolism |
16 |
3 |
9 |
2 |
2 |
| Biodegradation of xenobiotics |
4 |
1 |
0 |
1 |
2 |
| C1-metabolism |
6 |
1 |
4 |
0 |
1 |
| Miscellaneous |
73 |
20 |
30 |
14 |
9 |
| RNA |
177 |
55 |
57 |
37 |
28 |
| DNA |
29 |
5 |
17 |
5 |
2 |
| Protein |
157 |
41 |
64 |
17 |
35 |
| Signalling |
74 |
25 |
24 |
12 |
13 |
| Cell |
53 |
9 |
26 |
5 |
13 |
| Development |
37 |
9 |
12 |
10 |
6 |
| Transport |
81 |
40 |
19 |
15 |
7 |
| Not assigned | 471 | 159 | 148 | 67 | 97 |
Groups A, B, C, and D correspond to classifications in k-means clustering analysis presented in Figure 1.
Gene regulation network of starch metabolism in Arabidopsis leaves
To further investigate the transcriptional regulation of starch metabolic pathway, only metabolic genes in starch metabolism and all possible regulators (i.e. TF and clock genes) were focused in this research. The 133 significant genes —composed of 21 starch metabolic genes, 106 TF genes, and 6 clock genes— were subjected to GGM network construction [33]. Since GGM generates a conditional network depending on an input genes set, different sets of input genes are influent to the resulted network. Preservation of the TF-starch relationships in the networks constructed from expanded gene sets indicated the robustness of the network reconstructed by the focused set of starch metabolic and regulator genes (discussed in the following section).
The gene association network of starch metabolic pathway was constructed using small sample inference of GGM implemented in the R package ‘GeneNet’ [33]. The transcript profiles of 133 significant genes were retrieved from the original microarray data [36], and transformed to log-base 2 scales before inferring the gene association network. In the resulting network, a node represents a gene and an interaction between 2 genes is called an edge. Each edge indicates a correlated expression of any 2 genes after removing effects of other genes in a study set. The hypothesis testing of non-zero partial correlation and false discovery rate (FDR), multiple testing correction were employed to obtain significant correlation coefficients (i.e. significant edges) in the final network. From a total of 133 genes, the final network derived from GGM analysis at Q < 0.05 consisted of 117 nodes (or genes), corresponding with 16 starch (7 synthesis and 9 degradation), 97 TF, and 4 clock genes, and 266 edges (or, an association between 2 genes) (Figure 2). These edges could be the associations between 1) regulator and regulator genes (i.e. TF-TF, clock-TF, and clock-clock), 2) regulator and target genes (i.e. TF-starch and clock-starch), and 3) target and target genes (i.e. starch-starch). There are 215, 42, and 9 edges for the first, second, and third edge types, respectively.
Figure 2.
The gene association network of starch metabolism inferred from GGM (Q < 0.05). The network contains 117 nodes and 266 edges. A pink rectangle represents a TF gene; an orange diamond represents a clock gene; a green circle represent a starch gene; a black solid line represents an association with positive correlation; and a blue broken line represents an association with negative correlation.
Since 80% of the genes used in the network reconstruction are TF genes, predominant interactions in the final network represent those between TFs. To verify the significance of these TF-TF interactions and the robustness of the starch genes association network, especially the TF-starch relationships shown in Figure 2, another gene association network was reconstructed by expanding a set of metabolic genes from only genes in starch metabolism to genes in broader carbon-related metabolisms. The same set of 106 TFs, 6 clock genes, and 171 metabolic genes selected from 11 carbon-related functional groups categorized to be related to photosynthesis, major carbohydrate metabolism, minor carbohydrate metabolism, glycolysis, fermentation, gluconeogenesis/glyoxylate cycle, oxidative pentose phosphate pathway, TCA/organism transformation, mitochondrial electron transport/ATP synthesis, cell wall, and lipid metabolism by MapMan (Table 2) were utilized for network reconstruction. It should be noted that all 21 starch genes are present in the major carbohydrate metabolism category, thus included in the metabolic gene set. The gene association network of carbon-related metabolisms is shown in Additional file 1: Figure S1. In this expanded network, 151 out of 215 regulator- regulator edges (70%) and 26 out of 42 TF-starch or clock-starch relationships (62%) were identical with the starch network shown in Figure 2. All 6 TF candidates and their relationships to 5 starch genes that will be further experimentally validated (discussed in the following section) exist in the expanded network. The result indicates that most of the regulatory relationships of the starch gene association network are robust and preserved even when expanding an input gene set to carbon-related metabolic genes.
According to Figure 3A, the node degree of TF and clock genes ranged from 1 to 14 connections. Among the starch metabolic genes, β-amylase 3 (b-AMY3: At4g17090) was detected as the most-connected node or a hub gene with 15 neighbors (Figure 3B). This result may indicate the importance of b-AMY3 in the starch metabolism. The b-AMY3 represents a chloroplastic β-amylase [21,36,42] with a possible role in the leaf starch degradation process [3,43-45]. In the b-AMY3 sub-network (Figure 3B), there are 2 starch metabolic genes, starch synthase 4 (SS4: At4g18240) and β-amylase 6 (b-AMY6: At2g32290), that showed positive correlations with b-AMY3. There are reports indicating that SS4 involves in the starch granule initiation process [6,11], and b-AMY6 is repressed under heat shock stress [46]. However, no functional correlation among these 3 starch metabolic genes has ever been reported.
Figure 3.
β-amylase 3 is the most frequently connected gene (a hub gene). (a) The node degree distribution of starch and TF genes in the gene association network. The gene with the highest degree of connection is β-amylase 3 (b-AMY3: At4g17090) as indicated by a black arrow. (b) Sub-network of the most-connected node (a hub gene), β-amylase 3 at Q < 0.05. A pink rectangle represents a TF gene; an orange diamond represents a clock gene; a green circle represents a starch gene; a black solid line represents an association with positive correlation; and a blue broken line represents an association with negative correlation.
In addition to gene-to-gene association with two starch metabolic genes, b-AMY3 was also observed to have positive correlation with various TF genes and a clock gene, TOC1. TOC1 is an evening gene expressed during the night period with a major role in circadian rhythm [47-49]. From the k-means clustering, expression of b-AMY3 increased slightly during the night period, decreased at the dark-to-light transition phase, then increased again a few hours later. Our findings are not only in agreement with the results indicating regulation of b-AMY3 expression under diurnal and circadian rhythms [36,50,51], but they also suggest the circadian control of b-AMY3 via TOC1.
From the GGM network construction, we additionally found that the sub-networks of two starch metabolic genes encoding GBSS (At1g32900) and disproportionating enzyme (DPE1: At5g64860) and their TF neighbours are entirely separated from the rest of the network (Figure 2). These isolated modules indicate specific correlation between starch metabolic genes and their connected TFs. The LIM domain-containing protein (At2g39900) associated with DPE1 in the network is in fact WLIM2a, an actin-bundling protein that functions in cytoskeleton organization [52]; its gene expression is regulated by pickle (PKL), a member of CHD3 chromatin- remodelling protein involved in seed germination of Arabidopsis[53]. The other isolated module represents the association between GBSS and two zinc finger family proteins, constans-like (COL: At2g21320) and constans-like 7 (COL7: At1g73870). In rice, MYC and EREBP are known to act synergistically in transcriptional regulation of the rice Wx gene [23]. Since these homologues were not included in the GBSS sub-network of Arabidopsis, the results possibly suggest a difference in the mechanisms controlling the biosynthesis of storage starch (i.e. in rice endosperm) and transitory starch (i.e. in Arabidopsis leaves).
Sub-networks for transcriptional regulation of starch metabolism
To identify candidate TFs that may play regulatory roles in starch metabolism, we focused on the relationships between starch metabolic genes and their immediately connected TFs. The starch sub-network excerpted accordingly contained 49 nodes (16 starch, 1 clock, and 32 TF genes) and 79 edges (Figure 4A). Details of the genes in the starch sub-network are summarized in Additional file 2: Table S1. Within the starch sub-network, we observed 5 types of gene-to-gene interactions or edges; those were interactions between starch metabolic genes (8 edges), between TF genes (26 edges), between starch metabolic and TF genes (42 edges), between clock and starch metabolic genes (2 edges), and between clock and TF genes (1 edge).
