Abstract
Human serum glycan profiling with mass spectrometry (MS) has been employed to study several disease conditions and is demonstrating promise for e.g. clinical biomarker discovery. However, the low glycan ionization efficiency and the large dynamic range of glycan concentrations in human sera hinder comprehensive profiling. In particular, large glycans are problematic because they are present at low concentrations and prone to fragmentation. Here we show that, following liquid chromatographic separation on graphite columns, the sub-ambient pressure ionization with nanoelectrospray (SPIN)-MS can expand the serum glycome profile when compared with the conventional atmospheric pressure electrospray ionization (ESI)-MS with a heated capillary inlet. Notably, the ions generated by the SPIN interface were observed at higher charge states for 50% of the annotated glycans. Out of a total of 130 detected glycans, 34 were only detected with the SPIN-MS, resulting in improved coverage of glycan families as well as of glycans with larger numbers of labile monosaccharides.
Over the past few years, there has been a surge of reports discussing the potential of glycomics as a tool for identifying biomarkers useful for detecting disease states.1–8 Success is in part attributable to method improvements leading to increased sensitivity and richer glycan profiles.9–13 Greater coverage of the glycome fills in previously undetected glycan species, allowing the discovery of new glycan networks involved in disease. Glycan biomarker network analysis leads to glycan diagnostic panels rather than individual glycans, resulting in higher degrees of specificity and sensitivity.14–16
Many of the candidate cancer biomarkers identified are large sialylated and fucosylated (tri- and tetra- antennary) N-glycans and members of families differing by one monosaccharide.8 Besides their relatively poor ionization efficiency, the large glycans are particularly difficult to analyze due to labile sialic acid and fucose monosaccharides. To stabilize the labile monosaccharides, increase the ionization efficiency and simplify their fragmentation analysis, many research groups chemically modify the glycans by methylation17 or permethylation.7, 12, 18
Human serum and plasma glycan profiling has evolved from a discovery methodology to a high-throughput operation. Earlier serum profiling methods involved offline fractionation and MALDI ionization19 while more recent efforts have also incorporated sample depletion and on-line liquid chromatography.20 Tandem mass spectrometry is often incorporated for composition annotation verification and large scale structural elucidation.21 Subsequent LCMS N-glycan structural library generation are made possible by combining LC elution times and glycan structural information.22 However, the comprehensiveness of the serum glycan profiles varies greatly (18–113 glycan compositions) due to amount of starting material, challenges in sample preparation and instrument sensitivity (see Table S1, supplementary information).
Recently we described the sub-ambient pressure ionization with nanoelectrospray (SPIN) interface,23 capable of improving the MS sensitivity when compared to the conventional heated capillary interface. The SPIN interface eliminates the losses associated with the ion transfer into the first vacuum chamber of the mass spectrometer24–31 by facilitating the ionization at sub-ambient (e.g. 10–40 Torr) pressures. The ions are then efficiently captured by a dual ion funnel32 and transferred into the sub-Torr region of the mass spectrometer. With sufficiently low flow rates (e.g., 50 nL/min), the SPIN interface can deliver 50% of the analyte molecules as gas-phase ions into the sub-Torr region of the mass spectrometer.33
Here we show that the SPIN interface can improve the coverage of human serum glycome when compared to the conventional heated capillary ESI interface. The increased sensitivity provided by the SPIN source results in a greater number of analytes detected and more ions selected for fragmentation and ultimately identified by MS/MS. We demonstrate improved coverage of glycan families, especially glycans with large numbers of labile monosaccharides. A number of large glycans were also detected with sensitivity gains much larger than the 5–10 fold increase typically observed with the SPIN interface, as well as a shift to higher glycan charge states.
EXPERIMENTAL
Sample Preparation
All the chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. N-glycans were enzymatic released and purified by the method developed by Kronewitter et al.,11 with the following modifications. Briefly, the PNGase F reaction was performed in 50 mM sodium phosphate buffer at pH 5.5. Aliquots of 100 μL human serum samples (Sigma-Aldrich, St. Louis, MO) were reduced and heat denatured with dithiothreitol for 2 minutes at 95 °C. The samples were cooled and digested in a microwave reactor (CEM Discover, Matthews, NC) at 20 W for 40 min with 2uL PNGase F purified from Flavobacterium meningosepticum (New England BioLabs, Ipswich, MA). The proteins were precipitated with the addition of 80% ethanol and chilled to −80°C for 60 minutes. The samples were centrifuged at 15,000 rpm for 30 minutes to isolate the supernatant. The supernatant was extracted and dried. The samples were reduced with sodium borohydride for 60 min at 60°C in 1M sodium borohydride. The glycans were desalted and enriched on an automated Gilson GX-274 ASPEC liquid handler (Middleton, WI) with serial C8 (Discovery 500 mg, Sigma-Aldrich, St. Louis, MO) and graphitized carbon cartridges (Carbograph 150 mg, Alltech Associates, Inc., Deerfield, IL). Elution of the glycans from column was performed with 10%, 20%, and 40% acetonitrile solutions. The 40% fraction contained 0.05% TFA. The three fractions were collected and pooled together prior to LC/MS.
