Abstract
The hemagglutinin (HA) proteins derived from avian influenza viruses bind specifically to the α2-3 sialoglycan (Sia glycan), whereas human-adapted influenza viruses prefer to bind to the α2-6 Sia glycan. A switch of glycan specificity from α2-3 Sia glycan to α2-6 Sia glycan appears to be critical for a virus to become pandemic, therefore, it is important to monitor the influenza virus adaptation to glycan binding. In this article, we described surface plasmon resonance (SPR) methodology for reliable analyses of HA-glycan interactions. The methodology explores the synthetic tetravalent glycans (α2-3 Sia glycan and α2-6 Sia glycans) which facilitates not only the surface capacity of the sensor chip for better SPR signal but also enhance the affinity to the HA resulting an improved sensitivity. To adopt this method routinely for multiple samples of HA or virus, CAP-chip was adopted so that the regeneration of the sensor chip can be achieved. By combining the above developments with BiacoreT100 device, it is possible to program for analyzing multiple samples in continuous fashion under closed environment. Taken together we believe the above methodology is useful in influenza surveillance to monitor the HA adaptations to glycans among influenza viruses.
Keywords: influenza viruses, hemagglutinin, glycan, surface plasmon resonance, surveillance
Introduction
Influenza A viruses cause major respiratory tract infections in humans, birds and lower mammals and are responsible for many deaths and economic losses worldwide every year. Influenza virus belongs to the Orthomyxoviridae family and is composed of eight different single-stranded RNA segments surrounded by a lipid membrane that harbors two membrane glycoproteins, hemagglutinin (HA) and neuraminidase (NA). Among these 16 subtypes, only three HA subtypes—H1N1, H2N2 and H3N2—have successfully adapted to humans.1-3 However, in the past four decades, a growing number of human cases of avian influenza virus infections have been identified, including H5N1, H7N2, H7N7 and H9N2 sub-types.4-6
The HA protein of influenza viruses binds to host cell surface complex glycans through a terminal sialic acid (Sia) with α2-3 and α2-6 linkages. This binding is essential for the infection, transmission and virulence of influenza viruses.7,8 The crystal structures of H1, H2, H3, H5, H7 and H9 HA subtypes and their complexes with α2-3 Sia and/or α2-6 Sia glycans have been reported,9-16 revealing specific interactions between HA and α2-3 Sia or α2-6 Sia glycans. HAs derived from avian influenza viruses bind specifically to the α2-3 Sia glycan, which is preferentially expressed in the intestinal tracts of waterfowl. By contrast, human-adapted influenza viruses prefer to bind to the α2-6 Sia glycan, which is extensively expressed on the epithelial cells of the human upper respiratory tract.17 It was suggested that the HA of avian influenza viruses must adapt to bind to the α2-6 Sia glycan for efficient transmission to humans.18 In pandemic sub-types of avian influenza viruses, the HA switches its glycan specificity from α2-3 Sia to α2-6 Sia by adjusting the glycan-binding pocket through a combination of mutations. Therefore, it is important to monitor the switching of influenza virus binding specificity from α2-3 Sia to α2-6 Sia glycans, especially viruses originating in birds and swine, to understand clearly their potential for interspecies transmission.
In the past, traditional methods such as antigenic assays after isolating HA and NA from the viruses and serological analyses were commonly used for rapid detection and surveillance. However, these assays are insufficiently sensitive and require large amounts of sample. Newer label-free biosensing technologies are emerging, including optical detection platforms, such as SPR, localized surface plasmon resonance, surface-enhanced Raman scattering, fluorescence and colorimetry and several interference methods, such as microelectromechanical system cantilevers, reflectometric interference spectroscopy, interferometry and ellipsometry. Among these technologies, SPR-based analyses have attracted the most attention for many types of biomolecular interactions. The first paper published on the interaction between whole influenza virus and an antibody based on SPR was reported by Schofield and Dimmock.19 In that study, they immobilized different antibodies on a CM5 chip, analyzed whole influenza virus interactions and reported the kinetic parameters of their interactions. As an alternative to this strategy, by exploiting the efficient glycan-binding property of HA, Hidari et al.20 reported whole virus interactions with glycans, which were initially immobilized on the surface of liposomes and subsequently immobilized on the L1 sensor chip. These two studies suggest that SPR-based methodology may also be used to analyze influenza viruses. Further development of SPR hardware and software and parallel analyses of multiple samples may enable the development of a standard approach for influenza surveillance based on the SPR system in the near future.
