Abstract
Two-component signaling pathways involve sensory histidine kinases (HK), histidine phosphotransfer proteins (HpT) and response regulators (RR). Recent advancements in genome sequencing projects for a number of plant species have established the TCS family to be multigenic one. In plants, HKs operate through the His–Asp phosphorelay and control many physiological and developmental processes throughout the lifecycle of plants. Despite the huge diversity reported for the structural features of the HKs, their functional redundancy has also been reported via mutant approach. Several sensory HKs having a CHASE domain, transmembrane domain(s), transmitter domain and receiver domain have been reported to be involved in cytokinin and ethylene signaling. On the other hand, there are also increasing evidences for some of the sensory HKs to be performing their role as osmosensor, clearly indicating toward a possible cross-talk between hormone and stress responsive cascades. In this review, we bring out the latest knowledge about the structure and functions of histidine kinases in cytokinin and ethylene signaling and their role(s) in development and the regulation of environmental stress responses.
Keywords: Cytokinin, ethylene, environmental stress, histidine kinase, osmosensor, two component signaling, receptor
Introduction
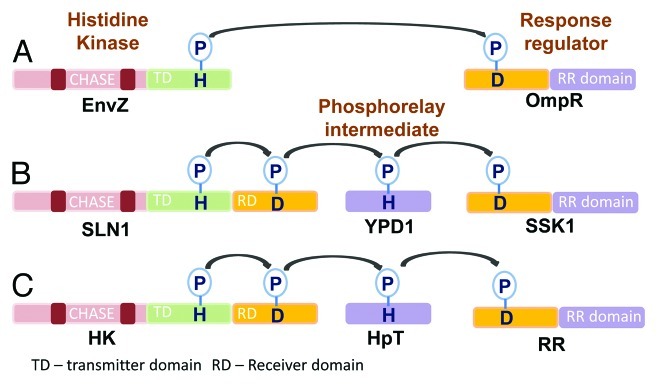
In all living organisms, responses to stimuli are mediated by signal transduction pathways which decide their physiological performance under a given set of conditions. In both eukaryotes and prokaryotes, protein phosphorylation is one of the major strategies by which intracellular signal-transduction proceeds. Protein kinases, using ATP as the phosphate donor, catalyze phosphorylation of either themselves (auto-phosphorylation) or other protein substrates (trans-phosphorylation) at specific serine, threonine, tyrosine or histidine residues. Based on this substrate specificity, protein kinases have been classified into three main categories: serine-threonine (ser-thr) kinases, tyrosine (tyr) kinases and histidine kinases (HKs). The ser-thr kinases and tyr kinases are predominant in eukaryotic systems. In bacteria, on the other hand, histidine kinases are the major signaling molecules and are involved in a plethora of responses such as osmosensing, chemotaxis and nutrient sensing. In this case, signal is perceived by the histidine kinases (HK) and signal transduction occurs through phosphotransfer to another group of signal transducers called the response regulators (RRs). This type of signal transduction has therefore been assigned the name, “Two Component System” or TCS, as it involves two types of highly conserved proteins- the HKs and their corresponding RRs. Two types of two-component signaling has been reported in literature; the first type is found primarily in bacteria and involves the direct transfer of the phosphoryl group from the histidine kinase (also called simple type HK) to the response regulator e.g., the E. coli EnvZ-OmpR two component system, which is responsible for osmotic stress response. The second type of two component signaling comprises a more complex histidine kinase which contains an extended C-terminal domain with a conserved aspartate residue. This type of histidine kinase is called a hybrid histidine kinase in which the phosphotransfer occours from the conserved histidine to the conserved aspartate residue, present within the same protein (Fig. 1). The phosphotransfer to the response regulator is mediated by an additional protein, the histidine-containing phosphotransfer protein (HpT), which itself has a conserved histidine phosphorylation site. An example of this type of phosphorelay is the Saccharomyces cerevisiae SLN1, YPD1 or SSK1 system1. Ever since the first reports for the existence of bacterial type HKs in yeast and Arabidopsis,1,2 many more genes encoding two component systems have not only been identified, but also well characterized in plants, yeast and Dictyostelium. However, only the second type of two component signaling has been reported in eukaryotes. In metazoa, the two component regulators (histidine kinases and response regulators) have not been identified so far. Furthermore, our understanding about the functioning of TCS members has been benefitted by the use of contemporary tools of genetics, molecular biology and microscopy techniques. This review will focus mainly on plant histidine kinases with a few cross references from their bacterial counterparts wherever deemed fit. For brevity sake, we have restricted our discussion to the functioning of HKs involved in ethylene and cytokinin signaling only, where major advancements have been made in the recent past. In addition, we have also tried to highlight the possible cross-talk between hormone signaling and stress responses, as mediated though HKs.