Figure 4.
The starch sub-network of 16 starch genes and their first step neighbours (Q < 0.05). (a) The graphical display of the starch sub-network. A pink rectangle represents a TF gene; an orange diamond represents a clock gene; a green circle represents a starch gene; a black solid line represents an association with positive correlation; and a blue broken line represents an association with negative correlation. (b) The metabolic display of the starch sub-network. A pink oval represents a TF gene; A red oval represents a TF gene predicted to have physical binding sites in target genes according to the comparative TF family analysis; an orange diamond represents a clock gene; a green rectangle represents a starch gene; a blue rectangle represents a metabolite; a black line represents an association with positive correlation; a red line represents an association with negative correlation; and a blue line represents a reaction in starch metabolism.
The Arabidopsis gene networks were previously reconstructed from global microarray conditions based on GGM [29] and co-expression analysis [32]. Interestingly, the sub-networks related to starch metabolism were extracted from both studies, even though different algorithms were applied. The sub-networks derived from GGM [29] and co-expression analysis [32] contain a total of 15 and 10 genes, and from these gene set 10 and 9 were identified as starch metabolic genes, respectively. There are 6 genes in starch degradation, i.e. disproportionating enzyme 1 and 2 (At5g64860 and At2g40840, respectively), glucan water dikinase 1 (starch excess1 - At1g10760), glucan phosphatase (starch excess 4 - At3g52180), starch phosphorylase 2 (At3g46970), and α-amylase 3 (At1g69830), and one gene in starch synthesis, branching enzymes 3 (At2g36390) that both of these networks have in common. From total 15 starch metabolic genes identified in the sub-networks of previous studies, 7 genes, which are starch synthase 4 (At4g18240), branching enzyme 3 (At2g36390), disproportionating enzyme 2 (At2g40840), phosphoglucan water dikinase (At5g26570), isoamylase 2 (At1g03310), cytosolic and plastidial starch phosphorylase (At3g46970 and At3g29320, respectively), were also observed in our starch sub-network (Additional file 2: Table S1). These results suggest coherent expressions of starch metabolic genes, especially those in the starch degradation process. It is worth to note that the starch sub-networks derived from those studies contain mainly metabolic genes, except one clock-regulated gene encoding pseudo-response regulator 3 (APRR3: At5g60100) which was identified in the starch sub-network derived from the GGM analysis [29]. However, this clock gene was not present in our starch sub-network.
Prediction of TF binding sites in promoter sequences of target genes
The 42 interactions between 12 starch and 32 TF genes were further analyzed by searching for the presence/absence of TF binding sites in the promoter region of starch metabolic genes (Table 3). The analysis conducted for prediction of physical binding of TF to target genes is TF family-based, thus, it stands on the assumption that different TFs of the same TF family most likely attach to the same TF-binding site on the promoter region of a target gene.
Table 3.
The TF-starch relationships from the starch sub-network
|
Starch Metabolic Gene (AGI code) |
Starch Metabolic Gene (Gene Name) |
pCor / Interaction1 |
TF (AGI code) |
TF (Gene Name) |
TF Family |
AthaMap2 |
Rank of TF-starch correlation in ATTED |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| 2-kb upstream | Correlation | Rank in all genes (total = 22,263 ) | Ranks in all TFs (total = 1,849) | Rank in specific TF family/No. of TFs in a family | ||||||
| At1g03310 |
ISA2/DBE1 |
-0.0756 / n |
At5g46710 |
zinc-binding |
PLATZ |
NA |
-0.05 |
11356 |
818 |
2/7 |
| At1g03310 |
ISA2/DBE1 |
0.0674 / p |
At2g34720 |
CBF-B/NF-YA |
CCAAT-HAP2 |
NA |
0.03 |
8424 |
557 |
6/10 |
| At1g03310 |
ISA2/DBE1 |
0.0733 / p |
At1g05805 |
bHLH |
bHLH |
N |
0.165 |
4598 |
241 |
27/114 |
| At1g32900 |
GBSS |
0.0736 / p |
At1g73870 |
COL7 |
C2C2-CO-like |
NA |
0.609 |
17 |
4 |
3/42 |
| At1g32900 |
GBSS |
0.0686 / p |
At2g21320 |
COL |
C2C2-CO-like |
NA |
0.608 |
18 |
5 |
4/42 |
| At1g76130 |
a-AMY2 |
0.0665 / p |
At2g31070 |
TCP |
TCP |
N |
0.174 |
3347 |
344 |
7/20 |
| At1g76130 |
a-AMY2 |
0.0758 / p |
At1g02340 |
HFR1/BHLH26 |
bHLH |
N |
-0.019 |
11812 |
1052 |
56/114 |
| At1g76130 |
a-AMY2 |
-0.1268 / n |
At5g65310 |
HB-5 |
HB |
N |
-0.372 |
21286 |
1809 |
87/89 |
| At2g32290 |
b-AMY6 |
0.0763 / p |
At1g22070 |
TGA3 |
bZIP |
Y |
0.111 |
3654 |
225 |
4/69 |
| At2g32290 |
b-AMY6 |
0.0676 / p |
At1g50420 |
SCL3 |
GRAS |
NA |
0.07 |
4806 |
328 |
10/32 |
| At2g32290 |
b-AMY6 |
-0.0707 / n |
At1g07520 |
scarecrow |
GRAS |
NA |
-0.125 |
18495 |
1527 |
30/32 |
| At2g32290 |
b-AMY6 |
0.0818 / p |
At2g31070 |
TCP |
TCP |
N |
0.204 |
2008 |
111 |
4/20 |
| At2g39930 |
ISA1 |
0.0917 / p |
At5g05090 |
myb |
GARP-G2-like |
Y |
0.242 |
3728 |
181 |
6/39 |
| At2g39930 |
ISA1 |
0.0763 / p |
At1g05805 |
bHLH |
bHLH |
Y |
0.356 |
2106 |
81 |
9/114 |
| At2g39930 |
ISA1 |
0.0783 / p |
At5g39860 |
bHLH |
bHLH |
Y |
0.255 |
3490 |
164 |
22/114 |
| At2g39930 |
ISA1 |
-0.0704 / n |
At1g51950 |
IAA18 |
AUX-IAA |
NA |
0.056 |
8010 |
542 |
14/28 |
| At3g46970 |
Stp,Cyt |
0.0670 / p |
At1g05805 |
bHLH |
bHLH |
Y |
0.442 |
511 |
37 |
1/114 |
| At4g17090 |
b-AMY3 |
0.0761 / p |
At2g35940 |
Homeo domain |
HB |
Y |
0.233 |
2173 |
127 |
12/89 |
| At4g17090 |
b-AMY3 |
0.0674 / p |
At1g19700 |
HB-ZIP |
HB |
Y |
0.009 |
7815 |
644 |
41/89 |
| At4g17090 |
b-AMY3 |
0.0916 / p |
At4g39780 |
AP2 domain |
AP2-EREBP |
Y |
-0.179 |
18803 |
1537 |
110/130 |
| At4g17090 |
b-AMY3 |
0.0810 / p |
At3g58680 |
ethylene-responsive |
MBF1 |
NA |
-0.009 |
8743 |
713 |
1/3 |
| At4g17090 |
b-AMY3 |
0.1093 / p |
At1g76590 |
zinc-binding |
PLATZ |
NA |
0.015 |
7522 |
608 |
2/7 |
| At4g17090 |
b-AMY3 |
-0.0871 / n |
At1g77850 |
B3 |
ARF |
NA |
0.026 |
7013 |
569 |
7/18 |
| At4g17090 |
b-AMY3 |
0.0684 / p |
At5g48250 |
B-box |
C2C2-CO-like |
NA |
0.285 |
1615 |
74 |
9/42 |
| At4g17090 |
b-AMY3 |
0.0671 / p |
At3g07650 |
B-box |
C2C2-CO-like |
NA |
0.264 |
1805 |
92 |
10/42 |
| At4g17090 |
b-AMY3 |
0.1051 / p |
At5g61380 |
TOC1 |
C2C2-CO-like |
NA |
0.181 |
2903 |
191 |
14/42 |
| At4g17090 |
b-AMY3 |
-0.0941 / n |
At5g05090 |
myb |
GARP-G2 |
NA |
0.015 |
7574 |
611 |
19/39 |
| At4g17090 |
b-AMY3 |
-0.0774 / n |
At1g07520 |
scarecrow |
GRAS |
NA |
-0.139 |
16919 |
1385 |
30/32 |
| At4g17090 |
b-AMY3 |
-0.0757 / n |
At4g39410 |
WRKY |
WRKY |
N |
0.094 |
4649 |
365 |
16/63 |
| At4g18240 |
SS4 |
0.0928 / p |
At2g02070 |
AtIDD5 |
C2H2 |
Y |
0.542 |
158 |
