Abstract
The development of an effective malaria vaccine has been hampered by the genetic diversity of commonly used target antigens. This diversity has led to concerns about allele-specific immunity limiting the effectiveness of vaccines. Despite extensive genetic diversity of circumsporozoite protein (CS), the most successful malaria vaccine is RTS/S, a monovalent CS vaccine. By use of massively parallel pyrosequencing, we evaluated the diversity of CS haplotypes across the T-cell epitopes in parasites from Lilongwe, Malawi. We identified 57 unique parasite haplotypes from 100 participants. By use of ecological and molecular indexes of diversity, we saw no difference in the diversity of CS haplotypes between adults and children. We saw evidence of weak variant-specific selection within this region of CS, suggesting naturally acquired immunity does induce variant-specific selection on CS. Therefore, the impact of CS vaccines on variant frequencies with widespread implementation of vaccination requires further study.
Although the estimated annual numbers of malaria infections (approximately 225 million) and deaths (approximately 781 000) remain high, recent successes in reducing transmission have triggered discussions about global malaria elimination [1–3]. However, our current tools are unlikely to achieve this goal. An effective Plasmodium falciparum vaccine would add an important tool to our armamentarium.
A major challenge to developing a broadly useful malaria vaccine has been the diversity of the falciparum parasite population [4]. P. falciparum is a highly genetically diverse organism with potentially hundreds of unique genotypes at key loci circulating within a population and infections commonly containing multiple variants [5]. Recent studies have shown that one potential vaccine target, apical membrane antigen-1, may have >200 haplotypes within a single town in Mali, and this diversity has caused a candidate vaccine to be broadly ineffective [6, 7].
To date, the most successful malaria vaccine has been RTS,S. RTS,S AS01E is a monovalent circumsporozoite protein (CS) malaria vaccine undergoing a phase III trial in 7 countries at 11 sites. Initial data from this trial has shown an estimated efficacy of 55% [8]. The molecular biology of CS and the development of RTS,S have recently been reviewed [9, 10]. The antigen covers the entire surface of sporozoites and is coded for by a single-copy gene (csp). The structure of CS is immunologically dominated by a central tandem amino acid repeat and a series of T-cell epitopes in the carboxy terminal of the molecule (Figure 1A). The primary T-cell epitopes, TH2 and TH3, are known to be highly polymorphic in natural parasite populations [11–15]. This diversity may in part explain why RTS,S is only partially effective.
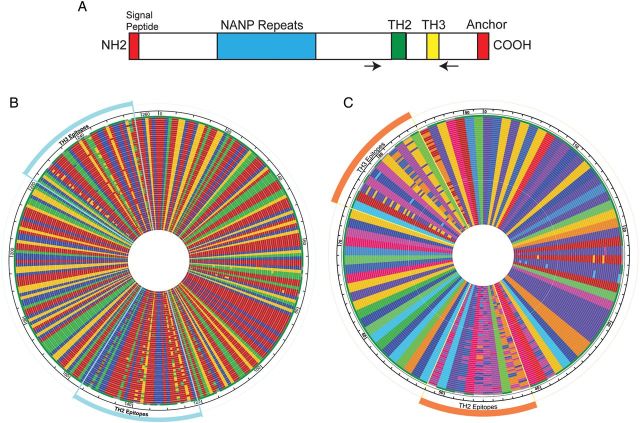
Figure 1.
TH2 and TH3 polymorphisms and haplotypes of csp variants. A, Schematic structure of CS (not to scale) with the locations of the NANP repeat (blue), TH2 epitope (green), and TH3 epitope (yellow) highlighted. The signal peptide and anchor sequences are also shown (red). The locations of the primers are marked by the arrows. B, DNA alignment of all 57 unique variants detected in the population. Nucleotides are represented by different colors (adenine, red; thymine, blue; cytosine, green; and guanine, yellow). The position of the TH2 and TH3 epitopes are marked. C, Amino acid alignment of all 57 unique variants detected in the population. Amino acids are represented by different colors (alanine, brick red; arginine, yellow ochre; asparagine, violet; aspartic acid, orange yellow; cysteine, forest green; glutamic acid, blue; glutamine, medium blue; glycine, dirty yellow; histidine, bright red; isoleucine, red; leucine, dark purple; lysine, pink; methionine, magenta; proline, indigo; serine, light blue; threonine, bright blue; tryptophan, medium green; tyrosine, parrot green; and valine, light green; phenylalanine is not seen). The position of the TH2 and TH3 epitopes are marked.
