Abstract
Islands are bounded areas where high endemism is explained either by allopatric speciation through the fragmentation of the limited amount of space available, or by sympatric speciation and accumulation of daughter species. Most empirical evidence point out the dominant action of allopatric speciation. We evaluate this general view by looking at a case study where sympatric speciation is suspected. We analyse the mode, tempo and geography of speciation in Agnotecous, a cricket genus endemic to New Caledonia showing a generalized pattern of sympatry between species making sympatric speciation plausible. We obtained five mitochondrial and five nuclear markers (6.8 kb) from 37 taxa corresponding to 17 of the 21 known extant species of Agnotecous, and including several localities per species, and we conducted phylogenetic and dating analyses. Our results suggest that the diversification of Agnotecous occurred mostly through allopatric speciation in the last 10 Myr. Highly microendemic species are the most recent ones (<2 Myr) and current sympatry is due to secondary range expansion after allopatric speciation. Species distribution should then be viewed as a highly dynamic process and extreme microendemism only as a temporary situation. We discuss these results considering the influence of climatic changes combined with intricate soil diversity and mountain topography. A complex interplay between these factors could have permitted repeated speciation events and range expansion.
Introduction
Diversity hotspots in islands are characterized by very high specific richness and endemism resulting from local diversification of groups which are “trapped” in very limited spatial areas [1], [2], [3]. The theory behind this diversity rests upon the simple idea that local diversification occurred either by allopatric speciation, through the fragmentation of the limited space available into even more restricted daughter areas (microendemism), or by accumulation of daughter species without vicariance, through sympatric speciation based on non geographic isolation, for example involving ecological specializations [4], [5], [6].
Empirical case studies showed or suggested that diversifications in islands most often result from repeated events of local allopatric speciation and subsequent establishment of microendemism [7] due to Plio-Pleistocene climatic fluctuations resulting in rainforest fragmentation [8], [9], [10], or by the barriers made by large rivers [11], [12]. Contrasting with this general pattern, sympatric speciation has also been documented in islands [4], [5], [13], [14] and a few cases have been suspected in a context where distribution of species is restricted to small areas, i.e. microendemism [8], [15], [16], [17], [18], [19].
New Caledonia, one of most diverse island biodiversity hotspots [20], is no exception to the largely dominant allopatric pattern of distribution among related species, as shown in plants [21], [22], insects [8], [9], [23], land snails [24] and lizards [25], [26]. This suggests a major impact of allopatric speciation during the set up of microendemism in New Caledonia [27]. Yet, the relative importance of allopatric and sympatric speciation has not been addressed in many cases because of the low number of opportunities and the difficulty to distinguish between primary and secondary sympatry [7], [14], [28].
Here we analyse the mode, tempo and geography of speciation in the microendemic species of the cricket genus Agnotecous [29], [30]. This insect group shows a remarkable and generalised pattern of sympatry involving up to three sympatric species per location throughout New Caledonia (see Figure S1). Despite previous evidence for a major impact of allopatric speciation in the island, this pattern of distribution makes sympatric speciation plausible in this group.
We use a dated species-level phylogeny, analyses of diversification time-course, and age-range correlation methods to investigate the mode of speciation and causes of microendemism. We aim at discriminating between two alternative scenarios:
Under a scenario of dominant allopatric speciation, the observed pattern of sympatry is caused by secondary sympatry with post-speciational movements.
Under a scenario of dominant sympatric speciation, the observed pattern of sympatry results directly from recent sympatric speciation, with little or no post-speciational movements.
According to the dominant mode of speciation, we will compare and discuss the pattern of microendemism and tempo of species diversification in New Caledonia with respect to the influence of climatic changes combined with intricate soil distribution and mountain topography. This study will allow us understanding the origin of microendemic distributions in an evolutionary and biogeographical perspective.
Materials and Methods
Sampling and DNA Sequencing
The molecular sampling consists of 37 individuals corresponding to 17 out of the 21 known extant species of Agnotecous. Each species is represented by specimens known from 1–7 localities (mean = 1.8). Three species from different eneopterine tribes (Nisitrus vittatus, Eneoptera guyanensis, Lebinthus santoensis) and one species belonging to another cricket subfamily (Acheta domesticus, Gryllinae) were used as outgroups [30], [31]. For additional information on taxonomic sampling and sequencing protocols, see Tables S1 and S2, and Text S1 and S2.
Phylogenetic and Dating Analyses
DNA sequences were aligned under Muscle with default parameters [32]. Individual datasets were constructed for the entire mitochondrial data, the entire nuclear data, and for the 28S and EF1α genes separately. Congruence tests between nuclear and mitochondrial datasets revealed a large extent of mitochondrial introgression (see Table S3, Figure S2 and Text S3), so only the nuclear data were used in the phylogenetic analyses. To obtain a species-level tree for Agnotecous, we generated a consensus sequence per Agnotecous species using IUPAC ambiguity codes under BIOEDIT v 7.0.9.0 [33]. Except slight differences in node supports, the phylogeny of the 16 consensus sequences does not reveal a different tree topology from the 37 specimens’ phylogeny (see Figure S3). For comparative purpose, we conducted Bayesian and Parsimony analyses on each dataset. Bayesian analyses used substitution models of evolution determined using the software MrModeltest v 2.3 [34], and selected using the Akaike Information Criteria [AIC; 35,36], and were performed in MrBayes 3.1.2 [37]. Parsimony analyses were performed under TNT [38]. For details about phylogenetic analyses, see Text S4.
Likelihood-ratio test [39] rejected rate homogeneity among taxa, advocating for a relaxed molecular clock model. To calibrate the trees, we used the results of a previous study [30] based on different combinations of five geological calibrations applied to a larger taxonomic scale. This study dated Agnotecous at 10.5 Myr ago, and its divergence with L. santoensis at 15 Myr ago, with standard deviations of 1 Myr for each calibration point [30]. We applied the Bayesian relaxed uncorrelated log normal approach implemented in BEAST v1.4.8 [40] to estimate the relative age of divergence of the lineages. We ran four Markov chains simultaneously for 15 million generations, sampling every 1000 generations to ensure the independence of samples. After checking for convergence a 10% burnin was applied, and the remaining posterior distribution of trees was summarized using TreeAnnotator [41], which retained the Maximum Clade Credibility (MCC) tree with branch length equal to the median lengths of all the trees (see Text S5 for additional BEAST parameters and details of calibrations).
Analysis of Temporal Diversification
Lineage Through Time (LTT) plot derived from the BEAST MCC tree was used to visualise the time-course of diversification. We tested the null hypothesis of equal diversification rates across the Agnotecous phylogeny using the software SymmeTREE [ver. 1.1.; 42], which implements a whole-tree topological approach to the study of diversification rates. Diversification rate variation within the phylogeny is reported by a range of test statistics that vary in their sensitivity to nodal depth scales [42]. We then looked for the shift points by testing diversification rate change along branches using the function shift.test in the R package apTreeshape [43]. This test is based on the Δ1 statistic computed by SymmeTREE [44], and returns the probability of a diversification rate shift along the internal branch.
