Abstract
We have developed a novel, silicon-based peptide array for broad biological applications, including potential for development as a real-time point-of-care platform. We employed photolithography on silicon wafers to synthesize microarrays (Intel arrays), containing every possible overlapping peptide within a linear protein sequence covering the N-terminal tail of human histone H2B. Arrays also included peptides with acetylated and methylated lysine residues reflecting post-translational modifications of H2B. We defined minimum binding epitopes for commercial antibodies recognizing modified and unmodified H2B peptides. We further demonstrated that this platform is suitable for highly sensitive methyltransferase and kinase substrate characterization. Intel arrays also revealed specific H2B epitopes recognized by autoantibodies in individuals with systemic lupus erythematosus (SLE) that have increased disease severity. By combining emerging nonfluorescence-based detection methods with an underlying integrated circuit, we are now poised to create a truly transformative proteomics platform with applications in bioscience, drug development, and clinical diagnostics.
INTRODUCTION
The development of highly multiplex, inexpensive proteomics tools is critical to deliver on the promise of individualized medicine and point-of-care diagnosis1. We have described a diverse group of array technologies to profile autoantibodies in autoimmune human and animal model samples2,3, as well as antibodies generated by vaccination4, to detect intracellular signaling states5, and to perform multiplex characterization of soluble and cell-surface analytes6. Other groups have described array-based approaches that have led to discoveries in cancer7, neurodegenerative disease8, aging9 and allergy10. While these and other innovative array techniques have developed at a rapid pace, technological innovation has not produced adequate tools for proteome-level assessment of biological and clinical samples on the small and rapid scale that point-of-care medicine demands11.
Many studies have used an “in silico”, or bioinformatics, approach to characterize pathways or disease states. We hypothesized that creating peptide arrays on a silicon surface (“on silico” arrays) could be transformative in the field because an integrated semiconductor circuit could be created beneath each peptide feature, allowing for real-time measurements and computations, neither of which are possible using current fluorescence-based approaches. Here we describe a novel, high-density, scalable platform using microprocessor-grade silicon wafers as a support surface (Supplementary Fig. 1 and Supplementary Methods). We used a maskless photolithographic approach to create arrays of overlapping peptides comprising every possible combination of contiguous amino acids within a naturally linear human polypeptide sequence. The synthesis process also permits the incorporation of functional group-modified amino acids, e.g. acetylated and methylated lysine, into a growing peptide sequence, allowing for subsequent analysis of protein interactions with modified and unmodified peptides in parallel on the same microarray. In previous published reports, photolithography has been used in a variety of peptide conjugation strategies, involving both mask-based12 and maskless13,14 techniques to generate location-specific peptides on microarray platforms using photo-labile or acid-labile Di-tert-butyl dicarbonate (t-BOC) protective groups; however, these approaches, with glass slides as the synthesis substrate, are difficult and expensive to scale up. In addition, these approaches are limited in the number of unique and overlapping sequences generated, raw number of discrete peptide addresses, and the ability to compare modified and unmodified peptide epitopes in parallel. Silicon provides a surface highly amenable to fluid-phase detection of peptide-reactive antibodies and other protein-protein interactions. Surprisingly and unexpectedly, there is no intrinsic background fluorescence of the surface when imaged in conventional microarray laser scanning systems, and there is no requirement for a pre-blocking step due to the near absence of non-specific biological molecule binding to the surface (data not shown). We have successfully leveraged mature semiconductor fabrication technology to produce 68 individual arrays on six-inch silicon wafers. The platform combines advances in two exciting fields, semiconductor manufacturing and multiplexed biochemical detection, potentially enabling analysis of multiplex protein-protein interactions in real time. Herein we describe the utility of silicon-base peptide arrays, which we have termed “Intel arrays”, for high-resolution epitope mapping of diverse commercial monoclonal and polyclonal antibody probes, characterization of specific protein lysine methyltransferase and kinase activity at single-amino acid precision, and identification of patterns of autoantibody reactivity that correlate significantly with disease severity in a cohort of individuals with systemic lupus erythematosus (SLE).
RESULTS
H2B peptide array design and synthesis
To demonstrate the power of the Intel array platform, we fabricated overlapping peptide arrays of a naturally linear region at the N-terminus of human histone H2B. H2B is a highly conserved protein that interacts with DNA and other histone proteins making up the intricate chromatin structures that organize the eukaryotic genome15. H2B is a major target for epigenetic regulation mediated by posttranslational modifications by kinases, peptidyl arginine deiminases, and methyl- and acetyl-transferases, among others, and manipulation of H2B via lysine acetylation and methylation within the linear N-terminal tail of the protein is critical for regulation of chromatin homeostasis16. Histone epitopes, including the N-terminus of H2B, are also clinically important autoantigens in SLE, drug-induced lupus, rheumatoid arthritis, and other diseases2,17,18.
