Abstract
Structural DNA nanotechnology is directed at building objects, lattices and arrays from cohesive interactions between DNA molecules. The predominant means of doing this takes advantage of the information inherent in Watson-Crick base pairing in duplex formation and in sticky-ended cohesion. Nevertheless, other forms of nucleic acid cohesion are also known, particularly paranemic edge-sharing interactions (PX). Here we report the formation of a triangular species that has four strands per edge, held together by PX interactions. We demonstrate by non-denaturing gel electrophoresis and by atomic force microscopy (AFM) that we can combine a partial triangle with other strands to form a four-stranded molecule that is robust. By combining them with a new mixed-fusion type of three-domain (TX) molecule, we demonstrate by AFM that these triangles can be self-assembled into a linear array.
Keywords: Unusual DNA Motifs, PX DNA, DNA Triangles, DNA self-assembly, Atomic Force Microscopy
The Watson-Crick base pairing of DNA not only provided the chemical basis for genetics but also has enabled the self-assembly of unusual DNA branched structures.1 This approach has led to the construction of DNA objects, such as a DNA cube2, a truncated octahedron3, an octahedron4 and tetrahedra.5 Several types of DNA triangles have been built6–10 and their self-assembly into 1D6 or 2D7–10 arrays has been demonstrated. All of these triangles have been built by using double helices or DNA double crossover molecules11 as the edges, and they have been connected to other triangles by sticky ended cohesion, using either individual sticky ends or doubly cohesive motifs. Motifs have also been described that contain triangles that cohere with each other via edge-sharing cohesion based on DX molecules.12 The diversity of DNA structures and types of cohesion could be increased if it were possible to build units based on edge-edge recognition.
Recently, we have reported a new DNA motif, known as the PX motif, wherein two double helices cohere in a paranemic manner.13 Here, we use that type of cohesion to produce a novel DNA triangle that is built in this fashion. The paranemic PX motif is derived theoretically by fusing two parallel double helices by reciprocal exchange between strands of the same polarity at every possible point where the two helices come in contact.14 All of the interactions between the nucleotides are of the Watson-Crick form. The distinguishing feature of this type of crossover structure is that the strands of the two interwound helices are completely unlinked and can be built from two closed DNA double helices; for example, the red and purple strands in Figure 1a could be separated from the blue and green strands even if all four ends of the two helices were closed by hairpins. Although we discuss the red and purple strands as one pair of strands and the green and blue strands as another pair, this is arbitrary: There is pairing between the blue and purple strand and between the red and green strand; thus, if they are separated into pairs, the restoration of the blue-purple and red-green pairing leads to the appropriate recognition; likewise, we could just as easily talk about the red-green strand pair wrapping around the blue-purple strand pair. PX DNA molecules have been used to link two large DNA objects,15 to work in nanomechanical devices,16–18 and to form a DNA octahedron in combination with DX molecules.4
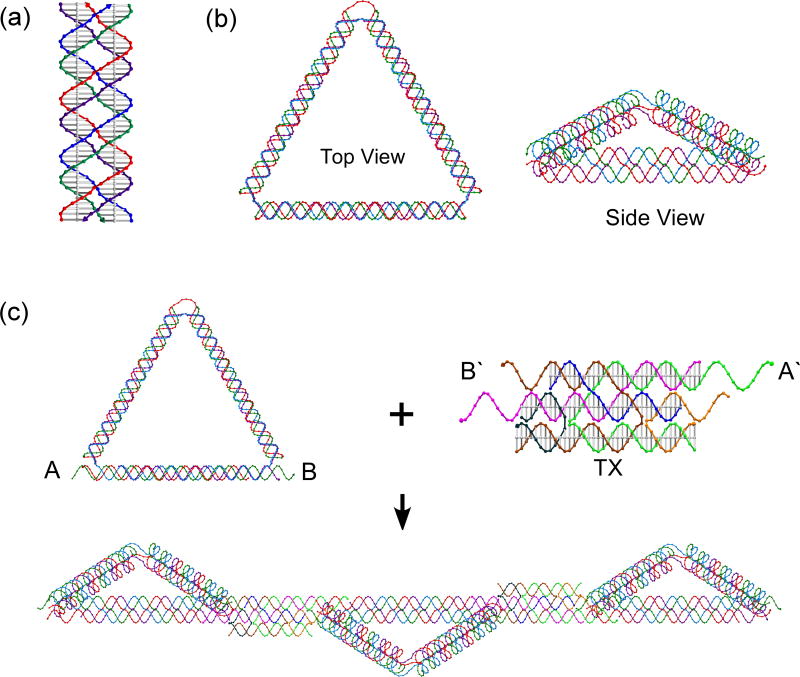
Figure 1.