We also compare the fit of rate-variable models to the null hypothesis of constant diversification by using likelihood methods based on birth-death models and model-fitting approach (BDL) implemented in the R package LASER [45]. We tested for rate variation using (1) two rate-constant models: pure birth (pb) and birth death (bd), as null hypotheses and (2) one multiple-pure-birth rate model (y-2-rate), which detect 2 shift points, in otherwise constant speciation rates. Model selection was accomplished by comparing the difference in Akaike Information Criteria corrected for small sample sizes (AICrc) scores between the best rate constant and rate variable models as test statistic (ΔAICrc) [46] for the full data combined BEAST chronogram. For the bd and each non rate-constant model, the ΔAICrc statistic was compared with a distribution generated under the null hypothesis of rate constancy by the mean of 50000 trees generated under pure birth (yuleSim command in LASER), using the same number of extant and collected taxa as in the Agnotecous data set and using the estimated pure speciation value.
To compare diversification rates, we used APE [47], LASER [45] and GEIGER [48] packages for R environment. Diversification rates were also compared on all trees of the posterior distribution for different clades using the Yule model in APE. Four species are not included in the phylogeny but were taken into account with the rate.estimate function in GEIGER (details in Text S6). According to morphological studies [49], all these species belong to clade B: A. petchekara is close to A. azurensis, A. humboldti and A. nekando are close to A. chopardi, and A. novaecaledoniae is close to A. sarramea (within clade B2).
Analysis of the Geographical Pattern of Speciation
Data on geographic distribution of the species were obtained from the collections of the following institutions: Muséum national d’Histoire naturelle (Paris), Natural History Museum (London), Queensland Museum (Brisbane), Osaka Museum of Natural History (Osaka), Naturhistorisches Museum (Vienna) and National Museum of Natural History (Leiden). The material examined in previous taxonomic studies of the genus was taken into account [29], [49], [50], [51], [52], [53], [54]. Species were recorded from a total of 52 localities for all the species (mean = 3.5; Table S4).
Distribution and range size of species were measured on a grid divided into quadrats of 0.025 degree intervals of latitude and longitude. For each spot from which a species was recorded, we considered an 8 km diameter circular area as the range size. This diameter was chosen based on the distribution of the most sampled species, Agnotecous azurensis, aimed at generating a continuous distribution area for this species. Considering these range sizes, our sampling covers 70% of Agnotecous main potential habitat (rainforest) (ca. 2670 km2 out of 3815 km2; after data available in DIVA-GIS v.7.1 [55] (see Figure S1).
To infer the predominant mode of speciation, and to account for post-speciational range changes, we applied the age-range correlation (ARC) method test [56]. A presence/absence matrix was used to calculate the degree of sympatry at each node of the tree. For pairs of terminal species, the degree of sympatry was calculated as the number of quadrats in which two species co-occur, divided by the number of quadrats occupied by the species with the smallest range [56], [57], [58], [59]. For deeper nodes in the tree we followed an alternative methodology using independent contrasts between pairs of taxa, which does not bring to artefactual increase of degree of sympatry through time [60], [61], [62], by acknowledging that unlike a phenotypic trait, distribution is an extrinsic feature of species that should not be interpreted only by mapping on a tree [63]. Average pairwise overlaps were calculated for each node. The degree of sympatry was plotted against the node ages estimated from BEAST analysis, and a linear regression was used to estimate intercept and slope. A positive slope with intercept <0.5 means that allopatric speciation predominates in the clade and that species become more sympatric as time since divergence increases, whereas a negative slope with intercept >0.5 means that sympatric speciation is the predominant mode and that overlap between species decreases over time [56], [57], [59], [60]. The statistical significance of the resulting slopes and intercepts was assessed via Monte Carlo simulation by random permutation of the overlap matrix to estimate a distribution under the null hypothesis of no phylogenetic signal. The fraction of randomized datasets with intercept and slopes greater than the observed data was calculated [60]. To conduct these analyses, we used the R package “phyloclim” v. 0.6 [64] with a modified function ARC provided by B. Fitzpatrick (pers. comm.).
Although Agnotecous is not characterised by high mobility (short-winged species unable to fly), we also computed two modified indicators following [65], known to be less sensitive to change in range size than the other indices: the proportion of cases where range overlap was 0 and the proportion of cases where range overlap was complete. These indicators were plotted against the node ages estimated from BEAST analysis, and a linear regression was used to estimate intercept and slope. A positive slope with intercept <0.5 means that allopatric speciation dominates, whereas a negative slope with intercept >0.5 means that sympatric speciation is the predominant mode [56], [60].
Finally, we computed the relationship between areas and ages of species to investigate post-speciational range expansion, and test whether microendemism is mostly found in younger clades.
Distribution areas and ages of clades were compared with the map of soil diversity using the GIS layers provided by Direction des Infrastructures, de la Topographie et des Transports Terrestres of the Gouvernement de la Nouvelle-Calédonie, and past climatic events with data in Chevillotte et al. [66].