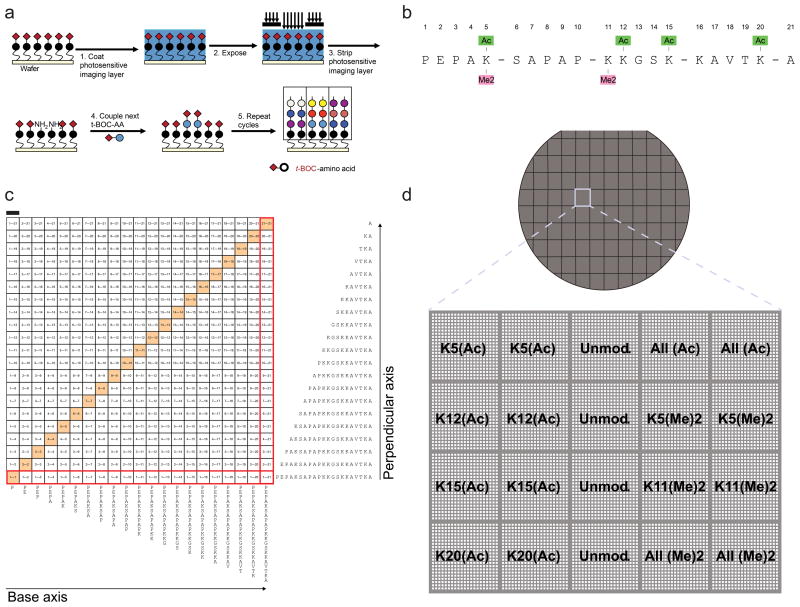
Using a photolithographic process (Fig. 1a), we constructed arrays of peptides corresponding to the N-terminal tail of H2B, ranging in length from 1–21 amino acids (sequence: NH3-PEPAKSAPAPKKGSKKAVTKA-COOH). Individual arrays were comprised of unmodified peptides, peptides with acetylated lysine at position 5, 12, 15 and 20, and peptides with dimethylated lysine at position 5 and 11 (Fig. 1b). Each sub-array is a square grid containing every possible peptide fragment within the 21-mer H2B sequence arranged in sequential order (Fig. 1c). The H2B array contains 20 square grids that individually contain all possible contiguous H2B peptide lengths either unmodified or with acetyl and methyl groups incorporated at specific lysine residues within the sequence, totaling 8,820 spatially addressable features (Fig. 1d). Intel array peptide yield, purity and sequence fidelity were routinely assessed using on-chip staircase fluorescence assays, off-chip fluorescence high-performance liquid chromatography, mass spectrometry, and highly sensitive kinase substrate assays, and were found to be equivalent or better than conventional peptide synthesis approaches (Supplementary Figs. 2–4 and data not shown)19,20.
Figure 1. Intel array design and construction.
(a) Schematic depicting photolithographic process to synthesize peptide microarrays on silicon wafers; see materials and methods for full details. (b) 21-mer H2B peptide sequence synthesized on the array, with acetylation and methylation sites annotated. (c) Layout of features from a representative block on the array. Peptides spanning the 21-mer histone H2B sequence increase by one amino acid to the C-terminus along the base axis from left to right, and decrease by one amino acid from the N-terminus along the perpendicular axis from bottom to top, generating an array of every possible contiguous H2B peptide fragment tiled at single-amino acid increments within the 21-mer sequence. The grid is symmetric along the diagonal axis (highlighted). Peptide sequences for outermost array features (boxed red) are shown along base and perpendicular axes. Scale bar (upper left) = 50 μm. (d) Above, schematic depicting diced six-inch silicon wafer containing 68 individual Intel arrays. Below, each array contains 20 square grids organized as depicted in (c) with either unmodified 21-mer H2B sequence or a sequence containing acetylated or dimethylated lysine at indicated positions.
Epitope mapping of commercial antibodies using Intel arrays
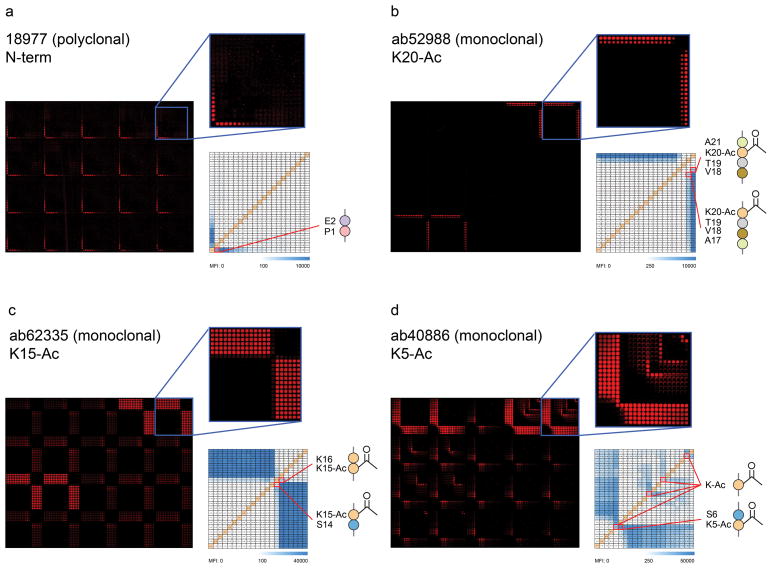
To determine the ability of Intel arrays to resolve epitopes recognized by H2B-reactive antibodies, we probed arrays with commercial antibodies with demonstrated H2B-binding capacity. Arrays probed with polyclonal antibody (pAb) 18977 (Abcam), directed against the N-terminus of H2B, displayed a minimum binding epitope of two N-terminal amino acids (Fig 2a). Interestingly, at higher concentration, pAb 18977 also displayed moderate binding to peptides containing acetylated Lys5 and Lys12, with minimum binding epitopes of three amino acids surrounding Lys5-Ac (Ala4–Ser6) and a 5-mer peptide containing Lys12-Ac (Lys11–Lys15) (data not shown). Monoclonal antibody (mAb) 52988 (Abcam) recognizes the H2B epitope surrounding Lys20-Ac. Intel arrays displayed reactivity only at peptide features containing C-terminal Lys20-Ac and indicate that a minimum peptide length of four amino acids containing Lys20-Ac is required for binding of mAb 52988 (Fig 2b). mAb 62335 (Abcam) is directed against an H2B epitope surrounding Lys15-Ac. Arrays probed with this antibody revealed reactivity at minimal binding epitopes composed of only three amino acids, Gly13–Lys15-Ac or S14–K16, surrounding Lys15-Ac; however, mAb 62335 also displayed cross reactivity with unmodified and dimethylated H2B peptides (Fig. 2c). Intel arrays probed with mAb 40886 (Abcam), which recognizes H2B Lys5-Ac, displayed reactivity to Lys5-Ac, but binding was also observed at acetylated lysine at positions 12, 15 and 20 (Fig. 2d). Titrations of antibodies 52988 and 62335 displayed a concentration-dependent decrease in signal on the array, and in comparison with indirect ELISA, the array platform demonstrated only slightly decreased sensitivity for antibody 52988, and when using dilutions of antibody 62335 ≥ 2500X (Supplementary Fig. 5). The specific epitopes for each antibody described were verified in pre-clearing experiments using unmodified and acetylated H2B peptides conjugated to sepharose beads (Supplementary Fig. 6a–e).