Schematic Drawings of the motifs used here. (a) The PX motif. The four strands are colored red, purple, green and blue; base pairs and helix axes are indicated. (b) Top and side views of the PX triangle are shown, using the same colors as in (a). Only the blue strand is a cyclic molecule. (c) The formation of a linear array. The left side of the upper panel shows the sticky ends on the triangle as A and B; the right side shows the PATX motif (base pairs and helix axes indicated); complementary sticky ends, A′ B′ are shown; the top two domains are parallel, the bottom two antiparallel. The array incorporating both motifs is shown at the bottom of the panel. Figures produced by the program GIDEON.29
The structure of the PX triangle built here is illustrated in Figure 1b. It is composed of four DNA strands (red, purple, blue and green). As in Figure 1a, the red and purple strands and are wrapped around the blue and green strands to form the edges of the DNA triangle, as well as forming the flexible single-stranded DNA loops or nicks that act as the vertices. The side view in Figure 1b showing the front edge of the triangle shows this point most clearly: red and purple strands wrap around blue and green strands to yield the edge. Each edge of PX triangle is 76 nucleotide pairs long, which contains two turns of a 6:5 PX molecule,13 where the major groove spacing contains six nucleotide pairs, the minor groove spacing contains five nucleotide pairs. Note that this length is composed from 76 pairs of pairs that are inter-wrapped. These edges are flanked by 1.5 turns of conventional double helix on either side. The blue strand is a long circular strand which is ligated from three short ones, and each of the other strands (red, purple and green) is composed of three short strands with the nicking positions in either vertices or edges. The detailed strand structures and sequences are shown in the Supporting Information.
Fusing triangle molecules into oligomeric units presents a further problem. We have found that intermolecular PX cohesion15 is much more difficult than intramolecular PX cohesion;4 although possible, it entails much higher concentrations of Mg2+ than does ordinary sticky-ended cohesion. As a solution to this problem, we have developed a new form of three-domain DNA motif (TX), which contains a mixture of helix pairs (DX components); one DX formed by fusion of strands of the same polarity (often called parallel) and the other DX is formed by fusion of strands of opposite polarity (often called antiparallel). Parallel DX molecules are less tractable than antiparallel DX components,11 but they are required to fuse PX triangles. This is because the phasing of strands is the same in PX and parallel DX molecules, but different in antiparallel DX molecules. The robust new TX motif (shown on the right of Figure 1c) solves this problem: The top two helical domains are parallel, and the bottom two are antiparallel, so we call it the PATX motif. Note that each of the top two domains contains two strands that participate in the bottom domain, although two strands (magenta and blue) are restricted to the parallel DX portion of the molecule. The parallel DX component of the PATX motif used here has a separation of one major groove spacing between its crossovers. This new motif makes it possible to connect PX molecules by double sticky-ended cohesion, which has been shown to be more robust than single sticky-ended cohesion.7 Figure 1c shows the combination of the PX triangle and the PATX motif to produce a linear array; this array will be described below.
