Abstract
Motivation: When working with large-scale protein interaction data, an important analysis task is the assignment of pairs of proteins to groups that correspond to higher order assemblies. Previously a common approach to this problem has been to apply standard hierarchical clustering methods to identify such a groups. Here we propose a new algorithm for aggregating a diverse collection of matrix factorizations to produce a more informative clustering, which takes the form of a ‘soft’ hierarchy of clusters.
Results: We apply the proposed Ensemble non-negative matrix factorization (NMF) algorithm to a high-quality assembly of binary protein interactions derived from two proteome-wide studies in yeast. Our experimental evaluation demonstrates that the algorithm lends itself to discovering small localized structures in this data, which correspond to known functional groupings of complexes. In addition, we show that the algorithm also supports the assignment of putative functions for previously uncharacterized proteins, for instance the protein YNR024W, which may be an uncharacterized component of the exosome.
Contact: derek.greene@ucd.ie
Supplementary information: Supplementary data are available at http://mlg.ucd.ie/nmf.
1 INTRODUCTION
Large biological datasets comprising thousands of protein–protein interactions have been assembled in recent years (Uetz and Finley, 2005). Proteins associate with each other in cells in order to carry out biological tasks, for example as enzyme and substrate, or as components of a protein complex. Cataloguing and analyzing such interaction data is therefore a first step toward understanding the biological basis of the interactions and the role of any network structure that underlies them. Subsequent steps may layer additional data onto these networks, for instance the strength of an interaction, or when and where it occurs within the life cycle of the cell. Therefore it is highly important that the organization of the data represents the state of the proteins as fully as possible.
In recent years, the size and density of these datasets has presented a barrier to analysis, even by individuals with extensive knowledge of the proteins. For instance, 18 324 physically interacting protein pairs are described from two-hybrid, large-scale affinity pulldown, and small-scale experiments for the Saccharomyces cerevisiae proteome alone (Salwinski et al., 2004). Fundamentally, two challenges exist. First, the data must be organized according to some metric, so that proteins that behave similarly are classed together. Second, the organized datasets must be visualized in a way that simplifies them and reveals any underlying structure. Traditionally, cluster analysis methods such as hierarchical agglomerative clustering have been applied to identify functional groupings in this kind of data (Collins et al., 2007). However, a distinct drawback of such methods lies in the fact that each data object can only reside in a single branch of the tree at a given level, and can only belong to a single leaf node. In many applications it may be the case that an object will belong to multiple distinct branches in the data's natural cluster hierarchy. For instance, while in large biological datasets proteins may be associated with multiple biological processes, the output of a traditional agglomerative clustering algorithm would fail to reflect this.
As an alternative, matrix decomposition techniques such as non-negative matrix factorization (NMF) (Lee and Seung, 1999) have been recently employed in the analysis of data where overlapping structures may exist, such as in cancer class discovery and gene expression analysis (Kim and Park, 2007). NMF produces a low-dimensional approximation of a high-dimensional data matrix, in the form of non-negative factors. The non-negativity of these factors allow them to be interpreted as a flat ‘soft’ clustering of the data, where cluster assignments are not mutually exclusive. However, biologists are frequently accustomed to exploring datasets via a tree-like organization, such as that produced by traditional hierarchical clustering algorithms. So while NMF can provide us with a powerful means of identifying cluster structures, the resulting flat clustering can potentially be difficult to interpret for a domain user. While it has been suggested that some hierarchical relations between NMF factors can be deduced via manual inspection (Brunet et al., 2004), such an approach may be impractical for larger datasets which contain a significant number of complex hierarchical structures.
When analyzing protein interaction networks, we would ideally like to combine both the ability of NMF techniques to accurately identify overlapping structures, with the interpretability and visualization benefits of hierarchical techniques. Towards this end, we consider the concept of ensemble machine learning techniques, the rationale for which is that the combined judgement of a group of experts will frequently be superior to that of an individual (Breiman, 1996). A variety of algorithms have been proposed to aggregate a collection of different clusterings to yield a more accurate, informative clustering of the data (Strehl and Ghosh, 2002). However, these algorithms have largely dealt with the problem of combining hard clusterings (i.e. sets of disjoint, non-overlapping clusters), produced by an algorithm such as k-means. Brunet et al. (2004) initially considered the possibility of combining factorizations produced by NMF. However, this involved thresholding each factor to become a hard clustering and subsequently producing a disjoint consensus solution, thereby losing the ability to represent overlapping groups.
