Abstract
Early development of the hippocampus, which is essential for spatial memory and learning, is controlled by secreted signaling molecules of the Wnt gene family and by Wnt/β-catenin signaling. Despite its importance, little is known, however, about Wnt-regulated genes during hippocampal development. Here, we used the Gli3 mutant mouse extra-toes (XtJ), in which Wnt gene expression in the forebrain is severely affected, as a tool in a microarray analyses to identify potential Wnt target genes. This approach revealed 53 candidate genes with restricted or graded expression patterns in the dorsomedial telencephalon. We identified conserved Tcf/Lef-binding sites in telencephalon-specific enhancers of several of these genes, including Dmrt3, Gli3, Nfia, and Wnt8b. Binding of Lef1 to these sites was confirmed using electrophoretic mobility shift assays. Mutations in these Tcf/Lef-binding sites disrupted or reduced enhancer activity in vivo. Moreover, ectopic activation of Wnt/β-catenin signaling in an ex vivo explant system led to increased telencephalic expression of these genes. Finally, conditional inactivation of Gli3 results in defective hippocampal growth. Collectively, these data strongly suggest that we have identified a set of direct Wnt target genes in the developing hippocampus and provide inside into the genetic hierarchy underlying Wnt-regulated hippocampal development.
Keywords: Dmrt3, Gli3, hippocampus, Wnt signal, ing, Wnt8b
Introduction
The hippocampus plays important roles in spatial navigation and in the consolidation of information from short-term memory to long-term memory. It can morphologically be subdivided into the CA1 and CA3 hippocampal fields and into the dentate gyrus (DG). Hippocampal development is initiated by embryonic day 10.5 (E10.5) in the mouse, when the dorsal midline starts to invaginate and to form the cortical hem, which acts as an organizer of hippocampal development (Mangale et al. 2008). By E12.5, the invaginated dorsal midline has morphologically differentiated into the choroid plexus, the cortical hem, and the progenitor zone of the hippocampal primordium. Neurons of the hippocampal fields will arise from these progenitors at specific positions such that progenitors close to the cortical hem will give rise to DG neurons, whereas CA3 and CA1 neurons are formed by progenitors located progressively further away from the cortical hem. Several findings strongly suggest that the cortical hem is both necessary and sufficient to control the formation of these different cell types. The formation of an ectopic cortical hem leads to the generation of extrahippocampal tissue in the neocortex with an exact arrangement of DG and hippocampal fields (Mangale et al. 2008). Moreover, the hem expresses several members of the Wnt gene family, including Wnt2b, 3a, 5a, 7a, 7b, and 8b (Grove et al. 1998), which mediate its function. Inactivation of Wnt3a results in the severe reduction of hippocampal tissue (Lee et al. 2000), and mice expressing a dominant negative form of the LEF1 transcription factor completely lack the hippocampus (Galceran et al. 2000) suggesting that the Wnt3a signal is transmitted by the Wnt/β-catenin signaling pathway. This idea is further supported by the recent finding that the ectopic expression of a constitutively active form of β-catenin is sufficient to ectopically induce the formation of hippocampal cell types in the neocortex (Machon et al. 2007; Mangale et al. 2008).
While it is clear from these studies that Wnts are important regulators of hippocampal development, relatively little is yet known about direct Wnt target genes during this process. The Sp5 transcription factor gene has recently been suggested to be a candidate Wnt target gene (Fujimura et al. 2007), but its role in hippocampal development has not been analyzed. Moreover, we and others showed that Wnt/β-catenin and Bmp signaling cooperatively regulate expression of the Emx2 homeobox gene (Theil et al. 2002; Suda et al. 2010), which has important functions in hippocampal development. Emx2 mutant mice lack the granule cells of the DG (Pellegrini et al. 1996; Yoshida et al. 1997; Oldekamp et al. 2004), and the CA1 and CA3 hippocampal fields are specified and correctly positioned but reduced in size (Tole and Grove 2001). Furthermore, Emx2 is required to maintain Wnt gene expression in the cortical hem suggesting the existence of a positive feedback loop between Wnts and their targets (Muzio et al. 2005). Finally, Emx2 cooperates with the Otx2 homeobox gene (Kimura et al. 2005), which is directly regulated by Wnt signaling in the forebrain (Kurokawa et al. 2004). However, the hippocampal defects in Emx2 mutant embryos are relatively mild compared with the severe phenotype of Wnt3a and dominant negative Lef1 mutants suggesting that, in addition to Emx2, Wnt/β-catenin signaling regulates other, hitherto unknown genes during hippocampal development.
The identification of such target genes, which is required to gain a better understanding of Wnt-mediated control of hippocampal development, is, however, impeded by the coexpression of several Wnt genes in the dorsal midline and potential redundancies between these factors (Fotaki et al. 2010). Here, we used extra-toes (XtJ) mutant mice, which carry a null mutation in the Gli3 gene (Büscher et al. 1998), as a tool to identify potential Wnt target genes in the developing hippocampus. XtJ/XtJ embryos lack the cortical hem and the hippocampus (Franz 1994; Grove et al. 1998; Theil et al. 1999) coinciding with a loss of Wnt3a expression and with a severe reduction in Wnt8b expression in the forebrain (Grove et al. 1998; Theil et al. 2002; Theil 2005). Using a microarray screen, we compared the gene expression profiles in the telencephalon of wild-type and XtJ/XtJ embryos at E10.5 when hippocampal development starts. We identified 53 genes with either restricted or graded expression in the dorsomedial telencephalon. Interestingly, the majority of these genes encode either transcription factors or components of the Wnt/β-catenin signaling cascade. Using DNA binding and reporter gene analysis and an ex vivo explant assay, we demonstrate that several of these genes, namely Dmrt3, Gli3, Nfia, and Wnt8b, are direct targets of Wnt/β-catenin signaling. Finally, we show that conditional inactivation of Gli3 in the dorsal telencephalon after E10.5 results in a size reduced and disorganized hippocampus indicating that Gli3 mediates aspects of Wnt/β-catenin signaling. Taken together, these findings provide insights into the genetic circuitry underlying Wnt-controlled hippocampal development.
Materials and Methods
Mice
XtJ/+ animals were kept on a mixed C57Bl6/C3H background. For microarray analysis, embryos were genotyped as described (Maynard et al. 2002). For in situ hybridization, heterozygous and wild-type embryos which did not show differences were used as control embryos, and forebrain morphology was used to distinguish them from XtJ/XtJ embryos (Theil et al. 1999). Emx1Cre and Gli3flox/flox mouse lines have been described previously (Gorski et al. 2002; Blaess et al. 2008). For Emx1Cre;Gli3flox/flox conditional embryos, Gli flox/flox, Gli3flox/+,Emx1Cre, and Gli3flox/+ embryos were used as controls. All experimental procedures involving mice were performed in accordance with local guidelines. For each marker and each stage, 3–5 embryos were analyzed.
Complementary Deoxyribonucleic Acid Microarray Analysis
The telencephali of 3 E10.5 XtJ/XtJ embryos or wild-type littermates were pooled and RNA was isolated using an RNeasy Micro Kit (Qiagen). Microarray analysis using Affymetrix GeneChip Mouse Genome 430 2.0 arrays was performed at the Sir Henry Wellcome Functional Genomics Facility (SHWFGF, Institute of Biomedical and Life Science, University of Glasgow). The resulting data were analyzed with the fully automated data analysis FunAlyse pipeline including a Robust Multichip Average (RMA) preprocessing step followed by the identification of differentially expressed genes using the RankProducts (RP) method (Breitling et al. 2004).
In Situ Hybridization and Immunohistochemistry
In situ hybridization and immunostaining on 12.5-μm coronal paraffin sections of E10.5 and E12.5 wild-type and XtJ/XtJ mouse brains were performed as described previously (Theil 2005). Digoxigenin-labeled antisense probes were generated from the following complementary deoxyribonucleic acid clones: Apcdd1 (Jukkola et al. 2004), Axin2 (Lustig et al. 2002), Bcl11a (IMAGE: 2631265), Cux2 (Zimmer et al. 2004), Dmrt3 (Smith et al. 2002), Efbn1 (Flenniken et al. 1996), E330013P04Rik (Genepaint riboprobe 1402), Gli3 (Hui et al. 1994), Lhx2 (Liem et al. 1997), Lrrn1 (IMAGE: 4972803), Nfib (Genepaint riboprobe 629), Nfix (Genepaint riboprobe 549), Nr4a2 (Quina et al. 2009), Nrp2 (Galceran et al. 2000), Rspo1 (IMAGE:1365431), Rspo2 (Genepaint riboprobe 844), Rspo3 (IMAGE: 40131232), Scip1 (Frantz et al. 1994), Sfrp1 (Genepaint riboprobe 418), Sp5 (Harrison et al. 2000), Tnfsfr19 (Pispa et al. 2003), Vegfc (IMAGE: 5002720), and Wnt9a (Summerhurst et al. 2008).
For immunofluorescence, antibodies against green fluorescent protein (GFP) (1:1000; Abcam), Nf1a (1:1000; Active Motif), and Prox1 (1:1000; RELIATech) were used followed by a nuclear counterstain with TO-PRO-1 (1:3000; Invitrogen).
Plasmid Construction and Mutagenesis
All genomic DNA fragments were generated via polymerase chain reaction (PCR) using wild-type genomic DNA (for oligonucleotides, see Supplementary Table 1). Enhancer sequences were subcloned using a TOPO TA cloning kit (Invitrogen) and verified by sequencing. Putative TCF/Lef1-binding sites were mutated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) (for oligonucleotides used in mutagenesis, see Supplementary Table 2). All mutations were confirmed by sequencing. To test for enhancer activity, wild-type and mutant regulatory elements were subcloned into the lacZ reporter gene vector pGZ40 upstream of a human β-globin minimal promoter (Yee and Rigby 1993). For generating transgenic embryos, the enhancer/reporter fragment was released from the plasmid backbone by digestion with the restriction enzymes indicated in Supplementary Table 1 and gel purified.
Electrophoretic Mobility Shift Assay
Electrophoretic mobility assays were performed as described previously (Theil et al. 2002) using purified GST and GST-LEF1 protein. For oligonucleotides covering the wild-type or mutated Tcf/Lef-binding sites, see Supplementary Table 3. The exchanged nucleotides in the mutated forms (AA or TT → GG or CC) are underlined. Wild-type and Tcf/Lef-binding site mutant oligonucleotides were used as specific and unspecific competitors, respectively, in a 10- to 100-fold molar excess.
Transgenic Embryos
Transgenic embryos were generated by microinjection of fertilized eggs from B6CBAF1/Crl crosses (Charles River) and were identified by PCR using extraembryonic yolk sac or tail DNA. Expression of the transgene was analyzed by staining E10.5 or E11.5 embryos for β-galactosidase activity as described previously (Theil et al. 1998).
In Utero Electroporation
E12.5 pregnant mice were anesthetized with sodium pentobarbitone at 50 mg/g of body weight, and the uterine horns were exposed. LacZ reporter gene plasmids and a GFP expression plasmid were coinjected into the lateral ventricle at 1 mg/mL each with a glass micropipette. The embryo in the uterus was placed between CUY650 tweezer-type electrodes (Nepagene). A CUY21E electroporator (Nepagene) was used to deliver 6 pulses (30 V, 50 ms each) at intervals of 950 ms. The uterine horns were placed back into the abdominal cavity, and embryos were allowed to develop for 36 h before further processing for immunofluorescence. For each construct and time point, at least 4 different embryos were analyzed.