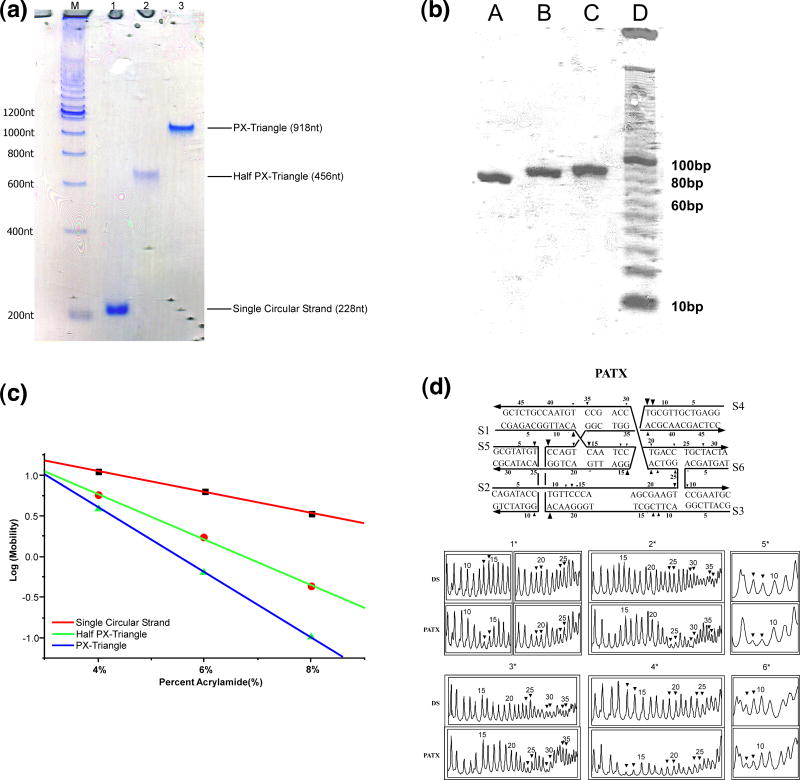
We first characterized the PX triangle by using non-denaturing gel electrophoresis. Figure 2a shows a 4% non-denaturing gel containing the single circular strand, the PX half-triangle and the whole PX triangle. The PX triangle appears as a single sharp band without any unwanted association or dissociation, indicating that the PX triangle constructed is a stable structure. Its mobility is in the expected range, relative to a ladder of linear duplex markers. We have also characterized the PATX by non-denaturing gel electrophoresis, shown in Figure 2b. Lanes A, B and C contain 4-strand, 5-strand and complete complexes, respectively. It is clear that the motif migrates as a single band. It is important that there is no multimerization of the molecule, as is often seen in parallel DX molecules.11
Figure 2.
Characterization of the Motifs Used. (a) A Non-Denaturing Gel of the PX Triangle. Lane M contains a 100 nucleotide duplex ladder (duplex sizes are indicated). Lane 1 contains the cyclic (blue) strand of the triangle, lane 2 contains a half-triangle (the blue strand plus the components of the green strand), and lane 3 contains the entire PX triangle. There is no dissociation of the PX triangle, nor is there multimerization. (b) A Non-Denaturing Gel of the PATX motif. Lane D contains a 10 nucleotide duplex marker. Lane C contains the complete PATX complex, while lane B lacks strand 6 and lane A lacks strands 5 and 6. Note that neither breakdown nor multimerization is seen for the complete complex; sticky ends have been deleted. (c) Ferguson Plot of the Triangle and its Substructures. The slope of the complete triangle does not scale directly with the number of nucleotides it contains, suggesting that nucleotides are buried by triangle formation. (d) Hydroxyl Radical Autofootprinting Analysis of the PATX Motif. The upper part of the panel is a schematic indicating protection sites by triangular markers; the extent of protection is reflected in the sizes of the triangles. The bottom part of the panel shows detailed scans that quantitate the autofootprinting. Labeled strands are indicated by an asterisk, nucleotide positions are numbered according to the schematic and sites of potential crossover are labeled with triangles.
The Ferguson plot is used to analyze electrophoretic mobility as a function of gel concentration. The slope of this plot yields information about the friction constant of the molecule.19 Figure 2c shows that the slopes for the single circular strand, the PX half-triangle, and the PX triangle are −0.257, −0.548, and −0.806 respectively. Thus the circular DNA and the half-triangle have friction constants roughly proportional to the number of nucleotides that they contain. By contrast, the friction constant of the triangle is only about 1.6 times that of the half-triangle. This finding suggests that the triangle is markedly more compact than the half-triangle. This can be ascribed to the burial of some of its residues as a consequence of the complete base pairing present in its edges; another contributing factor could also be the more compact nature of the triangle structure.
The most sensitive chemical method to characterize a small motif, such as the PATX molecule, is by hydroxyl radical autofootprinting experiments, 20 in which hydroxyl radicals are generated by Fenton chemistry involving Fe(II)EDTA2−.21 The strategy of such experiments is to compare the quantitative chemical attack pattern of each strand when it is part of an unusual motif, with the pattern obtained when a strand is hybridized with its normal Watson-Crick complement. From many experiments it has been concluded that strands exhibiting the same patterns in both pairing environments probably have double helical conformations near the junction, while strands whose patterns show protection at possible crossover points switch between helical domains. Crossover points and occluded points have evinced protection in virtually all analyses of unusual motifs.22 Figure 2d shows the hydroxyl radical autofootprinting data for a PATX molecule. The protection pattern is summarized in the schematic at the top of the panel, where protection is represented by triangular markers; the detailed scans of each of the gels is shown below this. Protection is seen at every expected crossover site. Weak protection is also visible on the bottom domain a half-turn from crossovers, likely resulting from occlusion of the site by the adjacent helix. The weak protection noted at sites opposite the crossover strands (nucleotides 35 and 36, strands S2 and S3) at the crossovers involving the two parallel domains are not readily explained, but could be the result of a small amount of crossover isomerization.23 Nevertheless, the structural description of the PATX molecule is in agreement with the target structure.