9 |
1/122 |
| At4g18240 |
SS4 |
-0.0671 / n |
At3g21175 |
GATTF_20_A |
ZIM |
NA |
0.225 |
2276 |
119 |
1/16 |
| At4g18240 |
SS4 |
0.1016 / p |
At5g61380 |
TOC1 |
C2C2-CO-like |
NA |
0.569 |
124 |
5 |
3/42 |
| At4g18240 |
SS4 |
0.0720 / p |
At3g07650 |
B-box |
C2C2-CO-like |
NA |
0.514 |
201 |
13 |
5/42 |
| At5g11720 |
AGLU-like4 |
0.0687 / p |
At3g50700 |
C2H2 |
C2H2 |
Y |
0.295 |
2024 |
87 |
5/122 |
| At5g11720 |
AGLU-like4 |
0.0837 / p |
At5g60850 |
Dof |
C2C2-Dof |
Y |
0.135 |
5244 |
310 |
6/32 |
| At5g11720 |
AGLU-like4 |
0.0910 / p |
At2g18280 |
TULP2 |
TLP |
NA |
0.213 |
3463 |
169 |
3/11 |
| At5g24300 |
SS1 |
0.0961 / p |
At5g06770 |
KH-CCCH |
C3H |
NA |
0.422 |
1194 |
31 |
3/172 |
| At5g24300 |
SS1 |
-0.0786 / n |
At3g55770 |
LIM |
LIM |
NA |
-0.079 |
13020 |
995 |
7/13 |
| At5g24300 |
SS1 |
0.0817 / p |
At3g06160 |
B3 |
ABI3-VP1 |
N |
0.241 |
2965 |
136 |
4/45 |
| At5g51820 |
PGM |
-0.0829 / n |
At3g55770 |
LIM |
LIM |
NA |
-0.07 |
11712 |
853 |
6/13 |
| At5g64860 | DPE1 | 0.0663 / p | At2g39900 | WLIM2a | LIM | NA | 0.715 | 30 | 1 | 1/13 |
Global-expression correlations extracted from ATTED database and the correlation ranks were shown in the table.
1. pCor is partial correlation of a gene pair under the diurnal condition predicted by GGM. p and n represent a positive and negative interactions, respectively.
2. Y = Physical binding of TFs to target genes was predicted by the comparative TF family analysis; N = No physical binding of TFs was predicted; NA = no TF binding site information in AthaMap.
In the TF family-based comparative analysis, all possible plant TF binding sites in the 2-kb upstream region of the 12 target genes (i.e. starch metabolic genes) were first obtained using a web-based tool from AthaMap database http://www.athamap.de/[54-57]. The names of known TFs and their relative binding locations predicted within the 2-kb upstream region of 12 starch metabolic genes are summarized in Additional file 3: Table S2. From the list of all possible TF binding sites, physical binding was predicted between 10 TFs and 6 starch metabolic genes, shown as 11 starch-TF interactions in the starch sub-network (Table 3). For easier visualization, we included information on the presence of putative TF-binding sites and re-drew the regulatory model of genes in the starch sub-network (Figure 4B). The results indicate that 10 TFs show only a positive correlation with 6 starch metabolic genes and can be classified into 7 families.
Predicted regulatory modules for a model validation
The robustness of the predicted regulatory network model of starch metabolism was further verified using a diverse set of “condition-independent” microarray data. This analysis has been implemented to find how the expression patterns of any 2 genes under various conditions correlate. The data representing the relationship of Arabidopsis co-regulated genes was obtained from the ATTED-II database http://atted.jp/[58,59]. In ATTED-II, the pair-wise correlation coefficients of 22,263 Arabidopsis genes were calculated from 58 experiments of GeneChip microarray (1,388 arrays in total) using weighted Pearson correlation. The pair-wise correlation coefficients of 12 starch metabolic genes with all Arabidopsis TF genes (1,849 genes listed in this database) were ranked from highest to lowest value (Table 3).
The pair-wise correlation coefficients between given target genes and all TFs were observed as normally distributed in this condition-independent analysis. The TF was thus considered significant when its correlation coefficient with its target starch metabolic gene was higher than the population mean with 97.5% confidence analysed by the single-sample t-test (one-tailed). From all the TFs in the starch sub-network, 8 TF genes passed this cut-off by being significantly and highly correlated with their target gene expression, and they were preliminarily chosen as candidates for experimental validation (discussed in the following section). However, among these 8 candidates, the expression patterns of 2 TF genes, constans-like 9 (COL9: At3g07650) and bHLH (At1g05805), did not adhere to the simple assumption that a TF should express at the same time, or earlier, than its target gene. The diurnal expression patterns of both COL9 and bHLH indicated that their induction took place after the expression of their potential target starch metabolic genes (Additional file 4: Figure S2). Therefore, by excluding these 2 TFs, the rest 6 candidate TF genes—AtIDD5 (At2g02070), C2H2 (At3g50700), COL (At2g21320), COL7 (At1g73870), WLIM2a (At2g39900), and KH-CCCH (At5g06770), each predicted as regulators for the expression of SS4, α-glucosidase-like 4 (AGLU-like 4: At5g11720), GBSS (co-regulated by COL and COL7), DPE1, and starch synthase 1 (SS1: At5g24300), respectively (Table 4)—were selected as final candidates and subsequently used in experimental validation. According to the TF family-based prediction of binding sequences (Tables 3 and 4) of these 6 candidates, putative binding sites for zinc finger C2H2 type TFs were located in the promoter regions of their target genes, SS4 and AGLU-like 4. Accordingly, the gene-to-gene associations between AtIDD5 and SS4, and between C2H2 and AGLU-like 4, gain strong support from both the TF binding site prediction and the global co-expression analysis.
Table 4.
Six candidate TF genes and their ranks based on the global-expression correlations
| Starch metabolic gene | TF gene | TF family | Binding Site1 | ATTED Correlation2 | TF rank3 | Family rank4 | ||
|---|---|---|---|---|---|---|---|---|
| At4g18240 |
SS4 |
At2g02070 |
AtIDD5 |
C2H2 |
Y |
0.542 |
9 |
1/122 |
| At5g11720 |
AGLU- like 4 |
At3g50700 |
C2H2 |
C2H2 |
Y |
0.295 |
87 |
5/122 |
| At5g64860 |
DPE 1 |
At2g39900 |
WLIM2a |
LIM |
n.a. |
0.715 |
1 |
1/13 |
| At1g32900 |
GBSS |
At1g73870 |
COL7 |
C2C2-CO-like |
n.a. |
0.609 |
4 |
3/42 |
| At1g32900 |
GBSS |
At2g21320 |
COL |
C2C2-CO-like |
n.a. |
0.608 |
5 |
4/42 |
| At5g24300 | SS1 | At5g06770 | KH-CCCH | C3H | n.a. | 0.422 | 31 | 3/172 |
1. Y = potential binding site of a TF was found at the 2,000-base upstream region of target gene by the comparative TF family analysis; n.a. = information of a binding site of that TF is not available in AthaMap http://www.athamap.de/.
2. Pearson correlation calculated from the 58 GeneChip experiments including 1,388 arrays using the weighted Pearson correlation in ATTED database http://atted.jp/.
3. Rank of TF based on the ATTED correlation of 1,849 TF genes in the database.
4. Rank of TF based on the ATTED correlation of TF genes in each specific TF family. The numerator indicates the rank and the denominator indicates the number of genes in a specific TF family.