Figure 1. Types of phosphorelay signaling in two-component systems. (A) The E. coli EnvZ-OmpR two component system includes a simple histidine kinase and a response regulator. Transfer of the phosphoryl group occurs from the conserved histidine at the transmitter domain of the HK to the conserved aspartate at the receiver domain of the response regulator. (B) The Yeast SLN1-YPD1-SSK1 two component system employs a hybrid histidine kinase which contains an additional receiver domain, a histidine containing phosphotransfer protein and a response regulator. It involves a multistep his to asp phosphorelay. (C) Plant two component systems have the same architecture as the yeast’s.
Histidine kinases in plants
The histidine kinases have been identified in increasing number of plant species, such as Arabidopsis, maize, rice, soybeans, poplar and Lotus japonicus besides a few others.3-8 The essentiality and importance of HKs can be judged by the level of conservation across plant species. In Arabidopsis, HKs are involved in different functions. These functions include ethylene signaling, osmosensing, cytokinin signaling, mega-gametophyte development, cold perception and more recently as regulating salt sensitivity and resistance against bacterial and fungal infection.1,9-14 Recently, several HK genes have been reported to be involved in drought response in Arabidopsis.10,15,16
Broadly speaking, HKs can be grouped into two categories; ethylene receptors and non-ethylene receptors. The latter includes cytokinin receptors, osmosensors and other histidine kinases whose functions are yet to be identified. Most of the plant genomes encode 4–5 ethylene receptors and 6–17 non-ethylene receptors. A list of HK genes identified in Arabidopsis, rice, maize, soybean, lotus and populus has been presented in Table 1. In literature, huge diversity has been reported for HKs as far as their structure, cellular localization and expression patterns are concerned, which are the topic of discussion in the next section.
Table 1. HK genes identified in various plant species. The numbers shown here represent the total number of genes identified in the particular genome.
| Organism | No. of HK Genes |
Reference | |
|---|---|---|---|
| Non-ethylene receptor | Ethylene receptor | ||
|
Aabidopsis thaliana |
6 |
5 |
(3) |
|
Oryza sativa |
6 |
5 |
(5) |
|
Zea mays |
11 |
- |
(4) |
|
Glycine max |
17 |
4 |
(8) |
|
Lotus japonicas |
11 |
5 |
(7) |
| Populus trichocarpa | 8 | 4 | (6) |
Diversity in Structure
Plant HKs have a high degree of structural similarity among different species. The majority of HKs are usually membrane anchored proteins with a sensory N-terminal domain which is highly variable and a highly conserved C-terminal kinase domain containing characteristic sequence motifs. The variability in the N-terminal is expected as different HKs are responsible for the perception of different types of signals. HKs have been found to exist as homodimers. Biochemical studies in bacteria have shown that when stimulated, the kinase domain of one subunit catalyzes the trans-phosphorylation of the conserved histidine residue of the other and subsequent transfer of the phosphoryl group occurs. The same mechanism appears to hold for plant HKs as well.
In Arabidopsis, non ethylene receptor HKs include the cytokinin receptors - AHK2, AHK3, AHK4 (CRE1/WOL); osmosensor (AHK1); AHK5 (CKI2) and CKI1. The role of CKI1 in cytokinin signaling has not been proved yet, although, it has been found to act upstream of histidine phosphotransfer (HpT) genes and activating cytokinin signaling when overexpressed in Arabidopsis.17 AHK5 (CKI2) has been shown to integrate endogenous and environmental signals via H2O2 homeostasis in guard cells.18 Most of these HKs have trans-membrane domains; CHASE domain and a receiver domain fused to the kinase domain, but in some cases, variations also exist. For example, AHK5 lacks trans-membrane domain and is predicted to be the only cytoplasmic HK. Likewise, an additional receiver-like domain is present in AHK2, AHK3 and AHK4.