In this study, we introduce a novel ensemble clustering algorithm, based on NMF, for the analysis and identification of localized structures in sparse data, represented in the form of a pairwise similarity matrix. This involves the construction of a soft hierar-chical clustering of the data, where data objects can be associated with multiple nodes in the tree to differing degrees. We focus on the application of this algorithm to a high-quality dataset of physically interacting proteins, and demonstrate that this approach can be used to group proteins according to function, and furthermore, to identify proteins linked to two or more functions. In addition to the proposed approach, we provide a new application for visualizing and exploring a soft hierarchy of clusters.
2 METHODS
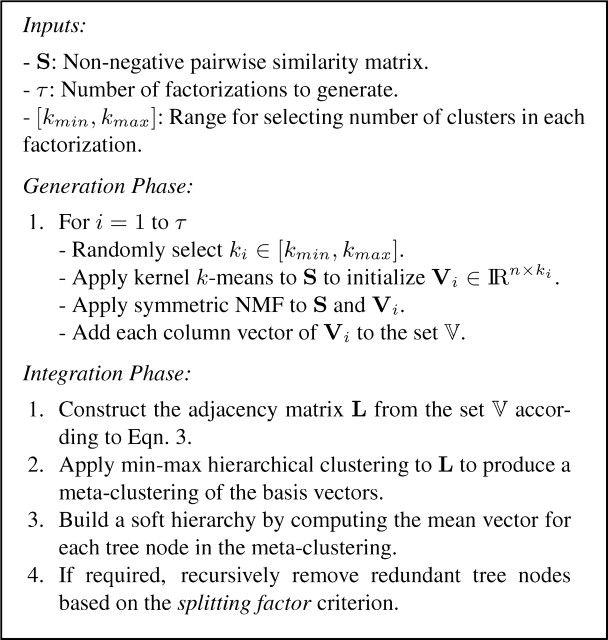
In this section, we describe an approach for producing ensemble factorizations of a symmetric matrix representing the pairwise associations in a given dataset. This approach consists of two distinct phases: a generation phase in which a collection of NMF factorizations is produced (i.e. the members of the ensemble), and an integration phase where these factorizations are aggregated to produce a final clustering of the dataset.
2.1 Ensemble generation
Given a dataset consisting of n data objects, the generation phase of the ensemble process involves the production of a collection of τ ‘base’ clusterings. These clusterings represent the individual members of the ensemble. Since we are interested in combining the output of multiple matrix factorizations, each member will take the form of a non-negative n×ki matrix factor Vi, such that ki is the number of basis vectors (i.e. clusters) specified for the i-th factorization procedure.
To generate the collection of base clusterings, we employ the symmetric NMF algorithm proposed by Ding and He, (2005). This algorithm decomposes a non-negative pairwise similarity matrix S∈IRnn to produce a factor V by minimizing the objective function:
| (1) |
The optimal factor can be approximated by starting with an initial randomly generated factor and repeatedly applying a single update rule until convergence
| (2) |
where 0<β≤1 is a user-defined parameter which controls the rate of convergence. We have observed that, not only is the algorithm efficient in comparison to other NMF algorithms, but it also has a tendency to produce relatively sparse factors representing localized clusters.
It has been demonstrated that supervised ensembles are most successful when constructed from a set of accurate classifiers whose errors lie in different parts of the data space (Opitz and Shavlik, 1996). Similarly, unsupervised ensemble procedures typically seek to encourage diversity with a view to improving the quality of the information available in the integration phase. A simple but effective strategy is to rely on the inherent instability of randomly initialized factorization algorithms. By employing a stochastic initialization scheme, symmetric NMF will generally converge to a variety of different local solutions when applied multiple times to the same matrix S. The level of diversity among the ensemble members can be increased by varying the number of clusters in each base clustering, such as by randomly selecting a value ki from a predefined range [kmin,kmax]. An important benefit of this strategy is that it ameliorates a model selection problem with NMF which is highly sensitive to the choice of the number of basis vectors ki.