Results
Phylogeny and Time-course of Diversification
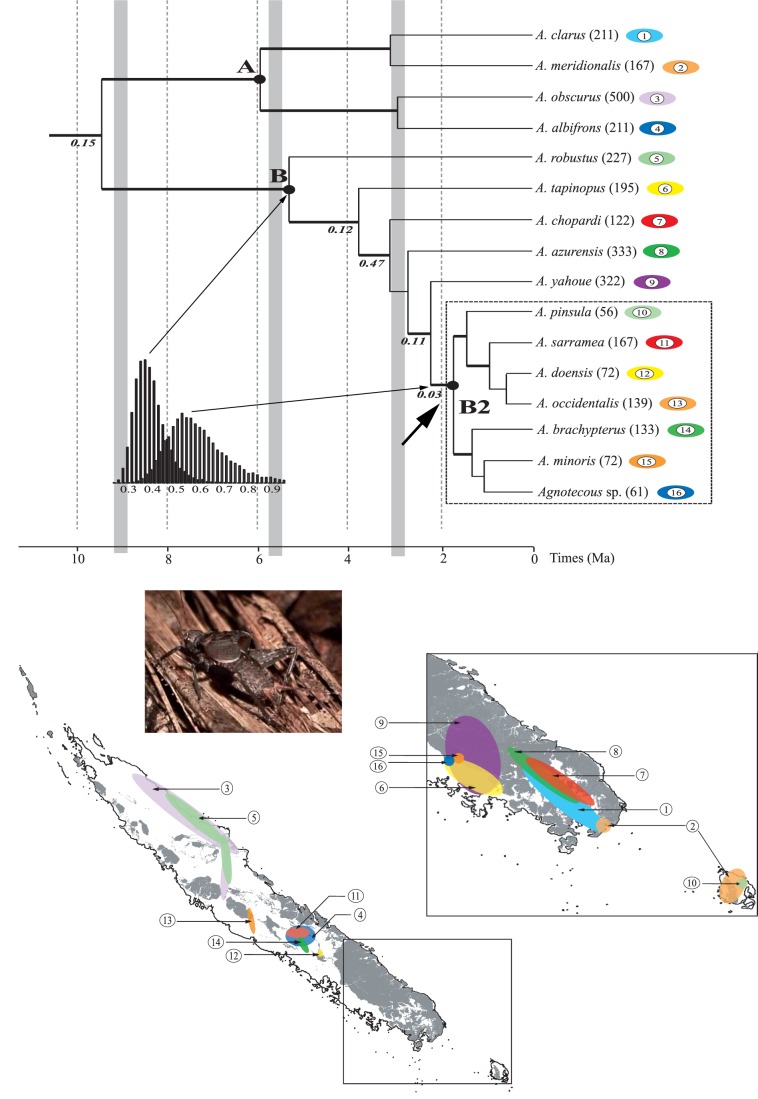
Two main lineages (A and B) of similar age (5.4 and 6 Ma, respectively) are recovered within the genus Agnotecous (Figure 1). Each clade consists of species distributed across New Caledonia. Major diversification events in Agnotecous are found around three major recent climatic events (Figure 1): The genus diversification started just before the first episode and was particularly high after the third episode, but the two main clades started diversifying during the second episode of dry and hot climate. In this context, clade B2 shows the highest diversification, which started just after the third episode of climatic changes and coincided with the peak of temperatures and the regular modifications of sea level.
Figure 1. Chronogram reconstructed under BEAST and distribution of Agnotecous species.
On the chronogram, branches shown as thick black lines indicate posterior probabilities ≥0.95. When different from 0, degrees of sympatry are indicated below branches. Grey vertical bars indicate morphogenesis episodes and dry periods (after [66]).The marginally significant case of diversification rate shifts detected with apTreeshape is indicate by an arrow, and the distribution of yule estimator of diversification for all trees the genus and clade B2 is shown. Hypothetical areas (in km2) are given on the right of the taxa names, and species are colored and numerated according their respective distribution. On the maps, the distribution of ultramafic rocks and corresponding metalliferous soils is indicated in grey (after [9]).
The species-level LTT plots suggested two minor increases in diversification rates around 3.5 and 2 Myr ago (Figure 2), and a significant tree asymmetry was detected through the most to least sensitive test-statistics in SymmeTREE (0.0066<P<0.022), indicating that lineages diversified at significantly different rates. Although no significant diversification rate shifts were detected with apTreeshape, one marginally significant case was identified (p = 0.11) for the divergence of clade B2, i.e. ∼1.7 Myr ago. All details about these analyses are available in Tables S5–S6 and Figure S4.
Figure 2. Species-level log lineages-through-time (LTT) plot for Agnotecous (excluding infra-specific data).
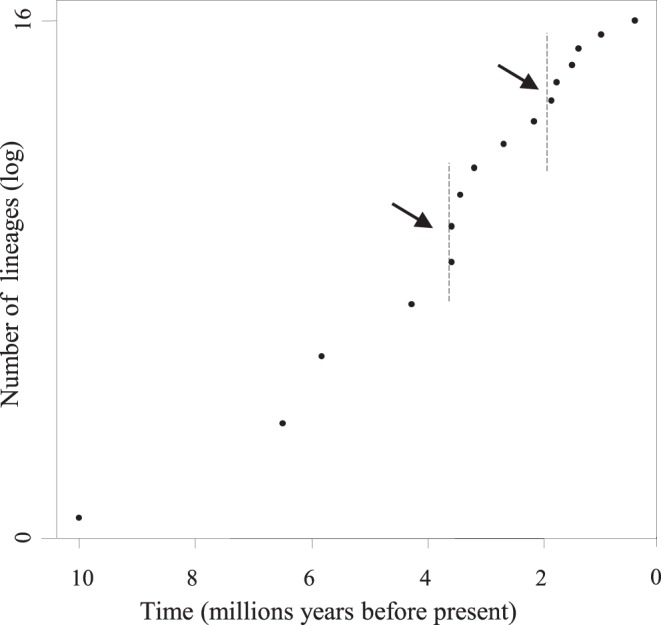
Two minor increases in diversification rates around 3.5 Ma and 2 Ma are showing, but likelihood analyses reject the null hypothesis that diversification rates in Agnotecous have been constant through time.
These results are not confirmed by BDL analyses: the best rate-constant model (pure birth) had an AIC of 13.16 versus 15.97 for the rate-variable model (yule-2-rate) (ΔAICrc = 3.19) (see Table S7). Although marginally significant, the observed ΔAICrc statistics do not indicate a departure from the null hypothesis of rate constancy (p = 0.066) under a pure birth model. There is a marked difference of diversification rate (measured with ratio speciation: extinction of 0–0.5) of the clade B (0.33–0.29 without missing species and 0.39–0.34 including missing species) and clade B2 (0.72–0.62 without missing species and 0.80–0.69 including missing species). This difference is also observed on all trees of the posterior distribution (Figure 1).
Geographical Pattern of Speciation
Geographical distributions reveal that sympatry is widespread amongst Agnotecous. Only three species belonging to clade B2 are not sympatric with any other (Figure 1). All highly microendemic species are located in the same clade (B2), which comprises 44% of the species and represents only 23% of the distribution area. Mann-Whitney tests show that these differences of area are highly significant: between clade B2 and other species (p = 0.004), and between clade B2 and all other species from clade B (p = 0.023). Species diversity and level of endemism are thus higher in the southern part of New Caledonia. Comparison of species distributions with soil diversity also shows that the southern region, where the highly endemic clade B2 occurs, is mostly characterized by metalliferous soils, while the rest of the species shows no relationship with the kind of soil.
In the age-range correlation analysis, the regression of range overlap against node age in each clade generated positive intercept (0.02170<0.5) and slope (0.01244>0), thus rejecting a scenario of fully sympatric speciation (Figure 3). Values were not statistically significant (P = 0.462), meaning that a fully allopatric speciation model was not supported either. Similarly, the test on the proportion of cases where ranges overlap did not show a clear pattern (P = 0.2746), although here again the observed trend is closer to a model of allopatric speciation (Figure 4). More importantly, except for A. minoris and Agnotecous sp., which are both microendemics, recently diverged sister species exhibit no geographical overlap, whereas more distantly related sister taxa do. This strongly suggests recent allopatric speciation. Finally, there is a negative relationship between area and age of species (Figure 5), indicating geographical range expansion.