Figure 2. Epitope mapping of polyclonal and monoclonal antibodies using H2B Intel arrays.
(a) Single-amino acid resolution map of the epitope recognized by H2B-reactive polyclonal antibody (pAb) 18977 (Abcam, Cambridge, MA) showing that a minimum of two N-terminal amino acids (Pro1–Glu2, inset) are required for antibody binding. (b) Epitope map for monoclonal antibody (mAb) 52988 (Abcam) directed against residues surrounding Lys20-Ac, showing a minimum binding requirement of either Ala17–Lys20-Ac or Val18–Ala21 (containing Lys20-Ac). (c) Epitope map and minimum peptide binding requirements for mAb 62335 (Abcam) directed against residues surrounding Lys15-Ac of H2B. The minimum binding requirements were determined to be Gly13–Lys15-Ac and Lys15-Ac–Lys16. (d) Epitope map for mAb 40886 (Abcam) directed against residues surrounding Lys5-Ac, showing binding of acetylated lysine (Lys-Ac), both at position 5 in the context of at least a 3-mer peptide Ala4-Ser6, but also Lys-Ac alone or in peptides at positions 12, 15 and 20. For each antibody, the “All (Ac)” subarray containing acetylated lysine at position 5, 12, 15 and 20 is enlarged. Grid schematic for each antibody corresponds to reactivity within the enlarged “All (Ac)” subarray. All primary antibodies were diluted 1:1000 from original stocks as received from vendors and reactive features were visualized with goat anti-rabbit IgG conjugated to Cy5. Color bars indicate observed median fluorescence intensity (MFI) at each peptide feature.
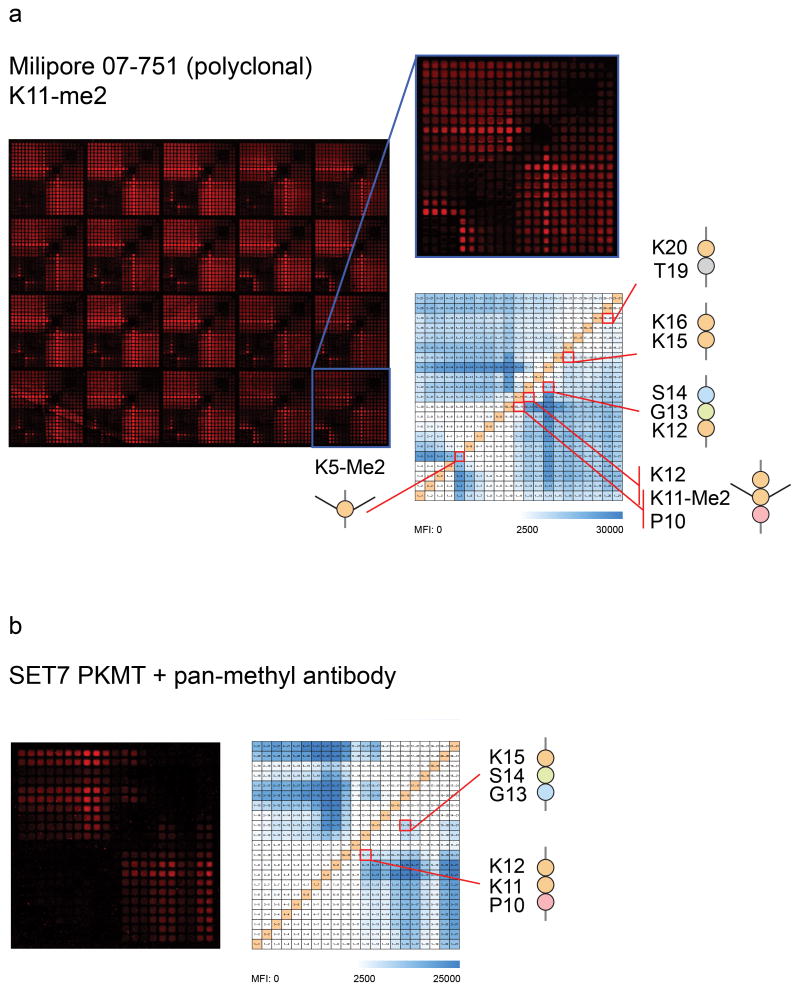
To verify the site-specific methylation of H2B peptides synthesized on Intel arrays we used a rabbit polyclonal antibody (07–751, Millipore), which is targeted to dimethylated Lys11 (Lys11-Me2). We characterized reactivity not only to a minimum peptide epitope containing Lys11-Me2 (Pro10–Lys12), but also to peptides containing Lys5-Me2 and unmodified peptides (Fig. 3a). To assess the nature of the polyclonal reactivity observed for this antibody, we performed a pre-clearing experiment using an H2B peptide dimethylated at Lys5. We observed loss of the reactivity seen against peptides containing Lys5-Me2, but not to peptides containing Lys11-Me2 or unmodified lysine (Supplementary Fig. 6f). This result indicates that within the polyclonal mixture of antibodies constituting pAb 07–751 there are at least two clones recognizing distinct epitopes of H2B, and further highlights the power of Intel arrays for deeply characterizing reactivity patterns of polyclonal antibody mixtures.