HPLC
Solvents A and B consisted of 0.1% formic acid (Sigma-Aldrich, St. Louis, MO) in deionized water (Barnstead Nanopure Infinity System, Dubuque, IA) and 0.2% trifluoroacetic acid (Sigma-Aldrich) in acetonitrile (Fisher Scientific, Pittsburgh, PA), respectively. An Agilent 1100 Series LC pump (Santa Clara, CA) was used to provide a 100 min linear gradient of solvent A up to 50% solvent B at 300 nL/min. The LC system was controlled with the LCMSNet automated control software. Graphitized carbon LC columns were prepared in-house by slurry packing 3-μm Hypercarb particles (Thermo Fisher, Madison, WI) into a 45-cm long and 75-μm-i.d./360-μm-o.d. fused silica capillary tubing (Polymicro Technologies, Phoenix, AZ).
Mass Spectrometry
Electrospray emitters were fabricated by chemically etching sections of 10-μm-i.d./150-μm-o.d. fused silica capillary tubing (Polymicro Technologies) as previously described.34 Mass spectra were acquired on an Agilent 6538 QToF mass spectrometer modified with a dual-ion funnel interface. A gate between the high- and low-pressure ion funnels allowed for easy exchange between a conventional heated capillary nanoESI interface and a SPIN interface without venting the instrument. Aliquots of the same sample were analyzed by HPLC/MS with ionization by both sources. Thermal energy for evaporation was provided by heating the conventional nanoESI inlet at 120°C. The temperature at the emitter tip in the SPIN source reaches ~55°C when the CO2 inlet chamber is heated at 160°C (typical operating temperature). The electrospray was operated at 1900 V with the conventional source and at 2300 V in the SPIN source. Different geometries and distances to the counter-electrode results in the larger voltage used for the SPIN source. To increase the confidence in glycan identifications, additional analyses were carried out with CID fragmentation enabled for ions exceeding 75k counts and a collision energy of E = −4.8 + 3.6V/(100Da). The spray stability in the SPIN interface was improved compared to that in the previous design35 by an additional CO2 sheath flow supplied around the emitter through a section of 200-μm-i.d./360-μm-o.d. fused silica capillary tubing (Polymicro Technologies) connected through a tee. Commercial instruments employ a similar approach at atmospheric pressure to deliver sheath gas around the ESI emitter.
Data Processing
Data was deisotoped using a glycan averagose16 in DeconTools,36 followed by internal mass calibration. Viper37 was used to find the LC/MS features (isomers, fragments, and charge states were counted as different features). Charged species with poor isotope fit scores or intensities below the limit of detection (3σ above the noise level) were not reported by the software. The glycans were then selected using the Glycolyzer16 with less than 10 ppm error and each assignment was manually validated by visually reviewing each isotopic profile.
RESULTS AND DISCUSSION
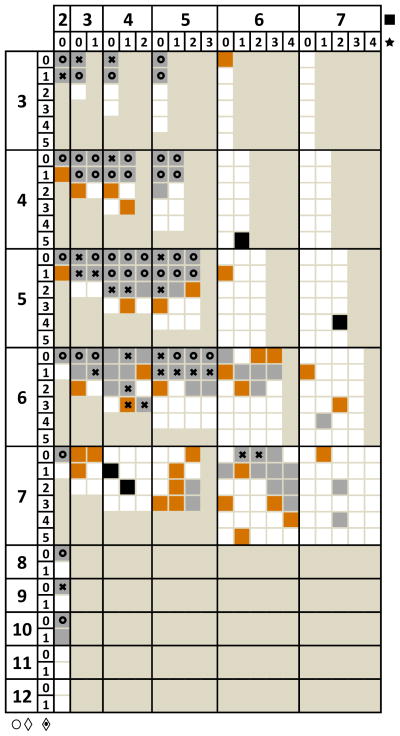
A comparison of glycan coverage by ESI- and SPIN-MS analyses of human serum is presented as a grid in Figure 1. The main grid indexes correspond to the number of hexose (H, left index: 3–12) and that of N-acetylglucosamine monosaccharides (N, top index: 2–7) in the glycan structure. Each element of the main matrix is an embedded matrix, whose indexes correspond to the number of fucose (F, left index: 0–5) and that of sialic acid monosaccharides (S, top index: 0–4). To make the matrix easier to read, we included standard symbols38 at the bottom of the header columns (○ = mannose, ⋄ = galactose, ⟐ = fucose) and at the right of the header rows (■ = N-acetylglucosamine, ★ = sialic acid). Each square in the matrix corresponds to a glycan composition, while the shaded regions correspond to compositions outside the search library.19 This representation provides additional information compared to Venn diagrams by categorizing glycans into families based on the number of H and N monosaccharides in their structure.