Figure 3. Degree of geographical overlap plotted as a function of relative age in the genus Agnotecous.
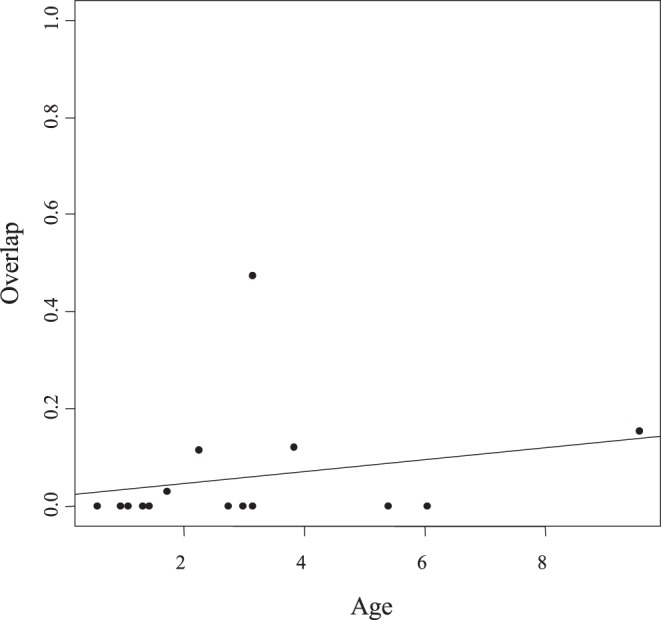
Each point represents a node in the species phylogeny in Figure 1. Linear regression yielded low intercept (0.02170<0.5) and positive slope (0.01244), suggestive of allopatric speciation, but values are not statically significant (P-value = 0.462>>0.05).
Figure 4. Correlation between the proportion of cases showing zero overlap and the proportion of range overlap.
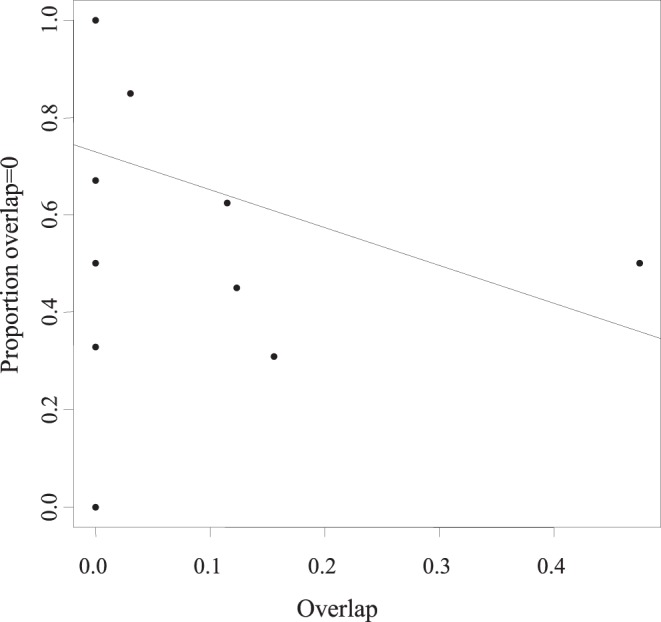
Using the Phillimore et al., 2008 method, allopatric speciation is suggestive but values are not statically significant (P-value = 0.2746>>0.05).
Figure 5. Relationship between areas and ages of species.
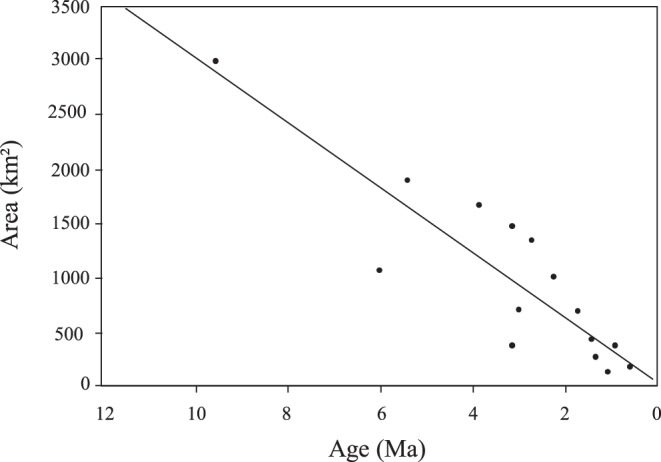
The negative relationship between area and age of species indicates a geographical range expansion.
Discussion
In Agnotecous, a cricket genus characterized by restricted areas of distribution (microendemism; Figure 1) and complex patterns of sympatric species, the molecular phylogeny supports a hypothesis of dominant and recent allopatric speciation. ARC and range overlap tests clearly rule out a dominant sympatric speciation model (slope and intercept values typical of allopatric speciation), but they do not significantly support a fully allopatric speciation model. ARC tests are often non-conclusive [65], especially when assumptions of the test are not met, i.e. limited post-speciational range change and a single speciation mode. Still, this method highlights the relative importance between sympatric versus non-sympatric speciation and allows discussing them in relation with biological features and temporal diversification [62], [67]. In the Agnotecous case study (Figures 3 and 4), dominant allopatric speciation appears more likely than dominant sympatric speciation, at least in the last millions of years, when most of the diversification and microendemism took place. Closely related species are clearly allopatric and only two sister species show partially overlapping ranges (A. minoris and Agnotecous sp.). However, this case of partial sympatry is mostly a consequence of our definition of estimated range sizes, and these species are indeed not syntopic, but were found at different altitudes on the same mountain (Mont Mou: 375 m for A. minoris versus 1010–1140 m for Agnotecous sp.).
All the studies analysing the mode of speciation in New Caledonia have concluded that both the faunal and floral diversity originated from dominant allopatric speciation [27]. Despite the observed pattern of sympatry which first suggested sympatric speciation, our study is finally consistent with this general picture. What is then the origin of the microendemism and sympatric distributions observed in Agnotecous?
Recent events may have affected the post-speciational distributions of the species, resulting in secondary contacts between them. This kind of pattern was already found in other microendemic taxa showing lower levels of sympatry. For example, sympatric species occupy different microhabitats as a consequence of character displacement in Australian freshwater snails [68], or New-Caledonian Heteroptera [23].