Figure 3. Detection of dimethlyated Intel array peptides and direct monomethylation of lysine-containing Intel array peptides by SETD7 protein lysine methyltransferase.
(a) Epitope map for pAb directed against Lys11-Me2 (07–751, Millipore, Billerica, MA) displaying minimum binding requirements of Lys11-Me2 within a three-mer Pro10–Lys12 as well as Lys5-Me2, Lys12–Ser14m Lys15–Lys16 and Thr19–Lys20. Subarray containing dimethylated lysine at position 5 and 11, “All (Me)2”, is enlarged. Grid schematic displays reactivity within the enlarged “All (Me)2” subarray. (b) Scanned image of an Intel array after PKMT reaction with SETD7 and subsequent detection with a pan-methyl antibody (ab23366, Abcam). Image and grid schematic display reactivity within an unmodified “Unmod.” subarray. Minimum peptide content required for methylation on Intel arrays by SETD7 is Pro10–Lys12 or Ser14–Lys15. Color bars indicate observed median fluorescence intensity (MFI) at each peptide feature.
Enzymatic modification of Intel array peptides
Protein lysine methyl transferases (PKMTs) are known to mediate the addition of methyl groups to lysine residues in the N-terminal tail of H2B16. To test whether site-specific methylation can be detected on Intel arrays, we incubated arrays with SETD7, a PKMT known to methlyate H2B21. SETD7 added a single methyl group to several lysine-containing peptides on Intel Arrays, detected by probing with a pan-methyl antibody following the PKMT reaction (Fig. 3b). Incubation with glutathione S-transferase or pan-methyl antibody alone did not result in the detection of peptide methylation (data not shown). The minimum epitopes required for SETD7 methylation of Intel array peptides were Pro10–Lys12 and Ser14–Lys15.
Phosphorylation of H2B is involved with chromatin regulation and is associated with the induction of apoptosis, specifically the phosphorylation of serine at position 14 (Ser14)22. To determine whether specific kinase assays could be performed on Intel arrays, we synthesized two different microarrays: one with a series of blocks containing sequentially growing peptides spanning a known phosphorylation target peptide (kemptide: LRRASL) of protein kinase A (PKA) (Supplementary Fig. 7a) and one with tiled peptides surrounding H2B Ser14 (Supplementary Fig. 7e). Intel arrays incubated with PKA displayed site-specific phosphorylation of serine residues in both configurations (Supplementary Fig. 7b–e). The most optimal orientation for H2B Ser14 phosphorylation appeared to be when Ser14 was flanked by two C-terminal lysine residues; peptides with one lysine C-terminal to Ser14 or in which Ser14 is the most C-terminal amino acid displayed less phosphorylation (Supplementary Fig 7e,f). This may indicate that there are conformational requirements for phosphorylation of H2B by PKA.
Autoantibody profiling of SLE sera using Intel arrays
We next assessed the ability of Intel arrays to detect H2B-reactive autoantibodies in human samples. SLE is a heterogeneous disease that is characterized by the development of immunoglobulin G (IgG) autoantibodies directed against DNA and RNA-associated proteins of the cell nucleus23. Interferon alpha (IFN-α) is known to drive autoimmune processes in SLE, and the level of IFN-α-driven pathology can be assessed by determining the extent of IFN-α-dependent gene transcription, the so-called “interferon signature”, with a high interferon signature correlating with increased SLE disease severity24,25. The presence of serum autoantibodies directed against a variety of nuclear antigens, including histone proteins, is coincident with overabundance of serum IFNα in SLE26. Furthermore, several reports have described autoantibody reactivity to post-translationally modified histone epitopes in mouse models of SLE and human SLE18,27,28.
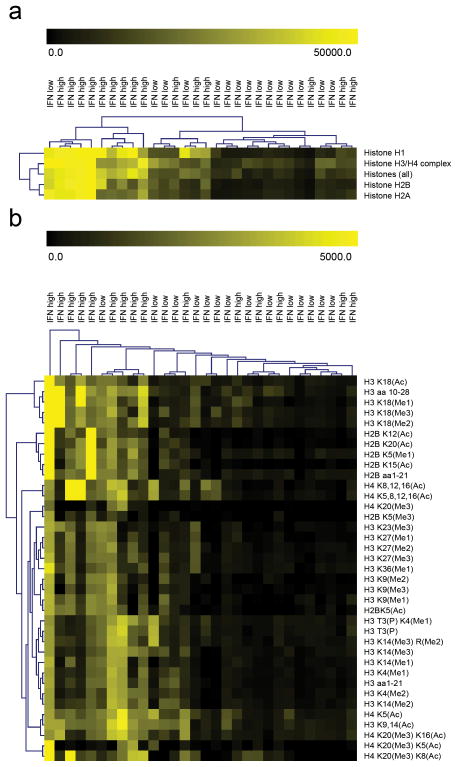
Using 30 sera from a cohort of SLE study participants collected through the Autoimmune Biomarkers Collaborative Network29 and profiled for IFN-α-induced gene expression25, we analyzed IgG reactivity to nucleosomes, individual purified H1, H2A, H2B and H3/H4 proteins, and peptide fragments with several acetylation and methylation modifications, using traditional spotted protein and peptide microarrays2. Using Significance Analysis of Microarrays (SAM)30, we observed that “IFN-high” SLE serum IgG autoantibodies were significantly more reactive with whole-protein histone antigens (q value = 0; Fig. 4a), as well as unmodified full-length H2B peptide and H2B peptides containing acetylated lysine at positions 5, 12, 15 and 20 (q value = 0; Fig. 4b).