It is still unclear what mechanisms are responsible for maintaining polymorphisms in the TH2 and TH3 epitopes of CS. Good et al suggested that they were maintained by natural selection favoring immune evasion (allele-specific immunity) [12]. This hypothesis was supported by the observation that the number of nonsynonymous nucleotide substitutions was higher than the number of synonymous nucleotide substitutions in some populations [16, 17]. On the other hand, recent evidence suggests that among CS isolates in the Gambia, there is only limited evidence of balancing selection, implying minimal allele-specific immunity in CS [18]. Whether there is allele-specific immunity generated against CS is critical to predicting the long-term usefulness of a monovalent CS vaccine.
To understand the diversity of csp within the falciparum parasite population in Lilongwe, Malawi, we conducted an observational study in 2010 involving 100 parasitemic participants. A region spanning the TH2 and TH3 epitopes of csp was amplified from each participant in duplicate and ultra-deep sequenced, using massively parallel pyrosequencing (MPP). To our knowledge, this is the first population genetic study of malaria diversity reported using MPP. By using the sequence data, we identified the existing parasite haplotypes (also termed “variants”) currently in the population and, for the first time, their relative frequency within individuals. By use of these metrics, we compared children and adults in terms of haplotype diversity and richness, using several ecological and molecular indexes. We then assessed the haplotypes for evidence of selection on the TH2 and TH3 epitope region.
MATERIALS AND METHODS
Study Population
Informed consent as approved by the University of North Carolina Institutional Review Board and Malawian National Health Sciences Research Committee was received from all study participants in this observational study. One hundred participants were enrolled at the outpatient clinics at Kamuzu Central Hospital in Lilongwe, Malawi, between March and June 2010. In total, 50 adults (age range, 19–61 years; mean age [±SD], 30 ± 10 years) who presented to the clinic with symptoms consistent with uncomplicated malaria were enrolled sequentially. In addition, 50 children (age range, 1–5 years; mean age [±SD], 3 ± 0.7 years) were enrolled in parallel. There was no difference in sex distribution between adults and children. Children <5 years of age routinely have malaria smears done in the clinic as part of the vast majority of evaluations, so some children may have been asymptomatic at the time of enrollment. All smears were reviewed by 2 microscopists, and patients who were smear positive for falciparum malaria with a parasitemia of ≥2+ (discussed in the Supplementary Materials) were approached about participation in the study. Any person with signs of severe malaria was excluded from the study. Dried blood spots were collected and stored in individual packets with desiccant at room temperature until shipment.
Amplification and Sequencing of csp
The region of csp containing the TH2 and TH3 epitopes was amplified from extracted DNA by use of previously described primers [19], which we modified for 454 sequencing by inclusion of a linker, tag, and a multiplex identifier (MID) sequence. Each patient sample was amplified in duplicate, using unique MIDs. Amplicons were pooled and sequenced on a 454 Life Sciences sequencer, using the Titanium chemistry at the University of North Carolina High Throughput Sequencing Facility. Further details are provided in the Supplementary Materials.
Haplotype Determination From MPP
Haplotypes of csp variants were determined using a combination of Bayesian and heuristic clustering that is detailed in the Supplementary Materials. In brief, sequence reads were separated on the basis of MIDs from the pooled data into amplicon-specific data, resulting in 2 independent amplicon data sets for each participant. The sequences were trimmed of MIDs, tags, and primers and were culled of low-quality reads on the basis of previously determined cutoffs for given characteristics (eg, length and quality scores). These resultant high-quality read sets were further clustered by ShoRAH to predict the most likely haplotypes within the patient [20]. The Bayesian model in this program provides a posterior probability that the predicted haplotype is a true haplotype. This was used to remove unlikely low-frequency haplotypes from further analysis.
ShoRAH is limited in that it models a uniform error rate across the sequence. This potentially leads to spurious haplotypes because of increased differences called within the error prone 3′ sequence. To correct for this, we employed a second heuristic clustering step to further collapse the combined ShoRAH haplotypes from both amplicons that differed by no more than 1 substitution but up to 5 small (1 or 2 bases) indels and to predict, by consensi, the likely true haplotypes for each participant. As each sample was amplified in duplicate and sequenced independently, we required that the final haplotypes from a participant be composed of only haplotypes identified from both independent amplicons and represent ≥1% of the total reads for a participant. By requiring the haplotype to occur independently in 2 polymerase chain reaction (PCR) assays, we limit the potential for false haplotypes due to PCR or sequencing error. To examine haplotypes at the population level, a similar clustering (allowing for only small indels) and consensus determination was performed as described above across the combined haplotypes from all individuals. The resulting weighted consensi provided the final haplotypes for analysis and was assigned a unique population identifier (pUID).