For ethylene perception, five receptors (ETR1, ETR2, EIN4, ERS1, and ERS2) have been identified in Arabidopsis.2,19-21 The members of ethylene receptor family contain highly conserved membrane bound N-terminal domain which is responsible for binding with ethylene. The histidine kinase and receiver domains are not conserved across these five family members. In fact, ETR1 and ERS2 are the only members containing a canonical histidine kinase moiety and the receiver domain is not present in ERS1 and ERS2.19
Diversity in sub cellular localization
Analysis of protein sequences of histidine kinases using bioinformatics tools predicted the presence of trans-membrane domains. ATHK1 has two hydrophobic regions in the N-terminal half and function as an osmosensor in yeast suggesting that it is a plasma membrane-bound HK.10,22 Experimentally, AHK5 is shown to be localized in both the cytoplasm and the plasma membrane.18 Interestingly, it contains putative N-myristoylation sites, indicating its possible association with the plasma membrane. Other HKs appear to be present in plasma membrane as revealed by subcellular localization of HK- GFP fusion proteins.23-25 But several new evidences, however, support their localization in the membrane of endoplasmic reticulum (ER) as well.26-28 Cell biological and biochemical evidences have been provided using transiently transformed tobacco and Arabidopsis cells as well as stable transgenic Arabidopsis plants.26 Cytokinin-binding studies with separated membrane fractions of plasma membrane and ER, fluorescence-labeled cytokinin receptors and biochemical fractionation confirmed the ER localization of cytokinin receptors.28 In addition, a cytokinin receptor, ZmHK1 has been reported to be ER localized in maize.27
Based on these new evidences, the old model of cytokinin perception at the plasma membrane has now been challenged. According to the new model,29 the cytokinin-binding CHASE domains of the cytokinin receptors (AHK2, AHK3 and AHK4) are exposed to the ER lumen, whereas their C-terminal histidine kinase domains are oriented to the cytoplasm, implying cytokinin perception to be occurring at the ER. In this context, either active cytokinins should be able to permeate the cell membrane or there should be carrier proteins present in plasma membrane as well as in ER membrane to transport cytokinin, first into the cell and further, into the lumen of ER. The first probability has not yet been investigated in detail, but several plasma-membrane bound cytokinin carriers have been identified.30-33 No intracellular transporter for movement of cytokinin into ER lumen has yet been identified.32 Studies on intracellular transport and distribution of cytokinin are still awaited. Biochemical studies show maximal hormone binding activity of cytokinin receptors at pH close to 6.534 - a pH characteristic for the ER lumen35 but not at the apoplastic pH of 5.5.36 Therefore, the current model of cytokinin signal perception at the plasma membrane should be reconsidered.
Interestingly enough, just like the cytokinin receptors, initially the ethylene receptors were also thought to be localized in the plasma membrane. Recent findings, however, provide convincing evidence that the ethylene receptors are in fact, localized at the endoplasmic reticulum membrane.37,38 Ethylene is presumed to enter the cell through passive diffusion as no ethylene transporters have been identified as yet.
Expression patterns of HKs indicate their developmental control
Expression patterns of HK genes have been determined using various techniques such as qRT-PCR, northern blotting, GUS assays and in situ hybridization. Expression of various HK genes has been reported in various organs of a plant, but at varying levels. For example, ATHK1 transcript shows more abundance in roots and its expression is also induced in response to various stresses such as high salinity, dehydration or cold.22 GUS reporter driven by ATHK1 promoter shows that transcription of ATHK1 is regulated by external osmolarity.22 AHK5 predominantly expresses in roots and regulates root elongation through an ETR1- dependent abscisic acid signaling pathway.39 Through the use of semi-quantitative RT-PCR, AHK5 transcript has also been detected in flowers, siliques and to a lower extent, in stems and leaves.39 Transient expression of GFP under the control of AHK5 promoter shows its presence in guard cells as well as in epidermal cells and its expression increases in guard cell by H2O2 treatment of leaves in Arabidopsis.18 In Arabidopsis, CKI1 predominantly expresses in flowers, where it is required for female gametophyte development. Cytokinin receptors predominantly express in meristematic tissues (such as, shoot and root tips, lateral root primordia and growing leaf), whereas, AHK4 shows high level of expression in the root of Arabidopsis.24,40,41 During the early stages of embryogenesis, its expression was restricted to procambium only, which is the precursor of the vascular tissue.24 Detailed analysis at cellular level by in situ hybridization showed that its expression is further confined to the vascular cylinder and pericycle of the root at the seedling stage. In Arabidopsis, AHK2 expresses at higher levels in leaf veins, inflorescence axis, flower and siliques and relatively low levels in roots. In contrast, AHK3 expresses in both root and shoot tissues including leaves, inflorescences axis and flowers.40,41 Thus cytokinin receptors express ubiquitously in various tissues in a nearly overlapping manner.