Figure 4. Conventional spotted peptide microarray shows increased reactivity to whole histones and post-translationally modified histone peptides in IFN-high sera.
Heatmaps displaying hierarchical clustering of peptide features identified by Significance Analysis of Microarrays (SAM) algorithm as significantly more reactive (q value = 0) in “IFN-high” SLE sera. SAM-identified whole protein histone antigens (a) and peptides (b) statistically more reactive with autoantibodies in IFN-high sera. Unmodified H2B peptides, as well as H2B peptides with acetylated lysines at position 5, 12, 15 and 20 are among the array features identified by SAM to be significantly associated with IFN-high disease state (q value = 0).
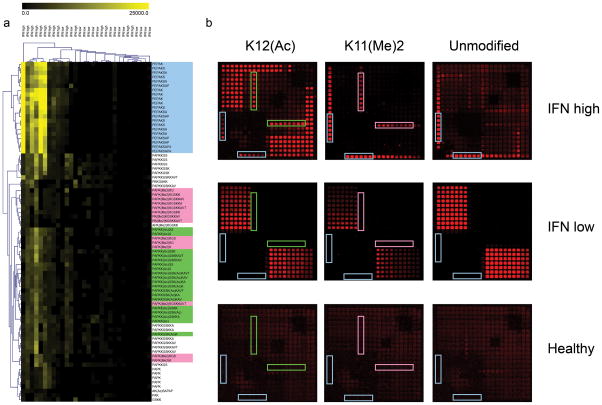
We tested whether Intel arrays could replicate our findings that IFN-high SLE sera is more reactive with unmodified and functional group-modified H2B peptides than IFN-low sera, and whether high-resolution mapping could define immunogenic regions of H2B in IFN-α-driven SLE. The SAM algorithm identified several classes of peptides on Intel arrays that were significantly more reactive with IgG from IFN-high SLE sera (q value = 0; Fig. 5a). These included an unmodified region near the N-terminus of the peptide with a minimum amino acid sequence of Pro1–Lys5; several peptides containing Lys12-Ac and Lys15-Ac; and peptides with Lys11-Me2 (Fig. 5a, b). Reactivity to peptides containing Pro1–Lys5, Lys12-Ac, Lys15-Ac and Lys11Me2 was largely absent in sera from IFN-low SLE and healthy controls (Fig. 5b). ELISA validation of Intel array reactivity was performed using full-length and truncated H2B peptides and confirmed that IFN-high sera were significantly more reactive with N-terminal H2B and H2B Lys12-Ac and Lys15-Ac (P <0.05; Supplementary Fig. 8). These data suggest that N-terminal reactivity to a minimum sequence Pro1-Lys5, as well as reactivity to epitopes surrounding the indicated acetylated and dimethylated lysine residues may be important for breaking tolerance to H2B and furthering autoantibody pathogenesis in IFN-α-driven SLE.
Figure 5. Differential reactivity to Histone H2B peptides in a cohort of individuals with systemic lupus erythematosus (SLE).
(a) Hierarchical clustering of peptide features identified by SAM as significantly more reactive (q-value = 0) in “IFN-high” SLE sera. Peptides containing the minimum epitope Pro1–Lys5 are highlighted in blue; peptides containing acetylated lysine at position 12 and 15 are highlighted in green; peptides with dimethlyated lysine at position 11 are highlighted in pink. Highlighted peptide sequences correspond with blue, green and pink-boxed array features in (b). (b) Representative images of individual grids from Intel arrays probed with serum from IFN-high and IFN-low SLE and healthy controls. Left, grids containing H2B peptides with acetylated lysine incorporated position 12. Center, grids containing H2B peptides with dimethylated lysine incorporated at position 11. Right, grids containing unmodified H2B peptides. Boxed peptide features correspond to corresponding sequences highlighted in (a).
DISCUSSION
Intel arrays constitute a considerable advance in proteomic analysis as the first highly multiplex, single-amino acid resolution array platform with utility in a variety of proteomic studies, such as epitope mapping and characterization of diverse protein-protein interactions. We have demonstrated the utility of Intel arrays in epitope mapping of commercial antibodies directed against H2B. The ability of Intel arrays to resolve commercial antibody reactivity to peptides as small as two amino acids speaks to the high degree of sensitivity and specificity of the platform, and demonstrates its potential utility in investigating basic immunological questions such as the precise epitopes recognized by antibodies. A high-resolution epitope mapping platform has application far beyond the characterization and development of chromatin-binding antibodies. Within the last decade, there has been an explosion of novel monoclonal antibody and other protein engineering technologies geared toward the development of disease therapeutics31. As the field of antibody and protein engineering evolves, it will be necessary to develop rapid, multiplex, and cost-effective testing platforms to determine precise target epitopes, to obtain information about interaction kinetics, and to determine whether there is cross-reactivity that may impede or enhance the activity of engineered molecules.