Data Analysis
DNA alignments and figures were generated using MegAlign and GeneVison software (DNAStar, Madison, WI). Ecological indexes of diversity and rarefaction curves were determined using EstimateS v8.2 [21]. Calculations of molecular diversity and evolution were done using Arlequin v3.5.1.2 and DnaSP v5.0 [22, 23]. The median-joining network was created using DNA Alignment v1.2.1.1 and Network v4.6.0.0 [24]. For this analysis, a multiplicity of infection (MOI) is defined as the number of unique CS haplotypes in an individual infection. Additional information about data analysis and MOI is provided in the Supplementary Materials.
RESULTS
Determination of Unique Haplotypes
The number of sequence reads passing the initial quality filter was 1 605 260. A total of 1 562 833 sequences (97.3%) are represented in the final data after construction of the unique variant consensus haplotypes. The average number of reads used to construct haplotypes in each patient was 15 682 (SD = 5301). Therefore, the majority of haplotypes at our lower limit, representing 1% of the parasite population within a patient, used a minimum of 100–200 sequencing reads to construct sequence of the haplotype. The final sequence length of all variants was 265 base pairs because of trimming of bases during haplotype building.
In total, 57 unique parasite haplotypes based upon DNA sequence spanning TH2 and TH3 were detected in the population (GenBank accession no. JN634586-JN634642) (Figure 1B). These variants represented frequencies of ≥1% within the individual participants and were supported by both independent amplifications for the given participant. Their frequencies within the entire population ranged from 0.01% to 12% and occurred in 1–25 samples. Population frequencies are estimated by the sum of the fractional representation across individuals and are not corrected for parasitemia because quantitative microscopy reads were not available. Amplicon sequencing by MPP has been used to determine frequencies of variants from clinical samples for other organisms [25]. However, it is possible that frequency estimates may be biased during PCR amplification, affecting any frequency-based estimates of diversity. Biases have been associated with variability in amplicon length, differences underlying the PCR primers, and extensive PCR cycles. This potential bias should be minimal in this study because the fragment is of uniform length with well-conserved primer regions and was amplified using a modest number of PCR cycles.
These 57 haplotypes also represented 57 unique amino acid sequences (Figure 1C). In total, 30 unique TH2 epitopes and 15 unique TH3 epitopes were identified. Among these, only 3 haplotypes (9%) and 1 haplotype (5%) carried the sample TH2 and TH3 epitopes, respectively, as falciparum strain 3d7 (the RTS,S strain). Only 1 variant contained the same TH2 and TH3 epitope haplotype as falciparum strain 3d7. This haplotype represented only 5% of the sequences from the population.
MOI and Ecological Indexes of Haplotype Diversity
Although several samples were highly complex (6 samples had a MOI of ≥5 variants), in general the average MOI was low (Table 1). The average MOI for the entire population was 2.31 (SD = 1.70) variants/infection (range, 1–12 variants/infection). Children and adults had similar MOIs (P = .7288, by the unpaired 2-tailed t test). There was no difference in mean MOI (SD) on the basis of parasitemia (2.5 [1.8], 2.4 [1.9], and 2 [1.2] in the 2 + , 3 + , and 4 + groups, respectively).
Table 1.
Multiplicity of Infection (MOI) and Indexes of Haplotype Diversity
| Variable | Total Population | Adults | Children |
|---|---|---|---|
| MOI | |||
| Mean (SD) | 2.31 (1.70) | 2.28 (1.81) | 2.34 (1.61) |
| Median | 2 | 2 | 2 |
| Chao2, mean (95% CI) | 77.34 (64.45–112.52) | 90 (58.31–183.23) | 65.34 (52.45–100.52) |
| Exponential Shannon, mean | 25.07 | 22.58 | 23.26 |
| θ(π) | 0.02 | 0.02 | 0.02 |
Species richness, or the number of species in a sample of a specified size, is an easily comprehensible expression of species “diversity.” To determine whether there were significant differences in species (or, in this case, haplotype) richness among the Lilongwe parasite population, we used EstimateS to evaluate 7 nonparametric estimators of species richness (Chao1, Chao2, incidence-based coverage estimator, abundance-based coverage estimator, Jack1, Jack2, and Michaelis-Menten mean) [21]. We looked at the total diversity of the sample, as well as the diversity of parasites within adults and within children. No differences in species richness were seen between any of these groups for any of these indexes. The Chao2 provides a 95% confidence interval for its estimate of species richness and gives an easily interpretable estimate of the expected richness in the population (Table 1) [26].