Expression analysis of target genes predicted in the regulatory modules
To experimentally verify the regulatory role of the 6 candidate TFs in the proposed TF-target model (Figure 4B; Table 4), the accumulation of starch metabolic gene transcripts was determined using homozygous knockout lines. T-DNA or transposon inserted knockout lines—Atidd5, c2h2, col, col7, wlim2a, and kh-ccch—were obtained from The Arabidopsis Biological Resource Centre (ABRC) [60] and The Nottingham Arabidopsis Stock Centre (NASC) [61]. The homozygous mutant plants were grown under the conditions described in [36] (See method). Rosette leaves at 3.90 developmental stage [62] were harvested 4 times within a 24- hour period, and used as materials for RNA extraction. The mRNA levels of TFs and target genes were quantified by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) analysis.
AtIDD5, C2H2, COL7, and KH-CCCH mRNAs were absent in Atidd5, c2h2, col7, and kh-ccch mutant lines, respectively, whereas COL and WLIM2a mRNAs were partially detected in col and wlim2a mutant lines, respectively (data not shown). The mRNA accumulations of target starch metabolic genes were then monitored in these mutant lines to determine the effect of disruption of TFs predicted to act as regulators. The results indicated that a starch metabolic gene, SS4, showed a decreased mRNA level in Atidd5 mutant (Figure 5A). Significant down-regulation of SS4 was observed at the end of the light period of both short and long days (Figures 5A and 5B). It appears that AtIDD5 plays an important role in the regulation of SS4 gene expression. By contrast, the mRNA levels of AGLU-like 4, GBSS, DPE1, and SS1 genes were not different between the wild type and mutant lines—c2h2, col &col7, wlim2a, and kh-ccch, respectively—during the time course of 12 L/12D condition (Additional file 5: Figure S3).
Figure 5.
SS4 gene expression in the wild type, Atidd5 and col mutants. The relative transcript level of SS4 mRNA was measured in Arabidopsis grown under (a) short (12 L/12D) and (b) long day (16 L/8D) conditions. Ubiquitin 2 (UBQ2) was used as a constitutive control for normalization.
In addition to the effect of direct TF-target relationships, the regulatory network can be affected by the associated modules particularly when the target genes are in the same metabolic process. Based on this assumption, the SS4 mRNA levels were determined in other TF mutant lines. Similar to the results obtained from the Atidd5 mutant, SS4 was down-regulated in the col mutant during the light period of both short and long day conditions (Figure 5A and 5B). Alteration of SS4 expression may have resulted from (i) the negative effect of disruption of COL on AtIDD5 gene expression or (ii) the direct control of SS4 by COL. Since the mRNA levels of AtIDD5 in the wild type and the col mutant were different only in the long day condition (Additional file 6: Figure S4), regulation of AtIDD5 expression by COL therefore remains inconclusive. Although the exact underlying mechanism is unknown, COL appears to be in part of the starch metabolic gene regulatory network, and its relevance is further evidenced by starch granule deformation (discussed in the following section).
According to in silico prediction of the Arabidopsis proteome, 176 proteins are classified in the C2H2 zinc finger family [63]. AtIDD5 belongs to this C2H2 gene family and is further classified into the same sub-family as the maize indeterminate 1 gene (ZmID1), which is a key regulator of flowering transition [63,64]. In Arabidopsis, there are 16 homologues of AtIDD genes. Among them, the biological functions of magpie (MGP/AtIDD3; At1g03840), nutcracker (NUC/AtIDD8; At5g44160), and jackdaw (JKD/AtIDD10; At5g03150) genes have been characterized [65,66]. To date, there are only three AtIDD genes—NUC/AtIDD8, AtIDD14 (At1g68130), and shoot graviropism 5 (SGR5/AtIDD15; At2g01940)—that have been reported to play roles, though indirectly, in sugar and starch metabolism [67-69]. Based on phylogenetic analysis, the AtIDD genes most closely related to AtIDD5 are AtIDD4 (At2g02080) and AtIDD6 (At1g14580). AtIDD4 is reported as a TF whose expression is affected by defects in chloroplast import machinery, and it is postulated to function as a transcriptional activator of nuclear-encoded photosynthetic gene expression [70]. In addition, AtIDD4 and AtIDD6 are identified as gibberellin-regulated genes [71]. Based on the evidence known to date, both AtIDD4 and AtIDD6 do not seem to have any function related to sugar and starch metabolism.
The database of global gene expression analysis provides evidence showing that AtIDD5 is abundantly expressed in leaf tissues. The global view of AtIDD5 gene expression was roughly examined using the Genevestigator web-based software [72]. The expression of AtIDD5 was observed ubiquitously in all stages, but its level was particularly high from stages of developed rosette leaves to developed flowers. In flowers, expression pattern of AtIDD5 was classified as ‘stamen-specific lack of expression’, suggesting that its expression disappears, especially in anthers of flowers from stage 7 to 11 [73,74].
Information on AtIDD5-interacting proteins further suggests that AtIDD5 is associated with other signalling components, such as radical-induced cell death 1 (RCD1: At1g32230) [75]. RCD1 is not only known as clone eighty-one (CEO1), which recovers the oxidative stress-sensitivity phenotype of the Yap1- mutant yeast [76], but also a major regulator of hormonal signalling and stress-response processes [77,78]. According to the AGRIS database http://arabidopsis.med.ohio-state.edu/REIN/, AtIDD5 is predicted to interact with a MADS-box domain TF, sepallata 3 (SEP3), which is a global moderator of multifunctional protein-complexes controlling flowering and hormonal signaling processes, especially responses to auxin stimuli [79].
Total starch content and granule morphology analysis
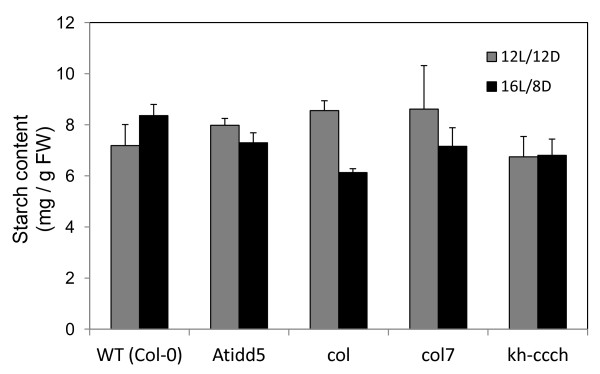
Total starch content was measured under both short and long day conditions. Starch was extracted from fully expanded leaves of all the mutants and the wild type, and analyzed in the form of glucose by capillary electrophoresis-diode array detector (CE-DAD) after enzymatic digestion (see method). Both the mutants and the wild type accumulated starch at relatively similar levels (Figure 6), even though SS4 was down-regulated in Atidd5 and col mutants (Figure 5). It has been reported that the size of starch granules can be significantly altered in ss4 mutant while only 35% reduction of the starch content could be observed [6]. Referring to these previous findings, we speculated that changes in starch granule morphology and number may occur in Atidd5 and col mutants as they show reduced levels of SS4 mRNA accumulation during the light period (Figure 5).
Figure 6.
Total starch content of leaves in the wild type and Atidd5, col, col7, and kh-ccch mutants.Arabidopsis plants were grown under short (12 L/12D; grey bar) and long day (16 L/8D; black bar) conditions, respectively.
Chloroplast and starch granule morphology of Atidd5 and col mutant lines were examined by transmitted electron microscopy (TEM). Transmission electron micrographs of Atidd5 and col mutant lines and wild type are shown in Figure 7. They were analyzed by the image processing software Image J (version 1.45) to obtain a group of data sets including (i) size measured by ‘Area’ and (ii) shape measured by ‘Width’, ‘Height’, and ‘Circularity’ of the chloroplast and starch granule cross-sections. Although these parameters might not represent the actual size and shape of chloroplasts and starch granules, we considered them suitable for a comparative purpose. Since the data was not normally distributed, a non-parametric statistic, the Mann–Whitney U test, was applied for testing significant differences between the wild type and the mutants. The relative mean ranks and P-values from the Mann–Whitney U test are described in Table 5. Descriptive statistics of chloroplast and starch granule morphology of Atidd5 and col mutants are summarized in Additional file 7: Table S3.
Figure 7.

Transmission electron micrographs of the wild type, Atidd5, and col chloroplast. Length of a scale bar indicated in the micrograph is 2 μm.
Table 5.