Our results have highlighted that distribution patterns are related to the age of species: relatively old species (>3–4 Ma) show a larger distribution (>200 km2), and younger species show more restricted distributions. This is especially true in the clade B2 (Figure 1), which contains 44% of the species in only 23% of the total area of distribution in the southeast of New Caledonia (Figure 1). Clade B2 is dated at 2.8 Myr ago only and includes 7 species showing no mutual sympatry, with putative range size comprised between 56–167 km2 (mean = 100 km2), compared to the 122–500 km2 for other species (mean = 253 km2). Highly microendemic species are thus clearly the most recent ones (Figure 1). In other words, correlation between size of distribution area and species age could be considered as a strong indication of range expansion after speciation events. In this view, extreme microendemism may be seen as a transitory step in the history of a species distribution, that is only observed in the case of the youngest species. The whole pattern, allopatric speciation and evolution toward secondary sympatry, is geographically very coherent, even though the most recent events of speciation date back to more than 1 Myr. It emphasizes once more that the geographical study of speciation depends on the stability of distributional patterns associated with speciation, that should not be obscured by too large and frequent events of range change or extinctions [69].
If generalised through relevant testing in other groups of organisms in other regions, this hypothesis may explain the extraordinary level of microendemism in some megadiverse hotspots. The question may then become not only why are there so many species with such small distribution areas, but why speciation and distributional patterns remain so stable without immediate range expansion. As a matter of consequence, range size of endemic species cannot only be assumed to be controlled by a given set of classical influential factors, but should also be considered a possible by-product of evolutionary time.
Several factors have been considered to explain high speciation rates and microendemism in biodiversity hotpots such as New Caledonia [27], Madagascar [28], Mexico [70] or Eastern North America [71]. Predominantly, altitude and climatic variations have been hypothesized to promote microendemic diversification as a consequence of climatic variations, through allopatric speciation with niche conservatism on different mountains [72]. Coupled with recent climatic changes, landscape complexity may be another factor involved in speciation. For example, in Malagasy endemic cophyline frogs, mountains constitute key areas for endemic diversity because of the presence of slopes, which increase community complexity and favour parapatric or sympatric speciation, while the altitudinal ranges promote allopatric speciation through topogeographic opportunities [28]. In New Caledonian cockroaches Lauraesilpha, speciation of microendemic species is explained by isolation on different mountains following climatic variations [9], [73].
In our study, there is no clear correspondence between climatic events [66] and species diversification (Figure 1). The initial diversification in Agnotecous took place during a climatic episode going from 16 to 4 Myr ago. This time lag, similar to that found in Trichoptera from New Caledonia [17], involves a period of long-term climate change and modification of the landscape with four successive planation episodes and dry periods, accompanied by oscillations of continental temperature and sea level [66]. In more recent taxa (after 3 Myr ago), we observe a marked difference of diversification rates (between clades B and B2) co-occurring with a period of climate change marked by frequent changes of sea level between −50 m and 60 m [66]. It suggests that the influence of climatic changes on speciation processes could be mediated by another factor.
Intense fragmentation of soil and habitats caused by climatic events may explain the pattern of distribution observed in Agnotecous. Soil diversity and especially metalliferous soils have often been seen as primarily important for speciation in New Caledonia [74], but the role of metalliferous soils has only been documented in plants [e.g. 75] and in one group of aquatic insects [76]. Metalliferous soils derived from ultramafic rocks almost covered the entire surface of New Caledonia after the Eocene obduction, and they still cover about one third of the territory today, mostly as a dense patchwork in the southeast, with addition of patches in the north-west [66], [77]. Ultramafic rocks are rich in ferromagnesian minerals (e.g. olivines, amphiboles and pyroxenes), which are easily altered by the action of water and wind [78]. Such alteration is responsible for the release of large quantities of iron, magnesium and nickel, and results in the complex landscape which is found in ultramafic massifs of New Caledonia [66]. This chemical process means that recent climatic changes may have affected these regions with metalliferous soils more intensely than nearby regions without these soils, especially during periods of alteration/erosion/peneplanation. This would result in range fragmentation with more complex landscape and habitat reduction, which in turn would favour recent allopatric speciation.
Agnotecous species are found both on metalliferous and non metalliferous soils, without a clear and contrasted radiation on one kind of soil (same conclusions as in [9]). Nevertheless, most species of the younger clade B2, i.e. species with restricted distribution areas, occur in the south, on or nearby metalliferous soils and ultramafic rocks (Figure 1). Species of clade A and of the first lineages of clade B mostly occur far from ultramafic rocks, and they have larger distribution ranges, especially in northern New Caledonia (Figure 2). As allopatry is the dominant mode of speciation in Agnotecous, soil heterogeneity may have an indirect effect on habitat fragmentation, which could in turn explain local microendemism concentrated in the south of the island.
Conclusion
Even in the Agnotecous lineage showing a generalized pattern of sympatry, speciation appeared predominantly allopatric. Therefore, our study brings new and conservative evidence in favour of the classical scenario of allopatric speciation leading to diversification in biodiversity hotspots. Our study also suggests that extreme local diversity may be explained by the recent establishment of microendemism through an environment-driven process. In this context, speciation studies help understanding that present-day microendemism is a first stage of a complex and dynamic process. Future studies should be conducted to specify the possible complex interplay between climatic changes, mountain complexity and soil-driven landscape changes, which could have permitted repeated speciation events and range changes.
Supporting Information
Geographic distribution of Agnotecous species showing the general pattern of sympatry throughout New Caledonia.
(PDF)
Topologies obtained in parsimony (a) and Bayesian inference (b) for mitochondrial dataset only.
(PDF)
Comparison between the phylogeny obtained from the 37 specimens (a, c) and the phylogeny from the 16 consensus sequences (b, d).
(PDF)
Repartition of p-values obtained by the analyses under apTreeshape.
(PDF)
Additional information on taxonomic sampling, voucher references (specimen numbers in MNHN collections) and GenBank accession numbers of the specimens included in this study.
(PDF)
Primers used in this study.
(PDF)
Results of the Bayesian test of monophyly for Agnotecous species.
(PDF)
Details of the geographic distribution of species used in the analysis of geographical pattern of speciation.
(PDF)
Results of the asymmetry tests conducted under SymmeTREE for seven test statistics.
(PDF)
Results of the diversification rate change test conducted under the R package apTreeshape.
(PDF)
Results of the BDL analyses and comparison of AIC values from different diversification models.
(PDF)
Additional information on sequencing protocols.
(PDF)
Characteristics of mitochondrial (a) and nuclear (b) datasets used in this study.
(PDF)
Congruence tests between nuclear and mitochondrial data sets.
(PDF)
Details about phylogenetic analyses.
(PDF)
Additional BEAST parameters and details of calibrations.
(PDF)
Details of the rate.estimate function used in GEIGER.
