Abstract
A comparative population genetics study revealed high levels of nucleotide polymorphism and intermediate-frequency alleles in an arcC gene of Staphylococcus epidermidis, but not in a homologous gene of the more aggressive human pathogen, Staphylococcus aureus. Further investigation showed that the arcC genes used in the multilocus sequence typing schemes of these two species were paralogs. Phylogenetic analyses of arcC-containing loci, including the arginine catabolic mobile element, from both species, suggested that these loci had an eventful history involving gene duplications, rearrangements, deletions, and horizontal transfers. The peak signatures in the polymorphic S. epidermidis locus were traced to an arcD-like gene adjacent to arcC; these signatures consisted of unusually elevated Tajima’s D and π/K ratios, which were robust to assumptions about recombination and species divergence time and among the most elevated in the S. epidermidis genome. Amino acid polymorphisms, including one that differed in polarity and hydropathy, were located in the peak signatures and defined two allelic lineages. Recombination events were detected between these allelic lineages and potential donors and recipients of S. epidermidis were identified in each case. By comparison, the orthologous gene of S. aureus showed no unusual signatures. The ArcD-like protein belonged to the unknown ion transporter 3 family and appeared to be unrelated to ArcD from the arginine deiminase pathway. These studies report the first comparative population genetics results for staphylococci and the first statistical evidence for a candidate target of balancing selection in S. epidermidis.
Keywords: Staphylococcus aureus, Staphylococcus epidermidis, Population genetics, Balancing selection, Genetic hitchhiking, Approximate Bayesian computation
Introduction
Staphylococcus aureus and Staphylococcus epidermidis are normal bacterial flora of humans and are opportunistic pathogens. These two species differ in the primary niche that they inhabit on the human body and in their ability to cause disease (Mathema et al. 2009). Staphylococcus epidermidis widely colonizes the skin of the entire human population, but is a mild opportunistic pathogen; infections are associated with indwelling medical devices such as intravenous catheters and prosthetic heart valves (von Eiff et al. 2002). By comparison, S. aureus asymptomatically colonizes the anterior nares of up to 30 % of the human population (van Belkum et al. 2009), while also causing infections that range in severity from relatively mild skin boils and food poisoning to life-threatening osteomyelitis, pneumonia, and endocarditis (Boucher et al. 2010). This difference in virulence between the two species has been credited to the greater number of virulence factors, including many toxins and immune evasion mechanisms, encoded within the genome of S. aureus (Foster 2005; Gill et al. 2005). The virulence factors of S. epidermidis are few and largely limited to colonization and persistence functions (Otto 2009).
In S. aureus, genetic variation is thought to occur primarily by point mutations that are only rarely recombined between strains, resulting in a number of well-defined clones and lineages of clones called clonal complexes (Feil et al. 2003; Ruimy et al. 2008; Vos and Didelot 2009). Far less is known about the population genetics of S. epidermidis. Multilocus sequence typing (MLST) of seven housekeeping gene fragments is able to define S. epidermidis clones that maintain some stability over space and time, but robust clonal complexes have not yet been identified (Miragaia et al. 2007; Wong et al. 2010). It is thought that recombination plays a greater role in producing genetic variation in S. epidermidis than in S. aureus (Kozitskaya et al. 2005; Miragaia et al. 2007). However, an analysis of linkage disequilibrium in MLST data from heterogenous samples deposited in sequence databases has indicated that the genetic variation of both species is equally impacted by recombination (Pérez-Losada et al. 2006). Understanding the relative roles of mutation and recombination in producing genetic variation and the relative roles of genetic drift and natural selection in modulating the frequency of polymorphisms is fundamental to understanding the natural history of pathogenic bacteria. To date, there has been no direct comparison of the population genetics of S. aureus and S. epidermidis based on sampling of both species from the same human population.
As a result of differences in ecology and pathogenicity, it could be hypothesized that the targets of natural selection in S. aureus and S. epidermidis differ, and that revealing these targets could provide unique insights into the adaptation of these species. Recently, a scan of the S. aureus genome identified candidate targets of balancing selection (Thomas et al. 2012), but no such candidates have been reported for S. epidermidis. Balancing selection is a rarely detected form of positive selection that maintains multiple favorable alleles at intermediate frequencies in a population, resulting in high levels of nucleotide polymorphism at the targeted loci (Charlesworth 2006; Kreitman and Di Rienzo 2004). Alleles that are maintained by balancing selection can persist for longer periods of time than neutral polymorphisms, even extending beyond speciation events in some cases (Wiuf et al. 2004). There are several reasons for studying these long-lived polymorphisms in pathogenic bacteria including their possible use for probing ancient population structure (Richman 2000), ecology (Brisson and Dykhuizen 2004; Wildschutte and Lawrence 2007), and the targets of host immunity (Weedall and Conway 2010).
In this study, we investigated the population genetics of S. aureus and S. epidermidis sampled from a single hospital over a period of 6 months. Our initial goal was to compare the mutation and recombination rates and levels of genetic variation in MLST genes of the two species. During this investigation, we identified statistically unusual molecular signatures that were consistent with balancing selection in an arcD-like gene of S. epidermidis.
Materials and Methods
Bacterial Isolates
The staphylococcal isolate collection described previously in Smyth et al. (2011) was used for this study. In brief, staphylococci from clinical specimens were collected weekly from January to June 2007 from the Microbiology Laboratory of Westchester Medical Center in Valhalla, NY. A total of 136 of 181 S. aureus isolates and 129 of 160 S. epidermidis isolates were selected for this study to provide ~130 isolates per species. Bacteria were grown overnight on tryptic soy agar plates at 37 °C. Isolates were stored long-term at −80 °C in a solution of tryptic soy broth and 15 % glycerol (v/v). Bacterial genomic DNA was isolated with the DNeasy kit (Qiagen), according to the manufacturer’s instructions. The genetic backgrounds of the isolates were determined using the multilocus sequence typing (MLST) schemes published for S. aureus (Enright et al. 2000) and S. epidermidis (Thomas et al. 2007). These MLST schemes consist of seven housekeeping gene fragments that are amplified by PCR and sequenced on both DNA strands. Multilocus sequence types (STs) for 86 of these isolates were reported in Smyth et al. (2011) in a study of the methicillin resistance genetic element, SCCmec type IV. Genetic data for all study isolates are provided in supplementary Table S1.