The statistics from Mann–WhitneyUtest of chloroplast and starch morphology, and granule number
| Arabidopsis Line | N1 | Area (μm2) | Width (μm) | Height (μm) | Circularity | |
|---|---|---|---|---|---|---|
| CHLOROPLAST |
|
|
Relative Mean Rank2 |
|||
|
Atidd5 |
259 |
35.52 |
41.96 |
45.06 |
42.75 |
|
| WT (Col-0) |
368 |
60.33 |
55.80 |
53.61 |
55.24 |
|
|
col |
176 |
38.81 |
53.70 |
33.94 |
48.20 |
|
| WT (Col-0) |
|
55.49 |
48.37 |
57.82 |
50.99 |
|
| |
|
P-value (Mann–Whitney U)3 |
||||
|
Atidd5 |
|
3.38E-26 |
3.52E-09 |
2.60E-04 |
9.86E-08 |
|
|
col |
|
2.99E-10 |
0.0442 |
1.90E-19 |
0.2919 |
|
| STARCH GRANULE |
|
|
Relative Mean Rank |
|||
|
Atidd5 |
890 |
45.63 |
47.58 |
47.35 |
47.66 |
|
| WT (Col-0) |
895 |
54.40 |
52.47 |
52.69 |
52.38 |
|
|
col |
505 |
48.82 |
54.72 |
38.91 |
60.53 |
|
| WT (Col-0) |
|
50.72 |
47.40 |
56.31 |
44.11 |
|
| |
|
P-value (Mann–Whitney U) |
||||
|
Atidd5 |
|
1.38E-10 |
3.45E-04 |
9.54E-05 |
5.54E-04 |
|
|
col |
|
0.2352 |
5.23E-06 |
2.49E-27 |
1.64E-24 |
|
| GRANULE NUMBER |
|
|
Number of starch granule / chloroplast |
|||
| |
|
Relative Mean Rank |
||||
|
Atidd5 |
283 |
53.57 |
||||
| WT (Col-0) |
408 |
47.65 |
||||
|
col |
177 |
54.18 |
||||
| WT (Col-0) |
|
48.31 |
||||
| |
|
P-value (Mann–Whitney U) |
||||
|
Atidd5 |
|
0.0067 |
||||
| col | 0.0213 | |||||
1. N is a number of observed chloroplast (or starch granule) from Transmission electron micrographs. For the granule number analysis, N is a number of observed chloroplast.
2. Relative Mean Rank was calculated from a ratio of mean rank to total rank and multiplied by 100 when the data was ranked in an ascending order.
3. The P-value calculated from the Mann–Whitney U test less than 0.05 was shown in bold letters indicating a significant difference of morphological parameters between the mutant and the wild type.
Area, width, height, and circularity of 259, 176, and 368 chloroplasts of Atidd5, col, and the wild type, respectively, were measured using the ImageJ software. According to the Mann–Whitney U analysis, chloroplast area of the wild type was significantly larger than that of Atidd5 and col mutants (P-value = 3.38E-26 and 2.99E-10, respectively) (Table 5) with the means of chloroplast areas at 13.18, 9.26, and 10.50 μm2 for the wild type, Atidd5, and col, respectively. In addition to the size, the shape of chloroplasts of both mutants also differed from the wild type. The width, height, and circularity of the chloroplasts were significantly smaller in Atidd5 than in the wild type. The small circularity values of Atidd5 chloroplasts indicate that they are in more oblong shapes relative to the wild type chloroplasts. In addition, the chloroplasts in the col mutant had longer width but less height than those in the wild type. The results, therefore, suggest that both mutants develop chloroplasts with altered morphology, which, particularly, appear smaller or thinner than the chloroplasts in the wild type (Figure 7 and Table 5).
Since chloroplasts of both mutants were altered with respect to their size and shape, we examined their effects on the morphology of accumulated starch granules. Reduction of starch granule size, inferred from the cross-section area, was significant in Atidd5 (P-value = 1.38E-10), but not in the col mutant. The means of starch granule areas of the wild type, Atidd5, and col were 0.54, 0.42, and 0.50 μm2, respectively. In contrast, the granule shape deformity was noticed in both col and Atidd5 mutants (Figure 7 and Table 5). The decrease in width, height, and circularity of Atidd5 starch granule most likely suggested that the granule was small and in oblong shapes. As compared to the wild type, the col starch granules were observed to have greater circularity, suggesting that they were relatively round in shape.
According to the work of Rolden and coworkers [6], a chloroplast of the ss4 mutant mostly contains one large starch granule. When examined under TEM—among 283, 177, and 408 chloroplasts of Atidd5, col, and the wild type, respectively—none of the chloroplasts were observed to contain a single large starch granule like the ss4 mutant. On the other hand, the majority of chloroplasts—corresponding to 25.4%, 27.1% and 27.5% of observed chloroplasts in Atidd5, col, and the wild type, respectively—normally contained 3 starch granules. We further investigated the distribution of starch granule number per chloroplast to find the difference between the mutants and wild type (Figure 8). In the Atidd5 mutant, 86.2% of the observed chloroplasts contained 2–5 starch granules, whereas 85.8% of chloroplasts from the wild type contained 1–4 starch granules. Interestingly, we observed that the number of chloroplasts containing 2 and 4 starch granules in the col mutant was lower than those in the Atidd5 and the wild type, whereas the number of chloroplasts containing more than 5 granules was higher than the other lines (Figure 8). Moreover, the col mutant was the only line that was observed to contain up to 10 starch granules per chloroplast. The relative mean rank and P-value from the Mann–Whitney U test of the mutants and the wild type shown in Table 5 indicated that both Atidd5 and col mutants had significantly higher numbers of starch granules per chloroplast than the wild type (P-value = 0.0067 and 0.0213, respectively). The results suggest that reduction of SS4 expression in the Atidd5 and col mutant lines leads to a significant increase in starch granule numbers, while their distributions of granule number per chloroplast are differently affected among the mutants (Figure 8).
Figure 8.
Distribution of granule number per chloroplast in the wild type, Atidd5, and col mutants. Means and P-value calculated from the Mann–Whitney U test are indicated.
The relationship between the size of chloroplast and the number of accumulated starch granules is shown in Figure 9. Our results indicate that the number of starch granules increased according to chloroplast size. Larger chloroplasts tended to contain greater numbers of starch granules; however the pattern of correlation was not uniform among the wild type and mutants. In the Atidd5 mutant, a positive correlation between chloroplast size and the number of starch granules was only observed in the chloroplast containing 1–4 granules. It appears that the chloroplasts in this mutant are unable to expand after reaching critical size; however, they continued to store higher numbers of starch granules without increasing their size. In col mutant, the positive correlation was observed in the chloroplast containing 1–6 granules, whereas the size of the chloroplast containing 6–8 starch granules tended to decrease in accordance with an increase in the number of starch granules. The average sizes of chloroplasts containing 7 and 8 granules were the same as those having 2 starch granules. In addition, the size of the col chloroplasts containing 10 starch granules was similar to the size of chloroplasts containing 6 granules, suggesting this might be the critical size limit of the col chloroplast.
Figure 9.
Relationship between granule number and chloroplast size in the wild type, Atidd5, and col mutants.
The results indicated that, in addition to having defects in their control of SS4 gene expression, both Atidd5 and col mutants are unable to increase the size of chloroplasts, although they may still retain the capability to expand their chloroplast to contain relatively small numbers of starch granules until the chloroplast reaches its critical size limit. Particularly in Atidd5, having relatively small starch granules can be another adaptive response caused by chloroplast deformity. The observed phenomena may suggest the alternative roles of AtIDD5 and COL in controlling chloroplast size limit, which may synchronize with transcriptional regulation of a starch biosynthetic enzyme, SS4. Our findings address a question of how starch biosynthesis and chloroplast development and/or functions are synergistically controlled in plant cells. The underlying mechanism of interaction awaits further investigation.