Figure 1.
N-Glycan coverage in human serum by both SPIN and ESI (
 ), SPIN only (
), SPIN only (
 ), ESI only (■) and glycans selected for fragmentation by both SPIN and ESI (
), ESI only (■) and glycans selected for fragmentation by both SPIN and ESI (
 ) and SPIN only (✖).
) and SPIN only (✖).
CID fragmentation validated 39 glycans in both ESI-MS and SPIN-MS, which are marked by (
 ) in Figure 1. These glycans are part of 18 glycan families as described above. For example, 6 out of 12 possible glycan compositions in the H5N4 family (block located at row 5, column 4) were validated by glycan diagnostic ions from MS/MS spectra. Additional 24 glycans, indicated by (✖) in Figure 1, were selected for CID fragmentation during the SPIN-MS analyses only. Most of the additional glycans extend the coverage of the same glycan families. For example, two extra glycans were validated in the H5N4 family; a significant extension of MS/MS validation (from 3 to 8 glycan compositions) can be observed in the H6N5 family. MS/MS validation by glycan diagnostic ions was obtained for three additional families (H9N2, H6N4, and H7N6); this was especially auspicious for H6N4 family, where 4 glycans (H6N4F0S1, H6N4F2S1, H6N4F3S1, and H6N4F3S2) were selected for fragmentation.
) in Figure 1. These glycans are part of 18 glycan families as described above. For example, 6 out of 12 possible glycan compositions in the H5N4 family (block located at row 5, column 4) were validated by glycan diagnostic ions from MS/MS spectra. Additional 24 glycans, indicated by (✖) in Figure 1, were selected for CID fragmentation during the SPIN-MS analyses only. Most of the additional glycans extend the coverage of the same glycan families. For example, two extra glycans were validated in the H5N4 family; a significant extension of MS/MS validation (from 3 to 8 glycan compositions) can be observed in the H6N5 family. MS/MS validation by glycan diagnostic ions was obtained for three additional families (H9N2, H6N4, and H7N6); this was especially auspicious for H6N4 family, where 4 glycans (H6N4F0S1, H6N4F2S1, H6N4F3S1, and H6N4F3S2) were selected for fragmentation.
The 92 glycans detected in both ESI-MS and SPIN-MS experiments were indicated by gray squares (
 ) in Figure 1. The glycans detected only in ESI-MS (4) and SPIN-MS (34) experiments were marked with black (■) and orange (
) in Figure 1. The glycans detected only in ESI-MS (4) and SPIN-MS (34) experiments were marked with black (■) and orange (
 ) squares, respectively. Out of the total of 130 glycans identified based on isotope distribution and exact mass (less than 10 ppm), 63 were validated by diagnostic ions in MS/MS spectra. The increase in glycome coverage and in the number of ions selected for fragmentation can be explained by a 5–10 sensitivity gain already demonstrated for ionization with the SPIN vs. the conventional ESI-MS interface.23, 35 Thus, many glycan ions with signal just below the fragmentation threshold in ESI-MS experiments will surpass it in SPIN-MS experiments.
) squares, respectively. Out of the total of 130 glycans identified based on isotope distribution and exact mass (less than 10 ppm), 63 were validated by diagnostic ions in MS/MS spectra. The increase in glycome coverage and in the number of ions selected for fragmentation can be explained by a 5–10 sensitivity gain already demonstrated for ionization with the SPIN vs. the conventional ESI-MS interface.23, 35 Thus, many glycan ions with signal just below the fragmentation threshold in ESI-MS experiments will surpass it in SPIN-MS experiments.