By use of EstimateS, we also calculated indexes of species diversity. Unlike indexes of species richness, indexes of species diversity combine information on species richness (ie number of variants) and species abundance (ie frequency of variants) in different ways. Again, no differences in species diversity were seen between these populations, using Shannon, Exponential Shannon, Alpha, and Simpsons Indexes.
Haplotype Discovery
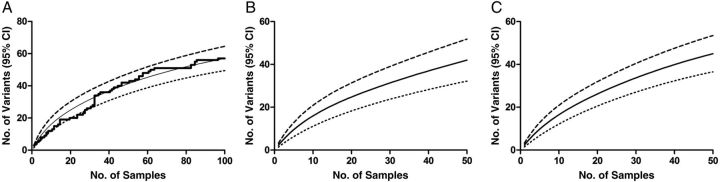
In general, the number of entities discovered is highly dependent on sampling effort. In the current context, this means that, as more sequences are analyzed, the number of variants discovered will increase. This is evident in our sample (Figure 2). The variant accumulation curve of our plotted data does not yet appear to be reaching its asymptote, suggesting that further sampling may result in the identification of additional haplotypes. This data was used to develop smoothed rarefaction curves for our population, which estimate the number of haplotypes that would be detected at any given sampling level. Figure 2A shows the rarefaction curve and raw data for the entire population. Figure 2B and 2C show the rarefaction curves for the adults and children, respectively. The curves for the 2 populations are similar, and neither reaches its asymptote.
Figure 2.
Accumulation and rarefaction curves of csp variants. A, Variant accumulation and rarefaction curves for the entire population. The thick solid line shows the raw data for the variant accumulation curve. The computed rarefaction curve (thin solid line) represents the expected average rate of variant accumulation that would be produced by repeated deep sequencing of the same population. Confidence intervals (CIs) for the species richness were also determined (dashed and dotted lines represent upper and lower 95% CIs, respectively). CIs for rarefaction curves allow for the comparison of diversity between populations, even when there are differences in sampling effort. B and C, Rarefaction curve (thin solid line) and 95% CIs (dotted lines) for the adult (B) and child (C) populations.
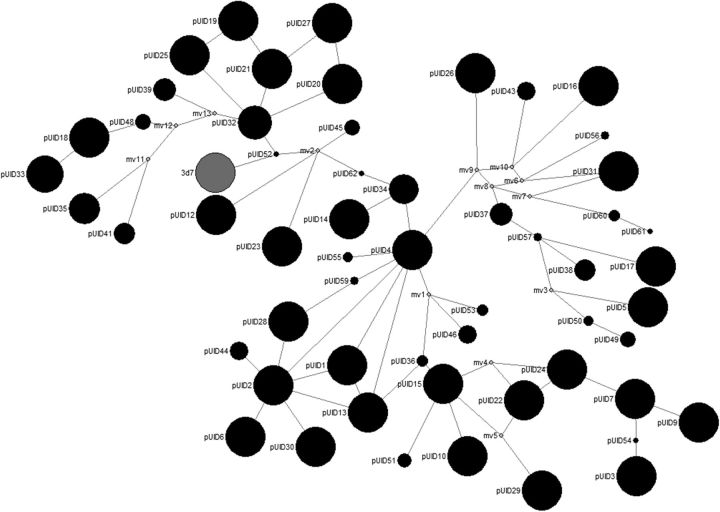
Median-Joining Network
A median-joining network was developed for the csp haplotypes (Figure 3). Similar approaches have been used previously in malaria research [27]. Figure 3 suggests that csp variants in Lilongwe are clustered into 3 major groups. All variants sharing the 3d7 TH2 epitope clustered in the arm of the network that contained 3d7 (including the 2 variants that shared TH2 and TH3). The variant that contained the same TH3 as 3d7, but a highly different TH2, clustered away from this arm (pUID10). Similar to previous reports, associations between the common haplotypes show that the TH2 and TH3 epitope sequences do not randomly assort and that the combinations that exist within a single CS molecule may be constrained (data not shown) [13].
Figure 3.