Explant Culture
Organotypic slice cultures of the E13.5 embryonic mouse telencephalon were prepared as previously described (Magnani et al. 2010). Brain slices were cultured on polycarbonate culture membranes (8-μm pore size; Corning Costar) in organ tissue dishes containing 1 ml of medium (Neurobasal/B-27 [Gibco] supplemented with glutamine, glucose, penicillin, and streptomycin) in the presence of either dimethyl sulfoxide (DMSO) or of 5, 25 or 50 μM CHIR99021 (CHIR) (Cambridge BioScience). Slices were cultured for 24 h, fixed with 4% paraformaldehyde, and processed for in situ hybridization as described above.
Results
Microarray Screening of Telencephali of Wild-Type and XtJ/XtJ Mutant Embryos
To identify new potential Wnt target genes in the developing hippocampus, we took advantage of the Gli3 mutant mouse extra-toes (XtJ) that shows severe downregulation of Wnt gene expression in the forebrain from E8.5 onwards (Grove et al. 1998; Theil et al. 2002; Theil 2005; Fotaki et al. 2011). To this end, we used microarrays to compare gene expression profiles between the telencephalon of wild-type and XtJ/XtJ E10.5 embryos. At this stage, the dorsal midline starts to invaginate thereby initiating hippocampal development. To define differentially expressed genes in our microarray experiment, we used the unique nonparametric RP statistical test, which includes an estimate of false discovery rates (FDRs) (Breitling et al. 2004). With statistically significant expression fold changes (meanFCnomN) of >±2.0 and an FDR of <2%, 300 downregulated, and 153 upregulated probe sets could be identified (Supplementary Tables 4 and 5). These probe sets are likely to contain genes that are directly regulated by the Gli3 transcription factor. Since Gli3 is believed to act predominantly as a repressor in the dorsal telencephalon (Fotaki et al. 2006), these direct Gli3 target genes are likely to be upregulated in XtJ/XtJ embryos. Gli3 can also affect gene regulation indirectly, for example, by regulating other transcription factors or by controlling the expression of signaling molecules such as Wnts and Bmps. Due to the downregulation of Wnt signaling in XtJ/XtJ embryos, we expected potential Wnt target genes to be among the downregulated probe sets and focused our further analyses on this group. For further refinement, the expression patterns of the downregulated probe sets were systematically analyzed using public databases for in situ hybridization data (http://www.genepaint.org; http://www.informatics.jax.org) under the condition that potential Wnt targets should show either restricted expression in dorsal midline tissues or graded expression in the dorsal telencephalon. These expression analyses helped to identify 53 genes, which meet these criteria (Table 1 and Supplementary Fig. 1). Noticeably, genes encoding proteins involved in 2 cellular functions are enriched among these candidate Wnt target genes. The first group consists of signaling molecules (n = 24; 45.3%) including several members of the Wnt/β-catenin signaling pathway (n = 12; 22.6%), while the second group codes for transcriptional regulators (n = 21; 39.6%).
Table 1.
Candidate Wnt target genes
| MeanFCn | Gene symbol | Gene title | Public ID | TFa | SMb | Wnt |
| −455.34 | Gli3 | GLI-Kruppel family member GLI3 | AW546010 | TF | ||
| −10.52 | Dmrt3 | Double sex and mab-3-related transcription factor 3 | AV298122 | TF | ||
| −10.26 | Lrrn1 | Leucine-rich repeat protein 1, neuronal | NM_008516 | |||
| −9.32 | Nfix | Nuclear factor I/X | AW049660 | TF | ||
| −6.81 | Wnt8b | Wingless-related MMTV integration site 8b | BG866612 | SM | Wnt | |
| −6.53 | Emx2 | Empty spiracles homolog 2 (Drosophila) | BG072869 | TF | ||
| −6.46 | Emx1 | Empty spiracles homolog 1 (Drosophila) | BB741819 | TF | ||
| −6.14 | Rspo1 | R-spondin homolog (Xenopus laevis) | NM_138683 | SM | Wnt | |
| −6.00 | Nr4a2 | Nuclear receptor subfamily 4, group A, member 2 | BB703394 | TF | ||
| −5.47 | Rspo2 | R-spondin 2 homolog (Xenopus laevis) | BG067392 | SM | Wnt | |
| −4.62 | Wnt9a | Wingless-type MMTV integration site 9A | AV273409 | SM | Wnt | |
| −4.55 | Bmp6 | Bone morphogenetic protein 6 | NM_007556 | SM | ||
| −4.44 | Wnt3a | Wingless-related MMTV integration site 3A | NM_009522 | SM | Wnt | |
| −4.20 | Sp5 | Transacting transcription factor 5 | NM_022435 | TF | ||
| −4.10 | E330013P04Rik | RIKEN cDNA E330013P04 gene | BG069958 | |||
| −4.08 | Msx2 | Homeobox, msh-like 2 | NM_013601 | TF | SM | |
| −3.81 | Otx1 | Orthodenticle homolog 1 (Drosophila) | BB438279 | TF | ||
| −3.80 | Efnb1 | Ephrin B1 | NM_010110 | SM | ||
| −3.70 | Tnfrsf19 | Tumor necrosis factor receptor superfamily, member 19 | NM_013869 | SM | ||
| −3.68 | Rspo3 | R-spondin 3 homolog (Xenopus laevis) | BG072958 | SM | Wnt | |
| −3.56 | Bcl11a | B-cell CLL/lymphoma 11A (zinc finger protein) | BB424718 | TF | ||
| −3.47 | Lhx2 | LIM homeobox protein 2 | NM_010710 | TF | ||
| −3.33 | Igfbpl1 | Insulin-like growth factor–binding protein-like 1 | BM935068 | SM | ||
| −3.21 | Ntrk3 | Neurotrophic tyrosine kinase, receptor, type 3 | BM245880 | SM | ||
| −3.34 | Dmrt4 (1a) | Double sex and mab-3-related transcription factor like family A1 | BB461344 | TF | ||
| −3.13 | Dkk2 | Dickkopf homolog 2 (Xenopus laevis) | NM_020265 | SM | Wnt | |
| −3.12 | Apcdd1 | Adenomatosis polyposis coli downregulated 1 | BB770932 | SM | Wnt | |
| −3.11 | Sfrp1 | Secreted frizzled-related sequence protein 1 | BI658627 | SM | Wnt | |
| −3.03 | Fzd1 | Frizzled homolog 1 (Drosophila) | BB259670 | SM | Wnt | |
| −2.92 | Rfx4 | Regulatory factor X, 4 (influences HLA class II expression) | AV255458 | TF | ||
| −2.91 | Dmrt5 (a2) | Double sex and mab-3-related transcription factor like family A2 | BB292639 | TF | ||
| −2.71 | Fgfr3 | Fibroblast growth factor receptor 3 | NM_008010 | SM | ||
| −2.70 | Cadps | Ca<2+>-dependent activator protein for secretion | NM_012061 | |||
| −2.70 | Tpbg | Trophoblast glycoprotein | BQ177165 | |||
| −2.63 | Nfib | Nuclear factor I/B | Y07687 | TF | ||
| −2.47 | Tgfb2 | Transforming growth factor, beta 2 | BF144658 | SM | ||
| −2.45 | Anp32a | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | AF022957 | |||
| −2.44 | Cachd1 | Cache domain containing 1 | BB730977 | |||
| −2.41 | Id4 | Inhibitor of DNA-binding 4 | BB121406 | TF | ||
| −2.40 | Dach1 | Dachshund 1 (Drosophila) | BG075820 | TF | ||
| −2.33 | Axin2 | Axin2 | BB398993 | SM | Wnt | |
| −2.32 | Hod | Homeobox only domain | BC024546 | TF | ||
| −2.28 | Kcnd2 | Potassium voltage-gated channel, Shal-related family, member 2 | BB051684 | |||
| −2.21 | Oprs1 | Opioid receptor, sigma 1 | BM220110 | SM | ||
| −2.19 | Emx2os | Empty spiracles homolog 2 (Drosophila) opposite strand | AV351746 | |||
| −2.18 | Elavl2 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) | BB105998 | |||
| −2.16 | Vegfc | Vascular endothelial growth factor C | AW228853 | SM | ||
| −2.15 | Zic2 | Zic finger protein of the cerebellum 2 | NM_009574 | TF | ||
| −2.15 | Cux2 (CutL2) | Cut-like 2 (Drosophila) | BB129488 | TF | ||
| −2.14 | Fzd8 | Frizzled homolog 8 (Drosophila) | BB086994 | SM | Wnt | |
| −2.13 | Bmp5 | Bone morphogenetic protein 5 | AV032115 | SM | ||
| −2.12 | Epha4 | Eph receptor A4 | AK013481 | SM | ||
| −2.10 | Nfia | Nuclear factor I/A | AW556192 | TF |
Transcription factor.
Signaling molecule.
Disrupted Gene Expression in XtJ/XtJ Mutants
Our initial expression pattern screen was mainly based upon public in situ hybridization data from wild-type E14.5 embryos. To gain further insights into the expression of our candidate genes during hippocampal development, we performed in situ hybridization and immunofluorescence analyses on coronal sections of wild-type and XtJ/XtJ embryos. While the dorsal midline has only just started to invaginate at E10.5, by E12.5 it has developed into morphologically distinct domains, including the choroid plexus, the cortical hem, and the hippocampal primordium allowing us to identify regionally restricted expression patterns. Therefore, we preferentially present in situ hybridization and immunofluorescence data from E12.5 embryos but also provide some data on E10.5 embryos. With these analyses, we mainly focused on the genes, which encode transcription factors or Wnt signaling components, which are overrepresented in our candidate group, but we have also included data on some of the most highly downregulated genes.
Genes Encoding Transcription Factors
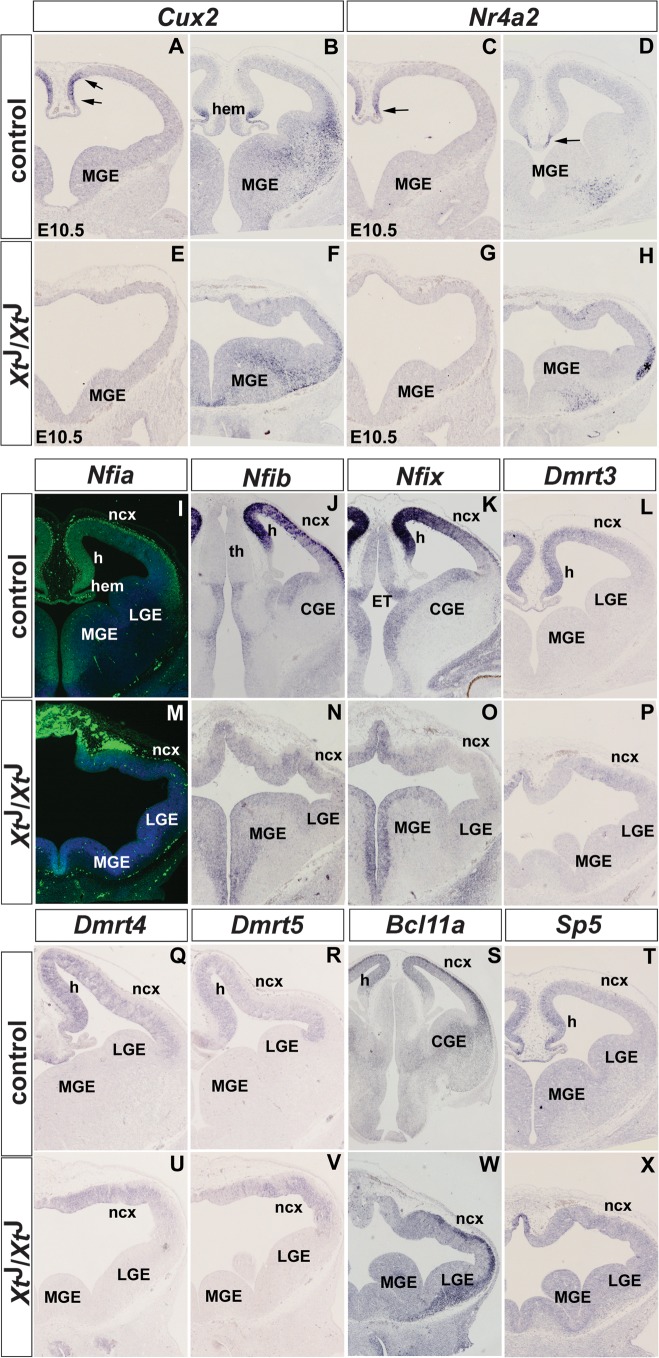
Based on their expression patterns, the transcription factor encoding genes fall into 2 subgroups. Genes in the first subgroup comprise the homeodomain transcription factor Cux2 and the orphan nuclear receptor Nr4a2 (Nurr1) and show restricted expression in the dorsal midline. Both of these genes are already expressed in the dorsomedial telencephalon at E10.5 and by E12.5 their expression becomes further restricted to the cortical hem (Cux2) and to the boundary between hem and choroid plexus (Nr4a2) (Fig. 1A–D). Consistent with the agenesis of midline structures in XtJ/XtJ embryos, these expression domains are lost at both stages, while Cux2 and Nr4a2 expression in basal progenitors and in the MGE mantle, respectively, remain unaffected (Fig. 1E–H).