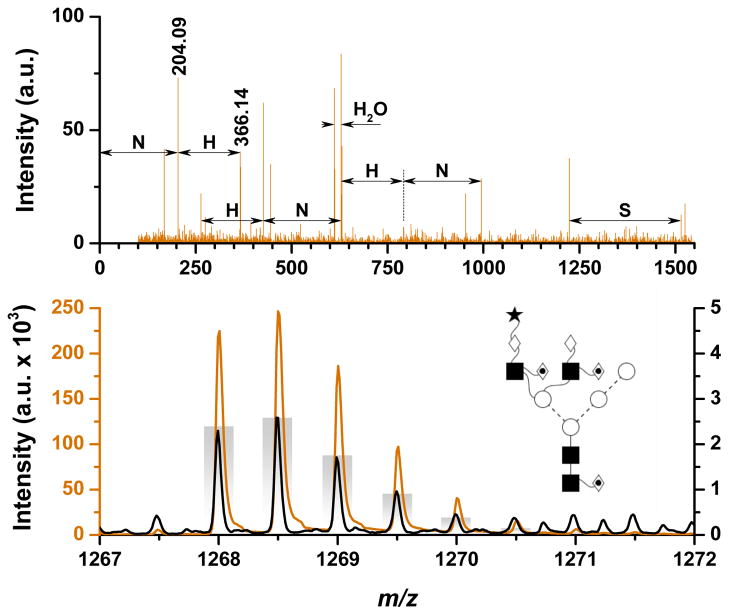
The glycan with composition H6N4F3S1 (2533.91 Da) provides an interesting case demonstrating how the increased sensitivity of the SPIN-MS experiments can improve the glycome coverage as well as glycan identification confidence. The bottom panel of Figure 2 compares the MS spectra (average of 40 scans) measured during the SPIN-MS (orange trace) and ESI-MS (black trace) experiments; note that the intensity scale is 50 times larger for the SPIN-MS (left, orange) than for the ESI-MS (right, black) data. Even though the presence of the analyte is clear in both SPIN and ESI data, the deconvolution software was not able to confidently assign the corresponding peaks in the ESI data. In this particular case, by switching from the ESI to the SPIN interface, the signal improved by almost two orders of magnitude, allowing the glycan to not only be confidently deisotoped in data processing, but also selected for fragmentation. The top panel of Figure 2 shows the MS/MS spectrum of H6N4F3S1 measured during the SPIN-MS experiment. This is the only case where an analyte not reported in the ESI data set had a large enough intensity to be selected for fragmentation in the SPIN-MS experiment. While the intensity gains are certainly helpful, they are also analyte dependent and will require additional research to understand and predict.
Figure 2.
Increased sensitivity in SPIN (
 ) vs. ESI (—) data (bottom) allows H6N4F3S1 (2533.91 Da) to be selected for fragmentation (top). Theoretical isotopic distribution provided as bar-graph and putative glycan structure provided as inset.
) vs. ESI (—) data (bottom) allows H6N4F3S1 (2533.91 Da) to be selected for fragmentation (top). Theoretical isotopic distribution provided as bar-graph and putative glycan structure provided as inset.
Several additional factors can augment the confidence in the identification of the 67 low abundant glycans not selected for fragmentation. We use composite information comprised of exact mass (10 ppm), high resolution (40,000), glycan relationships (monosaccharide mass differences) and isotope profiles (glycan averageose and fit scores). Furthermore, many of the same glycans were reported in similar samples analyzed on various platforms in other laboratories;7, 8, 10, 16, 21 grid views summarizing the corresponding glycan coverage mapped to the current working library are provided as supplementary information (Figures S1–S7). In addition, our sample preparation and liquid chromatography setup specifically targets the N-glycan separation and enrichment. Applying strict, multifaceted criteria for assignments decreases the probability for false positive assignments. Our confidence in glycan assignments increases when tandem mass spectra confirm parts of the glycan family.
The coverage of many glycan families (H4N3, H4N4, H4N5, H5N4, H5N5, and H6N5) was broadly validated by fragmentation data, with a small percentage of glycans identified by exact mass and isotope distribution only. For example, 8 out of 10 glycan compositions were validated by fragmentation in the H5N4 family, increasing the confidence in the identification of the two extra glycans (H5N4F3S1 and H5N4F2S2) that did not pass the fragmentation threshold. In several other families (H4N2, H5N2, H6N3, H6N4, and H10N2) the percentage of glycans validated by fragmentation is close to 50%.
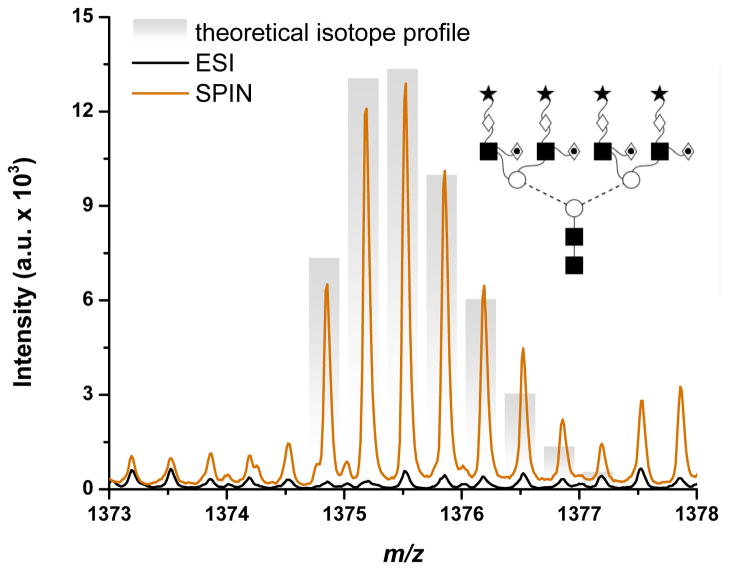
In the H7N6 family only two glycans were validated by fragmentation, while 13 other glycans were detected at relatively low concentration. Figure 3 shows the MS spectra corresponding to the largest glycan (4121.49 Da) identified in this family (H7N6F4S4). Because of the significantly larger signal-to-noise ratio, the deconvolution software was able to identify and assign the SPIN-MS profile (orange trace) based on the isotope distribution, but not the ESI-MS profile (black trace). In this particular case, the SPIN interface provided an increase in sensitivity of >20-fold.