(PDF)
Acknowledgments
We are grateful to the following institutions or programs that made our work possible: the Directions de l’Environnement of New Caledonia’s Province Nord and Province Sud for permits to work in natural parks and forest reserves; the Direction des Infrastructures, de la Topographie et des Transports Terrestres of the Gouvernement de la Nouvelle-Calédonie for kindly providing the GIS layers and electronic maps of New Caledonia (agreement number CS11-3170-865); and the IRD authorities for local support and hosting. We are also grateful to Geoff B. Monteith (QMF) for loan of specimens from the collections of Brisbane and Akihiko Ichikawa for loan of specimens from the collections of Osaka. Christian Mille and Fabrice Colin provided valuable help and are sincerely thanked. We are also grateful to Jérôme Murienne and Annabel Whibley for their critical reading of the paper.
Funding Statement
The following institutions or programs have supported the work: the French Agence Nationale de la Recherche (ANR) for funding via the Biodiversity research grant BIONEOCAL, to P.G.; the Muséum national d’Histoire naturelle (MNHN); the Gouvernement de Nouvelle-Calédonie and the Prévost Fund of the Ecole Doctorale of the Muséum for funding the PhD of R.N.; and the Consortium National de Recherche en Génomique, and the MNHN (CNRS UMS2700), under agreement number 2005/67 between Génoscope and the MNHN for the project ‘Macrophylogeny of life’ of Guillaume Lecointre. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wilson EO (1961) The Nature of the Taxon Cycle in the Melanesian Ant Fauna. American Naturalist 95: 169–193. [Google Scholar]
- 2. Losos JB, Ricklefs RE (2009) Adaptation and diversification on islands. Nature 457: 830–836. [DOI] [PubMed] [Google Scholar]
- 3. Losos JB, Schluter D (2000) Analysis of an evolutionary species-area relationship. Nature 408: 847–850. [DOI] [PubMed] [Google Scholar]
- 4. Schliewen U, Rassmann K, Markmann M, Markert J, Kocher T, et al. (2001) Genetic and ecological divergence of a monophyletic cichlid species pair under fully sympatric conditions in Lake Ejagham, Cameroon. Molecular Ecology 10: 1471–1488. [DOI] [PubMed] [Google Scholar]
- 5. Barluenga M, Stölting KN, Salzburger W, Muschick M, Meyer A (2006) Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature 439: 719–723. [DOI] [PubMed] [Google Scholar]
- 6. Papadopulos AST, Baker WJ, Crayn D, Butlin RK, Kynast RG, et al. (2011) Speciation with gene flow on Lord Howe Island. Proceedings of the National Academy of Sciences of the United States of America 108: 13188–13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vences M, Wollenberg KC, Vieites DR, Lees DC (2009) Madagascar as a model region of species diversification. Trends in Ecology & Evolution 24: 456–465. [DOI] [PubMed] [Google Scholar]
- 8. Murienne J, Grandcolas P, Piulachs MD, Bellés X, D’Haese C, et al. (2005) Evolution on a shaky piece of Gondwana: is local endemism recent in New Caledonia? Cladistics 21: 2–7. [DOI] [PubMed] [Google Scholar]
- 9. Murienne J, Pellens R, Budinoff RB, Wheeler WC, Grandcolas P (2008) Phylogenetic analysis of the endemic New Caledonian cockroach Lauraesilpha. Testing competing hypotheses of diversification. Cladistics 24: 802–812. [Google Scholar]
- 10. Raxworthy CJ, Nussbaum RA (1995) Systematics, speciation and biogeography of the dwarf chameleons (Brookesia; Reptilia, Squamata, Chamaeleontidae) of northern Madagascar. Journal of Zoology 235: 525–558. [Google Scholar]
- 11. Pastorini J, Thalmann U, Martin RD (2003) A molecular approach to comparative phylogeography of extant Malagasy lemurs. Proceedings of the National Academy of Sciences of the United States of America 100: 5879–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodman SM, Ganzhorn JU (2004) Biogeography of lemurs in the humid forests of Madagascar: the role of elevational distribution and rivers. Journal of Biogeography 31: 47–55. [Google Scholar]
- 13. Savolainen V, Anstett M-C, Lexer C, Hutton I, Clarkson JJ, et al. (2006) Sympatric speciation in palms on an oceanic island. Nature 441: 210–213. [DOI] [PubMed] [Google Scholar]
- 14.Grant PR, Grant V (2010) Sympatric speciation, immigration, and hybridization in island birds. In: Losos J, Ricklefs RE, editors. The theory of island biogeography revisited. Princeton (New Jersey): Princeton University Press. 326–357.
- 15. Balke M, Ribera I, Vogler AP (2004) MtDNA phylogeny and biogeography of Copelatinae, a highly diverse group of tropical diving beetles (Dytiscidae). Molecular Phylogenetics and Evolution 32: 866–880. [DOI] [PubMed] [Google Scholar]
- 16. Balke M, Pons J, Ribera I, Sagata K, Vogler AP (2007) Infrequent and unidirectional colonization of hyperdiverse Papuadytes diving beetles in New Caledonia and New Guinea. Molecular Phylogenetics and Evolution 42: 505–516. [DOI] [PubMed] [Google Scholar]
- 17. Espeland M, Johanson KA (2010) The effect of environmental diversification on species diversification in New Caledonian caddisflies (Insecta: Trichoptera: Hydropsychidae). Journal of Biogeography 37: 879–890. [Google Scholar]
- 18. Ratsoavina FM, Gehring P, Ranaivoarisoa FJ, Rafeliarisoa TH, Crottini A, et al. (2010) Molecular phylogeography of a widespread Malagasy leaf chameleon species, Brookesia superciliaris . Zootaxa 64: 62–64. [Google Scholar]
- 19. Vences M, Köhler J, Pabijan M, Glaw F (2010) Two syntopic and microendemic new frogs of the genus Blommersia from the east coast of Madagascar. African Journal of Herpetology 59: 133–156. [Google Scholar]
- 20. Kier G, Kreft H, Lee TM, Jetz W, Ibisch PL, et al. (2009) A global assessment of endemism and species richness across island and mainland regions. Proceedings of the National Academy of Sciences of the United States of America 106: 9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartish IV, Swenson U, Munzinger J, Anderberg AA (2005) Phylogenetic relationships among New Caledonian Sapotaceae (Ericales): Molecular evidence for generic polyphyly and repeated dispersal. American Journal of Botany 92: 667–673. [DOI] [PubMed] [Google Scholar]
- 22. Swenson U, Munzinger J, Bartish IV (2007) Molecular phylogeny of Planchonella (Sapotaceae) and eight new species from New Caledonia. Taxon 56: 329–354. [Google Scholar]
- 23.Murienne J, Guilbert E, Grandcolas P (2009) Species’ diversity in the New Caledonian endemic genera Cephalidiosus and Nobarnus (Insecta : Heteroptera : Tingidae), an approach using phylogeny and species’ distribution modelling. Biological Journal of the Linnean Society: 177–184.
- 24.Trewick S, Brescia F, Jordan C (2008) Diversity and phylogeny of New Caledonian Placostylus land snails; evidence from mitochondrial DNA. In: Grandcolas P, editor. Zoologia Neocaledonica 7 Biodiversity studies in New Caledonia. Paris: Mémoires du Muséum national d’Histoire naturelle. 421–436.