Further improvements in performance and accuracy can be achieved by seeding each NMF factorization using the output of the less computationally expensive kernel k-means algorithm (Schölkopf et al., 1998). Specifically, to seed the i-th base clustering, we randomly assign data objects to ki clusters and apply kernel k-means to the matrix S. The resulting disjoint clustering can be represented as an n × ki partition matrix, where the j-th column is a binary membership indicator for the j-th cluster. This partition matrix is subsequently used as the initial factor for symmetric NMF. The use of random cluster assignment and the tendency of kernel k-means to converge to a local solution ensures that sufficient diversity in the ensemble is maintained.
2.2 Ensemble integration
We now propose a novel approach for combining the factors produced during the generation phase to construct a soft hierarchical clustering of the original data.
2.2.1 Graph construction
From the generation phase, we have a collection of τ factors, giving a total of l=(k1+k2+···+kτ) individual basis vectors across all factors. We denote these vectors as the set 𝕍={v1…,vl}. This set can be modeled as a complete weighted graph consisting of l vertices, where each vertex represents a basis vector vi. The weight on each edge indicates the similarity between the pair of vectors associated with the two vertices. The value of the edge weight is computed as the [0,1]-normalized Pearson correlation (Strehl et al., 2000) between the pair of vectors (vi,vj):
| (3) |
The entire graph can be represented by its adjacency matrix L, where Lij=ncor(vi,vj).
2.2.2 Meta-clustering
Following the lead of the MCLA approach described by Strehl and Ghosh, (2002), we produce a ‘meta-clustering’ (i.e. a clustering of clusters) of the graph formed from the basis vectors in 𝕍. This is achieved by applying an agglomerative clustering algorithm to L, resulting in a disjoint hierarchy of ‘meta-clusters’ (i.e. tree nodes containing basis vectors from 𝕍). Rather than using a traditional linkage function such as average linkage during the agglomeration process, we compute the similarity between pairs of meta-clusters based on the min-max graph partitioning objective (Ding and He, 2002), as this linkage function has a tendency to produce clusters which are relatively balanced in size. Formally, given the matrix L, the min-max inter-cluster similarity between a pair of meta-clusters (Ma, Mb) is defined as:
| (4) |
such that
2.2.3 Soft hierarchy construction
The output of the meta-clustering procedure is a clustering of the basis vectors in 𝕍, in the form of a traditional disjoint hierarchical tree. We wish to transform this into a soft hierarchical clustering of the original dataset. That is, a binary tree structure, where each node Ma in the hierarchy is associated with an n-dimensional vector ya containing non-negative real values indicating the degree of membership for all n data objects. In practice, these node membership vectors will become increasingly sparse as we proceed further down the tree, representing more localized sub-structures.
To transform the meta-clustering into a soft hierarchy, we process each node Ma in the meta-clustering tree, computing the membership vector ya as the mean of all the basis vectors contained in Ma:
| (5) |
We associate the vector ya with the position held by the node Ma in the original meta-clustering tree. By preserving the parent–child relations from the previous tree, these vectors can be linked together to form a soft hierarchy as defined above.
2.2.4 Final model selection
A hierarchical meta-clustering of the l basis vectors in 𝕍 will yield a corresponding soft hierarchy containing l leaf nodes. However, a certain proportion of these nodes will be redundant, where the membership vectors of a pair of sibling nodes may be nearly identical to the membership vector of their parent node. This situation will arise when a tree node in the meta-clustering of 𝕍 contains basis vectors that are highly similar to one another. Ideally we would like to prune the soft hierarchy to remove all redundant leaf and internal nodes, thereby facilitating visualization and human interpretation. This problem is equivalent to the identification of an appropriate cut-off point in the tree. The concept of ensemble stability has previously been considered as a means of identifying an appropriate cut-off point in a disjoint hierarchy (Giurcaneanu and Tabus, 2004).