Figure 3.
Mass spectra of H7N6F4S4 (4121.49 Da) demonstrating a factor of more than 20 increase in sensitivity for SPIN (
 ) vs. ESI (—) data. Both traces are averages of 5 spectra. Theoretical isotopic distribution provided as bar-graph and putative glycan structure provided as inset.
) vs. ESI (—) data. Both traces are averages of 5 spectra. Theoretical isotopic distribution provided as bar-graph and putative glycan structure provided as inset.
Glycans from selected families (H6N6, H7N3, and H7N5) did not pass the fragmentation threshold; however, each family is well represented with all the identified glycans clustered together. Less confidently annotated glycans appear isolated in Figure 1 (see for example families H5N7, H6N7, and H7N7). Although detection by both ESI-MS and SPIN-MS (see for example H6N7F4S1, H7N7F2S2, and H7N7F4S2) improves identification confidence, the annotation certainty remains low.
The charge state distribution of analyte ions detected in ESI-MS experiments depends on various experimental parameters, some of which were associated with changes in analyte conformation:39 solvent composition,40–43 analyte basicity,44–46 gas-phase reactions,47, 48 temperature,49–52 and presence of additives.53–57 A shift to higher charge states is often highly desirable because it offers increased sensitivity and mass range, in particular with high resolution platforms.
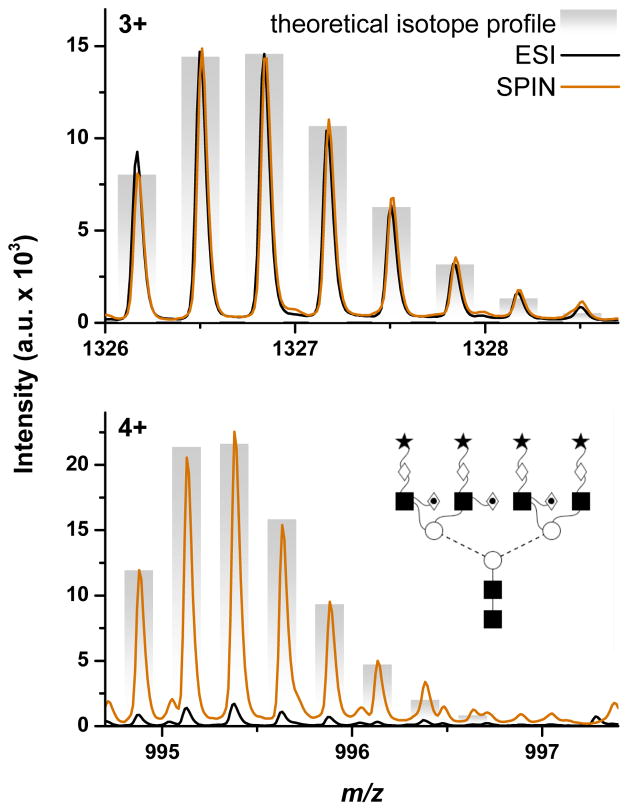
A notable shift to higher charge states was observed for many glycans observed by SPIN-MS vs. ESI-MS. Figure 4 compares averaged mass spectra corresponding to the glycan with the composition H7N6F4S3 observed by ESI-MS (black traces) and SPIN-MS (orange traces). The top panel shows that similar signal intensity for the 3+ charge state were measured for both interfaces, while the bottom panel demonstrates more than 10-fold signal increase in SPIN-MS for the 4+ charge state. In this particular case, the 4+ charge state in the ESI-MS data was missed by the automated data processing due to poor signal quality in individual spectra.
Figure 4.
Mass spectra of H7N6F3S4 (3975.43 Da) demonstrating a shift to higher charge states in SPIN (
 ) vs. ESI (—) data. Theoretical isotopic distribution provided as bar-graph and putative glycan structure provided as inset.
) vs. ESI (—) data. Theoretical isotopic distribution provided as bar-graph and putative glycan structure provided as inset.