Figure 1.
Expression of putative Wnt target genes encoding transcription factors in the dorsomedial telencephalon of wild-type and XtJ/XtJ embryos. A,C,E,G show expression in E10.5 embryos and B,D,F,H,I–X in E12.5 embryos. (A,B) Cux2 expression is confined to the dorsomedial telencephalon (arrows) and to the cortical hem in E10.5 and E12.5 embryos, respectively. (C,D) Nr4a2 is expressed ventrally to Cux2 at E10.5 (arrow), and its expression becomes restricted to the region between the hem and the choroid plexus (arrow) at E12.5. (E–H). These expression domains are lost in XtJ/XtJ embryos, while Cux2 and Nr4a2 expression in the medial ganglionic eminence (MGE) is not affected. (I–K) Nfia, Nfib, and Nfix show graded expression in the dorsomedial telencephalon and are strongly expressed in preplate neurons. (M–O) XtJ/XtJ embryos lack these expression domains, but Nfib and Nfix expression in the ventral telencephalon is not affected. (L,Q,R) Dmrt3, Dmrt4, and Dmrt5 show graded expression in the hippocampal anlagen and lower expression levels in the developing neocortex. (P,U,V) These genes have reduced expression levels in XtJ/XtJ embryos although expression remains in the mutant neocortex (ncx). (S,W) Bcl11a is expressed at high levels in the preplate and in a graded manner in the dorsomedial telencephalon. In XtJ/XtJ embryos, its dorsomedial expression is affected but not its expression in the preplate. (T,X) Sp5 expression in the dorsomedial telencephalon of wild-type embryos is lost in XtJ/XtJ embryos. Abbreviations: CGE, caudal ganglionic eminence; ET, eminentia thalami; h, hippocampus; th, thalamus.
The second subgroup of transcription factor genes show graded expression in the dorsomedial telencephalon and contains several genes, which were previously shown to be downregulated in XtJ/XtJ embryos, including Emx1/2 (Theil et al. 1999; Tole et al. 2000), Msx2 (Theil et al. 1999), Otx1 (Theil et al. 1999), Lhx2 (Fotaki et al. 2006), and Zic2 (Okada et al. 2008), while the expression of other transcription factors have not been reported in XtJ/XtJ embryos previously. Among this latter group, several members of the nuclear factor I (Nfi) and Dmrt families are downregulated in XtJ/XtJ embryos according to our microarray data (see Fig. 1). Nfi factors act in a cell type–specific and promoter-specific manner to activate or repress the expression of target genes (Gronostajski 2000). During development, all members of this gene family are expressed in unique but widely overlapping patterns (Chaudhry et al. 1997). Nfia, Nfib, and Nfix are expressed in the forebrain with a high medial to low lateral gradient in dorsal telencephalic progenitor cells and at high uniform level in preplate neurons (Fig. 1I–K). Both of these expression domains are completely abolished in XtJ/XtJ embryos, while Nfib and Nfix expression in the MGE is maintained (Fig. 1M–O). The Dmrt gene family has been implicated in sexual development in vertebrates and invertebrates, and several members show restricted expression patterns during murine embryogenesis suggesting that these genes might regulate other developmental processes (Hong et al. 2007). In our microarray screen, we identified Dmrt3, 4, and 5 as downregulated genes (Tab. 1). Indeed, all 3 genes show restricted expression in the dorsal telencephalon with Dmrt3 and 4 having a strongly graded expression in the hippocampal primordium (Fig. 1L,Q), while the Dmrt5 expression gradient is less pronounced (Fig. 1R). In XtJ/XtJ embryos, these genes show reduced expression although weak expression remains in the neocortex (Fig. 1P,U,V). In addition, we identified several other transcription factors with graded telencephalic expression. Bcl11a (Ctip1) is strongly expressed in preplate neurons and shows weaker but graded expression in the dorsomedial telencephalon (Fig. 1S and Leid et al. 2004). This expression in hippocampal progenitors is lost in XtJ/XtJ embryos, while neuronal Bcl11a expression is maintained (Fig. 1W). Finally, Sp5 transcripts are confined to the dorsomedial telencephalon (Fig. 1T) as described previously (Harrison et al. 2000; Fujimura et al. 2007). Here, we extend this finding by showing that XtJ/XtJ embryos lack this expression domain (Fig. 1X).
Genes Encoding Wnt Signaling Components
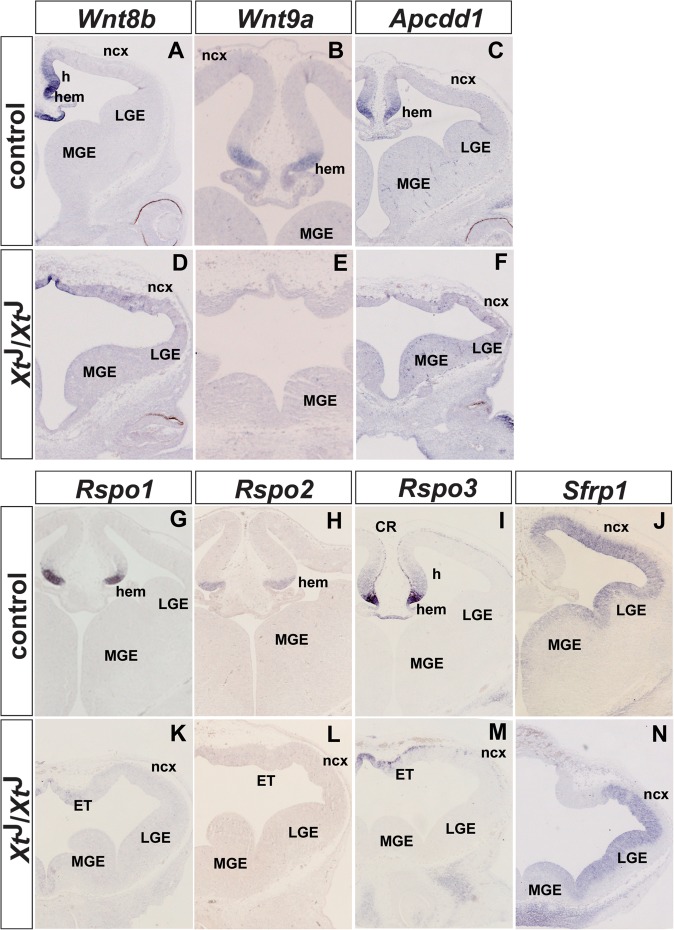
The second group of overrepresented genes that were found to be downregulated in our microarray analysis is associated with Wnt/β-catenin signaling. This group includes the Wnt ligands Wnt3a, Wnt8b, and Wnt9a (Table 1 and Fig. 2A,B,D,E) and several modulators of Wnt signaling, including Dkk2, Apcdd1, Rspo1-3, and Sfrp1. Except for Sfrp1, the expression of these genes is restricted to the cortical hem with Rspo3 showing additional weak expression in hippocampal progenitors adjacent to the hem and in Cajal–Retzius cells (Fig. 2C,G–J). Consistent with their restricted expression in the midline region, the expression of Wnt8b, Wnt9a, Apcdd1, Rspo1-3, and Sfrp1 is lost or severely reduced in XtJ/XtJ embryos (Fig. 2F,K–N). The expression of multiple Wnt modulators in the cortical hem strongly suggests a requirement for tight regulation of Wnt/β-catenin signaling during hippocampal development. Interestingly, Sfrp1 is not expressed in the hippocampal primordium and shows a lateral high to medial low gradient of expression in the neocortex (Fig. 2J,N) suggesting that Sfrp1 might have a role in ensuring graded Wnt/β-catenin signaling in the neocortex.
Figure 2.
Expression of Wnt genes and of several genes encoding Wnt signaling components in the dorsal telencephalon of E12.5 wild-type (A–C,G–J) and XtJ/XtJ (D–F,K–N) embryos. (A,D) Wnt8b is expressed in the cortical hem, the hippocampal anlagen, and in the roof plate of wild-type embryos, while Wnt8b transcripts are confined to the roof plate of the diencephalon. (B,C,E,F) Wnt9a and Apcdd1 expression are restricted to the cortical hem in wild-type embryos but are not expressed in XtJ/XtJ embryos. (G–I,K–M) Rspo1, Rspo2, and Rspo3 are strongly expressed in the cortical hem and Rspo3 mRNA is weakly expressed in hippocampal progenitors and in Cajal–Retzius (CR) cells. In XtJ/XtJ embryos, only Rspo3 expression could be detected in the diencephalic midline. (J,N) Wild-type embryos show strong Sfrp1 expression in the neocortex but lack Sfrp1 expression in the hippocampus.
Miscellaneous Genes
Finally, we analyzed the expression of some of the most highly downregulated genes, including the neuronal leucine-rich repeat protein 1 (Lrrn1) and E330013P04Rik. The tumor necrosis factor receptor superfamily member 19 (Tnfrsf19) was recently identified as a Wnt target gene in human mesenchymal stem cells where it mediates Wnt function in controlling osteoblast differentiation (Qiu et al. 2010). Vascular endothelial growth factor C (Vegfc) is inducible by Wnts in 3T3 fibroblasts (Longo et al. 2002). All these genes show graded expression in the dorsomedial telencephalon and their expression is lost in XtJ/XtJ embryos (Supplementary Fig. 2).
Taken together, our microarray and subsequent gene expression analyses have identified a large group of genes with graded or restricted expression patterns in the dorsomedial telencephalon. Among these genes, transcription factors and signaling molecules, in particular, Wnt signaling components, predominate.
Prediction of Tcf/Lef1-Binding Sites in Regulatory Elements of Candidate Wnt Target Genes
We next tested whether at least some of the genes, we identified in our microarray screen are directly regulated by Wnt/β-catenin signaling. The spatial and temporal expression of vertebrate developmental control genes is often regulated by enhancers located up to 1 Mb from the transcriptional start site (Williamson et al. 2011) significantly complicating the analyses of gene regulation during vertebrate development. Interestingly, a recent publication systematically described the systematic analyses of several hundred base pair long sequences, which are ultraconserved between vertebrate species and identified several of those as enhancer elements which can direct expression of a lacZ reporter gene in the dorsal telencephalon (Paparidis et al. 2007; Visel et al. 2008) (VISTA Enhancer Browser: http://enhancer.lbl.gov). We found that this database contained regulatory elements associated with 5 of our candidate Wnt-regulated genes that fell into the Wnt signaling or transcription factor groups, namely Cux2, Dmrt3, Gli3, Nfia, and Wnt8b.