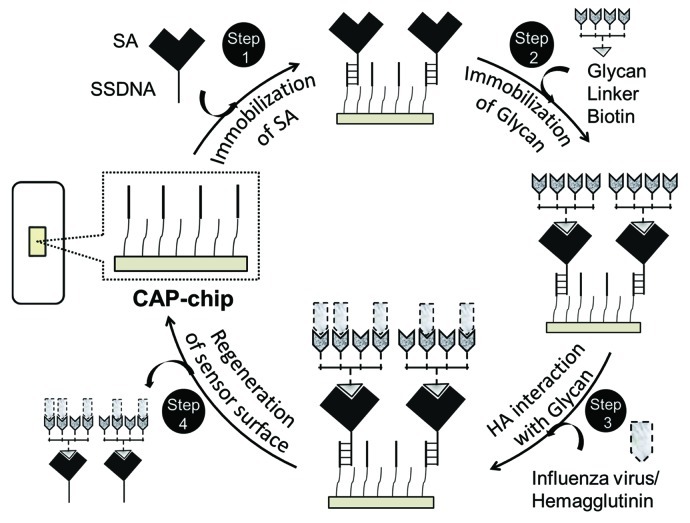
Toward this goal, we recently reported a protocol (Fig. 1) that allows the kinetic analysis of glycan-HA interactions by surface plasmon resonance (SPR),21 a technique that is commonly used to assess various bio-molecular interactions.22-24 In these analyses, first, chemically synthesized biotinylated tetravalent glycans was explored to facilitate the efficient recognition of HA. HA is a homotrimer. Based on the structure of the HA-glycan complex, each trimeric HA possesses three glycan-binding sites, which are estimated to be approximately 5 nm apart. The synthetic biotinylated tetravalent glycan has four Sia glycan moieties at the distal end of the biotinylated tetravalent glycan, and our building model predicts that the distance between the Sia glycan moieties is approximately 4 nm. Thus, the biotinylated tetravalent glycan would bind to HA monovalently but would capture other HA trimers using the three remaining unbound Sia glycan moieties. Thus, each biotinylated tetravalent glycan is expected to bind maximum of four HA trimmers, facilitating higher affinity compared with a monovalent glycan. Moreover, the application of mutlivalent glycans in our studies not only mimic the natural recognition of cell surface glycans for HA recognition but also increase the surface capacity of the sensor chip to facilitate the enhance binding of HA. Next, to expand this analysis for multiple HA samples derived from influenza viruses or whole influenza viruses we adopted the Biotin-CAP chip. The Biotin-CAP chip surface can be regenerated by incorporating a simple regeneration process (injecting regeneration solution), removing the whole complex (SA-Glycan-HA complex), for the next round of analyses. In terms of sensitivity, the SPR-based analyses known to provide, compared with other reported methods, specificity information with global rate constants [association rate (ka), dissociation rate (kd) and equilibrium dissociation constant (KD)], which allows us to monitor how well the HAs of different influenza viruses bind to α2-6 Sia glycans as compared with α2-3 Sia glycans. Importantly, combining the above developments with BiacoreT100 device, it is possible to program analyses for multiple samples continuously in a closed environment. The SPR-based methodology described here has potential applications not only in influenza surveillance to define a pandemic nature of influenza viruses but also useful for screening synthetic glycans and other compounds that may interfere with glycan-HA interactions.