Genetic Characterization of Loci that Encode arcC
Using reciprocal BLASTP searches, three different paralogs of the arcC gene, which mapped to three different loci, were identified among the published genome sequences of S. aureus strains FPR3757 and N315 and S. epidermidis strains RP62a and ATCC12228 (Diep et al. 2006; Gill et al. 2005; Kuroda et al. 2001; Zhang et al. 2003). These three loci included two native loci present in all of these genome sequences, which we named Locus 1 and Locus 2, as well as the variably present arginine catabolic mobile element (ACME) (Diep et al. 2006). All isolates in this study were screened for the arcC gene of these three loci by PCR, and amplicons were subsequently sequenced. The arcA gene of ACME was also screened by PCR as described previously (Diep et al. 2008). Full-length sequences of Locus 2 were obtained by PCR walking across arcB, arcC, and arcD-like genes for single, randomly selected isolates of each unique ST. In addition, a fragment of the arcD-like gene of Locus 2 was amplified by PCR and sequenced for all S. epidermidis isolates. PCR primer sequences are provided in supplementary Table S2. All sequencing was done on both DNA strands, and sequences were assembled, edited, and aligned using Lasergene software v7.2.1 (DNAStar, Madison, WI). Alignments were made on the translated amino acid sequences and back-translated to nucleotides. Gapped positions in alignments were excluded from all analyses. For phylogenetic analyses of ArcB, ArcC, and ArcD-like amino acid sequences, alignments were made with MUSCLE v3.7 (Edgar 2004) and curated with Gblocks v0.91b (Castresana 2000), using default settings. Maximum likelihood trees were constructed from the curated alignments using PhyML under a WAG model of amino acid substitution (Dereeper et al. 2008).
Analyses of Genetic Variation and Recombination
ST diversity was measured using Simpson’s index (Grundmann et al. 2001). Tajima’s and Watterson’s estimator of θ (Tajima 1989; Watterson 1975), Tajima’s D (Tajima 1989), the ratio of intraspecific nucleotide polymorphism (π) to interspecific nucleotide divergence (K) (Hudson et al. 1987), and Fay and Wu’s H (Fay and Wu 2000), were calculated using DnaSP v5.10 software (Librado and Rozas 2009). Mutation and recombination parameters were estimated from the MLST data of the two species separately using ClonalFrame v1.2 software (Didelot and Falush 2007). Each run of ClonalFrame used a Monte Carlo Markov chain (MCMC) of 500,000 iterations, discarding the first half as burn-in and saving every 500th iteration thereafter. The program was run independently five times for each dataset and the mixing and convergence of each MCMC was checked by manual comparisons and using the program’s built-in tools. The reported parameter estimates represent averages of the five runs.
RDP v.3.44 software (Martin et al. 2010) was used to detect recombinations in Locus 2 with the Chimaera (Posada and Crandall 2001), Geneconv (Sawyer 1989), MaxChi (Maynard Smith 1992), and RDP (Martin and Rybicki 2000) methods. Only events detected by three of the four methods were examined. Recombination was considered statistically significant at the P <0.05 level, and a Bonferroni correction for multiple testing was applied. Sequences were analyzed as linear fragments and recombination breakpoints were polished.
Analyses of Balancing Selection
Signatures of balancing selection can be detected in the allele frequency spectrum as summarized by Tajima’s D (Tajima 1989) and in the π/K ratio (Hudson et al. 1987). Under balancing selection, D is expected to be positive, reflecting intermediate-frequency alleles, and π/K is expected to be high, reflecting high levels of intraspecific polymorphism while controlling for local mutation rate (Andrés et al. 2009; Innan 2006; Ochola et al. 2010; Thomas et al. 2012). To test whether a focal region exhibits unusually high values of D and π/K that might have been caused by natural selection, a reference region representing a random sample of the genome and a null demographic model is needed (Innan 2006). All seven MLST genes were included in the reference region. As housekeeping genes, MLST genes may not represent a random sample of the genome, but they can capture similar phylogenetic and demographic information as that found in genome-wide data (Thomas et al. 2012; and described in “Results”). The standard neutral model (SNM), which assumes a constant-sized population, was used as the null model. Under the SNM, Tajima’s D is expected to be zero. However, D averaged across MLST genes and from concatenated MLST genes ranged from −0.64 to −0.89, when unique STs were considered. These data suggested that the SNM would be a conservative null model for testing for balancing selection in this sample.
To obtain expected π/K ratios under the SNM, it is necessary to estimate the time of divergence (Td) of the ingroup and outgroup species. An Approximate Bayesian computation (ABC) framework (Beaumont et al. 2002) was used to estimate Td in coalescent units of Ne generations, where Ne is the effective population size. Following Thomas et al. (2012), coalescent trees were simulated under a gene conversion model of recombination using ms software (Hudson 2002), sequences were simulated on those trees using Seq-Gen v1.3.2 software (Rambaut and Grassly 1997), and K was calculated from those sequences using VariScan v2.0.2 software (Vilella et al. 2005). K was used as the summary statistic because it was highly informative for Td (Spearman’s r >0.994 for all analyses). Sequences were simulated and summarized under a Jukes-Cantor model of nucleotide substitution. We simulated 100,000 datasets conditioned on the number of unique STs, length of the concatenated MLST genes, mutation and recombination parameters estimated from ClonalFrame, and Td values sampled from a uniform prior distribution between 3 and 30 coalescent units as described previously (Thomas et al. 2012). The distance between the summaries of simulated and observed data was measured with a standardized Euclidean metric (δ). The 100,000 simulations were ranked based on their δ’s, and the Td values from the top 100 simulations (corresponding to an acceptance rate of 0.001) were used as the approximate posterior distribution. The posterior was smoothed with an Epanechnikov kernel and the mode and 95 % highest posterior density intervals were calculated using the Locfit v1.5-6 module (Loader 1996) of the R v2.13.1 software package. Td was estimated with the 28 unique S. aureus STs as the ingroup and S. epidermidis strain RP62a as the outgroup, and with the 29 unique S. epidermidis STs as the ingroup and S. aureus strain N315 as the outgroup.
For each of the two species, null distributions of D and π/K were constructed from 10,000 simulations of the SNM, with each simulation conditioned on the number and length of sequences from the focal region, and the mutation, recombination, and Td parameter estimates from the reference region. To pinpoint statistically unusual signatures in the focal region, a sliding window approach was used as described previously (Thomas et al. 2012). In brief, D and π/K were calculated for consecutive, non-overlapping 100 bp windows, and the “unusualness” of each observed window in the focal region was measured as the proportion of simulated datasets that produced a window with both D and π/K as great or greater than the observed window.
To confirm the signatures detected with the above analyses, we performed an exploratory genome-wide scan for balancing selection in S. epidermidis. For this scan, the empirical distributions of D and π/K were generated from consecutive, non-overlapping 100 bp windows from an alignment of publicly available genome sequences of 13 S. epidermidis strains, selected to represent diverse STs, and S. aureus strain N315 as the outgroup (GenBank accession numbers are listed in supplementary Table S3). The genome sequences were aligned using the progressive Mauve algorithm of Mauve v2.3.1 software (Darling et al. 2010) with default parameters, followed by manual inspection of the alignment of the focal region. For this scan, DnaSP was used to calculate D and π/K, with a Jukes-Cantor correction of K.
Nucleotide Sequence Accession Numbers
The 57 full-length sequences of Locus 2 from S. aureus and S. epidermidis have been deposited in GenBank with accession numbers JQ031648-JQ031704.
Results
Genetic Variation in S. aureus and S. epidermidis
Similar-sized samples of S. aureus (n = 136 isolates) and S. epidermidis (n = 129 isolates) were drawn from the same hospital over the same time period. By MLST, the S. aureus isolates represented 28 STs and the S. epidermidis isolates represented 29 STs (supplementary Table S1). No significant differences in ST diversity were observed between these two samples as Simpson’s index of diversity (95 % confidence interval) was 0.874 (0.845, 0.903) for S. aureus and 0.894 (0.867, 0.921) for S. epidermidis.