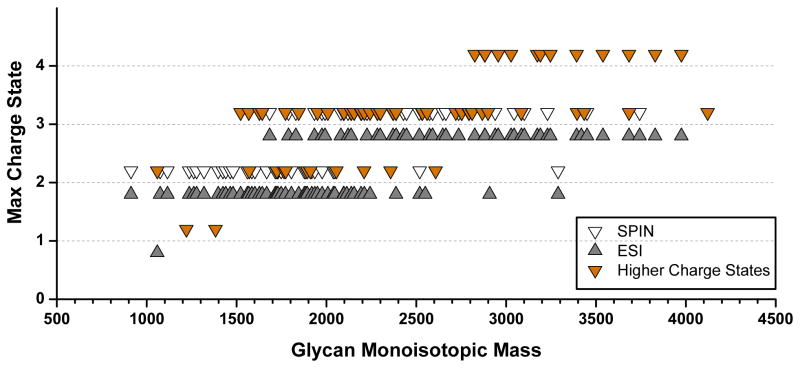
The shift to higher charge states occurred mostly for large glycans within the H6N4, H6N5, and H7N6 families. By adding the glycan compositions only detected by SPIN-MS (34), we conclude that almost half of the total number of glycans detected in these experiments (63 out of 130) were detected with higher charge states. Figure 5 plots the maximum charge state reported for each of the glycans detected in ESI-MS (
 ) and SPIN-MS (▽) experiments as a function of their molecular mass, emphasizing the glycans detected with at least one extra charge (
) and SPIN-MS (▽) experiments as a function of their molecular mass, emphasizing the glycans detected with at least one extra charge (
 ). We should note that even for the glycans reported with similar but multiple charge states, the relative contribution of higher charge states to the overall charge envelope was larger in SPIN-MS vs. ESI-MS data.
). We should note that even for the glycans reported with similar but multiple charge states, the relative contribution of higher charge states to the overall charge envelope was larger in SPIN-MS vs. ESI-MS data.
Figure 5.
Highest charge states of glycans detected by ionization with conventional ESI (
 ) and SPIN (▽) interfaces, emphasizing glycans detected with larger charge states in the latter (
) and SPIN (▽) interfaces, emphasizing glycans detected with larger charge states in the latter (
 ).
).
The early stages of electrospray ionization at atmospheric pressure and in the sub-ambient pressure environment are expected to be very similar, with only minor changes in droplet evaporation and temperature profiles. Additional charging of the ions after initial production in the SPIN interface is unlikely. Also, with glycans as analytes, increased charging cannot be explained by analyte conformation changes such as protein unfolding. Thus, the larger contribution of higher charge states in the SPIN-MS data can most likely be attributed to the additional time and likelihood of atmospheric pressure ion-molecule processes for the ESI interface. The difference between the ion residence times in the two ion sources is primarily in time spent at atmospheric pressure and inside the heated capillary. Gas-phase reactions, specifically in-source proton transfer reactions with e.g. solvent clusters and trace gas phase components, can reduce the charge state of analytes;47, 48 because the SPIN source operates at lower pressures and the residence time is much shorter, the probability of such reactions is greatly reduced. A preferential loss of higher mobility species during the transfer from atmospheric pressure into the first vacuum chamber of the mass spectrometer through a standard heated capillary inlet was recently demonstrated;58 this bias is effectively eliminated in the SPIN interface because the ion funnels provide efficient ion focusing prior to any conductance limiting aperture.
CONCLUSIONS
We compared the conventional ESI heated capillary inlet and SPIN interfaces for the analysis of human serum glycans and we demonstrated the SPIN interface expanded the coverage by >25%, while increasing the confidence in the glycan identifications. Our results indicate that the charged species MS detected from conventional electrospray ionization may be somewhat modified due to e.g. ion-molecule processes during the longer time at higher pressure, as well as during transfer through a heated capillary interface. We improved the coverage of large sialylated and fucosylated N-glycans, of possible interest for future biomarker candidate identification efforts.8 Additionally, nearly 50% of the glycans were detected with significant abundances in higher charge states, which could be used to improve the quality of tandem MS measurements and increase the mass range for their detection.
Supplementary Material
Acknowledgments
We thank Tom Fillmore, Danny Orton, Rui Zhao for help with the HPLC, and Brian LaMarche for the automated HPLC control software LCMSNet. Discussions with Dr. Erin Baker on the relationship between solution and gas-phase analyte charge states are gratefully acknowledged. The glycomics work was supported by the U. S. Department of Energy Office of Biological and Environmental Research (DOE/BER). SPIN source development was supported by grants from the National Institutes of Health: National Cancer Institute (1R33CA155252) and National Institute of General Medical Sciences (8 P41 GM103493-10). The experiments were performed in the Environmental Molecular Sciences Laboratory, a U.S. DOE national scientific user facility located at the Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RL01830.