Figure 1. The SPR-based surveillance of influenza virus or HA derived from influenza viruses.
Materials and Methods
Reagents
Proteins, glycans and buffers
The soluble form of HA of A/H5N1/Vietnam/1203/2004 (accession number AAW 80717) was obtained from Prospec-Tany TechnoGene Ltd. and was expressed in baculovirus. The purified HA of A/H3N2/Panama/2007/1999 (expressed in a mammalian system) was obtained from Immune Technology. The Biotinylated multivalent glycans; 01-077, [Neu5Acα2-3Gal β1-4GlcNAcβ1-PAA (polyacrylamide)-biotin]; 01-088, [Neu5Acα2-3Galβ1-3GalNAcα1-PAA-biotin]; 01-039a, [Neu5Acα2-6Galβ1-4GlcNAcβ1-PAA-biotin]; and 01-000, [HOCH2(HOCH)4 CH2NH-PAA-biotin] were purchased from GlycoTech. All the SPR-based analyses of HA-glycan interactions were performed in HBS-P+ buffer [0.01 M HEPES, 0.15 M NaCl, 0.05% (v/v) Surfactant P20, pH 7.4; GE Healthcare].
Equipment
We used Biacore T100 (GE Healthcare) instrument in evaluating HA and glycan interactions. This system explores the surface plasmon resonance (SPR) phenomena and exploited in the past for analyzing wide range of biomolecular interactions in a label-free environment. In this system the ligand is immobilized on the surface of the sensor chip and passed the cognate analyte (that binds with immobilized ligand) over the immobilized ligand. The binding of the passing analyte to the immobilized ligand generates a response corresponding to bound mass (down to picograms level) that depends on the bulk sample solution concentration down to nano to picomolar levels. Moreover, the interactions are monitored in real time fashion and thus possible to determine binding characteristics including rate constants.
Biotin CAP-chip (GE Healthcare) kit
The sensor chip is the main component in the biacore system which provides suitable condition for generating SPR signal. The glass surface of the sensor chip is coated with thin gold layer (~50 nM thickness) which facilitates the conditions for SPR signal. This signal is the basis for analyzing biomolecular interactions and thus, all biacore sensor chips bear this feature. To provide better environment on the surface of sensor chip, the chips are coated further with different polymers (alkanethiol or carbohydrate polymer). In the case of Biotin CAP-chip, the sensor surface was coated with carbohydrate polymer.
Reagent setup
The procedure for the SPR-based surveillance of influenza virus or HA derived from the A/H5N1/Vietnam/1203/2004 virus is summarized in Figure 1 and consists of four steps. The purified HA proteins were stored at 4°C and the diluted HA samples were used within 24 h of preparation. All the multivalent glycans stocks (33.3 μM) were stored at −80°C in 0.3 M sodium phosphate buffer pH 7.4 as small aliquots. For each analysis, the tetravalent glycans and control compounds were diluted freshly in 0.3 M sodium phosphate buffer, pH 7.4, to prepare 500 nM stocks. The diluted glycans stocks can be stored at 4°C up to three days. All buffers (10× stock solutions) were stores at room temperature and diluted freshly to 1× buffers followed by filtration (through 0.45 μM Millipore filters). The Biotin CAP-chip and supplied reagents with the kit are stored at 4°C and diluted freshly before use. Regeneration solution was prepared by mixing 3 parts of regeneration stock solution 1 (8 M guanidine-HCl) with 1 part regeneration solution stock 2 (1 M NaOH) provided by the supplier. The resulting stock should be used within 2 d when stored at 4°C. Upon completion of the analyses, the Biotin CAP-chip should be stored at 4°C.