Since Wnt/β-catenin signaling leads to the binding of β-catenin to Tcf/Lef transcription factors which in turn activate or repress Wnt target genes, we were interested to find out whether enhancer elements associated with these 5 candidate Wnt target genes contain Tcf/Lef-binding sites. To this end, we first used the probabilistic methods “Multiple Expectation Maximization for Motif Elicitation” (MEME) and “Motif Alignment and Scan Tool (MAST)” to predict a potential Tcf-binding motif from known in vivo Wnt target genes. This resulted in the identification of a 7 bp binding motif with strong resemblance to the previously identified Tcf/Lef consensus site (Bottomly et al. 2010) (Supplementary Fig. 3). Second, we used this binding motif to predict potential Tcf/Lef-binding sites within enhancer sequences of our candidate genes. To increase the probability for the functionality of these sites, we also analyzed their evolutionary conservation. Whenever available, sequences from human, mouse, chimpanzee, chicken, and zebra fish were included in this analysis. As a proof of principle, we also analyzed the Emx2 forebrain enhancer, which is regulated by Wnt/β-catenin signaling (Theil et al. 2002; Suda et al. 2010). This approach revealed, besides several nonconserved sites, one absolutely conserved putative Tcf/Lef binding in all 6 tested enhancers.
Recombinant Lef1 Protein Binds to the Tcf/Lef Sites Identified in the Telencephalon Enhancers
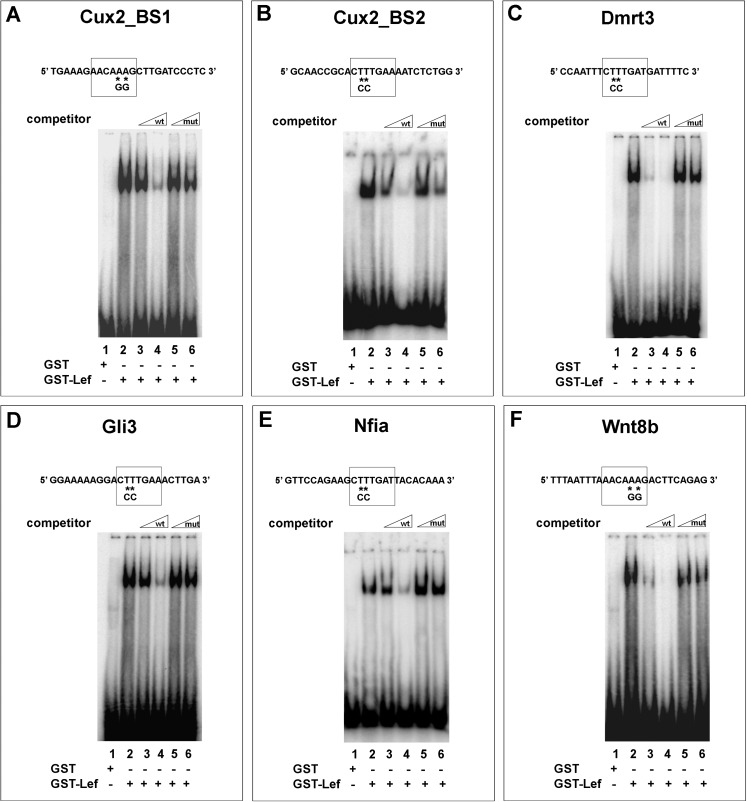
To start to analyze the functionality of these predicted Tcf/Lef-binding sites, we first performed electrophoretic mobility shift assays (EMSAs) using recombinant full-length Lef1 protein fused to glutathione-S-transferase (GST-Lef1) and radioactively labeled double-stranded oligonucleotides containing the predicted Lef/Tcf-binding motif. In this assay, GST-Lef1 fusion protein bound to all 6 Tcf/Lef-binding sites tested (Fig. 3A–F, lane 2). To further analyze the specificity and affinity of DNA binding, competition assays were conducted in the presence of an excess of unlabeled wild-type oligonucleotide (competitor). For all 6 Tcf/Lef-binding sites, these specific competition experiments resulted in progressively diminished binding of GST-Lef1 fusion protein with increasing amounts of competitor (Fig. 3A–F, compare lanes 2–4) and revealed binding with the highest affinity to sites in the Dmrt3 and Wnt8b enhancer elements (Fig. 3C,F). In contrast, competition with unlabeled oligonucleotides containing 2 point mutations in the central T stretch of the binding motif, which abolish Tcf/Lef1 binding (Tetsu and McCormick 1999), had no or little effect on the formation of the DNA protein complex (Fig. 3A–F, compare lanes 2, 5, and 6). Thus, all the 6 Tcf/Lef-binding sites can specifically bind to recombinant GST-Lef1 fusion protein in vitro.
Figure 3.
Electromobility shift assays showing in vitro binding of recombinant Lef1 protein to telencephalic enhancers. GST-Lef1 protein binds to oligonucleotides of the Cux2 (A,B), Dmrt3 (C), Gli3 (D), Nfia (E), and Wnt8b (F) telencephalic enhancers (lane 2). In each case, complex formation is progressively competed by increasing amounts of wild-type enhancer oligonucleotide (lanes 3 and 4) but not by oligonucleotides containing point mutations in the Tcf/Lef-binding site (lanes 5 and 6).
In Vivo Function of Tcf/Lef-Binding Sites in the Telencephalic Enhancers
Next, we analyzed whether these binding sites are functional in vivo. To this end, wild-type or Tcf mutant regulatory elements, which carried the same point mutations as used in the EMSA analyses to abolish Tcf/Lef binding, were linked to a lacZ reporter gene under the control of a human β-globin minimal promoter (Yee and Rigby 1993). To test for enhancer activity, we either used these reporter gene constructs to generate transient transgenic embryos or we electroporated them into the telencephalon of E12.5 embryos in utero.
Transcription Factor Genes
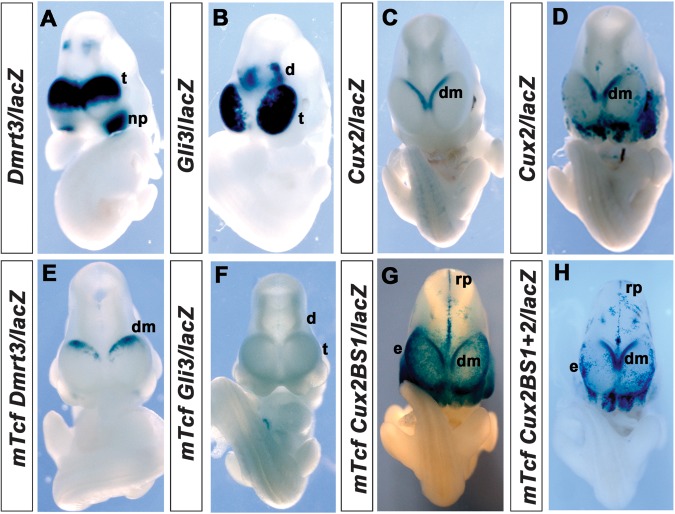
We first focused our analyses on the regulation of transcription factor genes identified in our screen. Dmrt3 represents one of the most highly downregulated genes in our microarray analysis. An ultraconserved element, which is located in the intergenic region between Dmrt1 and Dmrt3, directs lacZ reporter gene expression in the dorsal telencephalon and in the nasal placode (Visel et al. 2008). Since Dmrt1 is not expressed in the forebrain (Raymond et al. 1999), this element is likely to control telencephalic Dmrt3 expression. Moreover, the homologous mouse region also has strong enhancer activity in the dorsal telencephalon and in the nasal placode (n = 3/6 transgenic embryos) (Fig. 4A). In contrast, point mutations in the single, conserved Tcf-binding site contained in this regulatory element abolish enhancer activity in the nasal placode and strongly reduce enhancer activity in the dorsal telencephalon where it remains confined dorsomedially (n = 5/8) (Fig. 4E) suggesting that the Tcf/Lef-binding site is required for full enhancer activity in the dorsal telencephalon.
Figure 4.
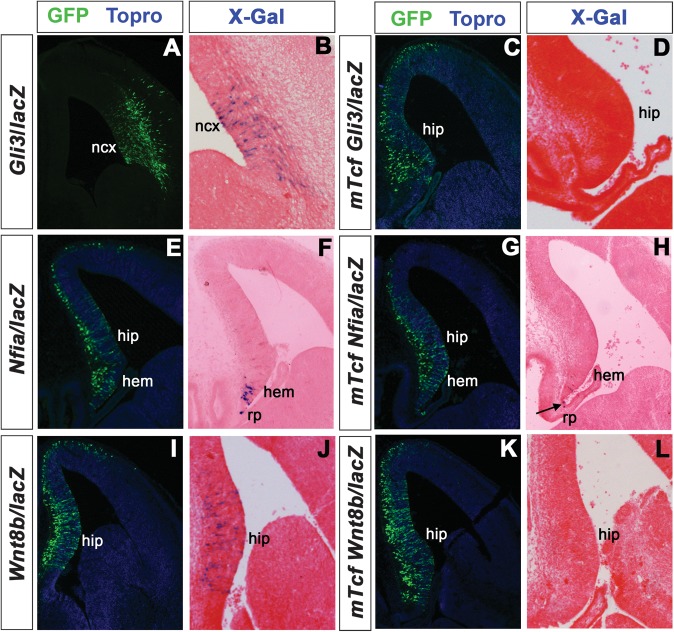
Role of the Tcf/Lef1-binding sites for in vivo enhancer activity in the dorsal telencephalon. (A) The Dmrt3 enhancer is active in the dorsomedial telencephalon (t) and in the nasal placode (np). (E) Mutation of the Tcf-binding site leads to weak lacZ staining in the dorsomedial telencephalon (dm). (B) Embryo transgenic for the Gli3/lacZ reporter gene construct shows enhancer activity in the dorsal telencephalon and in the diencephalon (d). (F) Mutation of the Tcf-binding site abolishes enhancer activity in the forebrain. (C,D) Staining in embryos transgenic for the Cux2/lacZ reporter gene is detected in the dorsomedial telencephalon. The enhancer also shows patchy activity in the ectoderm (e), roof plate (rp), and diencephalon. (G,H) Transgenic embryos carrying a mutation in the conserved Tcf/Lef-binding site (G) or mutations in both binding sites (H) consistently show strong expression in the cortical hem, ectoderm, roof plate, and diencephalon. Note that the embryos in G and H were stained for 3 h, while the embryos carrying the wild-type construct (C,D) were stained o/n.