Here we propose a stability-based approach to identifying an appropriate cut-off level, which is applicable to a soft hierarchy. Specifically, we consider a tree node to be stable if the basis vectors in the corresponding meta-cluster are highly similar, while an unstable node has a corresponding meta-cluster consisting of basis vectors that are dissimilar to one another. To numerically assess stability, we measure the extent to which an internal node can be split into diverse sub-nodes. Given a node Ma with child nodes (Mb,Mc), this can be quantified in terms of the weighted similarity between the membership vector ya and the pair of vectors (yb,yc) associated with the child nodes:
| (6) |
From this, we define the splitting factor of an internal node Ma as the minimum value for Equation 6 among Ma and all child nodes below Ma in the hierarchy. A lower value indicates a lower degree of stability for the branch beginning at Ma. Using this definition, we can prune a soft hierarchy by processing each internal node Ma in the tree, starting at the root node. The child nodes of Ma (together with all the nodes below them) are removed from the tree if the splitting factor of Ma is greater than or equal to a user-defined threshold λ. In practice we have observed that a threshold value of λ=0.9 frequently leads to the elimination of redundant nodes without removing those containing informative structures.
The pruning procedure outlined above allows us to construct a tree with k leaf nodes, where the value k does not need to be specified a priori. As with cut-off techniques used to convert a disjoint hierarchy to a flat partition, we can produce a flat soft clustering from the leaf nodes in the tree. Specifically, we construct a n × k matrix whose columns correspond to the vectors of the k non-redundant leaf nodes in the soft hierarchy. Unlike spectral dimension reduction procedures such as PCA, standard NMF techniques do not produce an ordering of the new dimensions in terms of importance. To produce an ordering of the columns in the flat soft clustering, the related k leaf nodes may be ranked based on their splitting factor, with the first column corresponding to the most stable node. The complete Ensemble NMF algorithm is summarized in Figure 1.
Fig. 1.
Summary of Ensemble NMF clustering algorithm.
3 RESULTS
3.1 Experimental setup
To examine the Ensemble NMF algorithm presented in Section 2, we focused on a real-life problem, the assignment of experimentally determined co-purifying protein pairs to groups that correspond to higher order protein assemblies. We used an extensive and high-quality assembly of binary interactions for 2390 proteins (Collins et al., 2007) derived from two proteome-wide studies in yeast (Gavin et al., 2006; Krogan et al., 2006). Both these studies used a genetically encoded affinity tag fused to a single protein (‘bait’) to pull down associated proteins (‘prey’), and each bait is linked to its respective preys by a confidence score measuring the evidence that the proteins do indeed co-purify, referred to as purification enrichment (PE). The resulting pairwise PE score matrix was normalized to the range [0,1] prior to the application of Ensemble NMF.
In total, an ensemble of 1000 factorizations was generated, which yielded a robust clustering. Each symmetric NMF factorization procedure was limited to a maximum of 150 iterations for practical purposes, although the algorithm frequently converged before this point was reached. When applying symmetric NMF, we set the convergence parameter β to 0.5 as recommended by Ding and He, (2005), which provides a good trade-off between accuracy and running time. As noted previously, we have proposed simplifying the NMF model selection process by allowing the use of a broad range for the number of basis vectors, rather than requiring a specific value for k. For the Collins dataset we use kmin=40 and kmax=60, and we observe that the clustering process was not particularly sensitive to variations of ±5 in these values. This range covers the expected number of functional groups in the data, so the user must have some prior expectations about that.
As a baseline for comparison, we also conducted experiments involving the application of traditional average-linkage agglomerative hierarchical clustering to the uncentered correlation matrix calculated from the PE interaction scores, as previously performed in Collins et al., (2007).
3.2 Discussion
In our experiments we wished to determine whether the Ensemble NMF algorithm satisfied any or all the features of an ideal procedure for assigning proteins to an appropriate complex. These are discussed in the following five subsections:
3.2.1 Similarity of groupings to known protein complex compositions
Cluster validation techniques are often employed to evaluate the output of cluster analysis algorithms in cases where some form of gold standard reference set is available. Although the catalogue of documented protein interactions for yeast is probably incomplete, we can make use of two resources to evaluate the biological relevance of the results produced by Ensemble NMF:
MIPS database: A database of stably interacting proteins supported by multiple experiments is maintained by the Munich Information Center for Protein Sequences1 (MIPS) and is often used to benchmark methods for defining protein complex composition from raw data (Mewes et al., 2004).
SGD database: Since MIPS data was used to estimate one of the constants for interaction confidence in the calculation of the PE score, we also used an additional independent reference set for validation. This set was generated from complexes given in the SGD database (Cherry et al., 1998) as described in (Collins et al. (2007).