Step 1
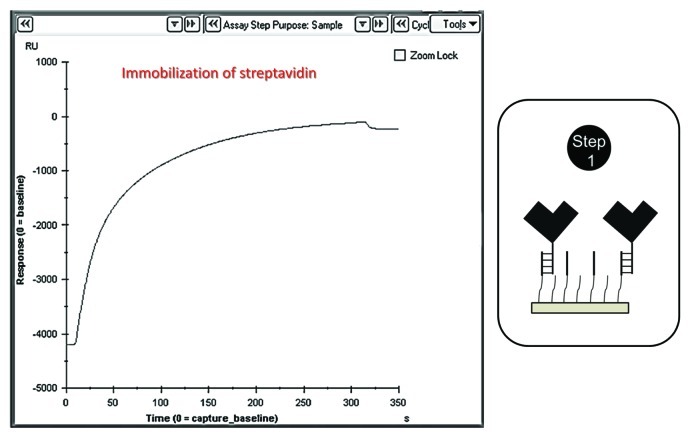
Immobilization of SA on the CAP chip using biotin CAPture reagent (provided by the supplier, do not dilute the reagent, GE Healthcare) at a flow rate of 2 μl/min for 5 min for all the flow cells (1–4) to facilitate the binding of biotinylated glycans (Fig. 2).
Figure 2. Injecting the biotin CAPture reagent into the flow cells (immobilization of streptavidin).
Step 2
Immobilization of synthetic α2-3 or α2-6 glycans on the surface through SA-biotin interactions.
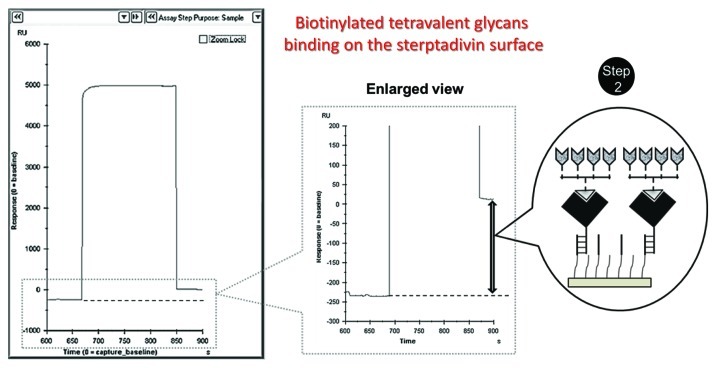
(1) Initially, the control 01-000 compound (500 nM stock) was injected into flow cell 1 and allowed to bind to the SA surface for approximately 3 min at a flow rate of 10 μl/min. This compound lacks the glycan but has all of the backbone structure (biotin and PAA chain). This compound is useful as a control to eliminate non-specific interactions of HA with the backbone chain in the absence of glycan (Fig. 3).
Figure 3. Immobilization of biotinylated tetravalent glycan on the streptavidin surface.
(2) Next, α2-3 glycan (01-077, 500 nM stock) was injected into flow cell 2 and allowed to bind to the SA surface at the same flow rate used above. This glycan is used for the analysis of influenza viruses that frequently infect birds.
(3) Another type of α2-3 glycan (01-088, muscine type 500 nM stock) was injected into flow cell 3 and allowed to bind to the SA surface at the same flow rate. This glycan is also used for the analysis of influenza viruses that frequently infect birds.
(4) Another type of α2-6 glycan (01-039a, 500 nM stock) was injected into flow cell 4 and allowed to bind to the SA surface at the same flow rate. This glycan is used for the analysis of influenza viruses that frequently infect humans and other mammals.
Step 3
Injection of either the virus or HA into the flow cells (1–4) as analytes (step 4) at a flow rate of 30 μl/min after the tetravalent glycan surface is generated on the sensor surface. For each sample, cycles of analyte injection for 120 sec and dissociation for 300 sec and analyte-free buffer injection were performed for 10 min. In general, this procedure is sufficient to obtain the response unit (RU). If analyses of the same virus or HA at increased concentrations are planned, continuous injection of samples is possible without surface regeneration (Fig. 4).