Conclusions
In this study, we proposed a transcriptional regulatory network of starch metabolism in Arabidopsis leaves, and examined the biological relevance of predicted network modules. The general workflow of data acquisition, refinement, and experimental validation provides a model case for reconstruction of transcriptional regulatory network. The present work widely utilizes publicly available biological information and resource databases, demonstrating how they can be integrated to find biological significance of predicted network modules. Construction of gene-to-gene association network models is based on diurnal regulation of starch metabolism in leaves where the transcriptomes oscillate during the day/night cycles. We first grouped time-series-dependent significant genes on transcriptome into four classes showing distinct patterns of co-regulation with starch biosynthesis or degradation. A particular focus has been placed on relationships between TFs, clock genes and starch metabolic genes, to obtain transcriptional regulatory network model of starch metabolism. The network constructed by the small sample inference of GGM suggests relationships between TFs and target starch metabolic genes. Gene-to-gene associations have been further refined by prediction of TF binding sites in target genes and by global co-expression analysis. Through these approaches, we finally showed the involvement of AtIDD5 and COL in transcriptional regulation of SS4. These regulatory networks were considered attributable to daytime starch biosynthesis by SS4. In addition, AtIDD5 and COL were shown to control chloroplast development and starch granule formation. The present work on TF network modelling and examination provides new insights into the regulatory mechanisms of starch biosynthesis and granule formation in the chloroplast.
Methods
Microarray data pre-processing
This study utilized Arabidopsis Affymetrix microarray data (CEL files) downloaded from the Nottingham Arabidopsis Stock Centre's microarray database (NASCArrays) [Experiment Reference Number: NASCARRAYS-60] http://affymetrix.arabidopsis.info/. This microarray experiment contains a set of 22 k Arabidopsis ATH-1 genome array transcriptome data of leaves at developmental stage 3.9 taken after 1, 2, 4, 8, and 12 hours in both darkness and light [36]. A ‘qspline’ normalization [80] and model-based expression index [81] were carried out in the microarray pre-processing, which was done using the Affy package in Bioconductor http://www.bioconductor.org.
Significant analysis of time-series data
The significant test for the time-series data was performed using the EDGE program (version 1.1.175) [39]http://faculty.washington.edu/jstorey/edge/. Hypothesis testing on time-series expression of each gene was performed to test whether an average expression constitutes a flat line. The gene expression profile was fitted under a model based on null and alternative hypotheses. The null hypothesis states that there is no differential gene expression over a time period. The alternative hypothesis states that a gene is differentially expressed over a time period. The goodness of fit of 2 models was compared by F-statistic using a significant cut-off based on a false discovery rate criterion [82,83].
The small-sample inference by graphical Gaussian model (GGM)
The R package “GeneNet” [33]—available at the R archive (CRAN) http://CRAN.R-project.org/—was used to construct the gene association network. The GeneNet was developed from the small-sample inference framework of graphical Gaussian model (GGM) to obtain a partial correlation coefficient, which is a correlation between 2 variables obtained when eliminating effects of other variables. In the case of 3 variables—x, y, and z—the partial correlation of x and y when eliminating the effect of z, prxy,z can be calculated as follows
| (1) |
where r is the correlation coefficient between 2 variables. In the case of more than 3 variables, the partial correlation can be calculated from the following equation.
| (2) |
The prxy,g is the partial correlation between x and y against variable 3 to g. The sxy = the xyth element of the inverse of variance matrix (S = V-1). The element in matrix V is vij (i, j = 1, … , n) corresponding to a covariance between variables i and j.
For microarray, the number of variables (i.e. genes) is much higher than the number of measurements (i.e. microarray conditions), thus making the inversion step of matrix V invalid. In the new framework of GGM, the parameter estimation techniques were used to obtain partial correlation of small sample size. In order to decide which edges are significant to be included in the resulting GGM network, statistical significance was further assigned to the edges in the GGM network by fitting a mixture model (as shown below) to the observed partial correlation coefficients [33].
The distribution of observed partial correlation coefficients is
| (3) |
where is the observed partial correlation, is the unknown proportion of null edges, is the distribution under the null hypothesis of zero-partial correlation, κ is the degree of freedom, and is the distribution of observed partial correlations assigned to actually existing edges. The two-sided p-values for each edge corresponding to the null distribution were subsequently calculated and followed by false discovery rate multiple testing [82,83] to obtain q-values. The edges with q-values are equal or lower than 0.05 were presented in the resulting GGM network in this study.
Arabidopsis lines and growth conditions
Arabidopsis ecotype Columbia-0 was used in this study. Arabidopsis mutant lines were obtained from the T-DNA or transposon inserted mutant collection (Col-0 background) of The Arabidopsis Biological Resource Centre (ABRC) [60] and The Nottingham Arabidopsis Stock Centre (NASC) [61]. Accession numbers of Atidd5, c2h2, col, col7, wlim2a, and kh-ccch are SALK_110990, SALK_070916, SALK_061956, SM_3_37788, SALK_067756, and SAIL_672_A10, respectively. Details of all mutant lines are shown in Additional file 8: Table S4. The seeds were vernalized in the dark for 3 days at 4°C before germination. Plants were grown on an equal mixture of sterile vermiculite and peat-based growing medium (PRO-MIX Bx/Microrise Pro, Premier) in a growth cabinet (SANYO) set at 60% humidity and 20-22°C with a light intensity of 100 μmol m-2 sec-1 and under 12 hr light/12 hr dark (short day) or 16 hr light/8 hr dark (long day) cycles. The trays of plant pots were sub-irrigated with a half-strength Arabidopsis liquid nutrient culture [84]. Leaves at a developmental stage of 3.90 [62] were harvested 4 times a day—1 hr before and after day break and night break. Leaves for starch analysis were harvested at the end of the light period.
Expression analysis by quantitative RT-PCR
Total RNA was extracted from 100–200 mg leaf material (3 biological replicates) using Plant RNeasy kit (Qiagen), treated with DNaseI (Invitrogen), and reverse transcribed by Omniscript Reverse Transcriptase (Qiagen). Subsequently, real-time PCR was carried out using SYBR® Premix Ex Taq ™ II (Perfect Real Time) (Takara) using ubiquitin 2 (UBQ2) as a constitutive internal control. Details of the primer pairs used in qRT-PCR experiments are shown in Additional file 9: Table S5.
Starch extraction and measurement
Starch from Arabidopsis leaves (3 biological replicates) was extracted using the method described by Smith and Zeeman [85]. Gelatinized starch was hydrolyzed to glucose by incubation for 4 hr at 37°C with α-amylase and α-amyloglucosidase. After the enzymatic digestion of starch to glucose, the amount of glucose was quantified by the capillary electrophoresis photodiode array detection (CE-DAD) system according to the manufacturer’s protocol (Agilent) [86]. Leaf starch content was calculated from the amount of glucose measured in this enzymatically-digested extract.
Starch granule morphology analysis by Transmission Electron Microscope (TEM)
Fully expanded Arabidopsis leaves were collected at end of day, cut into 2 x 2 mm2 pieces, and immediately fixed with a cold solution of glutaraldehyde. Various parameters describing starch granule morphology (i.e. area, perimeter, width, height, and circularity) and number of starch granules per chloroplast were measured from TEM micrographs using ImageJ software (version 1.45). It was noted that circularity is calculated by the following formula:
| (4) |
A value approaches 1.0 meaning a perfect circle and 0.0 meaning an elongated shape. The morphology data was tested for a statistically difference using a non-parametric Mann–Whitney U statistic (P-value < 0.05).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PI carried out the microarray data analysis, the model reconstruction, qRT-PCR experiments, and starch content and granule morphology analysis, and drafted the manuscript. PI, SC, and HT conceived and designed the research. SN and HT edited the manuscript. SP provided assistance in the statistical analysis of the research. JC provided assistance in R programming. HT provided assistance in growing Arabidopsis, qRT-PCR experiment, and starch measurement. AM, MT, and SB provided oversight of the work. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. The gene association network of 11 carbon-related metabolisms inferred from GGM (Q < 0.05).
Table S1. Genes in the starch sub-network.
Table S2. Prediction of TF binding sites in 12 starch metabolic genes (2 kb-upstream).
Figure S2. Expression patterns of 2 TFs, At3g07650 (COL9) and At1g05805 (bHLH), and their target genes.
Figure S3. Expression patterns of starch genes in other regulatory modules.
Figure S4. Expression pattern of C2H2 gene in the wild type, Atidd5, and col mutants quantified by qRT-PCR.
Table S3. Descriptive statistics of chloroplast morphology, starch granule morphology, and starch granule number in the wild type, Atidd5, and col mutants.