ClonalFrame was unable to estimate the recombination tract length (Δ) from the MLST data of either species, presumably because too few recombination events were detected with breakpoints occurring within the sequenced fragments (Didelot and Falush 2007). Thus, tract length was fixed to the biologically plausible values of 100, 1,000 and 10,000 bp and estimates were obtained in each case for the remaining parameters (Table 1). For each value of the tract length, both the recombination rate (ρ) and the rate at which nucleotides change by recombination events as opposed to mutation events (r/m) was higher for S. aureus than for S. epidermidis, but not significantly so (Table 1). These results indicated that if tract lengths are similar for these two species then recombination has a similar role in producing genetic variation.
Table 1.
ClonalFrame estimates of mutation and recombination parameters from MLST genes
| Species | Fixed parametera Δ | Estimated parameters (95 % credibility intervals)a
|
||
|---|---|---|---|---|
| θw | ρ | r/m | ||
| S. aureus | 100 | 0.0088 | 0.0059 (0.0021, 0.0127) | 0.8946 (0.3744, 1.6984) |
| 1,000 | 0.0088 | 0.0011 (0.0004, 0.0026) | 0.6752 (0.3054, 1.3544) | |
| 10,000 | 0.0088 | 0.00001 (0, 0.00003) | 0.0740 (0.0310, 0.1440) | |
| S. epidermidis | 100 | 0.0081 | 0.0032 (0.0014, 0.0067) | 0.7196 (0.3268, 1.3666) |
| 1,000 | 0.0081 | 0.0004 (0.0002, 0.0009) | 0.5948 (0.2896, 1.0540) | |
| 10,000 | 0.0081 | 0.000008 (0, 0.00001) | 0.0730 (0.0332, 0.1326) | |
Symbols recombination tract length (Δ), per-site mutation rate (θw), per-site recombination rate (ρ), rate at which nucleotides change by recombination events as opposed to mutation events (r/m)
Surprisingly, an examination of genetic variation at individual MLST genes revealed that arcC from S. epidermidis had high values of θπ, Tajima’s D, and π/K, compared to arcC from S. aureus and also compared to averaged and concatenated MLST data from both species (Table 2). These signatures were consistent with the effects of balancing selection, or a confounding process, in or near arcC from S. epidermidis. However, clone-corrected results, which were based on analyses with single representatives of each ST, presented weaker signatures in arcC (Table 2).
Table 2.
Genetic variation in MLST genes
| Species | Dataset | Sequence length (bp) | No. of alleles | All isolatesa
|
STs onlya
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| θπ | θw | D | π/K | θπ | θw | D | π/K | ||||
| S. aureus | arcC | 453 | 12 | 0.0041 | 0.0052 | −0.5813 | 0.0066 | 0.0059 | 0.0074 | −0.6735 | 0.0095 |
| MLST average | 456 | 11 | 0.0046 | 0.0063 | −0.5502 | 0.0173 | 0.0068 | 0.0089 | −0.6409 | 0.0258 | |
| MLST concatenate | 3,195 | 28 | 0.0043 | 0.0062 | −0.9793 | 0.0142 | 0.0068 | 0.0088 | −0.8921 | 0.0221 | |
| S. epidermidis | arcC | 465 | 9 | 0.0136 | 0.0063 | 3.1175 | 0.0527 | 0.0112 | 0.0088 | 0.9383 | 0.0433 |
| MLST average | 429 | 8 | 0.0053 | 0.0059 | −0.3734 | 0.0192 | 0.0063 | 0.0083 | −0.8041 | 0.0228 | |
| MLST concatenate | 3,003 | 29 | 0.0055 | 0.0059 | −0.2289 | 0.0193 | 0.0064 | 0.0081 | −0.8014 | 0.0227 | |
Symbols Tajima’s (θπ) and Watterson’s (θw) per-site mutation rate, Tajima’s D, ratio of intraspecific polymorphism to interspecific divergence (π/K)
Evolutionary History of Loci that Encode arcC in Staphylococci
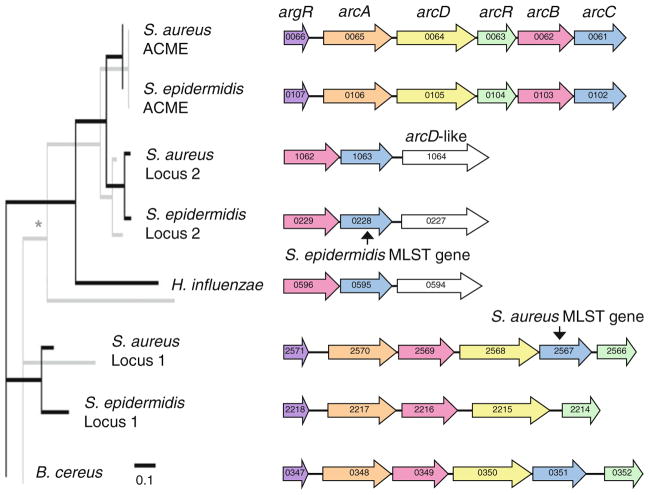
The potentially unusual signatures detected in arcC from S. epidermidis, but not from S. aureus, prompted further investigation of the loci that encode arcC in these two species. Reciprocal BLASTP searches of staphylococcal genome sequences revealed three paralogs of the arcC gene, which mapped to three different loci. The phylogeny of these loci was inferred from aligned ArcB and ArcC amino acid sequences. No species outside of the Staphylococcus genus was identified in GenBank database searches to contain all three loci, so outgroups were selected from Bacillus and Haemophilus because of their similarity to one of the different staphylococcal loci. Tip-mapping of the genetic architecture of these three loci onto the ArcB and ArcC phylogenies suggested an eventful history of gene duplications, rearrangements, and deletions (Fig. 1). Interestingly, these results indicated that the homologous arcC genes used in the MLST schemes of S. aureus and S. epidermidis were paralogs rather than orthologs. The arcC gene from the S. aureus MLST scheme mapped to the previously described native locus, which we named Locus 1, whereas the arcC gene from the S. epidermidis MLST scheme mapped to an undescribed native locus, which we named Locus 2 (Fig. 1). In fact, the arcC gene has been deleted from Locus 1 in S. epidermidis; this deletion was confirmed by PCR in all 129 S. epidermidis isolates studied here.
Fig. 1.