Cytokinin signaling and interactions of HKs within a cell
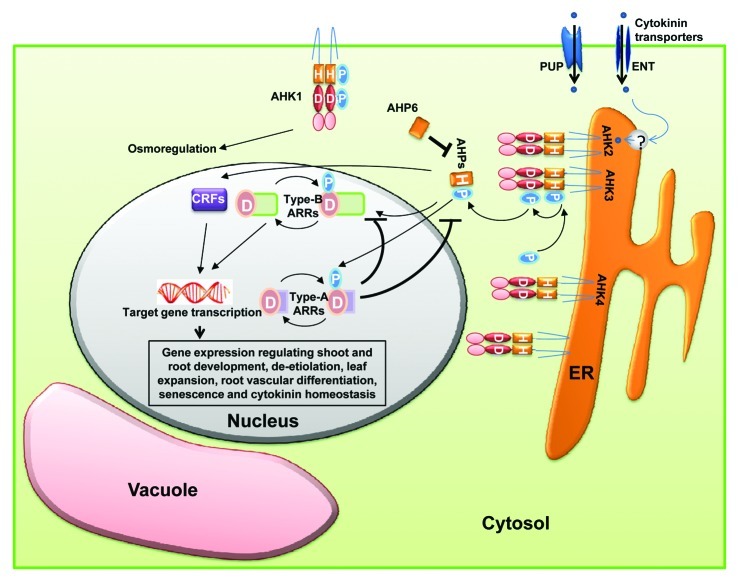
Binding of cytokinins to their HK receptors trigger their auto-phosphorylation. HpT proteins act as mediators receiving the phosphoryl group from HK and transmitting them to type-B RRs. Type-B RRs contain a receiver domain followed by a MYB like DNA binding motif at extended C-terminus.42 They activate transcription of target genes, including type-A RRs which are involved in negative feedback of cytokinin signaling.43 A schematic representation for cytokinin signaling is given in Figure 2 which shows that cytokinin signaling proceeds via two component system (histidine to aspartate phosphorelay) leading to the altered expression of cytokinin regulated genes. Out of the six non-ethylene receptor HKs in Arabidopsis; AHK2, AHK3 and AHK4 have been found to function as cytokinin receptors and act redundantly in cytokinin signaling. They bind cytokinins via the CHASE domain.44 Interestingly, AHK4 has dual activities, in the presence of cytokinins, it acts as kinase and phosphorylates HpT protein while in the absence of cytokinins, acts as a phosphatase and thus dephosphorylate Hpt.45 The cytokinin–receptor interactions (studied using bacterial, yeast and plant based bioassays), indicate that AHK2 and AHK4 have similar ligand preferences.46 Recently, the crystal structure of the AHK4 sensor domain has also been determined which has helped in elucidating the molecular basis for ligand-receptor interaction.47
Figure 2. Model for cytokinin signaling. Cytokinins are translocated by plasma membrane localized transporters and then by unknown mechanisms, enter into the ER lumen where their perception takes place. Cytokinin binding to their histidine kinase receptors triggers autophosphorylation at the conserved histidine residue. Then, the phosphoryl group is shuttled to the RR via the HpT. Type-B RRs are transcription factors which turn on the expression of cytokinin regulated genes as well as type-A RRs. The Type-A RRs are involved in repressing the cytokinin signaling via negative feedback loops. Alteration of target gene expression by type-B RRs results in cytokinin responses. AHK1, localized at the plasma membrane, perceives changes in osmolarity and via unknown mechanisms, mediate the corresponding physiological responses of the plant. For details, see text.
HpT proteins have been found to be present in both nucleus as well as cytoplasm. Contrary to previous assumptions, new studies show that their movements within the cell and their transcriptional regulation remain unaffected by cytokinin treatment.48 The authentic HpTs play a positive and redundant function in CK signaling,49 while the pseudo HpT viz. AHP6 which lacks a phosphorylation site, acts as negative regulator of CK signaling. AHP6 has been reported to be involved in xylem development.50
Apart from type-B RRs, HpT proteins also interact with a set of cytokinin-regulated transcription factors which are known as cytokinin response factors (CRFs).51 Their transcription is upregulated by cytokinin and they rapidly moves into nucleus. CRFs form a parallel branch of signaling with type-B RRs as many downstream targets of these two are found to be common.51 In addition, AHPs also interact with proteins of other signaling pathway.52 For example AHP1 has been shown to interact with ETR1, an ethylene receptor.52 As cytokinin receptors have been shown to be localized to ER membrane, proteins implicated in intracellular trafficking such as ADL1A and adaptin have been shown to interact with them.53
Histidine kinase activity and ethylene signaling
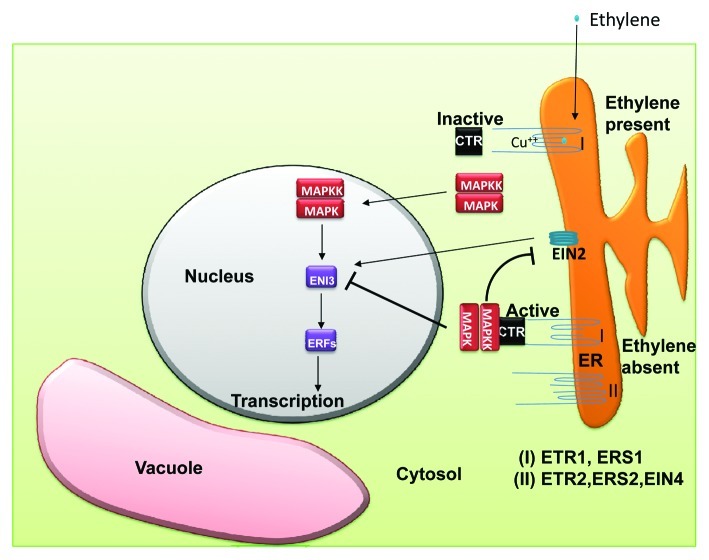