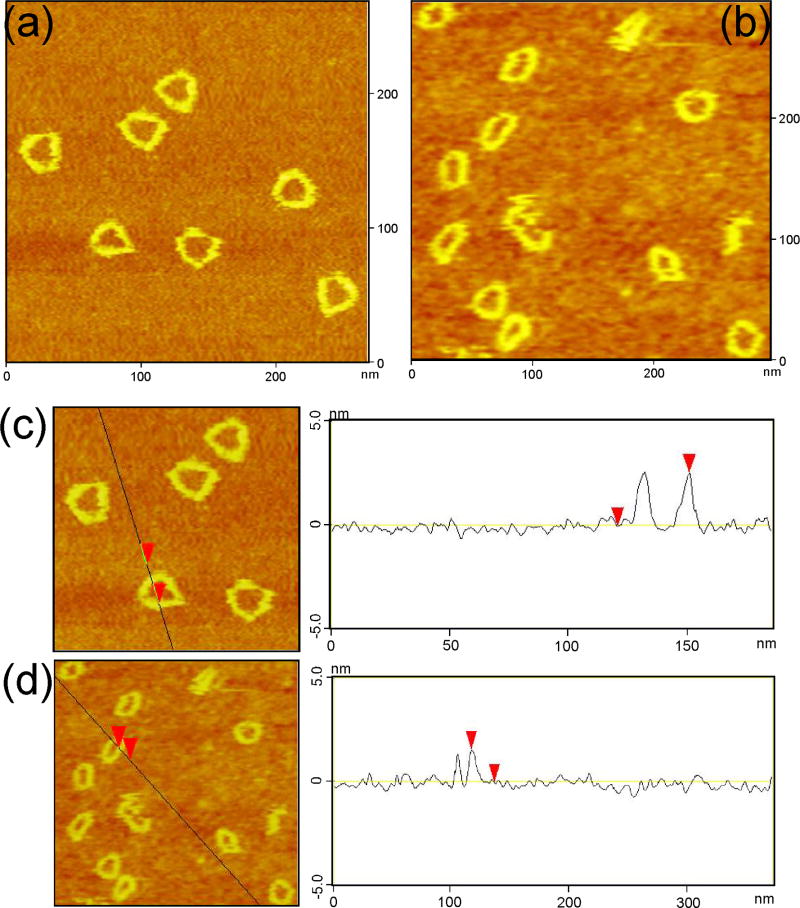
Direct visualization of the annealed products provides the strongest evidence for forming the PX DNA triangle structure. We have designed the PX triangle to be large enough that it can be seen in a conventional AFM imaging experiment. Figure 3a shows clearly the detailed triangular structure, including the triangular cavity. The length of each PX triangle edge is approximately 27.8 nm, which is consistent with the design; 76 nucleotide pairs in a double helix are expected to be about 25.8 nm in length.
Figure 3.
Atomic Force Micrographs of the PX Triangles. (a) contains images of the triangle, where the frequent appearance of a triangular cavity is evident. (b) shows the blue-green half-triangle; these images are more circular and less triangular than those in (a). (c) contains a cross-sectional analysis of the triangle, and (d) contains a cross-sectional analysis of the half-triangle; note the increased height in (c), relative to (d).
The blue-green triangle contains a cyclic strand, but the red-purple half-triangle is discontinuous, because its strands in the bottom edge (Figure 1b) are not attached to those in the other edges. Consequently, the two inter-wrapped triangles are unlinked, and the whole PX triangle can be formed directly from the components of two PX half-triangles. Figure 3b contains AFM images of the blue-green PX half-triangle. Most of the PX half-triangles fail to maintain the triangular shape and appear as round objects. Consistent with the Ferguson plot, this finding indicates that the PX half-triangle is less rigid than the whole PX triangle owing to the flexible unpaired regions in its edges. Figures 3c and 3d contain height analyses of the AFM images, which shows that the height of the PX triangle is ~2.6 nm, roughly twice of the height of the PX half-triangle (~1.7 nm), again consistent with the design.