Phylogeny of arcC-containing loci inferred from ArcB (black lines, foreground) and ArcC (gray lines, background) amino acid sequences. Non-parametric bootstrapping with 100 replicates revealed that all nodes had 100 % support except the node marked with an asterisk, which had 92 % support. Arrows represent open reading frames (ORFs), colors indicate homologous genes. Numbers on arrows indicate the ORF numbers in the corresponding strains. Sequences are from S. aureus strain FPR3757, S. epidermidis strain ATCC12228, Bacillus cereus strain NVH391-98, Haemophilus influenzae strain KW20Rd
ArcB sequences from the arginine catabolic mobile element (ACME) of the S. aureus and S. epidermidis genome sequenced strains were identical and their ArcC sequences differed by a single amino acid, consistent with the notion of recent cross-species spread of this element (Miragaia et al. 2009). We tested this hypothesis by sequencing a fragment of the arcC gene from ACME for all S. aureus (n = 18 isolates) and S. epidermidis (n = 54 isolates) that tested positive for the element. PCR amplification of arcA and arcC from ACME produced 100 % agreement regarding the presence or absence of the element in this population. The arcC gene from S. epidermidis ACME presented seven alleles and a θW of 0.003, whereas the arcC gene from S. aureus ACME presented two alleles, both of which were found among S. epidermidis, and a θW of 0.001. Thus, the frequency and diversity of this locus in this population was consistent with a recent cross-species spread of ACME, probably from S. epidermidis to S. aureus.
Functional Annotation of the arcD-like Gene from Locus 2
BLASTP searches of staphylococcal genome sequences using the ArcD amino acid sequences of both Locus 1 and ACME as queries failed to identify the Locus 2 ArcD-like sequences, suggesting that the Locus 2 arcD-like gene may encode an unrelated protein despite its annotation in some genome sequences and its association with the arcBC genes. BLASTP searches of the transporter classification database (TCDB; Saier et al. 2006) revealed that the Locus 2 ArcD-like protein belonged to the ion transporter superfamily and the unknown ion transporter 3 family (UIT3; 9.B.50), also known as the st313/AitC family (Lolkema and Slotboom 2003; Prakash et al. 2003). In contrast, both the Locus 1 and ACME ArcD proteins belonged to the amino acid/polyamine/organocation superfamily and the basic amino acid/polyamine antiporter family (APA; 2.A.3.2).
The Locus 2 ArcD-like amino acid sequences were subsequently aligned with the two UIT3 family reference sequences from the TCDB and 18 other sequences of this family previously used by Lolkema and Slotboom (2003) for the classification of secondary transport proteins. The phylogeny inferred from the ArcD-like sequences showed four distinct clades (Fig. 2). The two UIT3 family reference sequences clustered in clade 1, whereas the Locus 2 ArcD-like sequences clustered in clade 2 (Fig. 2, underlined). The Locus 2 ArcD-like sequences shared an average of 40.7 % identity with the sequences of clade 1, 65.5 % identity with clade 2, 36 % identity with clade 3, and 42.9 % identity with clade 4. By comparison, the Locus 2 ArcD-like sequences shared only 24.6 % and 15.2 % identity with the Locus 1 and ACME ArcD sequences, respectively. These results bring further support to the hypothesis that the Locus 2 ArcD-like protein is unrelated to, and is likely to be functionally distinct from, the Locus 1 and ACME ArcD proteins.
Fig. 2.
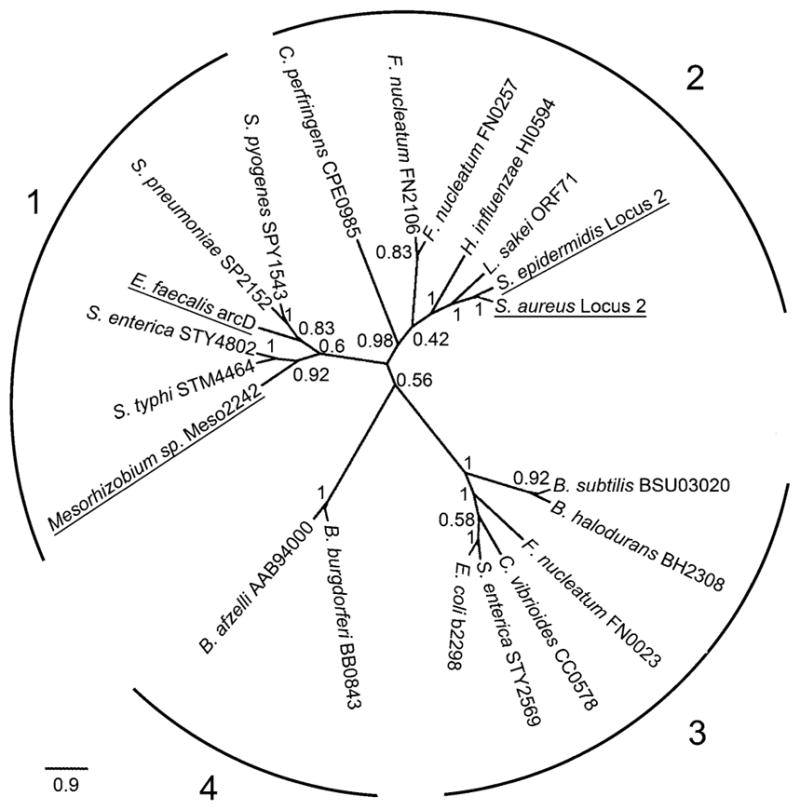
Clades within the unknown ion transporter 3 (UIT3) family. Numbers on branches indicate bootstrap proportions out of 100 replicates. Four clades are indicated. Underline indicates the two UIT3 reference sequences (in clade 1) and the two Staphylococcus Locus 2 sequences (in clade 2)
BLASTP searches of staphylococcal genome sequences with the Locus 2 ArcD-like sequences revealed a paralog encoded by a gene adjacent to the kdp potassium-sensing operon of the methicillin resistance genetic element, SCCmec type II. The SCCmec-borne paralog is shorter than the Locus 2 ArcD-like protein, 300 aa versus 521 aa, respectively, and the homology is restricted to the C-terminal half of the Locus 2 ArcD-like protein. The S. aureus and S. epidermidis versions of this SCCmec-borne paralog are identical, even at the nucleotide level, and they are more similar to the Locus 2 ArcD-like protein of S. epidermidis than S. aureus.
Further Investigation of the Signatures in Locus 2
Having identified Locus 2 as the focal region with potentially unusual signatures in S. epidermidis, we obtained full-length sequences of this locus from single representatives of each ST of both species. To test the unusualness of Tajima’s D and π/K from these sequences, the MLST genes were used as a reference region and the standard neutral model (SNM) was used as a null model. The SNM had one free parameter, the species divergence time (Td), which was estimated with Approximate Bayesian computation (ABC). The ABC estimates of Td were robust to the ingroup species and to assumptions about recombination, as none of the estimates differed significantly (Table 3).
Table 3.