Table S4. T-DNA insertion lines of 6 candidate TFs that were utilized in this experiment.
Table S5. Primer pairs for quantitative RT-PCR.
Contributor Information
Papapit Ingkasuwan, Email: papapit_ing@yahoo.com.
Supatcharee Netrphan, Email: supatchareen@biotec.or.th.
Sukon Prasitwattanaseree, Email: sprasitwattanaseree@gmail.com.
Morakot Tanticharoen, Email: morakot.tan@kmutt.ac.th.
Sakarindr Bhumiratana, Email: sakarindr.bhu@kmutt.ac.th.
Asawin Meechai, Email: asawin.mee@kmutt.ac.th.
Jeerayut Chaijaruwanich, Email: jeerayut.c@cmu.ac.th.
Hideki Takahashi, Email: htakaha@msu.edu.
Supapon Cheevadhanarak, Email: supapon.che@kmutt.ac.th.
Acknowledgements
We are grateful to Dr. Akinori Suzuki and Ms. Yumiko Tsuchiya, RIKEN Plant Science Center, for technical support in growing Arabidopsis and qRT-PCR experiment. We are also grateful to Prof. Dr. Maleeya Kruatrachue, with the assistance of Ms. Sombat Singhakaew, Mahidol University, for providing assistance in transmission electron micrographs analysis. TEM samples preparation and TEM analysis were carried out by service laboratories at Kasetsart University Research and Development Institute (KURDI) and Central Laboratory and Greenhouse complex, Kasetsart University (Kamphaengsaen Campus), respectively. PI is financially supported by Thailand Graduate Institute of Science and Technology (TGIST), Contract No. TGIST 01-47-048, and National Center for Genetic Engineering and Biotechnology (BIOTEC), Grant No. BT-B-02-PG-B5-4813. This work is supported by BIOTEC, Thailand; RIKEN Plant Science Center, Japan; and in part by the grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, and Bio-oriented Technology Research Advancement Institution (BRAIN).
References
- Smith AM. In: Regulation of Primary Metabolic Pathways in Plants. 42. Kruger NJ, Hill SA, Ratcliffe RG, editor. Kluwer Academic Publishers, Dordrecht; 1999. Regulation of starch synthesis in storage organs; pp. 173–193. Proceedings of the Phytochemical Society of Europe] [Google Scholar]
- Smith AM, Denyer K, Zeeman SC, Edwads A, Martin C. Plant Carbohydrate Biochemistry. BIOS Scienctific Publishers Ltd, Oxford; 1999. The synthesis of the starch granule; pp. 79–89. [Google Scholar]
- Smith AM, Zeeman SC, Thorneycroft D, Smith SM. Starch mobilization in leaves. J Exp Bot. 2003;54:577–583. doi: 10.1093/jxb/erg036. [DOI] [PubMed] [Google Scholar]
- Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC. A previously unknown maltose transporter essential for starch degradation in leaves. Science. 2004;303:87–89. doi: 10.1126/science.1091811. [DOI] [PubMed] [Google Scholar]
- Sokolov LN, Dominguez-Solis JR, Allary A-L, Buchanan BB, Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc Natl Acad Sci USA. 2006;103:9732–9737. doi: 10.1073/pnas.0603329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán I, Wattebled F, Mercedes Lucas M, Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D'Hulst C, Mérida A. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J. 2007;49:492–504. doi: 10.1111/j.1365-313X.2006.02968.x. [DOI] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco P, Gil M, Reinhold H, Eicke S, Messerli G. et al. β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell. 2008;20:1040–1508. doi: 10.1105/tpc.107.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmeier-Vogel E, Kerk D, Nimick M, Wrobel S, Vickerman L, Muench D, Moorhead G. Arabidopsis At5g39790 encodes a chloroplast-localized, carbohydrate-binding, coiled-coil domain-containing putative scaffold protein. BMC Plant Biol. 2008;8:120. doi: 10.1186/1471-2229-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, Comparot-Moss S, Chen J, Smith AM, Steup M. et al. STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell. 2009;21:334–346. doi: 10.1105/tpc.108.064360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Foster CM, Gan Q, Nettleton D, James MG, Myers AM, Wurtele ES. Identification of the novel protein QQS as a component of the starch metabolic network in Arabidopsis leaves. Plant J. 2009;58:485–498. doi: 10.1111/j.1365-313X.2009.03793.x. [DOI] [PubMed] [Google Scholar]
- Szydlowski N, Ragel P, Raynaud S, Lucas MM, Roldán I, Montero M, Muñoz FJ, Ovecka M, Bahaji A, Planchot V. et al. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell. 2009;21:2443–2457. doi: 10.1105/tpc.109.066522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Morell MK, Emes MJ. Recent developments in understanding the regulation of starch metabolism in higher plants. J Exp Bot. 2004;55:2131–2145. doi: 10.1093/jxb/erh248. [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Kolbe A, Tiessen A. Redox regulation of carbon storage and partitioning in response to light and sugars. J Exp Bot. 2005;56:1469–1479. doi: 10.1093/jxb/eri178. [DOI] [PubMed] [Google Scholar]
- Kötting O, Kossmann J, Zeeman SC, Lloyd JR. Regulation of starch metabolism: the age of enlightenment? Curr Opin Plant Biol. 2010;13:320–328. doi: 10.1016/j.pbi.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Ghosh HP, Preiss J. Adenosine diphosphate glucose pyrophosphorylase: a regulatory enzymein the biosynthesis of starch in spinach leaf chloroplasts. J Biol Chem. 1966;241:4491–4504. [PubMed] [Google Scholar]
- Fu Y, Ballicora MA, Leykam JF, Preiss J. Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J Biol Chem. 1998;273:25045–25052. doi: 10.1074/jbc.273.39.25045. [DOI] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farré EM, Geigenberger P. Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell. 2002;14:2191–2213. doi: 10.1105/tpc.003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler I, Renz A, Schmid FX, Beck E. Activation of spinach pullulanase by reduction results in a decrease in the number of isomeric forms. Biochim Biophys Acta Protein Struct Mol Enzymol. 2001;1548:175–186. doi: 10.1016/S0167-4838(01)00228-X. [DOI] [PubMed] [Google Scholar]
- Wu C, Colleoni C, Myers AM, James MG. Enzymatic properties and regulation of ZPU1, the maize pullulanase-type starch debranching enzyme. Arch Biochem Biophys. 2002;406:21–32. doi: 10.1016/S0003-9861(02)00412-5. [DOI] [PubMed] [Google Scholar]
- Mikkelsen R, Mutenda KE, Mant A, Schürmann P, Blennow A. α-Glucan, water dikinase (GWD): A plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc Natl Acad Sci USA. 2005;102:1785–1790. doi: 10.1073/pnas.0406674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P. Redox regulation of a novel plastid-targeted β-amylase of Arabidopsis. Plant Physiol. 2006;141:840–850. doi: 10.1104/pp.106.079186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Palmqvist S, Olsson H, Borén M, Ahlandsberg S, Jansson C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Cai X-L, Wang Z-Y, Hong M-M. An Interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J Biol Chem. 2003;278:47803–47811. doi: 10.1074/jbc.M302806200. [DOI] [PubMed] [Google Scholar]
- Tenorio G, Orea A, Romero JM, Mérida Á. Oscillation of mRNA level and activity of granule-bound starch synthase I in Arabidopsis leaves during the day/night cycle. Plant Mol Biol. 2003;51:949–958. doi: 10.1023/A:1023053420632. [DOI] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I. et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- Wang Y, Joshi T, Zhang X-S, Xu D, Chen L. Inferring gene regulatory networks from multiple microarray datasets. Bioinformatics. 2006;22:2413–2420. doi: 10.1093/bioinformatics/btl396. [DOI] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ. An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 2007;17:1614–1625. doi: 10.1101/gr.6911207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera J, Rodrigo G, Jaramillo A, Elena S. Reverse-engineering the Arabidopsis thaliana transcriptional network under changing environmental conditions. Genome Biol. 2009;10:R96. doi: 10.1186/gb-2009-10-9-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham C, Manfield I, Bulpitt A, Gilmartin P, Westhead D. From gene expression to gene regulatory networks in Arabidopsis thaliana. BMC Syst Biol. 2009;3:85. doi: 10.1186/1752-0509-3-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L, Van Hemert J, Dash S, Dickerson J. Arabidopsis gene co-expression network and its functional modules. BMC Bioinformatics. 2009;10:346. doi: 10.1186/1471-2105-10-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer J, Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics. 2005;21:754–764. doi: 10.1093/bioinformatics/bti062. [DOI] [PubMed] [Google Scholar]
- Opgen-Rhein R, Strimmer K. Infering gene dependency networks from genomic longitudinal data: a functional data approach. REVSTAT. 2006;4:53–65. [Google Scholar]