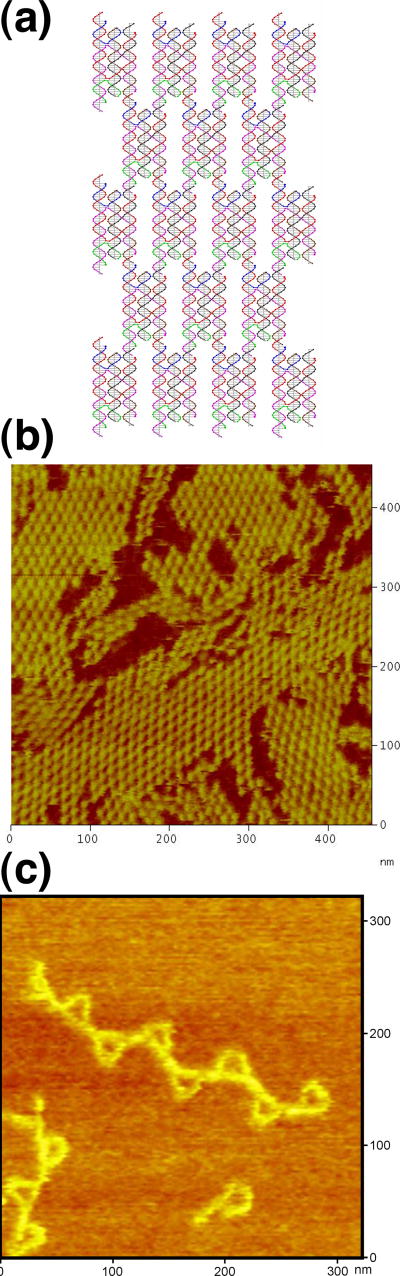
As with other DNA motifs containing parallel helix axes, the PATX motif can be formed into two dimensional arrays. In this case, we have used the top and bottom domains (which are antiparallel to each other) to form a 2D array connected 1–3, as done with a previous TX domain.24 Figure 4a contains a schematic illustrating this design, and Figure 4b shows an atomic force micrograph of such an array. The two large domains in the upper part of Figure 4b have their helix axes almost perpendicular to the top edge of the image, in an orientation similar to the drawing in Figure 4a; it is clear from those arrays how the 1–3 connectivity leads to the diamond-shaped appearance of the arrays. The gaps between the PATX components are also evident from this image.
Figure 4.
Arrays of the Motifs of the New Species Described Here. (a) A Schematic of the PATX 2-D Array. This is a GIDEON29 drawing showing that the antiparallel first and third domains are linked by sticky ends to product an array containing gaps. (b) An AFM image of the PATX 2-D Array. Note that the upper domains are oriented in almost the same direction as the schematic in (a). (c) A Linear Array of PX Triangles Assembled with PATX Motifs. The alternating orientation of the triangles conforms to the design of this arrangement.
Self-assembly of the PX triangles into arrays further demonstrates the nature of the PX triangle structure. The schematic drawing of a one dimensional (1D) array of the PX triangles fused with PATX linkers is shown in Figure 1c. A pair of the double sticky ends was added to both ends of the bottom edge of the PX triangle and connected by the PATX motif using sticky-ended DX cohesion.7 The PX motif is often poorly behaved, particularly as a triple helical motif.25 The relative twists between two consecutive triangles are designed to be 180°; this is achieved by separations between successive PX triangles that contain 48 nucleotide pairs. We arrived empirically at the separation of 48 nt pairs; PATX molecules containing 47, 49 and 50 base pairs were also examined. However, in those cases, the zigzag 1D arrays could not be formed properly, owing to phasing issues; we sometimes saw successive PX triangles on the same side of the 1D triangle arrays instead of on opposite sides. Besides the parallel TX molecules, DNA duplex and DX molecules were also tried unsuccessfully as linkers to form the zigzag 1D triangle arrays. It turned out that only the PATX connector provided the structural integrity and the double cohesion needed to form well-defined 1D PX triangle arrays.