ABC estimates of species divergence times from MLST genes under the standard neutral model
| Ingroup species | Δa | Td (95 % credibility intervals)b |
|---|---|---|
| S. aureus | 100 | 16.5 (15.1, 18.9) |
| 1,000 | 17.3 (14.9, 18.6) | |
| 10,000 | 17.0 (15.0, 18.7) | |
| S. epidermidis | 100 | 16.6 (15.3, 19.0) |
| 1,000 | 17.2 (14.9, 19.0) | |
| 10,000 | 17.2 (15.6, 18.3) |
Separate analyses using the recombination parameters (Δ, ρ) from Table 1
Species divergence time (Td) expressed in coalescent units of Ne generations
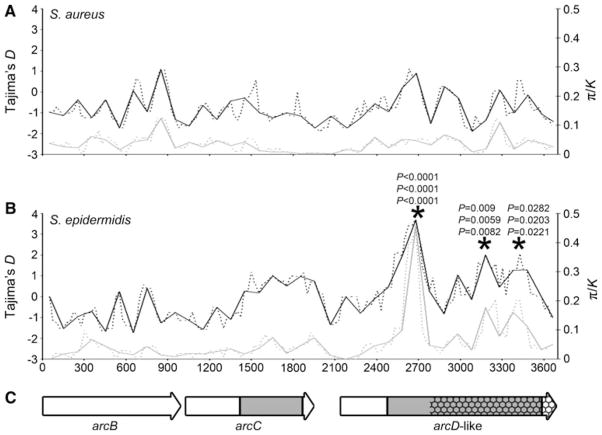
Sliding window analysis found no unusually high values of D and π/K in the S. aureus Locus 2 sequences (Fig. 3a). In contrast, the S. epidermidis Locus 2 sequences exhibited three windows that were unusual under the SNM when D and π/K were considered jointly (Fig. 3b, asterisks). The first unusual window was also unusual when D and π/K were considered separately (P <0.0001 for all tests). The unusualness of these signatures was robust to assumptions about recombination and species divergence time (Fig. 3b, asterisks). Notably, all three unusual windows were found within the arcD-like gene adjacent to arcC. These three unusual windows contained twenty, eight, and seven nucleotide polymorphisms, respectively, and all except one (in the third unusual window) was informative.
Fig. 3.
Analyses of full-length Locus 2 sequences, spanning 3,666 bp for S. aureus and 3,671 bp for S. epidermidis. a Sliding window analyses of Tajima’s D and π/K from 28 S. aureus STs. b Sliding window analyses of Tajima’s D and π/K from 29 S. epidermidis STs. Windows are 100 bp, X-axis indicates the midpoint of the windows. Black lines show D, gray lines show π/K. Solid lines show non-overlapping windows, dashed lines show windows with 25 bp steps. Asterisks indicate the three windows with the peak signatures of balancing selection. The three P values at each asterisk indicate the results assuming (top to bottom) 100, 1,000 and 10,000 bp recombination tract lengths and accompanying recombination rates and species divergence times from Tables 1 and 3. c Arrows represent open reading frames of arcBC and arcD-like genes, gray indicates the portions of the genes sequenced in all 129 S. epidermidis isolates, hatching indicates the portion of the arcD-like gene with homology to an SCCmec type II gene
The first unusual window contained three amino acid polymorphisms (M172A, A175S, L180I), which were perfectly associated with each other and defined two allelic lineages. The central A175S polymorphism results in a change in both the polarity and hydropathy of the residue. The second unusual window contained a single amino acid polymorphism (V347I) that was significantly (Fisher’s exact test, P = 2.48 × 10−12) but imperfectly associated with those in the first unusual window. The third unusual window contained no amino acid polymorphisms, suggesting that its signatures may be due to linkage with the other two windows. The same amino acid polymorphisms were identified when a 1,107 bp gene fragment, spanning all three unusual windows, was sequenced for all 129 S. epidermidis isolates (fragment indicated in Fig. 3c). The frequencies of the allelic lineages from this fragment were 72 % MAL, 28 % ASI when all isolates were considered and 55 % MAL, 45 % ASI when single representatives of each ST were considered.
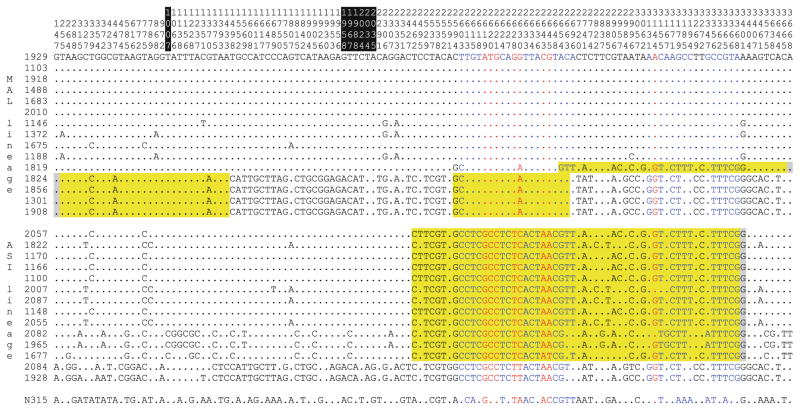
Four recombination events were detected in the S. epidermidis Locus 2 sequences (Fig. 4). All four cases involved putative members of the MAL allelic lineage mixing with putative members of the ASI allelic lineage. These results indicated that both allelic lineages have been in S. epidermidis populations long enough to mix with each other on multiple occasions. No recombination events were detected in the S. aureus Locus 2 sequences, and no recombination events were detected between the S. aureus and S. epidermidis Locus 2 sequences or between the Locus 2 and SCCmec-borne paralogs of the arcD-like gene. The first unusual window of the Locus 2 arcD-like gene has no homology with the SCCmec-borne paralog (Fig. 3c), indicating that its signatures cannot be explained by intragenomic recombinations.
Fig. 4.
Informative nucleotides across the full-length Locus 2 sequences of S. epidermidis. The corresponding nucleotides from S. aureus strain N315 are shown for comparison. The boundaries of arcB, arcC, and arcD-like genes are indicated at the top with black-and-white highlighting of the two intergenic regions. Four recombinations detected by RDP analysis are highlighted in yellow. Gray indicates uncertainty in the recombination breakpoints. Nucleotides colored blue correspond to the three unusual windows in the arcD-like gene, and nucleotides colored red correspond to the four amino acid polymorphisms within these three windows
Genomic Confirmation of the Signatures in Locus 2
To place the signatures in Locus 2 in a genome-wide context, we performed an exploratory scan of the genome sequences of 13 S. epidermidis strains and one S. aureus outgroup strain. The gap-free genome alignment was 1,614,681 bp in length, which covered an average of 65 % of these S. epidermidis genomes. A total of 47,600 SNPs distinguished the S. epidermidis strains from each other, and 413,548 SNPs distinguished S. epidermidis from S. aureus. Sliding window analysis of 16,147 consecutive, non-overlapping 100 bp windows, revealed that 14,687 windows had SNPs among the S. epidermidis strains. Across these variable windows, Tajima’s D ranged from −2.23 to 2.76, and π/K ranged from 0.002 to 0.394. Tajima’s D was −0.62 from the full-length S. epidermidis genomes and −0.42 when averaged across the windows. These genome-wide results were similar to results from the MLST genes for these strains: Tajima’s D was −0.61 from concatenated MLST genes and −0.31 when averaged across 100 bp windows of the MLST genes. Importantly, the three unusual windows in the Locus 2 arcD-like gene were all within the top 1.3 % of the empirical genome-wide distributions of both D and π/K (supplementary Table S4). These results demonstrate the strength of the signatures in the arcD-like gene relative to the rest of the S. epidermidis genome.