Median-joining network of csp variants. The network was developed using the complete 265–base pair sequence for each haplotype. These networks allow for a visual representation of the mutational paths that may have led to the observed data and assume that mutations are more likely to derive from a more frequent haplotype and proceed to a less frequent haplotype [27]. Parasite variants are shown in the black circles. The size of the circle is relative to the frequency of the variant in the population. Variant pUID8 (dark grey circle) shared the same 265–base pair sequence as strain 3d7 (labeled 3d7 in the figure). Median vectors are shown in light grey circles. Median vectors represent hypothetical ancestral haplotypes linking existent haplotypes or may represent haplotypes not sampled. The length of links is not proportional to the number of mutational steps separating haplotypes.
Sequence Polymorphisms and Molecular Evolution
In addition to the ecological evidence that there is not a difference in diversity between groups, our molecular evaluation supports this finding. With DNA sequences, diversity is described by variation in the nucleotide sequences among the parasites, since all the samples sequences are related through common ancestry. The total genetic variation is referred to as θ, and various statistics have been used to describe this diversity [23, 28–31]. In the population of parasites sampled in our study, several θ estimators (θHom, θπ, and θS) show no differences between the populations (Table 1) [23].
The overall genetic distance between parasite haplotypes was evaluated. The haplotypes on average had a mean number of pairwise differences (π) of 0.02 (Table 1). Differences of >50 single-nucleotide polymorphisms (across the whole gene) have been reported between some parasite variants. Individual haplotypic distance matrices for both adults and children are presented in Supplementary Figure 1, suggesting a uniform distribution of differences among the population. In total, there were 24 polymorphic loci within the 265–base pair fragment analyzed. The allele frequencies for these loci are presented in Supplementary Table 1, and the expected heterozygosity for these loci is shown in Supplementary Figure 1. On a population level, there was not a significant distance between parasites found in adults and children (population pairwise F statistic = 0.00022). This is lower than previously reported F statistic values from the Gambia for csp [18].
Interestingly, all 24 detected differences were nonsynonymous coding changes. This lack of synonymous changes suggests that our haplotypes contain minimal error, since we would expect that base changes would occur without difference to the coding potential, and thus we would expect approximately 1 of 3 errors to occur within a synonymous site.
Evidence of Selection on the C-terminus of CS
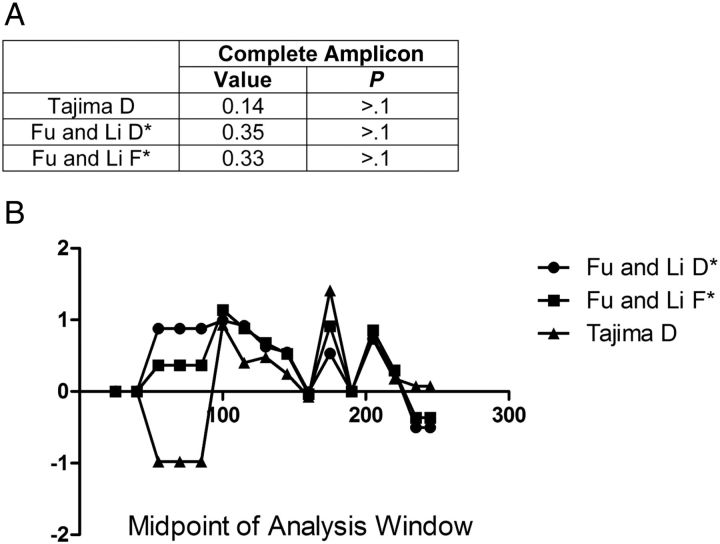
This study also showed some evolutionary selective pressure on csp within the parasite population sampled. Although all observed differences were nonsynonymous, nucleotide substitutions across the entire amplified region failed to show significant departures from neutrality (Figure 4). However, the regions containing TH2 and TH3 had weakly positive indexes for all 3 tests, suggesting an excess of intermediate-frequency alleles in a population. This is similar to previous reports in the Gambia [18]. Because of the lack of synonymous polymorphisms, dN/dS ratios were unable to be calculated. Finally, by use of a nonhierarchical island model to test for specific nucleotides under selection, we saw only 8 sites with evidence of selection (P < .05) (Supplementary Figure 1). Interestingly, all 8 loci fall within the TH2 and TH3 epitopes.
Figure 4.
Tests of neutrality to evaluate for selection on csp. A, Results for Tajima D and Fu and Li D* and F* statistics on the complete sequences of csp. B, Results of the same tests as in A, using a sliding window of 50–base pair size over the complete amplicon (slide of 15 base pairs). The y-axis is the statistical result for the index. The x-axis represents the midpoint base pair of the window evaluated. Despite peaks in the Tajima's D around TH2 and TH3, no significant evidence of selection is seen across this region (no value of the index >2). Fu and Li D* and F* tests also showed a positive trend in these regions. The positive nature of these indexes suggests an excess of intermediate-frequency alleles in a population and can result from either balancing selection or population bottlenecks.