We demonstrated the ability to use Intel arrays to detect the sequence-specific activity of SETD7 protein lysine methyltransferase and PKA. These proof of concept experiments demonstrate the power of Intel arrays for discovery of novel targets of these and other protein-modification enzymes. We have recently shown that a combination of methylation and phosphorylation regulates lymphocyte inflammatory responses through modifications to the NF-κB protein RelA by the PKMT SETD6 and PKC-ζ32. Peptide arrays have been instrumental in the discovery of these and other PKMT methylation targets21, suggesting that Intel arrays will prove useful in this field. Kinases and PKMTs are the targets of specific inhibitors, many of which have entered clinical trials for the treatment of cancer and other diseases33. Given the ability of Intel arrays to resolve specific activity of kinase and PKMT enzymes in parallel, we believe this platform is well suited for inhibitor screening.
We also demonstrated the utility of Intel arrays in defining epitopes within unmodified and post-translationally modified H2B associated with IFN-α-driven SLE. The identification of specific epitopes within this important antigen may help better define IFN-α-driven autoantibody pathogenesis in SLE. As Inhibitors of IFN-α and its receptor IFNAR have entered phase II clinical trials in SLE, these arrays may be useful in determining meaningful clinical outcomes. Our results corroborate reports showing that autoantibodies targeting post-translational modifications on histone proteins, notably acetylated and methylated epitopes, are associated with SLE pathogenesis18,27,28. The current results underscore that high-resolution mapping on the Intel array platform has great potential for identifying immunogenic regions relevant for the pathogenesis of SLE in a rapid and reliable way. While peptide-based approaches have not been developed sufficiently to replace gold-standard antibody detection tests in most clinics, several peptide platforms have been employed in the routine diagnosis of autoimmune diseases including celiac disease34 and rheumatoid arthritis35,36, and have been used to guide a phase II vaccine trial in multiple sclerosis37. Accordingly, the Intel array platform holds great promise in expanding the utility of peptide-based diagnostic technology for the detection of meaningful antibody reactivity in the clinical setting.
One potential limitation of peptide arrays is their reliance on linear peptide sequences, which precludes an assessment of conformational protein epitopes. This limitation in characterizing protein-protein interactions will diminish as technological advances improve the diversity of protein conformations “in miniature”. Combination of Intel array technology with emerging advances in the synthesis of peptide mimetics could directly address this limitation38.
Magnetic nanosensors are being developed to couple high-resolution Intel peptide arrays with underlying, in silico technology facilitating real-time detection of antibody-peptide interaction kinetics. Furthermore, we anticipate the rapid integration of the Intel peptide array platform into existing magnetic nanosensor point-of-care technology39. The inevitable combination of these technologies will yield a revolutionary tool for personalized, point-of-care proteomics, and allow for reactions on sub-micron array features to be detected in real time.
MATERIALS AND METHODS
Detailed methodology is available in the Supplementary Methods.
Wafer derivatization and linker attachment
We derivatized blank six-inch silicon wafers (Silicon Valley Microelectronics, Santa Clara, CA) by thermal oxidation followed by salination via immersion in a 0.5% v/v mixture of 3-aminopropyltriethoxysilane (Sigma-Aldrich, St. Louis, MO) in 95% ethyl alcohol (EtOH, Sigma) for 30 min at RT. We then cured wafers at 100 °C for 1 h under ultrapure and ultradry N2 (Airgas, Sacramento, CA) to crosslink silane groups to each other and to the silicon surface. Following salination, we added a PEG6 linker to the wafer surface to facilitate addition of custom peptides.
Array patterning by photolithography and amino acid coupling
For wafer-scale array patterning, we used a spin coater (Brewer Science, Rolla, MO) to apply a photosensitive imaging solution containing a photoacid generator and a sensitizer in propylene glycol methyl ether acetate (PGMEA, Sigma). In a class-10,000 clean room, we mounted wafers on a computer-controlled X–Y stage (Newport Corporation, Irvine, CA) and patterned array features by exposing selected areas of the wafer to UV light though a virtual mask with a digital exposure tool modified from a DLP device (Texas Instruments, Dallas, TX). At each coupling step, we treated UV-exposed wafers with coupling solution: [t-BOC amino acid of interest (Novabiochem, San Diego, CA), hydroxybenzotriazole (HOBt, CPC Scientific, Sunnyvale, CA), and N,N′-diisopropylcarbodiimide (DIC, CPC Scientific) dissolved in N-Methyl-2-pyrrolidone (NMP, Sigma) with a final concentration of 0.1 M each] for 30 min at room temperature (RT). After a brief rinse in N,N-dimethylformamide (DMF, Sigma), we transferred wafers into capping solution (5% Ac2O in DMF). We then washed the wafers extensively in DMF and 2-propanol (IPA, Sigma) in preparation for the subsequent photolithography step.
Antibody-binding assays
We equilibrated de-protected arrays by washing three times in phosphate-buffered saline (PBS, Bio-Rad, Hercules, CA) + 0.1% Tween-20 (Sigma) (PBST), once in PBS, and then once in diH20. We diluted commercial antibodies or patient serum in peptide binding buffer (PBB): [50 mM Tris (Mallinckrodt Baker, Phillipsburg, NJ) pH 7.5, 150 mM NaCl (Mallinckrodt AR, Phillipsburg NJ), 0.05% NP-40 (Sigma) + 2.5% fetal calf serum (FCS, Omega Scientific, Tarzana, CA)] and applied samples to arrays in 12-well tissue culture plates (Fisher Scientific, Pittsburg, PA) overnight at 4 °C on a rocking platform. After three washes in PBST, we applied secondary goat anti-mouse IgG, goat anti-rabbit IgG, or goat anti-human IgG Cy5 conjugated antibodies (Jackson Immunoresearch, West Grove, PA) diluted to 0.375 μg ml−1 in PBST + 20% FCS for 45 min at RT on a rocking platform. We then washed the arrays 3 × 5 min in PBST and briefly rinsed in PBS and diH20 before drying in microscope slide racks centrifuged at 300 ×g for 5 min at RT. We immediately scanned processed arrays using an Axon digital scanning system and performed analysis using Genepix Pro 6.1 software (Molecular Devices, Sunnyvale, CA).