To numerically compare the quality of the output produced by the two algorithms under consideration, we use two validation measures that have commonly been employed in a variety of machine learning tasks. First, we assess the precision of the clusters, which is defined as the fraction of proteins in each cluster that pertain to a specific complex in the MIPS or SGD database. Second, we assess the recall of the clusters, which refers to the fraction of the proteins from a given complex that were included in a cluster. High precision implies that most proteins in a given cluster belong to the same complex, while high recall suggests that most proteins from a single complex were assigned to the same cluster. Since both validation measures are applicable only to hard clusters, we produce hard overlapping clusters from the real-valued output of the Ensemble NMF procedure by assigning proteins to a cluster if their membership weight for that cluster exceeds a given threshold. While many cluster membership vectors will be naturally sparse, we found experimentally that a threshold of 0.1 proved suitable in this context.
MIPS-based validation scores2 a step to remove ribosomes were used to generate the raw data in Collins et al. (2007). for the most relevant clusters identified by Ensemble NMF are given in Table 1, with scores for average linkage hierarchical clustering listed in Table 2. Corresponding validation scores based on the SGD reference set are provided in Table 3 and Table 4, respectively. These tables display the 20 most significant clusters in the clustering trees, as ranked by their precision scores. Several notable differences are apparent when the consolidated protein interaction data was clustered using the Ensemble NMF algorithm, when compared to average-linkage clustering on the uncentered correlation matrix. First, Ensemble NMF, when combined with the sparse PE score matrix, lends itself to discovering small localized structures in the data (see subsequently). Notably the structures uncovered seem to be far more informative and interesting than those identified using the baseline approach. This is reflected in the substantially improved validation scores for both validation approaches, which illustrate that a higher number of significant structures are uncovered by Ensemble NMF. For instance, the algorithm successfully recovers all of the top 20 SGD complexes with perfect precision (1.0), while seven such complexes were perfectly identified by average-linkage clustering. Similarly, Ensemble NMF manages to recover nine MIPS complexes with perfect precision, compared to only two by the traditional clustering approach. Additional comparisons between the algorithms, based on cluster significance, can be found in the Supplementary Materials.
Table 1.
Validation scores for 20 most significant clusters identified by Ensemble NMF on Collins data, based on MIPS complexes
| MIPS complex | Precision | Recall |
|---|---|---|
| 20S proteasome | 1.00 | 0.88 |
| Anaphase promoting complex (APC) | 1.00 | 0.80 |
| H+-transporting ATPase vacuolar | 1.00 | 0.64 |
| Post-replication complex | 1.00 | 1.00 |
| Pre-replication complex (pre-RC) | 1.00 | 0.60 |
| Replication complex | 1.00 | 0.40 |
| Replication initiation complex | 1.00 | 0.75 |
| Septin filaments | 1.00 | 1.00 |
| TRAPP complex | 1.00 | 0.70 |
| RNA polymerase I | 0.93 | 0.59 |
| SWI/SNF activator complex | 0.89 | 0.89 |
| COPI | 0.88 | 1.00 |
| Exocyst complex | 0.88 | 1.00 |
| Kornbergs mediator (SRB) complex | 0.86 | 1.00 |
| Signal recognition particle (SRP) | 0.86 | 1.00 |
| Gim complexes | 0.83 | 1.00 |
| TFIIIC | 0.83 | 1.00 |
| 19/22S regulator | 0.78 | 1.00 |
| Arp2p/Arp3p complex | 0.71 | 1.00 |
| Class C Vps protein complex | 0.67 | 1.00 |
Table 2.
Validation scores for 20 most significant clusters identified by average-linkage clustering on Collins data, based on MIPS complexes
| MIPS complex | Precision | Recall |
|---|---|---|
| Geranylgeranyltransferase II | 1.00 | 0.67 |
| v-SNAREs | 1.00 | 0.33 |
| NEF3 complex | 0.50 | 0.14 |
| RNA polymerase I | 0.50 | 0.05 |
| RNase MRP | 0.50 | 1.00 |
| RNase P | 0.50 | 1.00 |
| Replication factor C complex | 0.50 | 1.00 |
| mRNA splicing | 0.50 | 0.04 |
| Other respiration chain complexes | 0.50 | 0.14 |
| RSC complex | 0.27 | 0.90 |
| SWI/SNF transcription activator complex | 0.27 | 1.00 |
| SAGA complex | 0.14 | 0.91 |
| rRNA splicing | 0.13 | 0.15 |
| Dam1 protein complex | 0.10 | 1.00 |
| 20S proteasome | 0.09 | 0.94 |
| RNA polymerase III | 0.08 | 0.92 |
| ADA complex | 0.07 | 0.83 |
| RNA polymerase II | 0.07 | 0.85 |
| TRAPP complex | 0.06 | 1.00 |
| Exocyst complex | 0.06 | 1.00 |
Table 3.