Figure 4. Biotinylated tetravalent glycan interaction with increasing concentrations of HA (single-cycle-kinetics mode).
Step 4
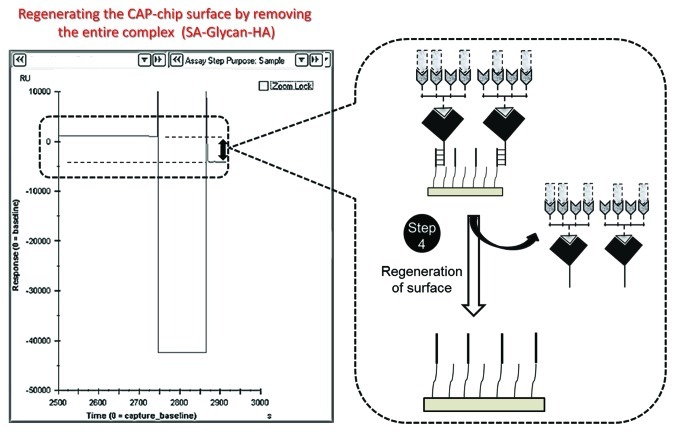
Regeneration of the surface for the next sample of viruses or HA (Fig. 5). Using this procedure, approximately two samples can be analyzed per hour, and the entire process can be programmed without interrupting the analyses. A caution for some errors during this analysis also included for ready reference (Table 1).
Figure 5. Regeneration of the sensor surface by stripping the entire complex.
Table 1. Potential problems and solution during the analyses of HA-glycan interaction using SPR-based analyses.
| Step | Observation | Probable reason | Solution |
|---|---|---|---|
| Step 1 |
No increase of response |
Biotin capture reagent is inactivated or CAP chip is damaged |
Prepare new capture reagent and change the CAP chip |
| Step 2 |
No increase of response |
Glycan binding failed |
Re-inject the freshly prepared glycans |
| Step 3 |
No increase of response |
HA failed form the trimeric structure |
Use higher HA conc. (up to 1 μM) |
| Step 4 | The response should decrease | Regeneration is not complete | Repeat the regeneration step |
Sensitivity of the SPR-based analyses
To evaluate the sensitivity of the SPR methodology suggested above, we performed analyses using different concentrations of purified HA derived from the H3N2/Panama virus (Fig. 6). Initially, the HA units in the purified HA were evaluated with an agglutination assay, and samples with different HA units/ml were prepared. The diluted samples were injected into the flow cell containing the glycan (01-039a), and continuous injection was performed with increasing concentrations of HA units (8–128 HA units/ml). The response unit was increased with increased concentrations of HA units, and a clear response was observed with 16 HA units/ml. However, the agglutination assay showed that 320 HA units/ml was required to obtain clear agglutination (data not shown).
Figure 6. SPR-based analyses of HA-glycan (01-039a) interactions on the CAP chip (HA derived from H3N2/Panama) using single-cycle kinetics.
Recent studies25-28 of H5N1 influenza viruses suggest that HA can evolve enhanced α2-6 glycan recognition by readjusting the glycan-binding region from α2-3 glycans. The HA variants of influenza viruses that attained enhanced binding to the α2-6 glycan may have significant effects on viral attachment to the human respiratory epithelium. This step seems to be important for adapting to humans, in addition to other supporting mutations in other influenza proteins. Thus, these studies suggest that it is important to evaluate the glycan-binding properties of HA derived from H5N1, H7N7 and H9N2 viruses. We believe that the protocol described above will aid in the evaluation of HA evolution among influenza viruses. Taken together, SPR-based analyses using either multivalent glycans or high-affinity ligands, such as anti-HA aptamers,29 may provide an efficient and rapid method for influenza surveillance, especially in a pandemic scenario.