DISCUSSION
The diversity of csp variants in Lilongwe, Malawi, was described using MPP, a novel method for a malaria population genetics study. This method is particularly useful in this context because it allows one to gain insights into infections that are polyclonal or contain minority variants (unlike Sanger sequencing, which provides only information on the dominant variant) and provides information on the frequency of each variant within an infection in a high-throughput manner [5, 20]. Multiple reports have applied MPP to the detection and characterization of rare drug-resistant human immunodeficiency virus (HIV) variants and to address questions on the dynamics of HIV quasispecies in response to selective pressure [32–36]. Here we describe a method that is capable of detecting variants that are ≥1% of the parasite population within an individual. The ability to accurately and quantitatively describe the in-host population of malaria parasite variants is critical for a more detailed understanding of the ecological interactions that selective pressures, such as drugs or vaccines, place on the parasite population.
By use of this method, we discovered 57 unique csp haplotypes in Lilongwe. However, the data suggest that this is only a portion of the actual number of circulating parasite haplotypes. It is clear from the variant accumulation curve and rarefaction curve that continued sampling would likely find additional haplotypes in the population. The extensive diversity of csp has clearly led to a significant number of different TH2 and TH3 alleles in the parasite population in Lilongwe. However, the impact of this diversity on the effectiveness of CS vaccines may be limited.
Tests for balancing selection were not significant across the complete region of CS in our study. This supports previous findings that showed no evidence of strong balancing selection for the complete csp gene and reports showing common csp alleles being maintained in an endemic Thai population for several years without declining in frequency [18, 37]. However, our study, along with previous studies, have shown peaks in Fu and Li D*, Fu and Li F*, and Tajima's D indexes around TH2 and TH3, suggesting an excess of intermediate-frequency alleles in a population [18]. This suggests that diversity at these regions is likely beneficial to parasite survival and that weak allele specific selective pressure, likely from naturally acquired immunity, does occur in these regions.
In this study, we saw no difference in the MOI between adults and children. We also saw no differences in the diversity of parasites between adults or children, using several ecological and molecular indexes of richness and diversity. However, we detected an extreme number of nonsynonymous polymorphisms, weak evidence of balancing selection at TH2 and TH3, and a high level of diversity consistent with previous African studies [38]. Given that this level of nonsynonymous substitution and diversity is far from the norm for the falciparum genome, this suggests that the selection may be a mixture of both balancing and directional selection, similar to the HLA loci [39], and only underscores the complexity of the region. Such mixed forces could maintain diversity over the long term while keeping certain alleles at higher frequency on the basis of inherently increased virulence. While this high-level of diversity in the T-cell epitopes suggests functional interplay with the human host immune system, it is not inconsistent with the hypothesis suggesting that this region may be under selective pressure in the mosquito host [37].
Variant replacement is a treatment-induced phenomenon in which vaccination drives the emergence and dominance of once-rare pathogen variants [40, 41]. This occurs because of a destabilization of the existing host-pathogen evolutionary equilibria or because of an acceleration of pathogen evolution [40, 41]. The standard explanation for treatment-induced pathogen variant replacement rests on the frequent observation that different strains respond differently to treatment (differential effectiveness) [40, 41]. However, some evidence suggests that vaccine-induced strain selection can also occur even in the case where a vaccine is equally effective against variants [40, 42]. The lack of a strong variant-specific effect by naturally acquired immunity between age groups suggests that strain selection by RTS/S may not occur quickly. To date, 3 studies have evaluated strain selection by RTS,S candidate vaccines by looking at the TH2 and TH3 epitopes [13, 15, 43]. None of these found evidence of selection occurring. However, 2 of the 3 studies showed a decrease in the MOI in vaccinated individuals. Unfortunately, these studies were somewhat methodologically limited because they used genotyping techniques that do not determine true frequencies and only detect the dominant strains in polyclonal infections. If the selective pressure of immunity on csp is weak, it is likely that the effects of selection or strain replacement may only be manifested in subtle shifts of frequency initially. In addition, these studies assessed time points shortly after initial vaccination and relatively small numbers of variants, and the time it takes to manifest strain selection may be quite long and require a larger population to detect. The observation of weak evidence of selection on TH2 and TH3 in population studies, as well as a decrease in MOI in several vaccine studies, suggests that close monitoring of strain selection by RTS/S should be conducted as more individuals receive the vaccine and duration of follow-up is extended.