Validation scores for 20 most significant clusters identified by Ensemble NMF on Collins data, based on SGD complexes
| SGD complex | Precision | Recall |
|---|---|---|
| Histone deacetylase complex | 1.00 | 0.24 |
| Vesicle coat | 1.00 | 0.28 |
| COPI vesicle coat | 1.00 | 1.00 |
| Ribonucleoprotein complex | 1.00 | 0.06 |
| Small nuclear ribonucleoprotein complex | 1.00 | 0.42 |
| Histone acetyltransferase complex | 1.00 | 0.46 |
| Preribosome | 1.00 | 0.18 |
| 90S preribosome | 1.00 | 0.21 |
| SAGA complex | 1.00 | 0.90 |
| Transcription factor TFIIIC complex | 1.00 | 1.00 |
| Exocyst | 1.00 | 1.00 |
| Ubiquitin ligase complex | 1.00 | 0.25 |
| U4 | 1.00 | 0.82 |
| COMPASS complex | 1.00 | 1.00 |
| Septin complex | 1.00 | 1.00 |
| DNA replication preinitiation complex | 1.00 | 0.50 |
| V-ATPase V1 domain | 1.00 | 0.88 |
| Small subunit processome | 1.00 | 0.27 |
| Rpd3L complex | 1.00 | 0.91 |
| Pre-replicative complex | 1.00 | 0.67 |
Table 4.
Validation scores for 20 most significant clusters identified by average-linkage clustering on Collins data, based on SGD complexes
| SGD complex | Precision | Recall |
|---|---|---|
| Ribonucleoprotein complex | 1.00 | 0.01 |
| NatB complex | 1.00 | 1.00 |
| NatC complex | 1.00 | 1.00 |
| Commitment complex | 1.00 | 0.18 |
| Transcription factor complex | 1.00 | 0.02 |
| Holo TFIIH complex | 1.00 | 0.20 |
| Spliceosome | 1.00 | 0.03 |
| Ctf18 RFC-like complex | 0.70 | 1.00 |
| Ribonuclease MRP complex | 0.63 | 1.00 |
| Elg1 RFC-like complex | 0.50 | 1.00 |
| Core TFIIH complex | 0.50 | 0.20 |
| SSL2-core TFIIH portion of NEF3 | 0.50 | 0.17 |
| SSL2-core TFIIH portion of holo TFIIH | 0.50 | 0.17 |
| Nucleolar ribonuclease P complex | 0.50 | 1.00 |
| DNA replication factor C complex | 0.50 | 1.00 |
| Histone acetyltransferase complex | 0.24 | 0.46 |
| SAGA complex | 0.24 | 0.90 |
| SLIK (SAGA-like) complex | 0.20 | 0.82 |
| Transcription factor TFIID complex | 0.20 | 0.93 |
| DASH complex | 0.11 | 0.89 |
3.2.2 Arrangement of the data should be presented in an intuitive visual format
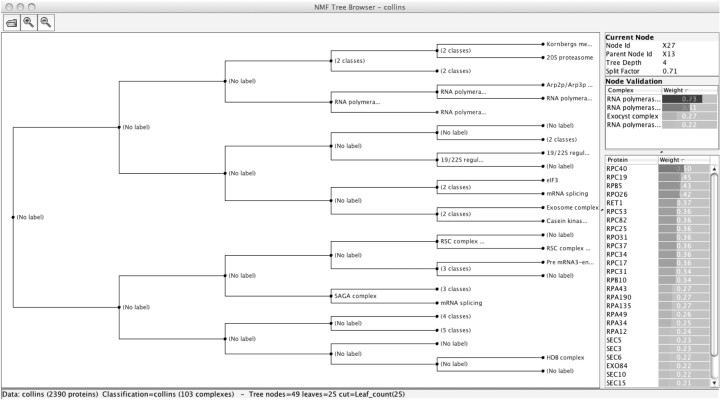
Visualization of high density biological data becomes more challenging as datasets increase in size and complexity (Uetz et al., 2002). As well as producing significant and accurate results, it is vital that the output of a cluster analysis procedure applied to biological data should be readily interpretable. Toward this end, we have developed the ‘NMF Tree Browser’, a cross-platform Java application for visually inspecting a soft hierarchy as produced by the Ensemble NMF algorithm. The output is graphically arranged in a hierarchical format where the user can click on any node to reveal its contents, in terms of membership weights for proteins together with any suggested complex annotations (e.g. from the MIPS or SGD reference set). A screenshot of the main window of the application is shown in Figure 2. Additionally, the user has two other options for viewing, one (‘Complex Browser’) focusing specifically on tree node labeling, revealing the protein components and their appropriate validation scores; the other (‘Protein Browser’) focuses on the individual proteins and revealing their association with different complexes. The application is freely available online3, together with an implementation of the Ensemble NMF algorithm itself.