Methylation assay
We incubated arrays in a 12-well plate overnight on a rocking platform at 30 °C in a reaction mixture containing 60 μg of SETD7 or GST purified proteins, and 0.1 mM S-adenosyl-methionine (AdoMet, Sigma) in a methylation buffer containing 50 mM Tris-HCl (pH 8.0), 10% glycerol, 20 mM KCl, 5 mM MgCl2, and 1 mM phenylmethanesulfonylfluoride (Roche Pharmaceuticals, San Francisco, CA) in a 500 μl total reaction volume. Following the methylation reaction, we washed arrays three times with PBST followed by three washes in PBST + 20% FCS. We then visualized array peptide monomethylation using a rabbit polyclonal pan-methyl antibody (ab23366, Abcam, Cambridge, MA).
Pre-clearing and peptide competition assay
For pre-clearing experiments, we incubated streptavidin-sepharose beads (GE Healthcare, Piscataway, NJ) with 0.2 mg ml−1 biotinylated histone peptide H2B 1–21 unmodified; H2B 1–21 acetylated Lys5,12,15,20 or H2B 1–21 Lys5-Me2 (W.M. Keck Foundation, Yale University) for 30 min at RT, washed twice in PBS + 10% FCS, and then incubated peptide-bound beads with commercial antibodies diluted 1:1000 in PBB + 2.5% FCS for 20 min at RT. We then centrifuged bead/Ab mixture at 850 xg for 1 min to pellet bead complexes. We repeated this procedure five times prior to incubation of the pre-cleared sample on arrays.
Sources of human samples and storage
We used serum samples from SLE patients collected as part of the Autoimmune Biomarkers Collaborative Network (ABCoN)29 maintained at −80 °C. Clinical information and research protocols for study enrollment, sample collection and processing are available as previously reported25.
Supplementary Material
Acknowledgments
We thank H. Lee, J. Zhu, J. Yu and the other members of the peptide array team at Biomedical Life Science at Digital Health, Intel, for development of early peptide array process and G. Credo for help with the M&M section of the manuscript. We thank D. Hall, K. C. Garcia, S. Sidhu, J. Haddon, J. Jarrell and other members of our laboratories for helpful discussion. J.V.P was funded by a US National Science Foundation Graduate Research Fellowship (NSF/GRFP) and is supported by the Stanford Genome Training Program (SGTP; US National Institutes of Health/US National Human Genome Research Institute). O.G. is a recipient of an Ellison Senior Scholar Award. P.J.U. is the recipient of a Donald E. and Delia B. Baxter Foundation Career Development Award and is supported by NHLBI Proteomics Contract- HHSN288201000034C, Proteomics of Inflammatory Immunity and Pulmonary Arterial Hypertension, 5 U19-AI082719, National Institutes of Health, 2 OR-92141, Canadian Institute of Health Research, 5 U19-AI050864, National Institutes of Health, FP Grant #261, Euro Commission, European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement no [261382], a gift from the Ben May Trust and a gift from the Floren Family Trust. C.L. is a recipient of an NIH National Research Service Award Fellowship (AI-080086-02).
Footnotes
AUTHOR CONTRIBUTIONS
J.V.P conducted antibody-binding experiments and ELISA, analyzed the data, made the figures and wrote the manuscript. S.T. performed antibody-binding experiments and contributed to data analysis and figure production. G.X. designed and fabricated Intel arrays. D.L. and O.G. contributed the methylation assay. E.C.B. characterized IFN signature and provided ABCoN patient samples. M.V. supervised the development team at Intel and edited the manuscript. P.J.U. and C.L. supervised the project and edited the manuscript.
References
- 1.Cortese DA. A vision of individualized medicine in the context of global health. Clin Pharmacol Ther. 2007;82:491–493. doi: 10.1038/sj.clpt.6100390. [DOI] [PubMed] [Google Scholar]
- 2.Robinson WH, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 3.Robinson WH, et al. Protein microarrays guide tolerizing DNA vaccine treatment of autoimmune encephalomyelitis. Nat Biotechnol. 2003;21:1033–1039. doi: 10.1038/nbt859. [DOI] [PubMed] [Google Scholar]
- 4.Neuman de Vegvar HE, et al. Microarray profiling of antibody responses against simian-human immunodeficiency virus: postchallenge convergence of reactivities independent of host histocompatibility type and vaccine regimen. J Virol. 2003;77:11125–11138. doi: 10.1128/JVI.77.20.11125-11138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan SM, Ermann J, Su L, Fathman CG, Utz PJ. Protein microarrays for multiplex analysis of signal transduction pathways. Nat Med. 2004;10:1390–1396. doi: 10.1038/nm1139. [DOI] [PubMed] [Google Scholar]
- 6.Kattah MG, Coller J, Cheung RK, Oshidary N, Utz PJ. HIT: a versatile proteomics platform for multianalyte phenotyping of cytokines, intracellular proteins and surface molecules. Nat Med. 2008;14:1284–1289. doi: 10.1038/nm.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haab BB. Antibody arrays in cancer research. Mol Cell Proteomics. 2005;4:377–383. doi: 10.1074/mcp.M500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Ray S, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 9.Britschgi M, et al. Neuroprotective natural antibodies to assemblies of amyloidogenic peptides decrease with normal aging and advancing Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106:12145–12150. doi: 10.1073/pnas.0904866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vigh-Conrad KA, Conrad DF, Preuss D. A Protein Allergen Microarray Detects Specific IgE to Pollen Surface, Cytoplasmic, and Commercial Allergen Extracts. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey RC. Grand challenge commentary: Informative diagnostics for personalized medicine. Nature chemical biology. 2010;6:857–859. doi: 10.1038/nchembio.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fodor SP, et al. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 13.Gao X, Zhou X, Gulari E. Light directed massively parallel on-chip synthesis of peptide arrays with t-Boc chemistry. Proteomics. 2003;3:2135–2141. doi: 10.1002/pmic.200300597. [DOI] [PubMed] [Google Scholar]