In this study, we report the first population genetic analysis in malaria using MPP. This technique provides a powerful tool for evaluating the ecological and evolutionary phenomenon induced by selective pressures, such as drugs or vaccines, on the parasite population. We saw extensive diversity in csp among the parasites in Lilongwe, Malawi. However, the selective pressures put on csp by naturally acquired immunity appear to be weak. This potentially bodes well for CS-based vaccines in terms of longevity and breadth of use. However, even weak levels of selection may lead to strain replacement, which will need to be monitored as CS vaccines become more widely used.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the National Institutes of Health (grants 1RO1AI089819, KL2RR025746, and UL1RR025747).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Das P, Horton R. Malaria elimination: worthy, challenging, and just possible. Lancet. 2010;376:1515–7. doi: 10.1016/S0140-6736(10)61551-6. [DOI] [PubMed] [Google Scholar]
- 2.Feachem RG, Phillips AA, Targett GA, Snow RW. Call to action: priorities for malaria elimination. Lancet. 2010;376:1517–21. doi: 10.1016/S0140-6736(10)61500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO) WHO: Geneva, Switzerland: 2010. World malaria report: 2010. [Google Scholar]
- 4.Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming ‘vaccine resistant malaria’. Parasite Immunol. 2009;31:560–73. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juliano JJ, Porter K, Mwapasa V, et al. Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc Natl Acad Sci U S A. 2010;107:20138–43. doi: 10.1073/pnas.1007068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thera MA, Doumbo OK, Coulibaly D, et al. A field trial to assess a blood-stage malaria vaccine. N Engl J Med. 2011;365:1004–13. doi: 10.1056/NEJMoa1008115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takala SL, Coulibaly D, Thera MA, et al. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci Transl Med. 2009;1:2ra5. doi: 10.1126/scitranslmed.3000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The RTS SCTP. First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N Engl J Med. 2011;20:1863–75. doi: 10.1056/NEJMoa1102287. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J, Nussenzweig V, Nussenzweig R, Vekemans J, Leach A. From the circumsporozoite protein to the RTS, S/AS candidate vaccine. Hum Vaccin. 2010;6:90–6. doi: 10.4161/hv.6.1.9677. [DOI] [PubMed] [Google Scholar]
- 10.Kappe SH, Buscaglia CA, Nussenzweig V. Plasmodium sporozoite molecular cell biology. Annu Rev Cell Dev Biol. 2004;20:29–59. doi: 10.1146/annurev.cellbio.20.011603.150935. [DOI] [PubMed] [Google Scholar]
- 11.Lockyer MJ, Marsh K, Newbold CI. Wild isolates of Plasmodium falciparum show extensive polymorphism in T cell epitopes of the circumsporozoite protein. Mol Biochem Parasitol. 1989;37:275–80. doi: 10.1016/0166-6851(89)90159-x. [DOI] [PubMed] [Google Scholar]
- 12.Good MF, Berzofsky JA, Miller LH. The T cell response to the malaria circumsporozoite protein: an immunological approach to vaccine development. Annu Rev Immunol. 1988;6:663–88. doi: 10.1146/annurev.iy.06.040188.003311. [DOI] [PubMed] [Google Scholar]
- 13.Waitumbi JN, Anyona SB, Hunja CW, et al. Impact of RTS,S/AS02(A) and RTS,S/AS01(B) on genotypes of P. falciparum in adults participating in a malaria vaccine clinical trial. PLoS One. 2009;4:e7849. doi: 10.1371/journal.pone.0007849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalloh A, van Thien H, Ferreira MU, et al. Sequence variation in the T-cell epitopes of the Plasmodium falciparum circumsporozoite protein among field isolates is temporally stable: a 5-year longitudinal study in southern Vietnam. J Clin Microbiol. 2006;44:1229–35. doi: 10.1128/JCM.44.4.1229-1235.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alloueche A, Milligan P, Conway DJ, et al. Protective efficacy of the RTS,S/AS02 Plasmodium falciparum malaria vaccine is not strain specific. Am J Trop Med Hyg. 2003;68:97–101. [PubMed] [Google Scholar]
- 16.Hughes AL. Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics. 1991;127:345–53. doi: 10.1093/genetics/127.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jongwutiwes S, Tanabe K, Hughes MK, Kanbara H, Hughes AL. Allelic variation in the circumsporozoite protein of Plasmodium falciparum from Thai field isolates. Am J Trop Med Hyg. 1994;51:659–68. doi: 10.4269/ajtmh.1994.51.659. [DOI] [PubMed] [Google Scholar]