Discussion
High mutation or recombination rates can make bacterial clones and clonal complexes ephemeral and of limited epidemiological use (Didelot and Maiden 2010). Genetic variation in S. aureus is thought to occur primarily by point mutations (Feil et al. 2003; Ruimy et al. 2008; Vos and Didelot 2009), and a limited role for recombination over the short-term is supported by the observations of high levels of multilocus linkage disequilibrium, congruence between gene trees, and well-defined clones and clonal complexes that are important units for infectious disease surveillance (Smyth and Robinson 2010). The relative roles of mutation and recombination in producing genetic variation in S. epidermidis are less clear, though a greater role for recombination has been suggested (Kozitskaya et al. 2005; Miragaia et al. 2007). In this study, S. aureus and S. epidermidis isolates were sampled from the same hospital over the same time period and found to have similar levels of genetic variation by MLST and similar recombination rates for a given recombination tract length. A negative correlation was apparent between recombination tract lengths and rates (Table 1), consistent with previous results showing the difficulty of distinguishing between high gene conversion rates with short tracts and lower rates with longer tracts (Padhukasahasram et al. 2004). Thus, in order for S. epidermidis to display a higher recombination rate than S. aureus, its tract length would need to be shorter on average than that of S. aureus, for which there is currently no evidence. To resolve this uncertainty, more sequence data than that provided by MLST genes will be needed.
The arcC-containing loci of staphylococci have experienced an eventful evolutionary history. Some of the duplication and rearrangement events required to produce the observed genetic architecture of these loci may predate the most recent common ancestor of staphylococci, as loci with similar architectures to those of staphylococci were found in distant species such as H. influenzae and B. cereus (Fig. 1). Alternatively, the examined non-staphylococcal outgroups may have acquired their arcC loci via horizontal transfer from staphylococci or from other species. The diversity of arcC-containing loci in bacteria and their inadequacy for inferring species relationships has been noted previously by Zúñiga et al. (2002). It was also noted previously that S. aureus and S. epidermidis both possessed a native arcC gene and that some strains possessed a second copy of arcC on a mobile element called ACME (Diep et al. 2006). ACME was discovered in the genome of the most prevalent methicillin-resistant S. aureus clone in the United States, called the USA300 clone, but its contribution to the ecological success of USA300 is unclear. Diep et al. (2008) reported a role for ACME in USA300 survival in a rabbit bacteremia model of infection, but Montgomery et al. (2009) found no role for ACME in USA300 virulence in a rat pneumonia model or in a mouse skin infection model. Joshi et al. (2011) recently made the observation that ACME provides resistance to antistaphylococcal polyamines, which might be encountered by the bacteria during human infection. We discovered that S. aureus and S. epidermidis both possessed two native arcC-containing loci, and that the arcC gene has been deleted from Locus 1 in all tested S. epidermidis isolates (Fig. 1). These observations raise the question of whether ACME is maintained more frequently in S. epidermidis than in S. aureus to compensate for the loss of arcC at Locus 1. In agreement with a previous conclusion that ACME flows from S. epidermidis to S. aureus (Miragaia et al. 2009), the frequency and diversity of ACME was higher in S. epidermidis than in S. aureus.
ArcD from the arginine deiminase pathway has been well-characterized in bacteria and functions as an arginine-ornithine antiporter (Liu et al. 2008; Makhlin et al. 2007; Verhoogt et al. 1992; Zúñiga et al. 1998). Under anaerobic conditions, this pathway converts arginine to ornithine, generates ammonia, carbon dioxide, and ATP, and can serve as the sole energy source for S. aureus when oxygen, glucose, and nitrates are all absent (Makhlin et al. 2007; Zuúñiga et al. 2002). However, several observations indicate that the Locus 2 arcD-like gene encodes a protein whose function is unrelated to that encoded by arcD. First, Zhu et al. (2007) reported that inactivation of Locus 1 arcD from S. aureus strain USA200, which lacks ACME, resulted in complete inhibition of arginine transport activity. These findings suggest that the Locus 2 arcD-like gene did not compensate for the loss of function of the Locus 1 arcD gene. Second, the genome sequence of S. haemolyticus strain JCSC1435 (Takeuchi et al. 2005) has a Locus 2 that contains arcBC genes but no arcD-like gene, indicating that these three genes are not necessarily clustered together in all staphylococci. Finally, the ArcD-like protein belongs to the unknown ion transporter 3 (UIT3) family (Fig. 2), which does not include the arginine-ornithine antiporter encoded by arcD.
Once the genetic architecture of the arcC-containing loci was understood, it was possible to investigate the causes of the potentially unusual signatures in the S. epidermidis Locus 2 arcC gene and to make comparisons with the orthologous S. aureus gene. When considered together, Tajima’s D and π/K provide powerful measures of balancing selection (Andrés et al. 2009; Innan 2006; Ochola et al. 2010; Thomas et al. 2012). However, demographic processes such as recent population contractions and population subdivisions can leave signatures similar to balancing selection (Ingvarsson 2004; Thornton and Andolfatto 2006). Since the signatures of demography are genome-wide and the signatures of selection are localized, the demographic and selective causes of genetic variation potentially can be disentangled (Kreitman 2000). By comparing genetic variation in a focal region with a reference region that is representative of the genome, the effects of demography potentially can be removed (Hiwatashi et al. 2010). Here, seven MLST genes were used as a reference region because they were capable of capturing similar phylogenetic and demographic information as found in genome-wide data (Thomas et al. 2012; and this study). In order to make statistical comparisons of π/K, even under the simplest demographic model, it is necessary to estimate the species divergence time. Our previous ABC estimates of divergence time between S. aureus and S. epidermidis (Thomas et al. 2012) overlapped with those found here (Table 3), which demonstrates that these estimates are robust to different sources of sequence (i.e., genome-wide data and MLST data), different ingroup and outgroup roles for the species, different null models, and different assumptions about recombination tract length and rate. The peak signatures in S. epidermidis Locus 2 were subsequently traced to three unusual windows in an arcD-like gene adjacent to arcC (Fig. 3b). These signatures were missing from the orthologous gene of S. aureus (Fig. 3a). Moreover, the strength of the signatures in these three windows was confirmed in an exploratory genome-wide scan of S. epidermidis (supplementary Table S4).