Figure 4c shows an AFM image of a zigzag 1D PX triangle array, demonstrating that the size and position of the PX triangles are in good agreement with our design. The alternating orientations of the triangles are evident in the image. Thus, it is possible not only to generate PX triangles, but it is possible to form linear arrays of them when the PATX motif is used as a connector providing DX sticky-ended cohesion.
We expect both the PX triangle and the PATX connector to be useful additions to the current DNA structure library for nano-construction. One possible application of the PX triangle is that it could be used as a marker to record motion in nanomechanical devices attached to solid supports.18 The half-triangle is much less likely than a rigid motif to perturb the formation of the device-containing cassettes involved; however, once the cassette is formed and in place, the other half-triangle could be added, leading to a distinct marker. A second application could be in the formation of face-sharing arrangements, a type of cohesion not yet seen in polyhedral DNA nanostructures.
EXPERIMENTAL
Design, Synthesis and Purification of DNA
Strands have been designed using the program SEQUIN.26 Oligonucleotides have been synthesized on an Applied Biosystems 394 synthesizer using routine phosphoramidite chemistry or have been purchased from the Integrated DNA Technology (www.idtDNA.com). DNA strands have been purified by gel electrophoresis: bands are cut out of 10–20% denaturing gels and eluted in a solution containing 500 mM ammonium acetate, 10 mM magnesium acetate, and 1 mM EDTA.
Formation of Hydrogen-Bonded Complexes
The PX triangles are formed by mixing a stoichiometric quantity of each strand, as estimated by OD260, in a solution containing 40 mM Tris-HCl, pH 8.0, 20 mM acetic acid, 2 mM EDTA, and 125 mM magnesium acetate (TAE/Mg). The mixture is cooled slowly from 90 °C to room temperature in about 2L water bath over the course of 48 h. The concentration of PX triangle is 0.1 μM.
Non-denaturing Polyacrylamide Gel Electrophoresis
Non-denaturing gels contain of 4% acrylamide (19:1, acrylamide: bisacrylamide) and the running buffer contains of 40mM Tris-HCl (pH 8.0), 20mM acetate acid, 2mM EDTA, and 125mM magnesium acetate (1XTAE/Mg). Tracking dye containing 1XTAE/Mg, 50% glycerol, and 0.02% each of Bromophenol Blue and Xylene Cyanol FF is added to the sample buffer. Gels are run on a Hoefer SE-600 gel electrophoresis unit at 4V/cm at 4°C and were stained with 0.01% Stains-all dye (Sigma), in 45% formamide.
Radioactive Phosphorylation
10 pmol of the each of the DNA strands was dissolved in 20 μL of a solution containing 50 mM Tris-HCl (pH 7.6), 10mM MgCl2, 10 mM 2-mercaptoethanol, and mixed with 1 μL of 1.25 mM γ32P-ATP (10 mCi/mL) and 2 unit of T4 polynucleotide kinase (USB) for 1 hour at 37°C. The reaction was stopped by ethanol precipitation of DNA.
Hydroxyl Radical Analysis
Individual strands of the PATX complex were 32P labeled and additionally gel purified from a 10% denaturing PAGE. Each of the labeled strands (approximately 10 pmol in 10 μL 1x TAE.Mg2+ buffer) was subjected to three different treatments: [1] It was annealed to an excess of the unlabeled complementary strand; [2] it was annealed to an excess of a mixture of the other strands in the complex; or [3] it was treated with sequencing reagents27 for a sizing ladder. The samples were annealed by heating to 90 °C for 5 min and then step-wised cooled at 65 °C for 10 min, 45 °C for 10 min, 37 °C for 10 min, room temperature for 10 min, and 4 °C for 10 min. Hydroxyl radical cleavage of the double-strand and complex samples took place at 4 °C for 2 min,28 with modifications noted by Churchill et al.20 The reaction was stopped by addition of thiourea. The samples were dried, dissolved in a formamide/dye mixture, and loaded directly on a 12% polyacrylamide/8.3 M urea sequencing gel. Autoradiograms were quantitated using a Storm 860 Gel and Blot Imaging System (Amersham Pharmacia Biotech, Piscataway, NJ).
Atomic Force Microscopy (AFM) Imaging
A 5 μL sample was spotted on freshly cleaved mica, and the sample was left to absorb for 2 minutes. Additional fresh 1XTAE/Mg (125mM) buffer was added to both the mica and to the liquid cell. All AFM imaging was performed on a NanoScope IV (Digital Instruments), using commercial cantilevers with Si3N4 tips.