References
- 1.Tanabe K, Deguchi A, Higashi M, Usuki H, Suzuki Y, Uchimura Y, Kuriyama S, Ikenaka K. Biochem Biophys Res Commun. 2008;374:219–225. doi: 10.1016/j.bbrc.2008.06.124. [DOI] [PubMed] [Google Scholar]
- 2.Kim YG, Jeong HJ, Jang KS, Yang YH, Song YS, Chung J, Kim BG. Anal Biochem. 2009;391:151–153. doi: 10.1016/j.ab.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Alley WR, Jr, Madera M, Mechref Y, Novotny MV. Anal Chem. 2010;82:5095–5106. doi: 10.1021/ac100131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saldova R, Fan Y, Fitzpatrick JM, Watson RW, Rudd PM. Glycobiology. 2011;21:195–205. doi: 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- 5.Arnold JN, Saldova R, Galligan MC, Murphy TB, Mimura-Kimura Y, Telford JE, Godwin AK, Rudd PM. J Proteome Res. 2011;10:1755–1764. doi: 10.1021/pr101034t. [DOI] [PubMed] [Google Scholar]
- 6.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Analyst. 2011;136:3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isailovic D, Plasencia MD, Gaye MM, Stokes ST, Kurulugama RT, Pungpapong V, Zhang M, Kyselova Z, Goldman R, Mechref Y, Novotny MV, Clemmer DE. Journal of Proteome Research. 2012;11:576–585. doi: 10.1021/pr200777u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alley WR, Vasseur JA, Goetz JA, Syoboda M, Mann BF, Matei DE, Menning N, Hussein A, Mechref Y, Novotny MV. Journal of Proteome Research. 2012;11:2282–2300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kita Y, Miura Y, Furukawa J, Nakano M, Shinohara Y, Ohno M, Takimoto A, Nishimura S. Mol Cell Proteomics. 2007;6:1437–1445. doi: 10.1074/mcp.T600063-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Bereman MS, Young DD, Deiters A, Muddiman DC. Journal of Proteome Research. 2009;8:3764–3770. doi: 10.1021/pr9002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kronewitter SR, de Leoz MLA, Peacock KS, McBride KR, An HJ, Miyamoto S, Leiserowitz GS, Lebrilla CB. Journal of Proteome Research. 2010;9:4952–4959. doi: 10.1021/pr100202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desantos-Garcia JL, Khalil SI, Hussein A, Hu Y, Mechref Y. Electrophoresis. 2011;32:3516–3525. doi: 10.1002/elps.201100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruhaak LR, Miyamoto S, Kelly K, Lebrilla CB. Anal Chem. 2012;84:396–402. doi: 10.1021/ac202775t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, Schnaper L, Hickey RJ, Malkas LH, Novotny MV. Clin Chem. 2008;54:1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- 15.Goldman R, Ressom HW, Varghese RS, Goldman L, Bascug G, Loffredo CA, Abdel-Hamid M, Gouda I, Ezzat S, Kyselova Z, Mechref Y, Novotny MV. Clin Cancer Res. 2009;15:1808–1813. doi: 10.1158/1078-0432.CCR-07-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kronewitter SR, De Leoz ML, Strum JS, An HJ, Dimapasoc LM, Guerrero A, Miyamoto S, Lebrilla C, Leiserowitz GS. Proteomics. 2012;12:2523–2538. doi: 10.1002/pmic.201100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell AK, Harvey DJ. Rapid Commun Mass Spectrom. 1996;10:1027–1032. doi: 10.1002/(SICI)1097-0231(19960715)10:9<1027::AID-RCM634>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 18.Zaia J. Mass Spectrom Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- 19.Kronewitter SR, An HJ, de Leoz ML, Lebrilla CB, Miyamoto S, Leiserowitz GS. Proteomics. 2009;9:2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bereman MS, Muddiman DC. Anal Bioanal Chem. 2010;396:1473–1479. doi: 10.1007/s00216-009-3368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stumpo KA, Reinhold VN. Journal of Proteome Research. 2010;9:4823–4830. doi: 10.1021/pr100528k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. J Proteome Res. 2012;11:1958–1968. doi: 10.1021/pr2011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page JS, Tang KQ, Kelly RT, Smith RD. Anal Chem. 2008;80:1800–1805. doi: 10.1021/ac702354b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith RD, Loo JA, Edmonds CG, Barinaga CJ, Udseth HR. Anal Chem. 1990;62:882–899. doi: 10.1021/ac00208a002. [DOI] [PubMed] [Google Scholar]
- 25.Lin BW, Sunner J. J Am Soc Mass Spectrom. 1994;5:873–885. doi: 10.1016/1044-0305(94)87012-8. [DOI] [PubMed] [Google Scholar]
- 26.Zook DR, Bruins AP. Int J Mass Spectrom Ion Process. 1997;162:129–147. [Google Scholar]
- 27.Geromanos S, Freckleton G, Tempst P. Anal Chem. 2000;72:777–790. doi: 10.1021/ac991071n. [DOI] [PubMed] [Google Scholar]