While these results are consistent with balancing selection, several alternative explanations need to be considered. Incomplete selective sweeps and sweeps acting on standing genetic variation rather than newly arisen genetic variation (i.e., soft sweeps) can temporarily result in intermediate-frequency alleles, but these processes ultimately result in reduced nucleotide polymorphism (Przeworski et al. 2005). Thus, the two-dimensional signatures of unusually elevated D and π/K should rule out these processes here. On the other hand, two selective sweeps from linked sites can interfere with each other and actually elevate both D and π (Chevin et al. 2008; Camus-Kulan-daivelu et al. 2008). However, interfering sweeps are expected to have the additional signature of a low Fay and Wu’s H, reflecting an excess of high-frequency derived alleles (Chevin et al. 2008). A scan of Locus 2 with Fay and Wu’s H indicated that the arcD-like gene has a generally higher H than the arcB and arcC genes (supplementary Figure S1), which makes interfering sweeps an unlikely explanation here. Diversifying selection and relaxed purifying selection can result in increased polymorphism, including increased amino acid polymorphism, but these processes are not expected to result in intermediate-frequency alleles (Andrés et al. 2009). A more plausible alternative explanation for the signatures observed here is genetic introgression from a diverged population or species (Evans et al. 2006; Plagnol and Wall 2006; Fumagalli et al. 2010). An allele that introgresses into S. epidermidis is likely to be lost by drift, so this explanation implies (though does not require) a period of more extensive genetic admixture or some selectable benefit of the introgressed allele. One signature used to detect introgression is an extended region of high linkage disequilibrium; however, since multiple recombinations were detected between the two allelic lineages of the arcD-like gene, and since both lineages were clearly identifiable as S. epidermidis (Fig. 4), our results are not entirely consistent with introgression. Formal statistical comparison of the two hypotheses of balancing selection and introgression would require a better understanding of patterns of linkage disequilibrium and recombination in S. epidermidis. Recent introgression can produce a negative Tajima’s D, similar to the dynamic of a new pathogen invading a susceptible host population (Gordo et al. 2009), and it can reduce K to the point that π/K is >1 (Castric et al. 2008; and unpublished results), and is therefore incompatible with our observations.
Candidate targets of balancing selection, identified by statistically unusual molecular signatures, can be validated with functional studies (Storz and Wheat 2010). Most of the functionally characterized members of the ion transporter superfamily transport anionic molecules (Prakash et al. 2003; Chen et al. 2011), but nothing specific is known about the substrate of the UIT3 family. Ion transport systems of all sorts are biologically important in staphylococci. It has been noted that both S. aureus and S. epidermidis are likely to encounter ionic and osmotic stresses in their natural habitats on human mucosal and skin surfaces, and that both species have at least 15 transport systems that provide physiological adaptation to these stresses (Gill et al. 2005). Ion transport systems in S. saprophyticus appear to have expanded via gene duplications, possibly as an adaptation to the ion contents of the urine environment (Kuroda et al. 2005). Moreover, inorganic ion transport systems in S. aureus are significantly overrepresented among the list of candidate genes with signatures of balancing selection compared to the remainder of the genome (Thomas et al. 2012). The possible selective maintenance of specific amino acid polymorphisms in the ArcD-like protein of S. epidermidis implies that the different alleles confer a fitness advantage under different conditions. Future investigations of the functions of these alleles may therefore provide unique insights into the adaptation of S. epidermidis.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health Grant GM080602 (to D.A.R.).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00239-012-9520-5) contains supplementary material, which is available to authorized users.
Contributor Information
Liangfen Zhang, Department of Microbiology, University of Mississippi Medical Center, 2500 North State Street, Jackson, MS 39216, USA.
Jonathan C. Thomas, Department of Microbiology, University of Mississippi Medical Center, 2500 North State Street, Jackson, MS 39216, USA
Xavier Didelot, Department of Infectious Disease Epidemiology, Imperial College London, London, UK.
D. Ashley Robinson, Email: darobinson@umc.edu, Department of Microbiology, University of Mississippi Medical Center, 2500 North State Street, Jackson, MS 39216, USA.
References
- Andrés AM, Hubisz MJ, Indap A, Torgerson DG, Degenhardt JD, Boyko AR, Gutenkunst RN, White TJ, Green ED, Bustamante CD, Clark AG, Nielsen R. Targets of balancing selection in the human genome. Mol Biol Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H, Miller LG, Razonable RR. Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2010;51:S183–S197. doi: 10.1086/653519. [DOI] [PubMed] [Google Scholar]
- Brisson D, Dykhuizen DE. ospC diversity in Borrelia burgdorferi. Genetics. 2004;168:713–722. doi: 10.1534/genetics.104.028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus-Kulandaivelu L, Chevin LM, Tollon-Cordet C, Charcosset A, Manicacci D, Tenaillon MI. Patterns of molecular evolution associated with two selective sweeps in the Tb1-Dwarf8 region in maize. Genetics. 2008;180:1107–1121. doi: 10.1534/genetics.108.088849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Castric V, Bechsgaard J, Schierup MH, Vekemans X. Repeated adaptive introgression at a gene under multiallelic balancing selection. PLoS Genet. 2008;4:e1000168. doi: 10.1371/journal.pgen.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Reddy V, Chen JH, Shlykov MA, Zheng WH, Cho J, Yen MR, Saier MH., Jr Phylogenetic characterization of transport protein superfamilies: superiority of SuperfamilyTree programs over those based on multiple alignments. J Mol Microbiol Biotechnol. 2011;21:83–96. doi: 10.1159/000334611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin LM, Billiard S, Hospital F. Hitchhiking both ways: effect of two interfering selective sweeps on linked neutral variation. Genetics. 2008;180:301–316. doi: 10.1534/genetics.108.089706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Maiden MC. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18:315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, Jones A, Palazzolo-Ballance AM, Perdreau-Remington F, Sen-sabaugh GF, DeLeo FR, Chambers HF. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PD, Mekel-Bobrov N, Vallender EJ, Hudson RR, Lahn BT. Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage. Proc Natl Acad Sci USA. 2006;103:18178–18183. doi: 10.1073/pnas.0606966103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, Peacock SJ, Smith JM, Murphy M, Spratt BG, Moore CE, Day NPJ. How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Cagliani R, Riva S, Pozzoli U, Biasin M, Piacentini L, Comi GP, Bresolin N, Clerici M, Sironi M. Population genetics of IFIH1: ancient population structure, local selection, and implications for susceptibility to type 1 diabetes. Mol Biol Evol. 2010;27:2555–2566. doi: 10.1093/molbev/msq141. [DOI] [PubMed] [Google Scholar]
- Gill SR, Fouts DE, Archer GL, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordo I, Gomes MG, Reis DG, Campos PR. Genetic diversity in the SIR model of pathogen evolution. PLoS One. 2009;4:e4876. doi: 10.1371/journal.pone.0004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol. 2001;39:4190–4192. doi: 10.1128/JCM.39.11.4190-4192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiwatashi T, Okabe Y, Tsutsui T, Melin AD, Oota H, Schaffner CM, Aureli F, Fedigan LM, Innan H, Kawamura S. An explicit signature of balancing selection for color-vision variation in new world monkeys. Mol Biol Evol. 2010;27:453–464. doi: 10.1093/molbev/msp262. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinfomatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson PK. Population subdivision and the Hudson–Kreitman–Aguade test: testing for deviations from the neutral model in organelle genomes. Genet Res. 2004;83:31–39. doi: 10.1017/s0016672303006529. [DOI] [PubMed] [Google Scholar]