Supplementary Material
Acknowledgments
This project was stimulated by a conversation with William DeGrado. This research has been supported by grants GM-29554 from NIGMS, grants DMI-0210844, EIA-0086015, CCF-0432009, CCF-0523290 and CTS-0548774, CTS-0608889, CCF-0726378 from the NSF, 48681-EL and MURI W911NF-07-1-0439 from ARO, DE-FG02-06ER64281 from DOE (Subcontract from the Research Foundation of SUNY), a grant from the W.M. Keck Foundation.
Footnotes
Supporting Information Available: The sequences of the molecules used. This material is available free of charge via the Internet at http://pubs.acs.org/.
Contributor Information
Wenyan Liu, Email: wl502@nyu.edu.
Xing Wang, Email: xw246@nyu.edu.
Tong Wang, Email: tw343@nyu.edu.
Ruojie Sha, Email: ruojie.sha@nyu.edu.
Nadrian C. Seeman, Email: ned.seeman@nyu.edu.
References
- 1.Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Seeman NC. J Am Chem Soc. 1994;116:1661–1669. [Google Scholar]
- 4.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 5.Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1664. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, Wenzler LA, Qi J, Li X, Seeman NC. J Am Chem Soc. 1998;120:9779–9786. [Google Scholar]
- 7.Ding B, Sha R, Seeman NC. J Am Chem Soc. 2004;126:10230–10231. doi: 10.1021/ja047486u. [DOI] [PubMed] [Google Scholar]
- 8.Constantinou PE, Wang T, Kopatsch J, Israel LB, Zhang X, Sherman WB, Wang X, Zheng J, Sha R, Seeman NC. Org Biomol Chem. 2006;4:3414–3419. doi: 10.1039/b605212f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu D, Wang M, Deng Z, Walulu R, Mao C. J Am Chem Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 10.Chelyapov N, Brun Y, Gopalkrishnan M, Reishus D, Shaw B, Adleman L. J Am Chem Soc. 2004;126:13924–13925. doi: 10.1021/ja0458120. [DOI] [PubMed] [Google Scholar]
- 11.Fu TJ, Seeman NC. Biochem. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 12.Yan H, Seeman NC. J Supramol Chem. 2001;1:229–237. [Google Scholar]
- 13.Shen Z, Yan H, Wang T, Seeman NC. J Am Chem Soc. 2004;126:1666–1674. doi: 10.1021/ja038381e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeman NC. NanoLett. 2001;1:22–26. [Google Scholar]
- 15.Zhang X, Yan H, Shen Z, Seeman NC. J Am Chem Soc. 2002;124:12940–12941. doi: 10.1021/ja026973b. [DOI] [PubMed] [Google Scholar]
- 16.Yan H, Zhang X, Shen Z, Seeman NC. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 17.Liao S, Seeman NC. Science. 2004;306:2072–2074. doi: 10.1126/science.1104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding B, Seeman NC. Science. 2006;314:1583–1585. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodbard D, Chrambach A. Anal Biochem. 1971;40:95–134. doi: 10.1016/0003-2697(71)90086-8. [DOI] [PubMed] [Google Scholar]
- 20.Churchill MEA, Tullius TD, Kallenbach NR, Seeman NC. Proc Nat Acad Sci (USA) 1988;85:4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tullius TD, Dombroski BA. Proc Nat Acad Sci (USA) 1986:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seeman NC. Curr Protocols Nucl Acid Chem. Unit 12.1. John Wiley & Sons; New York: 2002. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Wang H, Seeman NC. Biochem. 1997;36:4240–4247. doi: 10.1021/bi9629586. [DOI] [PubMed] [Google Scholar]
- 24.LaBean TH, Yan H, Kopatsch J, Winfree E, Liu F, Reif JH, Seeman NC. J Am Chem Soc. 2000;122:1848–1860. [Google Scholar]
- 25.Wang T. PhD Thesis. New York University; 2007. [Google Scholar]
- 26.Seeman NC. J Biomol Str & Dyns. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 27.Maxam AM, Gilbert W. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tullius TD, Dombroski B. Science. 1985;230:679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- 29.Birac JJ, Sherman WB, Kopatsch J, Constantinou PE, Seeman NC. J Mol Graphics & Modeling. 2006;25:470–480. doi: 10.1016/j.jmgm.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.