- 28.El-Faramawy A, Siu KWM, Thomson BA. J Am Soc Mass Spectrom. 2005;16:1702–1707. doi: 10.1016/j.jasms.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Schneider BB, Javaheri H, Covey TR. Rapid Commun Mass Spectrom. 2006;20:1538–1544. doi: 10.1002/rcm.2511. [DOI] [PubMed] [Google Scholar]
- 30.Page JS, Kelly RT, Tang KQ, Smith RD. J Am Soc Mass Spectrom. 2007;18:1582–1590. doi: 10.1016/j.jasms.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 31.Covey TR, Thomson BA, Schneider BB. Mass Spectrom Rev. 2009;28:870–897. doi: 10.1002/mas.20246. [DOI] [PubMed] [Google Scholar]
- 32.Ibrahim Y, Tang KQ, Tolmachev AV, Shvartsburg AA, Smith RD. J Am Soc Mass Spectrom. 2006;17:1299–1305. doi: 10.1016/j.jasms.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marginean I, Page J, Tolmachev A, Tang K, Smith R. Analytical Chemistry. 2010;82:9344–9349. doi: 10.1021/ac1019123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly RT, Page JS, Luo QZ, Moore RJ, Orton DJ, Tang KQ, Smith RD. Anal Chem. 2006;78:7796–7801. doi: 10.1021/ac061133r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang KQ, Page JS, Marginean I, Kelly RT, Smith RD. J Am Soc Mass Spectrom. 2011;22:1318–1325. doi: 10.1007/s13361-011-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. Bmc Bioinformatics. 2009;10 doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monroe ME, Tolic N, Jaitly N, Shaw JL, Adkins JN, Smith RD. Bioinformatics. 2007;23:2021–2023. doi: 10.1093/bioinformatics/btm281. [DOI] [PubMed] [Google Scholar]
- 38.Harvey DJ, Merry AH, Royle L, Campbell MP, Rudd PM. Proteomics. 2011;11:4291–4295. doi: 10.1002/pmic.201100300. [DOI] [PubMed] [Google Scholar]
- 39.Hall Z, Robinson CV. J Am Soc Mass Spectrom. 2012;23:1161–1168. doi: 10.1007/s13361-012-0393-z. [DOI] [PubMed] [Google Scholar]
- 40.Chowdhury SK, Katta V, Chait BT. J Am Chem Soc. 1990;112:9012–9013. [Google Scholar]
- 41.Loo JA, Udseth HR, Smith RD. Biomedical and Environmental Mass Spectrometry. 1988;17:411–414. [Google Scholar]
- 42.Wang GD, Cole RB. Anal Chem. 1994;66:3702–3708. [Google Scholar]
- 43.Iavarone AT, Jurchen JC, Williams ER. J Am Soc Mass Spectrom. 2000;11:976–985. doi: 10.1016/S1044-0305(00)00169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Covey TR, Bonner RF, Shushan BI, Henion J. Rapid Commun Mass Spectrom. 1988;2:249–256. doi: 10.1002/rcm.1290021111. [DOI] [PubMed] [Google Scholar]
- 45.Loo RRO, Smith RD. J Mass Spectrom. 1995;30:339–347. [Google Scholar]
- 46.Williams ER. J Mass Spectrom. 1996;31:831–842. doi: 10.1002/(SICI)1096-9888(199608)31:8<831::AID-JMS392>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLuckey SA, Stephenson JL. Mass Spectrom Rev. 1998;17:369–407. doi: 10.1002/(SICI)1098-2787(1998)17:6<369::AID-MAS1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 48.Pitteri SJ, McLuckey SA. Mass Spectrom Rev. 2005;24:931–958. doi: 10.1002/mas.20048. [DOI] [PubMed] [Google Scholar]
- 49.Le Blanc JCY, Beuchemin D, Siu KWM, Guevremont R, Berman SS. Org Mass Spectrom. 1991;26:831–839. [Google Scholar]
- 50.Mirza UA, Cohen SL, Chait BT. Anal Chem. 1993;65:1–6. doi: 10.1021/ac00049a003. [DOI] [PubMed] [Google Scholar]
- 51.Liu JL, Konermann L. J Am Soc Mass Spectrom. 2009;20:819–828. doi: 10.1016/j.jasms.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 52.Sterling HJ, Cassou CA, Susa AC, Williams ER. Anal Chem. 2012;84:3795–3801. doi: 10.1021/ac300468a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iavarone AT, Jurchen JC, Williams ER. Anal Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lomeli SH, Peng IX, Yin S, Loo RRO, Loo JA. J Am Soc Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hogan CJ, Loo RRO, Loo JA, de la Mora JF. Phys Chem Chem Phys. 2010;12:13476–13483. doi: 10.1039/c0cp01208d. [DOI] [PubMed] [Google Scholar]
- 56.Sterling HJ, Kintzer AF, Feld GK, Cassou CA, Krantz BA, Williams ER. J Am Soc Mass Spectrom. 2012;23:191–200. doi: 10.1007/s13361-011-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miladinović SM, Fornelli L, Lu Y, Piech KM, Girault HH, Tsybin YO. Anal Chem. 2012;84:4647–4651. doi: 10.1021/ac300845n. [DOI] [PubMed] [Google Scholar]
- 58.Page JS, Marginean I, Baker ES, Kelly RT, Tang KQ, Smith RD. J Am Soc Mass Spectrom. 2009;20:2265–2272. doi: 10.1016/j.jasms.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.