- 18.Weedall GD, Preston BM, Thomas AW, Sutherland CJ, Conway DJ. Differential evidence of natural selection on two leading sporozoite stage malaria vaccine candidate antigens. Int J Parasitol. 2007;37:77–85. doi: 10.1016/j.ijpara.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Alloueche A, Silveira H, Conway DJ, et al. High-throughput sequence typing of T-cell epitope polymorphisms in Plasmodium falciparum circumsporozoite protein. Mol Biochem Parasitol. 2000;106:273–82. doi: 10.1016/s0166-6851(99)00221-2. [DOI] [PubMed] [Google Scholar]
- 20.Zagordi O, Bhattacharya A, Eriksson N, Beerenwinkel N. ShoRAH: estimating the genetic diversity of a mixed sample from next-generation sequencing data. BMC Bioinformatics. 2011;12:119. doi: 10.1186/1471-2105-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colwell RK. EstimateS: statistical estimation of species richness and shared species from samples. 2011 http://viceroy.eeb.uconn.edu/estimates . Accessed 17 September 2011. [Google Scholar]
- 22.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 23.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 24.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 25.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199:693–701. doi: 10.1086/596736. [DOI] [PubMed] [Google Scholar]
- 26.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–91. [PubMed] [Google Scholar]
- 27.Hawkins VN, Auliff A, Prajapati SK, et al. Multiple origins of resistance-conferring mutations in Plasmodium vivax dihydrofolate reductase. Malar J. 2008;7:72. doi: 10.1186/1475-2875-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–60. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zouros E. Mutation rates, population sizes and amounts of electrophoretic variation of enzyme loci in natural populations. Genetics. 1979;92:623–46. doi: 10.1093/genetics/92.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tajima F. Measurement of DNA polymorphism. In: Takahata N, Clark AG, editors. Introduction to molecular paleopopulation biology. Tokyo: Japan Scientific Societies Press; 1993. pp. 37–59. [Google Scholar]
- 32.Hoffmann C, Minkah N, Leipzig J, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozera G, Abbate I, Bruselles A, et al. Massively parallel pyrosequencing highlights minority variants in the HIV-1 env quasispecies deriving from lymphomonocyte sub-populations. Retrovirology. 2009;6:15. doi: 10.1186/1742-4690-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitsuya Y, Varghese V, Wang C, et al. Minority human immunodeficiency virus type 1 variants in antiretroviral-naive persons with reverse transcriptase codon 215 revertant mutations. J Virol. 2008;82:10747–55. doi: 10.1128/JVI.01827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Archer J, Braverman MS, Taillon BE, et al. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS. 2009;23:1209–18. doi: 10.1097/QAD.0b013e32832b4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumkhaek C, Phra-Ek K, Renia L, et al. Are extensive T cell epitope polymorphisms in the Plasmodium falciparum circumsporozoite antigen, a leading sporozoite vaccine candidate, selected by immune pressure? J Immunol. 2005;175:3935–9. doi: 10.4049/jimmunol.175.6.3935. [DOI] [PubMed] [Google Scholar]
- 38.Jalloh A, Jalloh M, Matsuoka H. T-cell epitope polymorphisms of the Plasmodium falciparum circumsporozoite protein among field isolates from Sierra Leone: age-dependent haplotype distribution? Malar J. 2009;8:120. doi: 10.1186/1475-2875-8-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 2010;277:979–88. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martcheva M, Bolker BM, Holt RD. Vaccine-induced pathogen strain replacement: what are the mechanisms? J R Soc Interface. 2008;5:3–13. doi: 10.1098/rsif.2007.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gandon S, Day T. The evolutionary epidemiology of vaccination. J R Soc Interface. 2007;4:803–17. doi: 10.1098/rsif.2006.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iannelli M, Martcheva M, Li XZ. Strain replacement in an epidemic model with super-infection and perfect vaccination. Math Biosci. 2005;195:23–46. doi: 10.1016/j.mbs.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Enosse S, Dobano C, Quelhas D, et al. RTS,S/AS02A malaria vaccine does not induce parasite CSP T cell epitope selection and reduces multiplicity of infection. PLoS Clin Trials. 2006;1:e5. doi: 10.1371/journal.pctr.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.