A human 438 bp Gli3 intragenic enhancer directs strong reporter gene expression in the E11.5 telencephalon and in the dorsal thalamus (Paparidis et al. 2007; Visel et al. 2008). We show here that the corresponding murine regulatory element, which is completely conserved between humans and mice gives rise to an identical pattern of reporter gene activity in E10.5 embryos (n = 2/5) (Fig. 4B). In contrast, mutations in the single Tcf/Lef-binding site in this enhancer completely abolish enhancer activity (n = 4/4) (Fig. 4F). We also electroporated the Gli3 reporter gene construct together with a GFP expression plasmid as an electroporation control, into the telencephalon of E12.5 embryos in utero and analyzed electroporated embryos for enhancer activity 36 h later. Electroporation of the wild-type construct into the neocortex resulted in strong X-Gal staining in electroporated cells within the ventricular zone (n = 6 electroporated embryos) but not in differentiating neurons (Fig. 5A,B). This pattern reflects the expression of the endogenous Gli3 gene, which is expressed in cortical progenitor cells but not in differentiating neurons (Magnani et al. 2010). In contrast, embryos electroporated with the Gli3 reporter gene construct containing the mutated Tcf/Lef-binding site did not show enhancer activity (n = 4 electroporated embryos) (Fig. 5C,D). Taken together, the reporter gene analyses in transgenic mice and the in utero electroporation assay both show an absolute requirement for the Tcf/Lef-binding site in regulating the activity of the Gli3 forebrain enhancer.
Figure 5.
In utero electroporation of lacZ reporter gene constructs into the telencephalon. Shown are GFP immunofluorescences (A,C,E,G,I,K) to detect electroporated cells and X-Gal staining to reveal enhancer activity (B,D,F,H,J,L). (A–D) Electroporation of a Gli3/lacZ reporter gene construct results in reporter gene activation in cortical progenitor cells (A,B), while enhancer activity is abolished by mutations in the Tcf/Lef-binding site (C,D). (E–H) The Nfia telencephalic enhancer is active in the cortical hem and in roof plate (rp) cells (E,F). X-Gal staining is confined to roof plate cells (arrow) after mutating the Tcf/Lef-binding site (G,H). (I–L) Embryo electroporated with a Wnt8b/LacZ reporter gene construct shows X-Gal staining in the hippocampus (hip) (I,J). The Tcf/Lef mutant Wnt8b/LacZ reporter construct lacks enhancer activity in the dorsomedial telencephalon (K,L).
Next, we determined whether enhancers with more restricted activity in the dorsal telencephalon are dependent on Tcf/Lef binding. To this end, we analyzed the Cux2 and Nfia regulatory elements, which are specifically active in the dorsomedial telencephalon (Visel et al. 2008). Embryos carrying a transgene in which the murine Cux2 telencephalic enhancer is coupled to a lacZ gene (Cux2/lacZ) show restricted reporter gene expression in the dorsomedial telencephalon though with strong variations in staining intensity ranging from weak and patchy (n = 6/14 lacZ stained embryos; 43%) to strong staining (n = 8/14 lacZ stained embryos; 57%) (Fig. 4C,D). In contrast to the corresponding human element, this enhancer consistently gave rise to patchy and variable X-Gal staining in the roof plate, ectoderm, hindbrain, diencephalon, midbrain, and branchial arches. Bioinformatic analysis revealed 2 candidate Tcf/Lef1-binding motifs in the murine Cux2 telencephalic enhancer, both of which bound Tcf/Lef1 protein in vitro (Fig. 3A,B). As only one of these sites could also be identified in the human, chimpanzee, and chicken enhancer sequences, we initially focused on this conserved site. All transgenic embryos (mTcfCux2BS1/lacZ) carrying point mutations in this site showed strong X-Gal staining in the dorsomedial telencephalon (n = 10/10 lacZ stained embryos; 100%), in the roof plate as well as in the diencephalon, midbrain, hindbrain, ectoderm, and branchial arches (Fig. 4G). Moreover, strong and persistent cortical hem staining in mTcfCux2BS1/lacZ embryos was detected after 3 h staining compared with the overnight staining that was required to detect lacZ expression in Cux2/lacZ embryos suggesting that this site is not essential for enhancer activity but might be important to adjust levels of Cux2 expression. These findings also raise the possibility that the second, nonconserved Tcf/Lef1-binding site might compensate for the loss of the conserved site. Therefore, we analyzed reporter gene expression in transgenic embryos in which both Tcf/Lef1-binding sites had been mutated. mTcfCux2BS1+2/lacZ embryos showed as strong expression in the dorsomedial region of the telencephalon as mTcfCux2BS1/lacZ embryos (n = 7/7 lacZ stained embryos; 100%) and in the hindbrain but reduced and very patchy expression in the roof plate, diencephalon, midbrain, ectoderm, and branchial arches (Fig. 4H). Taken together, these findings suggest that neither Tcf/Lef1-binding site in the Cux2 telencephalic enhancer is required for its spatial activity in the dorsomedial telencephalon but that the conserved site might regulate levels of enhancer activity.
The Nfia telencephalic enhancer is located in the intergenic region between Nfia and Tm2d1 (Visel et al. 2008), which is not expressed in the telencephalon (http://www.genepaint.org). This enhancer drives reporter gene expression in the cortical hem partially overlapping with Nfia expression in the dorsal midline region suggesting that it controls a subset of the Nfia telencephalic expression domains. Electroporation of an Nfia/lacZ reporter gene construct containing the corresponding murine genomic sequences into the E12.5 telencephalon resulted in the robust activation of the reporter in cortical hem cells (n = 3 embryos) and in roof plate cells (Fig. 5E,F). In contrast, mutation of the single Tcf/Lef site in this enhancer abolished cortical hem enhancer activity despite efficient electroporation, while lacZ expression in the roof plate is not affected (n = 4 embryos) (Fig. 5G,H). Thus, in contrast to Cux2, cortical hem activity of the Nfia telencephalic enhancer critically depends on Tcf/Lef binding.
Wnt Signaling Components
Finally, we analyzed whether Tcf/Lef binding is also required for the activity of the Wnt8b telencephalic enhancer. For this purpose, we used an ultraconserved element located between Wnt8b and Sec31b (Visel et al. 2008). Since Sec31b is not expressed in the brain (Tang et al. 2000) and since this regulatory element faithfully reflects the expression of endogenous Wnt8b it likely controls Wnt8b forebrain expression. Moreover, the corresponding mouse genomic region has enhancer activity in hippocampal progenitor cells as demonstrated by electroporation of the Wnt8b/lacZ reporter gene construct (Wnt8b/lacZ) (n = 4 embryos) (Fig. 5I,J). Our bioinformatic and bandshift analyses indicated the presence of a single conserved Tcf/Lef-binding site within this enhancer. Electroporation of a reporter gene construct (mTcfWnt8b/lacZ) containing point mutations in this site did not result in lacZ expression in the developing hippocampus (n = 4 embryos) (Fig. 5K,L) suggesting that Tcf/Lef binding is essential for the activity of the Wnt8b dorsomedial telencephalon enhancer.
Ectopic Activation of Wnt/β-Catenin Signaling Results in Ectopic Activation of Putative Wnt Target Genes
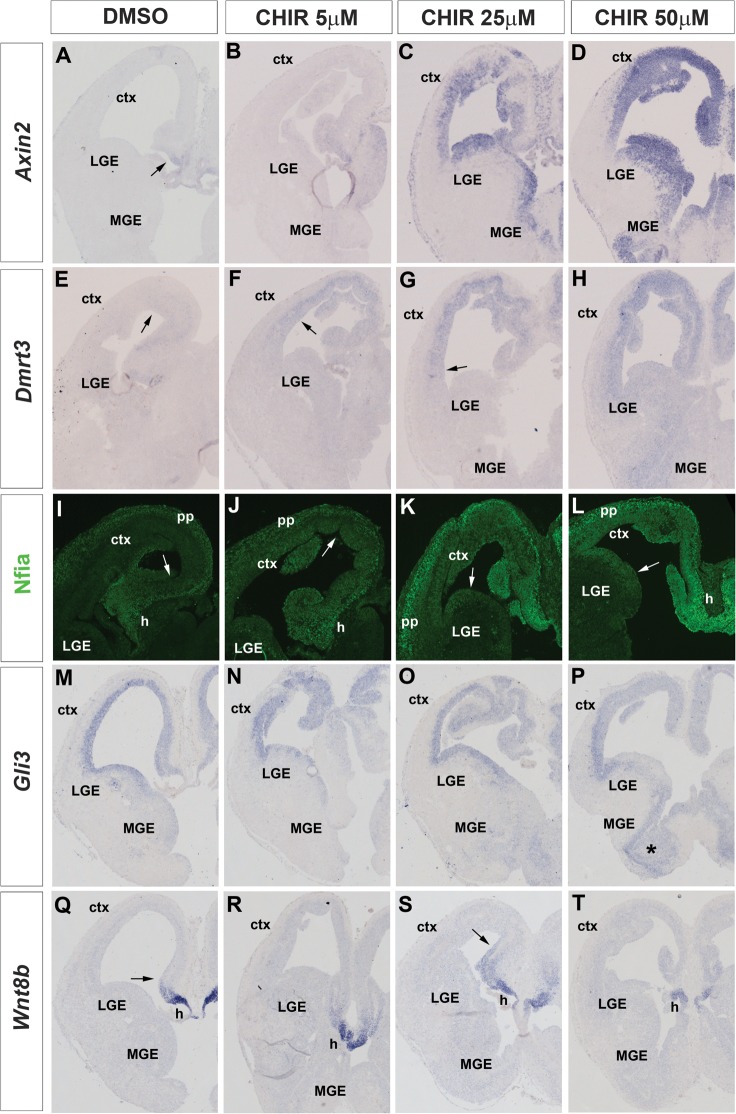
To obtain further evidence for the ability of the Wnt/β-catenin signaling to regulate the expression of these candidate target genes, we wanted to determine the consequences of ectopic activation of this pathway on the expression of these candidates. To this end, we employed an ex vivo explant assay in which we prepared coronal sections of the E13.5 mouse telencephalon and maintained these sections in culture for 24 h in the presence of DMSO or various concentrations of CHIR, which selectively inhibits GSK3β and thereby activates Wnt/β-catenin signaling (Ring et al. 2003). In control experiments, we first determined the effects of these treatments on the expression of Axin2, a general target of Wnt/β-catenin signaling (Jho et al. 2002; Lustig et al. 2002). Under control conditions, Axin2 expression is confined to the dorsomedial telencephalon, but the addition of CHIR led to a concentration dependent induction of Axin2 expression (Fig. 6A–D). Moreover, the Axin2 expression domain expands with increasing amounts of CHIR and at its highest levels, the whole telencephalic ventricular zone is positive for Axin2 indicating that CHIR treatment resulted in the activation of Wnt/β-catenin signaling in telencephalic progenitors. Next, we analyzed the expression of candidate target genes after CHIR treatment. In control experiments, Dmrt3 expression is restricted to the dorsomedial telencephalon, but its expression increases and expands into the neocortex and even into the lateral ganglionic eminence (LGE) at 50 μM CHIR (Fig. 6E–H) suggesting that Dmrt3 expression is regulated by Wnt/β-catenin signaling in a concentration dependent manner. Nfia is detected in the cortical hem, the hippocampal primordium, and in preplate neurons (Fig. 6I). CHIR treatment results in an Nfia upregulation in all these expression domains and to an expanded expression into the neocortex and even the LGE at the highest CHIR concentration levels (Fig. 6J–L). These findings indicate that Nfia expression not only in the dorsomedial telencephalon but also in preplate neurons is controlled by Wnt/β-catenin signaling, which is consistent with the presence of conserved Lef/Tcf-binding sites in the Nfia dorsomedial (Figs 3E and 5E–H) and preplate enhancers (Visel et al. 2008; K Hasenpusch-Theil, T Theil, unpublished data). In contrast, Wnt8b and Gli3 showed a more varied response to this pharmacological treatment. Gli3 expression on control sections is detected at high levels in the dorsal telencephalon and in the LGE but at low levels in the MGE. While this widespread expression makes it difficult to determine effects of CHIR treatment at low and intermediate concentrations, Gli3 expression was slightly downregulated in the neocortex but upregulated in the MGE at high CHIR levels (Fig. 6M–P). Similarly, addition of 25 μm CHIR resulted in an expanded Wnt8b expression in the dorsomedial telencephalon, while Wnt8b expression was reduced in samples treated with 50 μm CHIR (Fig. 6Q–T). These findings suggest that Gli3 and Wnt8b are regulated by Wnt/β-catenin signaling in a dose dependent manner.