- 25. Bauer AM, Todd J, Sadlier RA, Whitaker AH (2006) A Revision of the Bavayia validiclavis group (Squamata: Gekkota: Diplodactylidae), a Clade of New Caledonian Geckos Exhibiting Microendemism. Proceedings of the California Academy of Sciences 57: 503–547. [Google Scholar]
- 26. Smith SA, Sadlier RA, Bauer AM, Austin CC, Jackman T (2007) Molecular phylogeny of the scincid lizards of New Caledonia and adjacent areas: Evidence for a single origin of the endemic skinks of Tasmantis. Molecular Phylogenetics and Evolution 43: 1151–1166. [DOI] [PubMed] [Google Scholar]
- 27. Grandcolas P, Murienne J, Robillard T, Desutter-Grandcolas L, Jourdan H, et al. (2008) New Caledonia: a very old Darwinian island? Philosophical Transactions of the Royal Society B: Biological Sciences 363: 3309–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wollenberg KC, Vieites DR, van der Meijden A, Glaw F, Cannatella DC, et al. (2008) Patterns of endemism and species richness in Malagasy cophyline frogs support a key role of mountainous areas for speciation. Evolution 62: 1890–1907. [DOI] [PubMed] [Google Scholar]
- 29. Desutter-Grandcolas L, Robillard T (2006) Phylogenetic systematics and evolution of Agnotecous in New Caledonia (Orthoptera: Grylloidea, Eneopteridae). Systematic Entomology 31: 65–92. [Google Scholar]
- 30. Nattier R, Robillard T, Desutter-Grandcolas L, Couloux A, Grandcolas P (2011) Older than New Caledonia emergence? A molecular phylogenetic study of the eneopterine crickets (Orthoptera: Grylloidea). Journal of Biogeography 38: 2195–2209. [Google Scholar]
- 31. Robillard T, Desutter-Grandcolas L (2006) Phylogeny of the cricket subfamily Eneopterinae (Orthoptera, Grylloidea, Eneopteridae) based on four molecular loci and morphology. Molecular Phylogenetics and Evolution 40: 643–661. [DOI] [PubMed] [Google Scholar]
- 32. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- 34.Nylander JAA (2004) MrModeltest, version 2.3. Available: http://www.abc.se/~nylander/.
- 35.Akaike H (1973) Information theory and an extension of the maximum likelihood principle. In: Csaki BPaF, editor. Second International Symposium on Information Theory. Budapest: Akademiai Kiado. 267–281.
- 36. Akaike H (1974) New Look at Statistical-Model Identification. Ieee Transactions on Automatic Control Ac19: 716–723. [Google Scholar]
- 37. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 38. Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786. [Google Scholar]
- 39. Huelsenbeck JP, Crandall KA (1997) Phylogeny estimation and hypothesis testing using maximum likelihood. Annual Review of Ecology and Systematics 28: 437–466. [Google Scholar]
- 40. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. Bmc Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rambaut A, Drummond AJ (2007) Tracer v1.4, Available: http://beast.bio.ed.ac.uk/Tracer.
- 42. Chan KMA, Moore BR (2005) SymmeTREE: whole-tree analysis of differential diversification rates. Bioinformatics 21: 1709–1710. [DOI] [PubMed] [Google Scholar]
- 43. Bortolussi N, Durand E, Blum M, François O (2006) apTreeshape: statistical analysis of phylogenetic tree shape. Bioinformatics (Oxford, England) 22: 363–364. [DOI] [PubMed] [Google Scholar]
- 44.Moore BR, Chan KMA, Donoghue MJ (2004) Detecting diversification rate variation in supertrees. In: Bininda-Emonds ORP, editor. Phylogenetic Supertrees: Combining Information to Reveal the Tree of Life. Dordrecht, the Netherlands: Kluwer Academic. 487–533.
- 45. Rabosky DL (2006) LASER: A Maximum Likelihood Toolkit for Detecting Temporal Shifts in Diversification Rates From Molecular Phylogenies. Evolutionary Bioinformatics 2: 247–250. [PMC free article] [PubMed] [Google Scholar]
- 46. Rabosky DL (2006) Likelihood methods for detecting temporal shifts in diversification rates. Evolution 60: 1152–1164. [PubMed] [Google Scholar]
- 47. Paradis E, Claude J, Strimmer K (2004) APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- 48. Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W (2008) GEIGER: investigating evolutionary radiations. Bioinformatics 24: 129–131. [DOI] [PubMed] [Google Scholar]
- 49. Robillard T, Nattier R, Desutter-Grandcolas L (2010) New species of the New Caledonian endemic genus Agnotecous (Orthoptera, Grylloidea, Eneopterinae, Lebinthini). Zootaxa 2559: 17–35. [Google Scholar]
- 50. Gorochov AV (1985) Mesozoic crickets (Orthoptera, Grylloidea) of Asia. Paleontologicheskii Zhurnal 2: 59–68. [Google Scholar]
- 51. Otte D, Alexander RD, Cade W (1987) The crickets of New Caledonia (Gryllidae). Proceedings of the Academy of Natural Sciences of Philadelphia 139: 375–457. [Google Scholar]
- 52.Desutter-Grandcolas L (1997) Le peuplement de grillons du sous-bois (Orthoptères Grylloidea) de la Réserve de faune du Col d’Amieu (Nouvelle-Calédonie). I. Etude du peuplement. In: Najt J, Matile L, editors. Zoologia Neocaledonica Volume 4: Mémoires du Muséum national d’Histoire naturelle, Paris. 125–135.
- 53.Desutter-Grandcolas L (1997) Les grillons de Nouvelle-Calédonie (Orthoptères, Grylloidea): espèces et données nouvelles. In: Najt J, Matile L, editors. Zoologia Neocaledonica Volume 4: Mémoires du Muséum national d’Histoire naturelle, Paris. 165–177.
- 54. Gorochov AV (1986) New and little known crickets (Orthoptera, Gryllidae) from Australia and Oceania. Revue d’Entomologie de l’URSS 65: 692–708. [Google Scholar]
- 55. Hijmans RJ, Cruz M, Rojas E (2002) Computer tools for spatial analysis of plant genetic resource data: DIVA-GIS. Plant Genetic Resources Newsletter 127: 15–19. [Google Scholar]
- 56. Barraclough TG, Vogler AP (2000) Detecting the geographical pattern of speciation from species-level phylogenies. American Naturalist 155: 419–434. [DOI] [PubMed] [Google Scholar]
- 57.Lynch JD (1989) The gauge of speciation: on the frequency of modes of speciation. In: Endler JA, Otte D, editors. Speciation and its consequences. Sunderland, MA: Sinauer Associates. 527–553.