Ethylene is a major hormone which regulates various processes ranging from plant development to mediating cellular responses to various environmental signals.54 Hence, a lot has gone into identifying the mechanism through which ethylene functions, and in Arabidopsis, through the use of ethylene insensitive mutants, the ethylene receptors ETR1, ETR2, EIN4, ERS1 and ERS2 were thus isolated, cloned and sequenced.2,19-21 Interestingly, ETR1 and ERS1 displayed high sequence similarities with the hybrid HKs that function in bacterial two component systems but, as has been mentioned earlier, the remaining three, lack some of the consensus motifs that are characteristic of HKs.19 Mutations in the hydrophobic domains of any one of these genes led to dominant ethylene insensitivity of the mutant plants and moreover some of these mutations were responsible for disrupting the binding of ethylene to the receptor.19,20 These findings suggested that these receptors were in fact, negative regulators of the ethylene response in the absence of the hormone and the mutations were in fact ‘gain of function’ mutations. This negative regulatory role of the ethylene receptors was confirmed through the use of ‘loss of function’ mutants.55 Genetic analysis of single ‘loss of function’ mutants of ETR1, ETR2, EIN4 and ERS2 showed that these mutants have almost normal sensitivity to ethylene which explains the functional redundancy of the receptors.40 However, the double, triple and quadruple loss of function mutants displayed constitutive ethylene responses even in the absence of the hormone.55 A model for ethylene signaling is represented in Figure 3. According to this model, the ethylene receptors act as negative regulators of ethylene signaling and upon ethylene binding these proteins get inactivated and thereby permit signaling to proceed. Ethylene binding also causes the inactivation of the associated negative regulators (CTR 1 and RTE 1) which, in turn, leads toward the activation of a MAP kinase pathway which ultimately results in altered expression of the ethylene regulated genes. The ethylene receptors have been proposed to function as dimers. Ethylene binding requires a copper ion cofactor which is supplied by the copper transporter RAN1.56 Furthermore, it has been reported that the histidine kinase domains of ETR1 and ERS2 interacts and activates yet another negative regulator of the ethylene response: CTR1, a raf-like serine-threonine kinase.57 ETR1 also interacts with RTE1, another negative regulator of the ethylene response.58 Interestingly, enough RTE1 has been found to be co-localized with ETR1 at the golgi.59 Thus ETR1 is not exclusively located at the ER as was previously thought. It has also been shown that Arabidopsis ETR1 functions as a HK which was the definitive experiment which extended the known organisms in which HKs function from bacteria and yeast to plants.60 Since then, ethylene receptors of other plants such as tobacco have also been reported to possess HK activity.61 However, the role of the HK activity of the ethylene receptors in mediating the ethylene response has remained unclear for quite a long time. In fact, it was reported that the canonical HK activity of the transmitter domain of the Arabidopsis ETR1 was not required for signal transduction.62 Very recently, it was published that in Arabidopsis, the complementation of the etr1;ers1 double mutant with wild type-ETR1 or kinase-inactive ETR1 was able to rescue the dominant ethylene insensitive phenotype of the mutant lines in air but the mutant lines carrying the kinase-inactive ETR1 showed reduced sensitivity to ethylene as well as decreased levels of the CTR1 protein suggesting that the HK activity of ETR1, although not required for ethylene responses, plays a modulating role in its regulation.63
Figure 3. Model for ethylene signaling. Ethylene crosses the cell membranes presumably by passive diffusion and is perceived by receptors localized ER or Golgi (not shown in diagram). Two types of receptors, type-I and type-II, are present which are active and thus suppress the ethylene responses in the absence of the hormone. Binding of ethylene results in the inactivation of the type I receptors as well as the associated negative regulatory proteins, CTR1 and RTE1 (in Golgi, not shown in diagram). This activates a MAP kinase pathway which facilitates the ethylene response factors (ERFs) through the eventual activation of EIN3, EIL1. For details, see text.