Figure 6.
Ex vivo explant assay to determine the effects of ectopic activation of Wnt/β-catenin signaling on the expression of candidate target genes. (A–D) Axin2 expression is detected in the cortical hem region (arrow) of DMSO-treated control section (A) but is activated throughout the telencephalic ventricular zone after the addition of CHIR in a concentration dependent manner (B–D). (E–H) Dmrt3 expression is confined to the dorsomedial telencephalon but becomes activated in the neocortex at low (F) and intermediate (G) CHIR concentration levels and even in the LGE/MGE at 50 μM CHIR (H). The arrows in E–G mark the expression boundary in the neocortex. (I–L) Nfia is expressed in the cortical hem (h), dorsomedial telencephalon, and preplate (pp) neurons of control sections (I). CHIR treatment leads to an Nfia upregulation in all 3 expression domains, and the lateral expression boundary (arrows) shifts into the neocortex (J) and into the LGE (K,L). (M–P) Gli3 mRNA is detected at high levels in the cortex and the LGE but at low levels in the MGE (M). Treatment with 50 μM CHIR results in a slight downregulation in the medial neocortex but into a strong upregulation in the MGE (asterisks) (P). (Q–T) Wnt8b expression is confined to the cortical hem and to the ventral most hippocampal primordium (arrow) (Q). Wnt8b expression spreads dorsally in the hippocampus anlagen at 25 μM CHIR (arrow) but becomes downregulated after the addition of 50 μM CHIR.
Size Reduction and Disorganization of the Hippocampus in Gli3 Conditional Mouse Mutants
Finally, we started to test for possible functions of the newly identified Wnt target genes during hippocampus development. For this purpose, we focused on Gli3 since previous analyses of the Gli3 null mutant XtJ had shown a requirement for Gli3 in establishing/maintaining Wnt gene expression early in forebrain (E8.5) development (Grove et al. 1998; Fotaki et al. 2011), but it remained unclear whether Gli3 also has role(s) later in hippocampal development as a Wnt target gene. We therefore analyzed hippocampal formation in a Gli3 conditional mouse mutant (Blaess et al. 2008). Using an Emx1Cre driver line (Gorski et al. 2002), Gli3 mRNA expression is lost in the dorsomedial telencephalon of these mutants from E10.5 (Fig. 7A,B). Next, we performed a marker analysis for each structure of the medial pallium in E18.5 embryos. In control embryos, Nrp2 expression is found throughout the entire hippocampus (Galceran et al. 2000 and Fig. 7C), Scip1 is expressed in the CA1 field and in the neopallium (Frantz et al. 1994 and Fig. 7E), while Prox1 expression is confined to the DG (Oliver et al. 1993 and Fig. 7G). In Emx1Cre;Gli3 conditional mutants, these hippocampal structures are present but are severely reduced in size and severely disorganized (Fig. 7D,F,H). In particular, the DG does not form its typical blade like structure (Fig. 7H). Thus, Gli3 is required for hippocampal growth and organization after E10.5.
Figure 7.
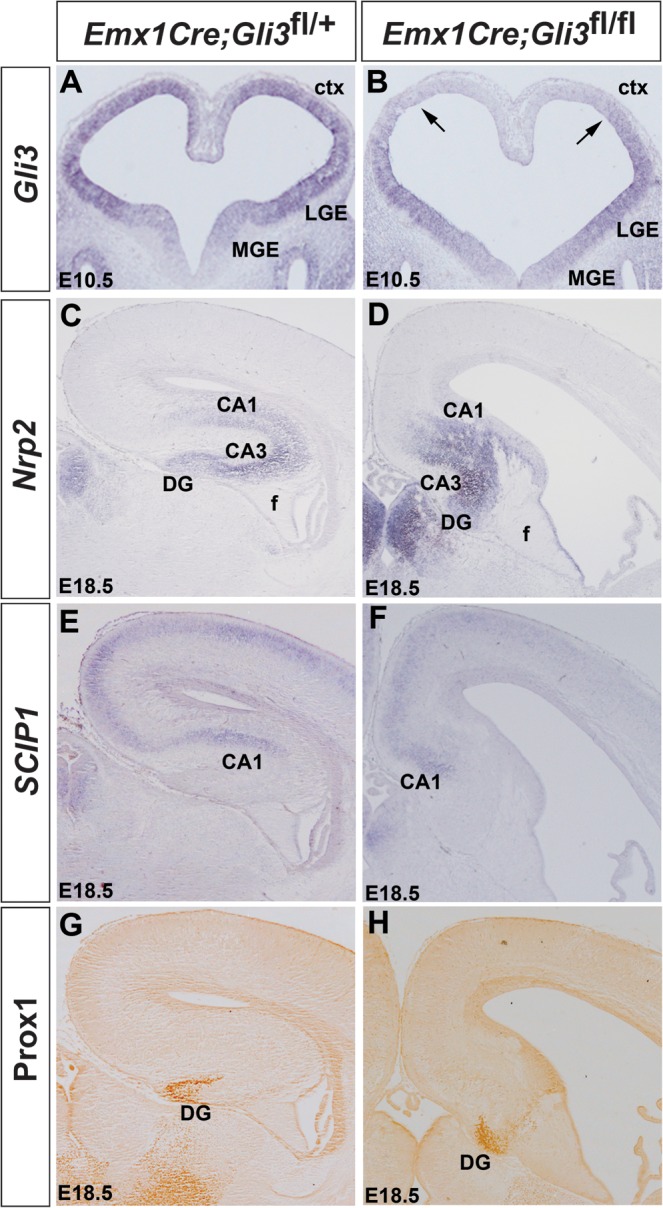
Hippocampus formation in Emx1Cre;Gli3 conditional mouse mutants. (A,B) Gli3 is expressed at high levels in the dorsal telencephalon and in the LGE but at lower levels in the MGE of E10.5 control embryos (A), but Gli3 expression is lost from the dorsomedial telencephalon of conditional mutants (B). (C–H) Hippocampal marker gene expression in E18.5 control (C,E,G) and Emx1Cre;Gli3 conditional mutants (D,F,H). Expression of Nrp2 labels the whole hippocampal formation (C), while Scip1 is expressed in CA1 and in the neocortex (E). Prox1 expression is confined to the DG (G). In Emx1Cre;Gli3 conditional mutants, these hippocampal markers are expressed but their expression domains are severely reduced (D,F,H). Note that the Prox1+ cells do not form the characteristic blades of the DG (G).
Discussion
Previous analyses have shown that Wnt/β-catenin signaling is essential for hippocampal development, but Wnt target genes remain largely unknown during this process. Here, we present a microarray analysis, which resulted in the identification of 53 genes, which according to their restricted or graded expression represent potential Wnt target genes. These genes encode proteins involved in a diverse range of cellular functions, but transcription factor and Wnt signaling components prevail. Using a combination of DNA-binding assays and reporter gene analyses, we have shown that telencephalic enhancers of several of these genes depend on Tcf/Lef binding. Moreover, the expression of these candidate genes can also be activated by ectopic activation of Wnt/β-catenin signaling. Given the nature of these genes and their interactions, our studies start to unravel a genetic circuitry underlying hippocampal formation.
XtJ Mutant Embryos Provide a Valuable Tool for the Identification of Candidate Wnt Target Genes
The identification of Wnt target genes in the developing hippocampus has been impeded by the presence of several Wnt genes, which show nested expression patterns (Parr et al. 1993) and by redundant Wnt functions (Fotaki et al. 2010). The fact that XtJ mutant embryos lack the expression of Wnt3a and have reduced Wnt8b expression in the dorsal telencephalon and phenocopy the hippocampal defects of Wnt3a mutants allowed us to overcome these difficulties and to use these embryos in a microarray analysis to identify candidate Wnt targets. Changes in telencephalic gene expression in these mutants are likely to result from cell-autonomous effects of the Gli3 transcription factor (Quinn et al. 2009) as well as from noncell-autonomous effects due to altered Wnt/Bmp signaling. Since the Gli3 repressor form predominates in the dorsal telencephalon (Fotaki et al. 2006), direct Gli3 target genes were expected to be upregulated, while Wnt targets were expected to be downregulated. Indeed, by focusing on the downregulated genes, our microarray and subsequent in situ hybridization analyses identified 53 genes which have restricted or graded expression in the dorsomedial telencephalon making them excellent candidates for being regulated by Wnt/β-catenin signaling. Moreover, this list contained previously identified Wnt target genes, such as Emx2 and Otx1 (Theil et al. 2002; Kurokawa et al. 2004; Suda et al. 2010). Our DNA binding and reporter gene analyses also showed that recombinant Lef1 binds to the telencephalic enhancers of several of these genes, including Dmrt3, Gli3, Nfia, and Wnt8b, and that this binding is essential for enhancer activity. Using an ex vivo explant culture system, we show that the expression of the corresponding endogenous genes is also regulated by Wnt/β-catenin signaling. Taken together with their reduced expression in the Wnt signaling deficient dorsal telencephalon of XtJ/XtJ embryos and with our observation that none of these enhancers contained a consensus Gli3-binding site (K Hasenpusch-Theil, T Theil, unpublished data), these findings suggest that Dmrt3, Gli3, Nfia, and Wnt8b are direct targets of Wnt/β-catenin signaling in the developing hippocampus.
Tight Regulation of Wnt Signaling Is Essential for Hippocampal Development
The specification of hippocampal field (CA1/3 and DG) fate crucially depends on Wnt signaling (Galceran et al. 2000; Lee et al. 2000). The cortical hem produces several Wnt proteins which are thought to act in a concentration dependent manner to specify different hippocampal cell fates (Machon et al. 2007) suggesting that levels of Wnt/β-catenin signaling have to be tightly controlled during hippocampal development. Consistent with this idea, we identified not only several Wnt genes in our microarray analysis but also several components of Wnt/β-catenin signaling, which act as modulators of this pathway. In contrast to Axin2 and Apcdd1, which inhibit Wnt/β-catenin signaling (Jho et al. 2002; Lustig et al. 2002), Rspo1/2/3 can enhance this pathway (Shimomura et al. 2010), while Dkk2 acts as an agonist or antagonist depending on the cellular context (Mao et al. 2002; Mao and Niehrs 2003). Moreover, the Wnt antagonist Dkk3 is expressed in the cortical hem from E11.5 onwards (Diep et al. 2004) further suggesting that enhancement of Wnt/β-catenin signaling and negative feedback regulation are integral parts of Wnt regulation in hippocampal development. Moreover, our findings add another level of complexity to this regulation by showing that the expression of at least one Wnt gene, Wnt8b, is directly controlled by Wnt/β-catenin signaling. The Wnt8b hippocampus enhancer contains a Tcf/Lef-binding site mutation of which results in loss of enhancer activity in hippocampal progenitors. Therefore, a negative feedback loop acts in conjunction with Wnt autoregulation to adjust levels of Wnt signaling in the dorsomedial telencephalon. This interaction between an autoregulatory and a negative feedback loop is also supported by our explant assay where intermediate concentrations of the Gsk3β inhibitor CHIR led to an expanded Wnt8b expression but higher concentrations resulted in a downregulation. The Wnt autoregulation also suggests a possible mechanism for the transcriptional upregulation of several Wnt genes after Wnt8b inactivation (Fotaki et al. 2010). An autorepressive feedback loop that results in a change of the Wingless signaling profile has also been identified during fly development (Yu et al. 1998) suggesting an evolutionarily conserved mechanism to control Wnt signaling. In summary, our findings suggest a requirement for a complex regulation of Wnt signaling during hippocampal development.