Fig. 2.
Screenshot of the NMF Tree Browser application displaying the output of Ensemble NMF, when applied to the Collins protein interaction dataset.
3.2.3 Provision of meaningful hierarchical structure
An additional feature of real life protein complexes is that the number of constituent proteins is highly variable. For example, the ribosome contains approximately 150 different proteins, while enolase, which catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate, is a dimer. It can be difficult for an algorithm using a single set of parameters to accurately identify such complexes, particularly as it is now evident that the larger complexes are composed, sometimes transiently, from smaller subcomplexes (Gavin et al., 2006; Krogan et al., 2006). In addition, this kind of composition might not be revealed by algorithms that produce flat clusterings, such as standard NMF (Lee and Seung, 1999) or symmetric NMF (Ding and He, 2005). In contrast, the hierarchical nature of the output produced by the Ensemble NMF algorithm lends itself to this analysis, where for instance the COMA subcomplex (Ame1, Okp1, Mcm21, Ctf19) of the larger CTF19 central kinetochore complex can be resolved.
3.2.4 Identification of shared subunits
An analysis tool for protein interaction data should be able to accommodate proteins that are present in two or more groupings, a situation that may arise when a single protein carries out similar functions in different protein complexes (i.e. shared subunits). Complete lists of shared subunits defined in the MIPS and SGD reference sets, which were classified as such using Ensemble NMF, can be found in Tables 7 and 8, respectively in the Supplementary Materials.
A well-known example in this context is the sharing of subunits between the distinct protein complexes RNA polymerase I, II and III that specialize in the transcription of different RNA species (Shpakovski et al., 1995). This is accurately reflected in the soft hierarchy produced by Ensemble NMF, highlighting the flexibility and resolution of the approach for the analysis of protein interactions. Four subunits common to all three polymerases (RPB5, RPB8, RPB10, RPO26) are appropriately classified, as are RPC19 and RPC40, subunits shared between RNA polymerases I and III, but not found in RNA polymerase II (Lalo et al., 1993). Similarly, shared components of the ADA, SAGA and SLIK histone acetyltransferase complexes (ADA2, SPT3, SPT7, SPT20) are also appropriately classified (Sterner and Berger, 2000), as are ARP7 and ARP9, two proteins that form a stable heterodimer within the SWI/SNF and RSC chromatin remodeling complexes (Szerlong et al., 2003).
3.2.5 Assignment of putative protein function
Grouping proteins based on common properties has been used to assign putative functions to those proteins, using the principle of ‘guilt by association’ (Hazbun and Fields, 2001). That is, proteins tend to associate with other proteins involved in similar processes. In fact, the fraction of uncharacterized proteins, even for the intensively studied S.cerevisiae species, is still substantial (Pena-Castillo and Hughes, 2007). The NMF Browser application supports the discovery of such associations through the use of the ‘Protein Browser’ window, which lists all unannotated proteins associated with a given node in the hierarchy, and suggests putative functional groupings for those proteins.