- Zeeman SC, Tiessen A, Pilling E, Kato KL, Donald AM, Smith AM. Starch synthesis in Arabidopsis. granule synthesis, composition, and structure. Plant Physiol. 2002;129:516–529. doi: 10.1104/pp.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 2004;136:2687–2699. doi: 10.1104/pp.104.044347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible W-R, Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Bläsing OE, Palacios -Rojas N, Pankovic D, Hendriks JHM, Fisahn J, Höhne M, Günther M, Stitt M. Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 2004;39:847–862. doi: 10.1111/j.1365-313X.2004.02173.x. [DOI] [PubMed] [Google Scholar]
- Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. Mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Lao NT, Schoneveld O, Mould RM, Hibberd JM, Gray JC, Kavanagh TA. An Arabidopsis gene encoding a chloroplast-targeted β-amylase. Plant J. 1999;20:519–527. doi: 10.1046/j.1365-313X.1999.00625.x. [DOI] [PubMed] [Google Scholar]
- Kossmann J, Lloyd J. Understanding and influencing starch biochemistry. Crit Rev Biochem Mol Biol. 2000;35:141–196. [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005;138:2280–2291. doi: 10.1104/pp.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidig A, Fröhlich A, Schulze S, Lloyd JR, Kossmann J. Downregulation of a chloroplast-targeted beta-amylase leads to a starch-excess phenotype in leaves. Plant J. 2002;30:581–591. doi: 10.1046/j.1365-313X.2002.01317.x. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Sung DY, Guy CL. Roles of beta-amylase and starch breakdown during temperatures stress. Physiol Plantarum. 2006;126:120–128. doi: 10.1111/j.1399-3054.2006.00604.x. [DOI] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T. Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:791–803. doi: 10.1093/pcp/41.6.791. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Ito S, Kawamura H, Niwa Y, Nakamichi N, Yamashino T, Mizuno T. A genetic study of the Arabidopsis circadian clock with reference to the TIMING OF CAB EXPRESSION 1 (TOC1) gene. Plant Cell Physiol. 2009;50:290–303. doi: 10.1093/pcp/pcn198. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Guy CL. RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J. 2005;44:730–743. doi: 10.1111/j.1365-313X.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- Papuga J, Hoffmann C, Dieterle M, Moes D, Moreau F, Tholl S, Steinmetz A, Thomas C. Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell. 2010;22:3034–3052. doi: 10.1105/tpc.110.075960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean Rider S, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J. Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 2003;35:33–43. doi: 10.1046/j.1365-313X.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bulow L, Hehl R. AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucl Acids Res. 2004;32:D368–372. doi: 10.1093/nar/gkh017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bulow L, Hehl R. AthaMap web tools for database-assisted identification of combinatorial cis-regulatory elements and the display of highly conserved transcription factor binding sites in Arabidopsis thaliana. Nucl Acids Res. 2005;33:W397–402. doi: 10.1093/nar/gki395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow L, Steffens NO, Galuschka C, Schindler M, Hehl R. AthaMap: from in silico data to real transcription factor binding sites. In Silico Biol. 2006;6:243–252. doi: 10.1007/3-540-28185-1_10. [DOI] [PubMed] [Google Scholar]
- Galuschka C, Schindler M, Bulow L, Hehl R. AthaMap web tools for the analysis and identification of co-regulated genes. Nucl Acids Res. 2007;35:D857–862. doi: 10.1093/nar/gkl1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucl Acids Res. 2009;37:D987–D991. doi: 10.1093/nar/gkn807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 2009;16:249–260. doi: 10.1093/dnares/dsp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Tissier AF, Marillonnet S, Klimyuk V, Patel K, Torres MA, Murphy G, Jones JDG. Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell. 1999;11:1841–1852. doi: 10.1105/tpc.11.10.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englbrecht C, Schoof H, Bohm S. Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics. 2004;5:39. doi: 10.1186/1471-2164-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Tremblay R, Wong A, Coneva V, Kozaki A, Mable B. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics. 2006;7:158. doi: 10.1186/1471-2164-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 2007;21:2196–2204. doi: 10.1101/gad.440307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU. et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto M, Tremblay R, Colasanti J. Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Mol Biol. 2008;67:57–69. doi: 10.1007/s11103-008-9301-0. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Ryu J-Y, Jeong E-Y, Park C-M. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat Commun. 2011;2:303. doi: 10.1038/ncomms1303. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Ryu J, Kang SK, Park C-M. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 2011;65:418–429. doi: 10.1111/j.1365-313X.2010.04432.x. [DOI] [PubMed] [Google Scholar]
- Kakizaki T, Matsumura H, Nakayama K, Che F-S, Terauchi R, Inaba T. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 2009;151:1339–1353. doi: 10.1104/pp.109.145987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang Z-L, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, Sun T-P. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptome. Adv Bioinformatics. 2008. p. 5. [DOI] [PMC free article] [PubMed]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama N, Arroyo JM, Simorowski J, May B, Martienssen R, Irish VF. Gene trap lines define domains of gene regulation in Arabidopsis petals and stamens. Plant Cell. 2005;17:2486–2506. doi: 10.1105/tpc.105.033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P, Blomster T, Brosché M, Salojärvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F. et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 2009;60:268–279. doi: 10.1111/j.1365-313X.2009.03951.x. [DOI] [PubMed] [Google Scholar]
- Belles-Boix E, Babiychuk E, Van Montagu M, Inzé D, Kushnir S. CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 2000;482:19–24. doi: 10.1016/S0014-5793(00)02016-0. [DOI] [PubMed] [Google Scholar]
- Overmyer K, Tuominen H, Kettunen R, Betz C, Langebartels C, Sandermann H, Kangasjarvi J. Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell. 2000;12:1849–1862. doi: 10.1105/tpc.12.10.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlfors R, Lang S, Overmyer K, Jaspers P, Brosche M, Tauriainen A, Kollist H, Tuominen H, Belles-Boix E, Piippo M. et al. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein–protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell. 2004;16:1925–1937. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E. AGRIS: the Arabidopsis gene regulatory information server, an update. Nucl Acids Res. 2011;39:D1118–D1122. doi: 10.1093/nar/gkq1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild H-H, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3:research0048.0041–research0048.0016. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.98.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J Royal Stat Soc Ser B. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- Fujiwara T, Hirai MY, Chino M, Komeda Y, Naito S. Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 1992;99:263–268. doi: 10.1104/pp.99.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC. Quantification of starch in plant tissues. Nat Protoc. 2006;1:1342–1345. doi: 10.1038/nprot.2006.232. [DOI] [PubMed] [Google Scholar]
- Sato S, Soga T, Nishioka T, Tomita M. Simultaneous determination of the main metabolites in rice leaves using capillary electrophoresis mass spectrometry and capillary electrophoresis diode array detection. Plant J. 2004;40:151–163. doi: 10.1111/j.1365-313X.2004.02187.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The gene association network of 11 carbon-related metabolisms inferred from GGM (Q < 0.05).
Table S1. Genes in the starch sub-network.
Table S2. Prediction of TF binding sites in 12 starch metabolic genes (2 kb-upstream).
Figure S2. Expression patterns of 2 TFs, At3g07650 (COL9) and At1g05805 (bHLH), and their target genes.
Figure S3. Expression patterns of starch genes in other regulatory modules.
Figure S4. Expression pattern of C2H2 gene in the wild type, Atidd5, and col mutants quantified by qRT-PCR.
Table S3. Descriptive statistics of chloroplast morphology, starch granule morphology, and starch granule number in the wild type, Atidd5, and col mutants.
Table S4. T-DNA insertion lines of 6 candidate TFs that were utilized in this experiment.
Table S5. Primer pairs for quantitative RT-PCR.