- 58. Chesser RT, Zink RM (1994) Modes of speciation in birds: a test of Lynch’s method. Evolution 48: 490–497. [DOI] [PubMed] [Google Scholar]
- 59. Berlocher SH, Feder JL (2002) Sympatric speciation in phytophagous insects: Moving beyond controversy? Annual Review of Entomology 47: 773–815. [DOI] [PubMed] [Google Scholar]
- 60. Fitzpatrick BM, Turelli M (2006) The geography of mammalian speciation: Mixed signals from phylogenies and range maps. Evolution 60: 601–615. [PubMed] [Google Scholar]
- 61. Jiggins CD, Mallarino R, Willmott KR, Bermingham E (2006) The phylogenetic pattern of speciation and wing pattern change in neotropical Ithomia butterflies (Lepidoptera : Nymphalidae). Evolution 60: 1454–1466. [DOI] [PubMed] [Google Scholar]
- 62. Linnen CR, Farrell BD (2010) A test of the sympatric host race formation hypothesis in Neodiprion (Hymenoptera: Diprionidae). Proceedings of the Royal Society B: Biological Sciences 277: 3131–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Grandcolas P, Nattier R, Legendre F, Pellens R (2011) Mapping extrinsic traits such as extinction risks or modelled bioclimatic niches on phylogenies: does it make sense at all? Cladistics 27: 181–185. [DOI] [PubMed] [Google Scholar]
- 64.Heibl C (2011) Phyloclim: integrating phylogenetics and climatic niche modelling. R package accessed: September-20–2010.
- 65. Phillimore AB, Orme CDL, Thomas GH, Blackburn TM, Bennett PM, et al. (2008) Sympatric speciation in birds is rare: Insights from range data and simulations. American Naturalist 171: 646–657. [DOI] [PubMed] [Google Scholar]
- 66. Chevillotte V, Chardon D, Beauvais A, Maurizot P, Colin F (2006) Long-term tropical morphogenesis of New Caledonia (Southwest Pacific): Importance of positive epeirogeny and climate change. Geomorphology 81: 361–375. [Google Scholar]
- 67. Perret M, Chautems A, Spichiger R, Barraclough TG, Savolainen V (2007) The geographical pattern of speciation and floral diversification in the neotropics: The tribe Sinningieae (Gesneriaceae) as a case study. Evolution 61: 1641–1660. [DOI] [PubMed] [Google Scholar]
- 68. Ponder WF, Colgan DJ (2002) What makes a narrow-range taxon? Insights from Australian freshwater snails. Invertebrate Systematics 16: 571–582. [Google Scholar]
- 69. Losos JB, Glor RE (2003) Phylogenetic comparative methods and the geography of speciation. Trends in Ecology & Evolution 18: 220–227. [Google Scholar]
- 70. Ochoa-Ochoa LM, Rodríguez P, Mora F, Flores-Villela O, Whittaker RJ (2012) Climate change and amphibian diversity patterns in Mexico. Biological Conservation 150: 94–102. [Google Scholar]
- 71. Hollingsworth PR Jr, Near TJ (2009) Temporal patterns od fiversification and microendemism in eastern highland endemic barcheek darters (Percidae: Etheostomatinae). Evolution 63: 228–243. [DOI] [PubMed] [Google Scholar]
- 72. Wiens JJ, Harrison R (2004) Speciation and Ecology Revisited: phylogenetic niche conservatism and the origin of species. Evolution 58: 193–197. [DOI] [PubMed] [Google Scholar]
- 73.Murienne J, Pellens R, Grandcolas P (2008) Short-range endemism in New Caledonia. New species and distribution of the genus Lauraesilpha Grandcolas, 1997 (Insecta, Dictyoptera, Blattidae, Tryonicinae). In: Grandcolas P, editor. Zoologia Neocaledonica 6 Biodiversity studies in New Caledonia. Paris. 261–271.
- 74.Lowry PP (1998) Diversity, endemism, and extinction in the flora of New Caledonia: a review. In: Peng CI, Lowry PP, editors. Proc Int Symp on Rare, Threatened, and Endangered Floras of Asia and the Pacific. Monograph ed. Taipei, Taiwan: Institute of Botany, Academica Sinica. 181–206.
- 75. Pillon Y, Munzinger J, Amir H, Hopkins HCF, Chase MW (2009) Reticulate evolution on a mosaic of soils: diversification of the New Caledonian endemic genus Codia (Cunoniaceae). Molecular ecology 18: 2263–2275. [DOI] [PubMed] [Google Scholar]
- 76. Espeland M, Johanson KA, Hovmöller R (2008) Early Xanthochorema (Trichoptera, Insecta) radiations in New Caledonia originated on ultrabasic rocks. Molecular Phylogenetics and Evolution 48: 904–917. [DOI] [PubMed] [Google Scholar]
- 77. Paris JP (1981) Géologie de la Nouvelle-Calédonie. Un essai de synthèse (Mémoire pour servir notice explicative à la carte géologique de la Nouvelle-Calédonie à l’échelle du 1/200000): 1–278. [Google Scholar]
- 78. Sánchez-Marañón M, Gámiz E, Delgado G, Delgado R (1999) Mafic-ultramafic soils affected by silicic colluvium in the Sierra Nevada Mountains (southern Spain). Canadian Journal of Soil Science 79: 431–442. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographic distribution of Agnotecous species showing the general pattern of sympatry throughout New Caledonia.
(PDF)
Topologies obtained in parsimony (a) and Bayesian inference (b) for mitochondrial dataset only.
(PDF)
Comparison between the phylogeny obtained from the 37 specimens (a, c) and the phylogeny from the 16 consensus sequences (b, d).
(PDF)
Repartition of p-values obtained by the analyses under apTreeshape.
(PDF)
Additional information on taxonomic sampling, voucher references (specimen numbers in MNHN collections) and GenBank accession numbers of the specimens included in this study.
(PDF)
Primers used in this study.
(PDF)
Results of the Bayesian test of monophyly for Agnotecous species.
(PDF)
Details of the geographic distribution of species used in the analysis of geographical pattern of speciation.
(PDF)
Results of the asymmetry tests conducted under SymmeTREE for seven test statistics.
(PDF)
Results of the diversification rate change test conducted under the R package apTreeshape.
(PDF)
Results of the BDL analyses and comparison of AIC values from different diversification models.
(PDF)
Additional information on sequencing protocols.
(PDF)
Characteristics of mitochondrial (a) and nuclear (b) datasets used in this study.
(PDF)
Congruence tests between nuclear and mitochondrial data sets.
(PDF)
Details about phylogenetic analyses.
(PDF)
Additional BEAST parameters and details of calibrations.
(PDF)
Details of the rate.estimate function used in GEIGER.
(PDF)