Wnt Signaling Regulates the Expression of Several Transcription Factors
Consistent with the idea that the interplay between signaling cascades and transcription factors underlies patterning of the telencephalon, transcription factors are the second class of proteins, which are highly overrepresented in our list of potential Wnt target genes. Several of these factors show highly restricted expression patterns thereby delineating subdivisions of the dorsal midline region, while others show graded expression in the hippocampal primordium. Here, we present evidence that genes of both groups are regulated by Wnt signaling. The Cux2 and the Nfia telencephalic enhancers show restricted activity in the cortical hem and contain Tcf-binding sites. However, while mutation of this site within the Nfia telencephalic enhancer led to a loss of enhancer activity, Tcf/Lef-binding sites within the Cux2 telencephalic enhancer are not required but may be important to regulate expression levels. Since Cux2 regulates neural precursor proliferation in the developing spinal cord (Iulianella et al. 2008, 2009), the regulation of Cux2 expression levels might have important implications for the cortical hem as a source of Cajal–Retzius cells. Interestingly, Axin2 and a Tcf/lacZ reporter transgene (Maretto et al. 2003), which have been used previously to monitor Wnt signaling in the dorsal telencephalon, are differentially regulated in the cortical hem (Machon et al. 2007). While Axin2 is strongly expressed in the hem, the Tcf/lacZ reporter is only activated in a few isolated cells. Taken together, regulation of genes in the cortical hem by Wnt signaling appears complex and is likely to depend on a context specific combination of transcription factors. Possible candidates include Bmp signaling and Smad transcription factors which are known to cooperate with Wnt signaling in regulating gene expression in the dorsomedial telencephalon (Theil et al. 2002; Suda et al. 2010). Indeed, the Cux2 but not the Nfia telencephalic enhancer contains an evolutionary conserved Smad-binding site adjacent to the Tcf/Lef sites similar to the Emx2 forebrain enhancer (K Hasenpusch-Theil, T Theil, unpublished data). It will therefore be interesting to analyze whether Bmp signaling is involved in regulating the Cux2 telencephalic enhancer.
Amongst the transcription factor genes with graded expression in the dorsal telencephalon, Dmrt3 attracted our attention as one of the most strongly regulated genes. The Dmrt genes encode a family of transcription factors whose function in sexual development has been well studied in invertebrates and vertebrates, but emerging evidence indicates that these genes might also regulate other developmental processes (Hong et al. 2007). In this respect, it is intriguing that our microarray screen revealed a reduced expression for 3 members of this gene family, Dmrt3, 4, and 5, which in addition show graded expression in the wild-type dorsal telencephalon. While the C. elegans Dmrt homolog mab-3 is directly regulated by the Gli homolog Tra-3 (Yi et al. 2000), we were unable to identify a consensus Gli3-binding site in the Dmrt3 telencephalic enhancer (K Hasenpusch-Theil, T Theil, unpublished data). In contrast, we showed that this enhancer contains an essential Tcf/Lef-binding site and that the endogenous gene is positively regulated by ectopic activation of Wnt/β-catenin signaling in a concentration dependent manner. Therefore, Dmrt3 represents a direct Wnt target gene. Moreover, a recently identified Dmrt4 enhancer also contains an evolutionarily conserved, Tcf/Lef-binding sequence (Visel et al. 2008), the functionality of which still needs to be demonstrated. In addition, in Dmrt5 mutant mouse embryos, the cortex is strongly reduced in size and posterior midline structures fail to develop (E Bellefroit, personal communication) similar to Wnt3a and XtJ/XtJ mutants suggesting that these genes represent important downstream mediators of Wnt/β-catenin signaling during telencephalic development. In the future, it will be interesting to further investigate the regulatory relationship between Dmrt genes and other genes identified in our screen.
Given its crucial role in telencephalic development, we were also interested in the transcriptional regulation of Gli3 expression and indeed we were able to identify an essential Tcf/Lef-binding site within the Gli3 forebrain enhancer (Paparidis et al. 2007; Visel et al. 2008). A previous bioinformatic analysis on the regulation of Gli3 expression in the spinal cord identified conserved potential Tcf/Lef-binding sites (Alvarez-Medina et al. 2008) in a region distinct from the forebrain enhancer suggesting that Gli3 forebrain and spinal cord expression is controlled by different regulatory elements, but the functionality of these sites for Gli3 expression in the spinal cord was not tested. These findings taken together with the results from our ex vivo explant assay present the first evidence that Gli3 is a direct target of Wnt/β-catenin signaling. Moreover, our analysis of Gli3 conditional mouse mutants shows a requirement for Gli3 after E10.5 in controlling the growth and organization of the hippocampus and therefore indicates a novel role for Gli3 in mediating aspects of Wnt/β-catenin signaling in hippocampal development.
These findings taken together with previous studies suggest a complex regulatory relationship between Gli3 and Wnts. First, Gli3 is required for the expression of several Wnt genes from earliest stages of forebrain development (Fotaki et al. 2011). Since the Wnt8b telencephalic enhancer lacks canonical Gli3-binding sites and since the Gli3 repressor form predominates in the dorsal telencephalon, this regulation is likely to be indirect. Second, Gli3 repressor negatively regulates Wnt/β-catenin signaling in the spinal cord by binding to β-catenin thereby inhibiting its transcriptional activity (Ulloa and Briscoe 2007). Finally, we show here that the expression of Gli3 is directly regulated by Wnt/β-catenin signaling.
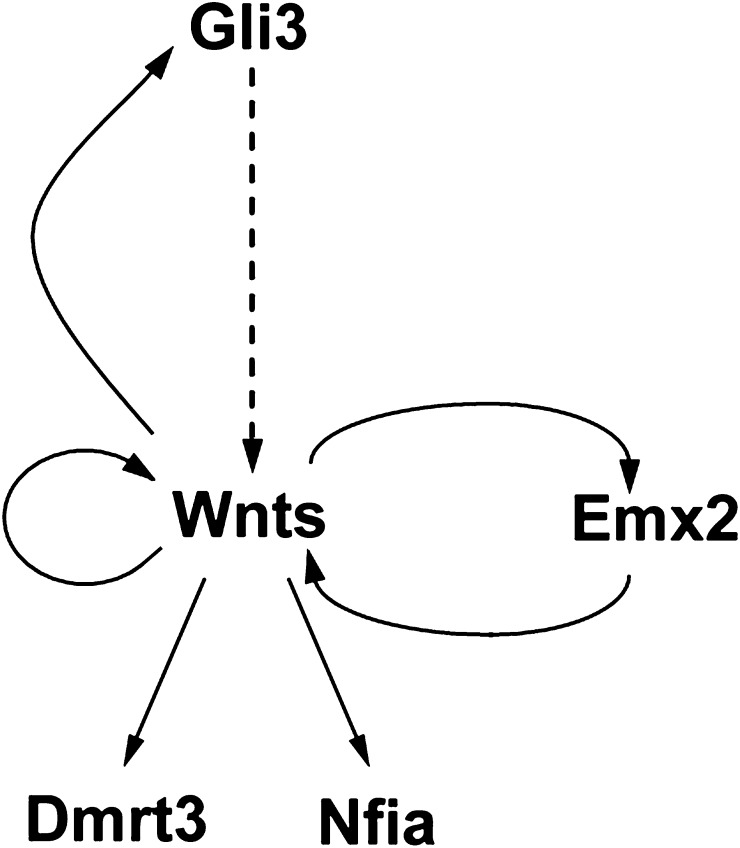
In summary, our study begins to unravel the complexity of the interactions between Gli3, Wnts, and Wnt targets and provides a model whereby several feedbacks and an autoregulatory loop control early hippocampal development (Fig. 8). Initially, Gli3 is required to establish/maintain Wnt gene expression in the dorsal midline. Since the Gli3 repressor form predominates in the dorsal telencephalon, this regulation is likely to be indirect and could involve a second, hitherto unknown transcriptional repressor, expression of which is derepressed by Gli3. Conversely, Wnts directly control Gli3 expression. This feedback loop to maintain expression of Wnts and Gli3 acts in parallel to a similar positive feedback loop between the Wnts and the Emx2 homeobox gene (Muzio et al. 2005). Moreover, levels of Wnt gene expression and Wnt signaling are controlled by an autoregulatory mechanism and by the expression of several modulators in the cortical hem, respectively. Finally, Wnt signaling regulates the expression of several transcription factors, which control hippocampal development. In the future, it will be interesting to further characterize potential interactions between these transcription factors and Wnt signaling to gain a better understanding of the genetic programs regulating formation of the hippocampus.
Figure 8.
A simple model of the genetic circuitry underlying hippocampal development. Gli3R controls Wnt gene expression presumably by a derepression mechanism. Wnts in turn activate Gli3 expression and autoregulate their own expression. In addition to this Gli3/Wnt feedback loop, a second positive feedback loop maintains expression of Wnts and the hippocampal determinant Emx2. Dmrt3 and Nfia represent additional Wnt target genes in the hippocampal primordium and in the cortical hem, respectively.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
Wellcome Trust (078100).
Supplementary Material
Acknowledgments
We thank John Mason, Tom Pratt, and David Price for critical comments on the manuscript. We are grateful to Sandra Blaess and Alex Joyner for providing the Gli3 floxed mouse line and Gonzalo Alvarez-Bolado, Harold Cremer, Denis Houzelstein, Juha Partanen, Irma Thesleff, and David Wilkinson for providing in situ hybridization probes. Conflict of Interest: None declared.