Histidine kinases and response to environmental stresses
Plants are subject to a variety of environmental stresses. These stresses, whether biotic or abiotic, more often have detrimental effects on overall development of the plant. Therefore, plants have evolved intricate strategies to recognize process and respond to these stresses. In plants, HKs have been reported to play an integral role in the response to environmental stresses (Table 2). The first such report came when an Arabidopsis HK -ATHK1 was reported to function as a putative osmosensor in yeast.22 ATHK1 expression was found to be regulated by changes in external osmolarity.22 ATHK1 was also able to functionally complement SLN1 in a yeast osmosensing deficient mutant SLN1-ts.22 However, when either of the putative phosphorylation sites (conserved his or asp) were substituted with other amino acid residues, ATHK1 was unable to complement the SLN1 mutant suggesting that signaling proceeds through a his to asp phosphorelay.22 Also in Arabidopsis, the ATHK1 null mutants show increased sensitivity to osmotic stress and 35S:ATHK1 overexpressing transgenic lines show more tolerance to various types of water stress as compared with wild type plants which confirms that ATHK1 functions as an osmosensor in planta.10 A maize histidine kinase gene, ZmHK9, was found to mediate drought tolerance through the regulation of stomatal opening in Arabidopsis.15 In rice, a multi-stress inducible histidine kinase, OsHK3b, also seems to function as an osmosensor (our unpublished data). Thus sensing of changes in osmolarity seems to be mediated by HKs in plants.
Table 2. Summary of HK genes involved in abiotic stress responses in Arabidopsis.
| Gene | Type | Regulatory function in stress response | stresses | References |
|---|---|---|---|---|
|
AHK1 |
Osmosensor |
positive |
drought, salt, osmotic stress |
(10, 22) |
|
AHK2 |
CK receptor |
negative |
cold, drought, salt |
(10,13) |
|
AHK3 |
CK receptor |
negative |
cold, drought, salt |
(10,13) |
|
AHK4 |
CK receptor |
negative |
salt, drought |
(10) |
| AHK5 | Intracellular sensor | negative for abiotic, positive for biotic | salt, bacterial and fungal infection | (14) |
It has been reported that HKs are involved in responses to other stresses besides osmotic stress. In Arabidopsis, it was shown that the cytokinin receptors AHK2 and AHK3 play a negative regulatory role in cold stress signaling via inhibition of the ABA response, occurring independently of the cold acclimation pathway.13 In another report, the loss of function ahk2, ahk3 single mutants and ahk2-ahk3 double mutants displayed strong tolerance toward drought and salinity indicating that AHK2 and AHK3 function as negative regulators of salt and osmotic stress.10 Furthermore, Arabidopsis ahk4 mutants also displayed salinity and drought tolerance but only in the presence of cytokinin.10 Yet another HK in Arabidopsis, AHK5 was found to regulate salt sensitivity.14 The ahk5 mutants displayed higher root and shoot weight than the wild type and AHK5 complemented mutant lines when grown under salt stress. Moreover, this difference was not observed when the plants were subjected to osmotic stress using sorbitol or KCl as the osmoticum which suggests that the differences in root length and seed germination were the result of the ionic component of salt stress.14 This suggests that AHK5 is in fact, a negative regulator of salt tolerance. Besides this, AHK5 is also involved in resistance to biotic stress. It has been shown that AHK5 has a role in providing resistance to bacterial and fungal infections.14 Another interesting finding is that, although there is no direct evidence of it, the ethylene receptors may also be involved in salt tolerance. This derivation can be made simply from the fact that the Arabidopsis EIN2, which is a positive regulator of the ethylene response downstream of the ethylene receptors, has been shown to play a role in providing salinity tolerance thereby providing a link between ethylene signaling and salinity stress tolerance.64 The ein2 mutant plants displayed extreme sensitivity to salinity whereas overexpression of the C-terminus of EIN2 in the mutant line suppressed the salt sensitivity.64 Since EIN2 is a positive regulator of the ethylene response and the ethylene receptors negatively regulate EIN2, it can be reasoned that the ethylene receptors negatively regulate salinity tolerance. This is an interesting thought as it implies that ethylene may have a positive role in regulation of salinity and drought stress through the activation of EIN2. But at this point, this can only be considered as a speculation. However, cross talk between hormones and salinity stress has been also predicted based on bioinformatics based analysis of transcriptome of roots of Arabidopsis.65
Work from our own laboratory has shown that the TCS members (including sensory HKs) are differentially expressed in contrasting salinity responsive genotypes of rice under non stress conditions indicating their role in salinity stress tolerance.66 Further support to this hypothesis is obtained from the observation that quite a number of these TCS members are co-localized within salinity related QTLs in rice. Moreover, the salinity tolerant genotype Pokkali maintains relatively higher transcript levels for all TCS members under normal conditions as compared with the sensitive genotype IR64, in anticipation of stress. However, the trend is seen to be reversed upon salinity treatment with strong upregulation of TCS members in IR64, though to differential levels. We have picked up several of these sensory HKs and currently, the work is in progress related to their validation as ‘candidate gene’ for improving stress tolerance through transgenic approach.