- Innan H. Modified Hudson–Kreitman–Aguadé test and two-dimensional evaluation of neutrality tests. Genetics. 2006;173:1725–1733. doi: 10.1534/genetics.106.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi GS, Spontak JS, Klapper DG, Richardson AR. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol. 2011;82:19–20. doi: 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J Clin Microbiol. 2005;43:4751–4757. doi: 10.1128/JCM.43.9.4751-4757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M. Methods to detect selection in populations with application to the human. Annu Rev Genomics Hum Genet. 2000;1:539–559. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- Kreitman M, Di Rienzo A. Balancing claims for balancing selection. Trends Genet. 2004;20:300–304. doi: 10.1016/j.tig.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Ohta T, Uchiyama I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Yamashita A, Hirakawa H, Kumano M, Morikawa K, Higashide M, Maruyama A, Inose Y, Matoba K, Toh H, Kuhara S, Hattori M, Ohta T. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc Natl Acad Sci USA. 2005;102:13272–13277. doi: 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dong Y, Chen YY, Burne RA. Environmental and growth phase regulation of the Streptococcus gordonii arginine deiminase genes. Appl Environ Microbiol. 2008;74:5023–5030. doi: 10.1128/AEM.00556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loader CR. Local likelihood density estimation. Ann Stat. 1996;24:1602–1618. [Google Scholar]
- Lolkema JS, Slotboom DJ. Classification of 29 families of secondary transport proteins into a single structural class using hydropathy profile analysis. J Mol Biol. 2003;327:901–909. doi: 10.1016/s0022-2836(03)00214-6. [DOI] [PubMed] [Google Scholar]
- Makhlin J, Kofman T, Borovok I, Kohler C, Engelmann S, Cohen G, Aharonowitz Y. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J Bacteriol. 2007;189:5976–5986. doi: 10.1128/JB.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathema B, Mediavilla JR, Chen L, Kreiswirth B. Evolution and taxonomy of staphylococci. In: Crossley KB, Jefferson KK, Archer GL, Fowler VG, editors. Staphylococci in human disease. 2. Wiley-Blackwell; Oxford Maynard: 2009. [Google Scholar]
- Smith J. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J Bacteriol. 2007;189:2540–2552. doi: 10.1128/JB.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One. 2009;4:e7722. doi: 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery CP, Boyle-Vavra S, Daum RS. The arginine catabolic mobile element is not associated with enhanced virulence in experimental invasive disease caused by the community-associated methicillin-resistant Staphylococcus aureus USA300 genetic background. Infect Immun. 2009;77:2650–2656. doi: 10.1128/IAI.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochola LI, Tetteh KK, Stewart LB, Riitho V, Marsh K, Conway DJ. Allele frequency-based and polymorphism-versus-divergence indices of balancing selection in a new filtered set of polymorphic genes in Plasmodium falciparum. Mol Biol Evol. 2010;27:2344–2351. doi: 10.1093/molbev/msq119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhukasahasram B, Marjoram P, Nordborg M. Estimating the rate of gene conversion on human chromosome 21. Am J Hum Genet. 2004;75:386–397. doi: 10.1086/423451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Losada M, Browne EB, Madsen A, Wirth T, Viscidi RP, Crandall KA. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect Genet Evol. 2006;6:97–112. doi: 10.1016/j.meegid.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plagnol V, Wall JD. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Cooper G, Singhi S, Saier MH., Jr The ion transporter superfamily. Biochim Biophys Acta. 2003;1618:79–92. doi: 10.1016/j.bbamem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Richman A. Evolution of balanced genetic polymorphism. Mol Ecol. 2000;9:1953–1963. doi: 10.1046/j.1365-294x.2000.01125.x. [DOI] [PubMed] [Google Scholar]
- Ruimy R, Maiga A, Armand-Lefevre L, Maiga I, Diallo A, Koumaré AK, Ouattara K, Soumaré S, Gaillard K, Lucet JC, Andremont A, Feil EJ. The carriage population of Staphylococcus aureus from Mali is composed of a combination of pandemic clones and the divergent Panton-Valentine leukocidin-positive genotype ST152. J Bacteriol. 2008;190:3962–3968. doi: 10.1128/JB.01947-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SA. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Smyth DS, Robinson DA. Population genetics of Staphylococcus. In: Robinson DA, Falush D, Feil EJ, editors. Bacterial population genetics in infectious disease. 1. Wiley-Blackwell; New Jersey: 2010. [Google Scholar]
- Smyth DS, Wong A, Robinson DA. Cross-species spread of SCCmecIV subtypes in staphylococci. Infect Genet Evol. 2011;11:446–451. doi: 10.1016/j.meegid.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Wheat CW. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 2010;64:2489–2509. doi: 10.1111/j.1558-5646.2010.01044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol. 2005;187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC. An improved multilocus sequence typing scheme for Staphylococcus epidermidis. J Clin Microbiol. 2007;45:616–619. doi: 10.1128/JCM.01934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, Godfrey PA, Feldgarden M, Robinson DA. Candidate targets of balancing selection in the genome of Staphylococcus aureus. Mol Biol Evol. 2012;29:1175–1186. doi: 10.1093/molbev/msr286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Melles DC, Nouwen J, van Leeuwen WB, van Wamel W, Vos MC, Wertheim HFL, Verbrugh HA. Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol. 2009;9:32–47. doi: 10.1016/j.meegid.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Verhoogt HJ, Smit H, Abee T, Gamper M, Driessen AJ, Haas D, Konings WN. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine–ornithine exchanger. J Bacteriol. 1992;174:1568–1573. doi: 10.1128/jb.174.5.1568-1573.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Blanco-Garcia A, Hutter S, Rozas J. VariScan: analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics. 2005;21:2791–2793. doi: 10.1093/bioinformatics/bti403. [DOI] [PubMed] [Google Scholar]
- von Eiff C, Peters G, Heilmann C. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect Dis. 2002;2:677–685. doi: 10.1016/s1473-3099(02)00438-3. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Watterson GA. Number of segregating sites in genetics models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Weedall GD, Conway DJ. Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol. 2010;26:363–369. doi: 10.1016/j.pt.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Wildschutte H, Lawrence JG. Differential Salmonella survival against communities of intestinal amoebae. Microbiology. 2007;41:10095–10104. doi: 10.1099/mic.0.2006/003616-0. [DOI] [PubMed] [Google Scholar]
- Wiuf C, Zhao K, Innan H, Nordborg M. The probability and chromosomal extent of trans-specific polymorphism. Genetics. 2004;168:2363–2372. doi: 10.1534/genetics.104.029488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Reddy SP, Smyth DS, Aguero-Rosenfeld ME, Sakoulas G, Robinson DA. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob Agents Chemother. 2010;54:742–748. doi: 10.1128/AAC.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Weiss EC, Otto M, Fey PD, Smeltzer MS, Somerville GA. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect Immun. 2007;75:4219–4226. doi: 10.1128/IAI.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga M, Champomier-Verges M, Zagorec M, Pérez-Martínez G. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J Bacteriol. 1998;180:4154–4159. doi: 10.1128/jb.180.16.4154-4159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zúñiga M, Pérez G, González-Candelas F. Evolution of arginine deiminase (ADI) pathway genes. Mol Phylogenet Evol. 2002;25:429–444. doi: 10.1016/s1055-7903(02)00277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.