References
- Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135:237–247. doi: 10.1242/dev.012054. [DOI] [PubMed] [Google Scholar]
- Blaess S, Stephen D, Joyner AL. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development. 2008;135:2093–2103. doi: 10.1242/dev.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomly D, Kyler SL, McWeeney SK, Yochum GS. Identification of {beta}-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010;38:5735–5745. doi: 10.1093/nar/gkq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Büscher D, Grotewold L, Rüther U. The Xt-J allele generates a Gli3 fusion transcript. Mamm Genome. 1998;9:676–678. doi: 10.1007/s003359900845. [DOI] [PubMed] [Google Scholar]
- Chaudhry AZ, Lyons GE, Gronostajski RM. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Diep DB, Hoen N, Backman M, Machon O, Krauss S. Characterisation of the Wnt antagonists and their response to conditionally activated Wnt signalling in the developing mouse forebrain. Brain Res Dev Brain Res. 2004;153:261–270. doi: 10.1016/j.devbrainres.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Flenniken AM, Gale NW, Yancopoulos GD, Wilkinson DG. Distinct and overlapping expression patterns of ligands for Eph-related receptor tyrosine kinases during mouse embryogenesis. Dev Biol. 1996;179:382–401. doi: 10.1006/dbio.1996.0269. [DOI] [PubMed] [Google Scholar]
- Fotaki V, Larralde O, Zeng S, McLaughlin D, Nichols J, Price DJ, Theil T, Mason JO. Loss of Wnt8b has no overt effect on hippocampus development but leads to altered Wnt gene expression levels in dorsomedial telencephalon. Dev Dyn. 2010;239:284–296. doi: 10.1002/dvdy.22137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotaki V, Price DJ, Mason JO. Wnt/beta-catenin signaling is disrupted in the extra-toes (Gli3(Xt/Xt) ) mutant from early stages of forebrain development, concomitant with anterior neural plate patterning defects. J Comp Neurol. 2011;519:1640–1657. doi: 10.1002/cne.22592. [DOI] [PubMed] [Google Scholar]
- Fotaki V, Yu T, Zaki PA, Mason JO, Price DJ. Abnormal positioning of diencephalic cell types in neocortical tissue in the dorsal telencephalon of mice lacking functional Gli3. J Neurosci. 2006;26:9282–9292. doi: 10.1523/JNEUROSCI.2673-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz GD, Bohner AP, Akers RM, McConnell SK. Regulation of the POU domain gene SCIP during cerebral cortical development. J Neurosci. 1994;14:472–485. doi: 10.1523/JNEUROSCI.14-02-00472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz T. Extra-toes (Xt) homozygous mutant mice demonstrate a role for the Gli-3 gene in the development of the forebrain. Acta Anat (Basel) 1994;150:38–44. doi: 10.1159/000147600. [DOI] [PubMed] [Google Scholar]
- Fujimura N, Vacik T, Machon O, Vlcek C, Scalabrin S, Speth M, Diep D, Krauss S, Kozmik Z. Wnt-mediated down-regulation of Sp1 target genes by a transcriptional repressor Sp5. J Biol Chem. 2007;282:1225–1237. doi: 10.1074/jbc.M605851200. [DOI] [PubMed] [Google Scholar]
- Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–482. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- Harrison SM, Houzelstein D, Dunwoodie SL, Beddington RS. Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with Brachyury. Dev Biol. 2000;227:358–372. doi: 10.1006/dbio.2000.9878. [DOI] [PubMed] [Google Scholar]
- Hong CS, Park BY, Saint-Jeannet JP. The function of Dmrt genes in vertebrate development: it is not just about sex. Dev Biol. 2007;310:1–9. doi: 10.1016/j.ydbio.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev Biol. 1994;162:402–413. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- Iulianella A, Sharma M, Durnin M, Vanden Heuvel GB, Trainor PA. Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development. 2008;135:729–741. doi: 10.1242/dev.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iulianella A, Sharma M, Vanden Heuvel GB, Trainor PA. Cux2 functions downstream of Notch signaling to regulate dorsal interneuron formation in the spinal cord. Development. 2009;136:2329–2334. doi: 10.1242/dev.032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho E-h, Zhang T, Domon C, Joo C-K, Freund J-N, Costantini F. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukkola T, Sinjushina N, Partanen J. Drapc1 expression during mouse embryonic development. Gene Expr Patterns. 2004;4:755–762. doi: 10.1016/j.modgep.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Kimura J, Suda Y, Kurokawa D, Hossain ZM, Nakamura M, Takahashi M, Hara A, Aizawa S. Emx2 and Pax6 function in cooperation with Otx2 and Otx1 to develop caudal forebrain primordium that includes future archipallium. J Neurosci. 2005;25:5097–5108. doi: 10.1523/JNEUROSCI.0239-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa D, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development. 2004;131:3319–3331. doi: 10.1242/dev.01220. [DOI] [PubMed] [Google Scholar]
- Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Jr, Tremml G, Jessell TM. A role for the roof plate and its resident TGFbeta-related proteins in neuronal patterning in the dorsal spinal cord. Cell. 1997;91:127–138. doi: 10.1016/s0092-8674(01)80015-5. [DOI] [PubMed] [Google Scholar]
- Longo KA, Kennell JA, Ochocinska MJ, Ross SE, Wright WS, MacDougald OA. Wnt signaling protects 3T3-L1 preadipocytes from apoptosis through induction of insulin-like growth factors. J Biol Chem. 2002;277:38239–38244. doi: 10.1074/jbc.M206402200. [DOI] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machon O, Backman M, Machonova O, Kozmik Z, Vacik T, Andersen L, Krauss S. A dynamic gradient of Wnt signaling controls initiation of neurogenesis in the mammalian cortex and cellular specification in the hippocampus. Dev Biol. 2007;311:223–237. doi: 10.1016/j.ydbio.2007.08.038. [DOI] [PubMed] [Google Scholar]
- Magnani D, Hasenpusch-Theil K, Jacobs EC, Campagnoni AT, Price DJ, Theil T. The Gli3 hypomorphic mutation Pdn causes selective impairment in the growth, patterning, and axon guidance capability of the lateral ganglionic eminence. J Neurosci. 2010;30:13883–13894. doi: 10.1523/JNEUROSCI.3650-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, et al. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 2008;319:304–309. doi: 10.1126/science.1151695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Niehrs C. Kremen2 modulates Dickkopf2 activity during Wnt/LRP6 signaling. Gene. 2003;302:179–183. doi: 10.1016/s0378-1119(02)01106-x. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard TM, Jain MD, Balmer CW, LaMantia AS. High-resolution mapping of the Gli3 mutation extra-toes reveals a 51.5-kb deletion. Mamm Genome. 2002;13:58–61. doi: 10.1007/s00335-001-2115-x. [DOI] [PubMed] [Google Scholar]
- Muzio L, Soria JM, Pannese M, Piccolo S, Mallamaci A. A mutually stimulating loop involving emx2 and canonical wnt signalling specifically promotes expansion of occipital cortex and hippocampus. Cereb Cortex. 2005;15:2021–2028. doi: 10.1093/cercor/bhi077. [DOI] [PubMed] [Google Scholar]
- Okada T, Okumura Y, Motoyama J, Ogawa M. FGF8 signaling patterns the telencephalic midline by regulating putative key factors of midline development. Dev Biol. 2008;320:92–101. doi: 10.1016/j.ydbio.2008.04.034. [DOI] [PubMed] [Google Scholar]
- Oldekamp J, Kraemer N, Alvarez-Bolado G, Skutella T. bHLH gene expression in the Emx2-deficient dentate gyrus reveals defective granule cells and absence of migrating precursors. Cereb Cortex. 2004;14:1045–1058. doi: 10.1093/cercor/bhh064. [DOI] [PubMed] [Google Scholar]
- Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- Paparidis Z, Abbasi AA, Malik S, Goode DK, Callaway H, Elgar G, deGraaff E, Lopez-Rios J, Zeller R, Grzeschik KH. Ultraconserved non-coding sequence element controls a subset of spatiotemporal GLI3 expression. Dev Growth Differ. 2007;49:543–553. doi: 10.1111/j.1440-169X.2007.00954.x. [DOI] [PubMed] [Google Scholar]
- Parr BA, Shea MJ, Vassileva G, McMahon AP. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development. 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Mansouri A, Simeone A, Boncinelli E, Gruss P. Dentate gyrus formation requires Emx2. Development. 1996;122:3893–3898. doi: 10.1242/dev.122.12.3893. [DOI] [PubMed] [Google Scholar]
- Pispa J, Mikkola ML, Mustonen T, Thesleff I. Ectodysplasin, Edar and TNFRSF19 are expressed in complementary and overlapping patterns during mouse embryogenesis. Gene Expr Patterns. 2003;3:675–679. doi: 10.1016/s1567-133x(03)00092-9. [DOI] [PubMed] [Google Scholar]
- Qiu W, Hu Y, Andersen TE, Jafari A, Li N, Chen W, Kassem M. Tumor necrosis factor receptor superfamily member 19 (TNFRSF19) regulates differentiation fate of human mesenchymal (stromal) stem cells through canonical Wnt signaling and C/EBP. J Biol Chem. 2010;285:14438–14449. doi: 10.1074/jbc.M109.052001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Mason JO, Price DJ. Gli3 is required autonomously for dorsal telencephalic cells to adopt appropriate fates during embryonic forebrain development. Dev Biol. 2009;327:204–215. doi: 10.1016/j.ydbio.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Quina LA, Wang S, Ng L, Turner EE. Brn3a and Nurr1 mediate a gene regulatory pathway for habenula development. J Neurosci. 2009;29:14309–14322. doi: 10.1523/JNEUROSCI.2430-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999;215:208–220. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, Slabiak T, et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Agalliu D, Vonica A, Luria V, Wajid M, Baumer A, Belli S, Petukhova L, Schinzel A, Brivanlou AH, et al. APCDD1 is a novel Wnt inhibitor mutated in hereditary hypotrichosis simplex. Nature. 2010;464:1043–1047. doi: 10.1038/nature08875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Hurley TM, McClive PJ, Sinclair AH. Restricted expression of DMRT3 in chicken and mouse embryos. Gene Expr Patterns. 2002;2:69–72. doi: 10.1016/s0925-4773(02)00360-x. [DOI] [PubMed] [Google Scholar]
- Suda Y, Kokura K, Kimura J, Kajikawa E, Inoue F, Aizawa S. The same enhancer regulates the earliest Emx2 expression in caudal forebrain primordium, subsequent expression in dorsal telencephalon and later expression in the cortical ventricular zone. Development. 2010;137:2939–2949. doi: 10.1242/dev.048843. [DOI] [PubMed] [Google Scholar]
- Summerhurst K, Stark M, Sharpe J, Davidson D, Murphy P. 3D representation of Wnt and Frizzled gene expression patterns in the mouse embryo at embryonic day 11.5 (Ts19) Gene Expr Patterns. 2008;8:331–348. doi: 10.1016/j.gep.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL, Zhang T, Low DY, Wong ET, Horstmann H, Hong W. Mammalian homologues of yeast sec31p. An ubiquitously expressed form is localized to endoplasmic reticulum (ER) exit sites and is essential for ER-Golgi transport. J Biol Chem. 2000;275:13597–13604. doi: 10.1074/jbc.275.18.13597. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Theil T. Gli3 is required for the specification and differentiation of preplate neurons. Dev Biol. 2005;286:559–571. doi: 10.1016/j.ydbio.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Theil T, Alvarez-Bolado G, Walter A, Rüther U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 1999;126:3561–3571. doi: 10.1242/dev.126.16.3561. [DOI] [PubMed] [Google Scholar]
- Theil T, Aydin S, Koch S, Grotewold L, Rüther U. Wnt and Bmp signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development. 2002;129:3045–3054. doi: 10.1242/dev.129.13.3045. [DOI] [PubMed] [Google Scholar]
- Theil T, Frain M, Gilardi-Hebenstreit P, Flenniken A, Charnay P, Wilkinson DG. Segmental expression of the EphA4 (Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development. 1998;125:443–452. doi: 10.1242/dev.125.3.443. [DOI] [PubMed] [Google Scholar]
- Tole S, Grove EA. Detailed field pattern is intrinsic to the embryonic mouse hippocampus early in neurogenesis. J Neurosci. 2001;21:1580–1589. doi: 10.1523/JNEUROSCI.21-05-01580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tole S, Ragsdale CW, Grove EA. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J) Dev Biol. 2000;217:254–265. doi: 10.1006/dbio.1999.9509. [DOI] [PubMed] [Google Scholar]
- Ulloa F, Briscoe J. Morphogens and the control of cell proliferation and patterning in the spinal cord. Cell Cycle. 2007;6:2640–2649. doi: 10.4161/cc.6.21.4822. [DOI] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Hill RE, Bickmore WA. Enhancers: from developmental genetics to the genetics of common human disease. Dev Cell. 2011;21:17–19. doi: 10.1016/j.devcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Yee SP, Rigby PW. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Suda Y, Matsuo I, Miyamoto N, Takeda N, Kuratani S, Aizawa S. Emx1 and Emx2 functions in development of dorsal telencephalon. Development. 1997;124:101–111. doi: 10.1242/dev.124.1.101. [DOI] [PubMed] [Google Scholar]
- Yu X, Riese J, Eresh S, Bienz M. Transcriptional repression due to high levels of Wingless signalling. EMBO J. 1998;17:7021–7032. doi: 10.1093/emboj/17.23.7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.