Interestingly, using the application we find that the uncharacterized protein YNR024W is grouped within a tree node that contains all 12 members of the exosome complex. In the original data, YNR024W was experimentally co-purified with Rrp4, Rrp6, Rrp42, Rrp45, Rrp46, Cls4 and Lrp1 (Krogan et al., 2006). The exosome mediates 3′$ processing and degradation of eukaryotic RNA (Mitchell et al., 1997). This suggests that YNR024W may be a previously undescribed component of this complex, and/or participate in these processes. Indeed, we have detected a genetic interaction between Lrp1 and YNR024W, providing independent evidence that this is the case (N.Krogan, data not shown).
4 CONCLUSION
In this article, we have presented a new clustering approach that involves aggregating a collection of matrix factorizations generated using NMF-like techniques. In evaluations performed on the high-quality protein interaction dataset produced by (Collins et al., 2007), we have observed that the proposed algorithm can improve our ability to identify groupings that accurately reflect known protein complex compositions. Unlike traditional agglomerative algorithms that have previously been used in this context, the soft hierarchical clustering produced by Ensemble NMF facilitates the discovery of overlapping groups and multifunction proteins, while still providing the user with an intuitive, tree-like organization of the data. To visualize the output of the algorithm and to assist in the labeling of previously uncharacterized proteins, we have developed the ‘NMF Tree Browser’ application. We suggest that this application, combined with the Ensemble NMF algorithm, may also prove useful in other data exploration tasks, such as the extraction of meaningful structures from genetic interaction data.
Conflict of Interest: none declared.
Footnotes
REFERENCES
- Breiman L. Bagging predictors. Mach. Learn. 1996;24:123–140. [Google Scholar]
- Brunet J-P, et al. Metagenes and molecular pattern discovery using matrix factorization. Proc. Nat. Acad. Sci. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SRR, et al. Towards a comprehensive atlas of the physical interactome of saccharomyces cerevisiae. Mol. Cell. Proteomics. 2007 doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Ding C, He X. Cluster merging and splitting in hierarchical clustering algorithms. In. 2002:139–146. Proceedings of the IEEE International Conference on Data Mining (ICDM'02) [Google Scholar]
- Ding C, He X. On the equivalence of nonnegative matrix factorization and spectral clustering. In. 2005:606–610. Proceedings of the SIAM International Conference on Data Mining (SDM'05) [Google Scholar]
- Gavin A-C, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Giurcaneanu C, Tabus I. Cluster structure inference based on clustering stability with applications to microarray data analysis. EURASIP J. Appl. Sign. Process. 2004;1:64–80. [Google Scholar]
- Hazbun TR, Fields S. Networking proteins in yeast. Proc. Natl Acad. Sci. USA. 2001;98:4277–4278. doi: 10.1073/pnas.091096398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Park H. Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics. 2007;23:1495–1502. doi: 10.1093/bioinformatics/btm134. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, et al. Global landscape of protein complexes in the yeast saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Lalo D, et al. Interactions between three common subunits of yeast RNA polymerases i and iii. Proc. Natl Acad. Sci. USA. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- Mewes H, et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32(Database Issue):D41. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, et al. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3'–5' exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Opitz DW, Shavlik JW. Generating accurate and diverse members of a neural-network ensemble. Neural Inf. Process. Syst. 1996;8:535–541. [Google Scholar]
- Pena-Castillo L, Hughes TR. Why are there still over 1000 uncharacterized yeast genes? Genetics. 2007;176:7–14. doi: 10.1534/genetics.107.074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwinski L. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölkopf B, et al. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 1998;10:1299–1319. [Google Scholar]
- Sterner D, Berger S. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. R. 2000;64:435. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehl A, Ghosh J. AAAI/MIT Press; 2002. Cluster ensembles – a knowledge reuse framework for combining partitionings. In; pp. 93–98. Proceedings of the Conference on Artificial Intelligence (AAAI'02) [Google Scholar]
- Strehl A, et al. AAAI/MIT Press; 2000. Impact of similarity measures on web-page clustering. In; pp. 58–64. Proceeding of the AAAI Workshop on AI for Web Search. [Google Scholar]
- Szerlong H, et al. The nuclear actin-related proteins arp7 and arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. . EMBO J. 2003;22:3175–3187. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Finley R. From protein networks to biological systems. FEBS Lett. 2005;579:1821–1827. doi: 10.1016/j.febslet.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Uetz P, et al. Visualization and integration of protein-protein interactions. In Protein-Protein Interactions–a Molecular Cloning Manual. 2002 [Google Scholar]