The SPR system used in the above studies (BiacoreT100) is programmable and can analyze a number of samples automatically and continuously, and the CAP chip surface can be regenerated at least 90 times. More importantly, the SPR-based analysis is a closed system (samples are not exposed to the environment) and requires minimal time and small sample quantities for analysis, which is important when analyzing pandemic influenza viruses. The above procedure may be readily adjusted for portable SPR devices or commercially available systems to expand the application of SPR-based point-of-care influenza surveillance.
Glossary
Abbreviations:
- SPR
surface plasmon resonance
- HA
hemagglutinin
- NA
neuraminidase
- PCR
polymerase chain reaction
- KD
equilibrium dissociation constant
- SA
streptavidin
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/21822
References
- 1.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 3.Kawaoka Y, Bean WJ, Webster RG. Evolution of the hemagglutinin of equine H3 influenza viruses. Virology. 1989;169:283–92. doi: 10.1016/0042-6822(89)90153-0. [DOI] [PubMed] [Google Scholar]
- 4.de Jong JC, Claas EC, Osterhaus AD, Webster RG, Lim WL. A pandemic warning? Nature. 1997;389:554. doi: 10.1038/39218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taubenberger JK, Morens DM, Fauci AS. The next influenza pandemic: can it be predicted? JAMA. 2007;297:2025–7. doi: 10.1001/jama.297.18.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012;2:160–7. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 8.Russell RJ, Stevens DJ, Haire LF, Gamblin SJ, Skehel JJ. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj J. 2006;23:85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- 9.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–42. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 10.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr., Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–60. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen MB, Sabesan S, Skehel JJ, Wiley DC. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology. 1997;232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 12.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci U S A. 2001;98:11181–6. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology. 2003;309:209–18. doi: 10.1016/S0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 2010;6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, et al. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci U S A. 2009;106:17175–80. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–10. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 17.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–6. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 18.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–12. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schofield DJ, Dimmock NJ. Determination of affinities of a panel of IgGs and Fabs for whole enveloped (influenza A) virions using surface plasmon resonance. J Virol Methods. 1996;62:33–42. doi: 10.1016/0166-0934(96)02086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hidari KIPJ, Shimada S, Suzuki Y, Suzuki T. Binding kinetics of influenza viruses to sialic acid-containing carbohydrates. Glycoconj J. 2007;24:583–90. doi: 10.1007/s10719-007-9055-y. [DOI] [PubMed] [Google Scholar]
- 21.Suenaga E, Mizuno H, Penmetcha KK. Monitoring influenza hemagglutinin and glycan interactions using surface plasmon resonance. Biosens Bioelectron. 2012;32:195–201. doi: 10.1016/j.bios.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Kodoyianni V. Label-free analysis of biomolecular interactions using SPR imaging. Biotechniques. 2011;50:32–40. doi: 10.2144/000113569. [DOI] [PubMed] [Google Scholar]
- 23.Cooper MA. Label-free screening of bio-molecular interactions. Anal Bioanal Chem. 2003;377:834–42. doi: 10.1007/s00216-003-2111-y. [DOI] [PubMed] [Google Scholar]
- 24.Misono TS, Kumar PKR. Selection of RNA aptamers against human influenza virus hemagglutinin using surface plasmon resonance. Anal Biochem. 2005;342:312–7. doi: 10.1016/j.ab.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Chen LM, Blixt O, Stevens J, Lipatov AS, Davis CT, Collins BE, et al. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology. 2012;422:105–13. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–8. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maines TR, Chen LM, Van Hoeven N, Tumpey TM, Blixt O, Belser JA, et al. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011;413:139–47. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, de Wit E, Munster VJ, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–41. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gopinath SCB, Misono TS, Kawasaki K, Mizuno T, Imai M, Odagiri T, et al. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J Gen Virol. 2006;87:479–87. doi: 10.1099/vir.0.81508-0. [DOI] [PubMed] [Google Scholar]