- 14.Shin DS, et al. Automated maskless photolithography system for peptide microarray synthesis on a chip. J Comb Chem. 2010;12:463–471. doi: 10.1021/cc100009g. [DOI] [PubMed] [Google Scholar]
- 15.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Monestier M, Decker P, Briand JP, Gabriel JL, Muller S. Molecular and structural properties of three autoimmune IgG monoclonal antibodies to histone H2B. J Biol Chem. 2000;275:13558–13563. doi: 10.1074/jbc.275.18.13558. [DOI] [PubMed] [Google Scholar]
- 18.van Bavel CC, et al. Apoptosis-associated acetylation on histone H2B is an epitope for lupus autoantibodies. Mol Immunol. 2009;47:511–516. doi: 10.1016/j.molimm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Barone AD, et al. Photolithographic synthesis of high-density oligonucleotide probe arrays. Nucleosides Nucleotides Nucleic Acids. 2001;20:525–531. doi: 10.1081/NCN-100002328. [DOI] [PubMed] [Google Scholar]
- 20.Lee MS, Kerns EH. LC/MS applications in drug development. Mass Spectrom Rev. 1999;18:187–279. doi: 10.1002/(SICI)1098-2787(1999)18:3/4<187::AID-MAS2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 21.Dhayalan A, Kudithipudi S, Rathert P, Jeltsch A. Specificity Analysis-Based Identification of New Methylation Targets of the SET7/9 Protein Lysine Methyltransferase. Chemistry & Biology. 2011;18:111–120. doi: 10.1016/j.chembiol.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Cheung WL, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 23.von Muhlen CA, Tan EM. Autoantibodies in the diagnosis of systemic rheumatic diseases. Semin Arthritis Rheum. 1995;24:323–358. doi: 10.1016/s0049-0172(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 24.Baechler EC, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer JW, et al. Elevated serum levels of interferon-regulated chemokines are biomarkers for active human systemic lupus erythematosus. PLoS Med. 2006;3:e491. doi: 10.1371/journal.pmed.0030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obermoser G, Pascual V. The interferon-alpha signature of systemic lupus erythematosus. Lupus. 2010;19:1012–1019. doi: 10.1177/0961203310371161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dieker JW, et al. Apoptosis-induced acetylation of histones is pathogenic in systemic lupus erythematosus. Arthritis & Rheumatism. 2007;56:1921–1933. doi: 10.1002/art.22646. [DOI] [PubMed] [Google Scholar]
- 28.van Bavel CC, et al. Apoptosis-induced histone H3 methylation is targeted by autoantibodies in systemic lupus erythematosus. Annals of the rheumatic diseases. 2011;70:201–207. doi: 10.1136/ard.2010.129320. [DOI] [PubMed] [Google Scholar]
- 29.Petri M. Hopkins Lupus Cohort. 1999 update. Rheum Dis Clin North Am. 2000;26:199–213. v. doi: 10.1016/s0889-857x(05)70135-6. [DOI] [PubMed] [Google Scholar]
- 30.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker M. Upping the ante on antibodies. Nat Biotechnol. 2005;23:1065–1072. doi: 10.1038/nbt0905-1065. [DOI] [PubMed] [Google Scholar]
- 32.Levy D, et al. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat Immunol. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 34.Volta U, et al. Deamidated Gliadin Peptide Antibodies as a Routine Test for Celiac Disease: A Prospective Analysis. Journal of Clinical Gastroenterology. 2010;44:186–190. doi: 10.1097/MCG.0b013e3181c378f6. 110.1097/MCG.1090b1013e3181c1378f1096. [DOI] [PubMed] [Google Scholar]
- 35.Lee DM, Schur PH. Clinical utility of the anti-CCP assay in patients with rheumatic diseases. Annals of the rheumatic diseases. 2003;62:870–874. doi: 10.1136/ard.62.9.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffman IE, et al. Diagnostic performance and predictive value of rheumatoid factor, anti-citrullinated peptide antibodies, and the HLA shared epitope for diagnosis of rheumatoid arthritis. Clin Chem. 2005;51:261–263. doi: 10.1373/clinchem.2004.034728. [DOI] [PubMed] [Google Scholar]
- 37.Garren H, et al. Phase 2 trial of a DNA vaccine encoding myelin basic protein for multiple sclerosis. Ann Neurol. 2008;63:611–620. doi: 10.1002/ana.21370. [DOI] [PubMed] [Google Scholar]
- 38.Liskamp RMJ, Rijkers DTS, Kruijtzer JAW, Kemmink J. Peptides and Proteins as a Continuing Exciting Source of Inspiration for Peptidomimetics. Chembiochem. 2011;12:1626–1653. doi: 10.1002/cbic.201000717. [DOI] [PubMed] [Google Scholar]
- 39.Gaster RS, Hall DA, Wang SX. nanoLAB: An ultraportable, handheld diagnostic laboratory for global health. Lab on a Chip. 2011;11:950–956. doi: 10.1039/c0lc00534g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.