Conclusion
Almost, 20 y have passed since the identification and cloning of the first plant HK. A huge amount of research has been performed not only on plant HKs but on two-component systems, in general. The advances in technology have enabled us to decode and decipher various signaling pathways and mechanisms of their action. But this is just the tip of the iceberg. There is a lot of work which needs to done in this regard. For example, the mechanism by which ATHK1 recognizes changes in osmolarity is still to be elucidated; the regulation of expression of most of these genes is still unclear; crystal structures of proteins, which can clear many doubts, are still elusive; complete interactomes are lacking. Furthermore, when there are reports about multiple roles of a single protein cropping ever so frequently, it makes easier to speculate that there are many more processes in which the plant HKs identified so far could be involved in.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Authors would like to thank Department of Biotechnology, Department of Science and Technology (Government of India) and Jawaharlal Nehru University (through Capacity building, CAS and PURSE) for supporting research work in the laboratory. Award of research fellowship from Council of Scientific and Industrial Research (CSIR) to RN and PS is also thankfully acknowledged.
Glossary
Abbreviations:
- HK
histidine kinase
- HpT
histidine phospho-transfer protein
- RR
response regulator
- TCS
two component system
Footnotes
†Current address: School of Plant, Environmental and Soil Sciences; Louisiana State University Agricultural Center; Baton Rouge, LA USA
Metabolomics Research and Core Laboratories; UC Davis Genome Center; Davis, CA USA
Previously published online: www.landesbioscience.com/journals/psb/article/21516
References
- 1.Ota IM, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–9. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 2.Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–44. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 3.Hwang I, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–15. doi: 10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol. 2004;134:1654–61. doi: 10.1104/pp.103.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pareek A, Singh A, Kumar M, Kushwaha HR, Lynn AM, Singla-Pareek SL. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006;142:380–97. doi: 10.1104/pp.106.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh G, Kumar R. Genome-wide in silico analysis of plant two component signaling system in woody model plant Populus trichocarpa. Research in Plant Biology. 2012;2:13–23. [Google Scholar]
- 7.Ishida K, Niwa Y, Yamashino T, Mizuno T. A genome-wide compilation of the two-component systems in Lotus japonicus. DNA Res. 2009;16:237–47. doi: 10.1093/dnares/dsp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, et al. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011;18:17–29. doi: 10.1093/dnares/dsq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urao T, Yamaguchi-Shinozaki K, Shinozaki K. Plant Histidine Kinases: An emerging picture of two-component signal transduction in hormone and environmental responses, Science. STKE 2001 (109). [DOI] [PubMed]
- 10.Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:20623–8. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci U S A. 2004;101:8821–6. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hejátko J, Pernisová M, Eneva T, Palme K, Brzobohatý B. The putative sensor histidine kinase CKI1 is involved in female gametophyte development in Arabidopsis. Mol Genet Genomics. 2003;269:443–53. doi: 10.1007/s00438-003-0858-7. [DOI] [PubMed] [Google Scholar]
- 13.Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem. 2010;285:23371–86. doi: 10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pham J, Liu J, Bennett MH, Mansfield JW, Desikan R. Arabidopsis histidine kinase 5 regulates salt sensitivity and resistance against bacterial and fungal infection. New Phytol. 2012;194:168–80. doi: 10.1111/j.1469-8137.2011.04033.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Guo B, Xie X, Yao Y, Peng H, Xie C, et al. A novel histidine kinase gene, ZmHK9, mediate drought tolerance through the regulation of stomatal development in Arabidopsis. Gene. 2012;501:171–9. doi: 10.1016/j.gene.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Muñiz LM, Royo J, Gómez E, Baudot G, Paul W, Hueros G. Atypical response regulators expressed in the maize endosperm transfer cells link canonical two component systems and seed biology. BMC Plant Biol. 2010;10:84. doi: 10.1186/1471-2229-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng Y, Dong H, Mu J, Ren B, Zheng B, Ji Z, et al. Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell. 2010;22:1232–48. doi: 10.1105/tpc.108.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desikan R, Horák J, Chaban C, Mira-Rodado V, Witthöft J, Elgass K, et al. The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS One. 2008;3:e2491. doi: 10.1371/journal.pone.0002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, et al. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–32. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, et al. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci U S A. 1998;95:5812–7. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall AE, Findell JL, Schaller GE, Sisler EC, Bleecker AB. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 2000;123:1449–58. doi: 10.1104/pp.123.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, et al. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–54. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, et al. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–3. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 24.Mähönen AP, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000;14:2938–43. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, et al. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:814–9. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caesar K, Thamm AMK, Witthöft J, Elgass K, Huppenberger P, Grefen C, et al. Evidence for the localization of the Arabidopsis cytokinin receptors AHK3 and AHK4 in the endoplasmic reticulum. J Exp Bot. 2011;62:5571–80. doi: 10.1093/jxb/err238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomin SN, Yonekura-Sakakibara K, Romanov GA, Sakakibara H. Ligand-binding properties and subcellular localization of maize cytokinin receptors. J Exp Bot. 2011;62:5149–59. doi: 10.1093/jxb/err220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wulfetange K, Lomin SN, Romanov GA, Stolz A, Heyl A, Schmülling T. The cytokinin receptors of Arabidopsis are located mainly to the endoplasmic reticulum. Plant Physiol. 2011;156:1808–18. doi: 10.1104/pp.111.180539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012;17:172–9. doi: 10.1016/j.tplants.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Bürkle L, Cedzich A, Döpke C, Stransky H, Okumoto S, Gillissen B, et al. Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 2003;34:13–26. doi: 10.1046/j.1365-313X.2003.01700.x. [DOI] [PubMed] [Google Scholar]
- 31.Wormit A, Traub M, Flörchinger M, Neuhaus HE, Möhlmann T. Characterization of three novel members of the Arabidopsis thaliana equilibrative nucleoside transporter (ENT) family. Biochem J. 2004;383:19–26. doi: 10.1042/BJ20040389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirose N, Makita N, Yamaya T, Sakakibara H. Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 2005;138:196–206. doi: 10.1104/pp.105.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cedzich A, Stransky H, Schulz B, Frommer WB. Characterization of cytokinin and adenine transport in Arabidopsis cell cultures. Plant Physiol. 2008;148:1857–67. doi: 10.1104/pp.108.128454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romanov GA, Lomin SN, Schmülling T. Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J Exp Bot. 2006;57:4051–8. doi: 10.1093/jxb/erl179. [DOI] [PubMed] [Google Scholar]
- 35.Kim JH, Johannes L, Goud B, Antony C, Lingwood CA, Daneman R, et al. Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc Natl Acad Sci U S A. 1998;95:2997–3002. doi: 10.1073/pnas.95.6.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, et al. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science. 2005;310:121–5. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- 37.Chen YF, Randlett MD, Findell JL, Schaller GE. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem. 2002;277:19861–6. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- 38.Grefen C, Städele K, Růzicka K, Obrdlik P, Harter K, Horák J. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Mol Plant. 2008;1:308–20. doi: 10.1093/mp/ssm015. [DOI] [PubMed] [Google Scholar]
- 39.Iwama A, Yamashino T, Tanaka Y, Sakakibara H, Kakimoto T, Sato S, et al. AHK5 histidine kinase regulates root elongation through an ETR1-dependent abscisic acid and ethylene signaling pathway in Arabidopsis thaliana. Plant Cell Physiol. 2007;48:375–80. doi: 10.1093/pcp/pcl065. [DOI] [PubMed] [Google Scholar]
- 40.Ueguchi C, Koizumi H, Suzuki T, Mizuno T. Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:231–5. doi: 10.1093/pcp/pce015. [DOI] [PubMed] [Google Scholar]
- 41.Higuchi M, Pischke MS, Mähönen AP, Miyawaki K, Hashimoto Y, Seki M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci U S A. 2004;101:8821–6. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–10. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 43.Müller B, Sheen J. Advances in cytokinin signaling. Science. 2007;318:68–9. doi: 10.1126/science.1145461. [DOI] [PubMed] [Google Scholar]
- 44.Heyl A, Wulfetange K, Pils B, Nielsen N, Romanov GA, Schmülling T. Evolutionary proteomics identifies amino acids essential for ligand-binding of the cytokinin receptor CHASE domain. BMC Evol Biol. 2007;7:62. doi: 10.1186/1471-2148-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mähönen AP, Higuchi M, Törmäkangas K, Miyawaki K, Pischke MS, Sussman MR, et al. Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr Biol. 2006;16:1116–22. doi: 10.1016/j.cub.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 46.Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 2011;67:157–68. doi: 10.1111/j.1365-313X.2011.04584.x. [DOI] [PubMed] [Google Scholar]
- 47.Hothorn M, Dabi T, Chory J. Structural basis for cytokinin recognition by Arabidopsis thaliana histidine kinase 4. Nat Chem Biol. 2011;7:766–8. doi: 10.1038/nchembio.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Punwani JA, Kieber JJ. Localization of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin. Plant Signal Behav. 2010;5:896–8. doi: 10.4161/psb.5.7.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell. 2006;18:3073–87. doi: 10.1105/tpc.106.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–8. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 51.Cutcliffe JW, Hellmann E, Heyl A, Rashotte AM. CRFs form protein-protein interactions with each other and with members of the cytokinin signalling pathway in Arabidopsis via the CRF domain. J Exp Bot. 2011;62:4995–5002. doi: 10.1093/jxb/err199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urao T, Miyata S, Yamaguchi-Shinozaki K, Shinozaki K. Possible His to Asp phosphorelay signaling in an Arabidopsis two-component system. FEBS Lett. 2000;478:227–32. doi: 10.1016/S0014-5793(00)01860-3. [DOI] [PubMed] [Google Scholar]
- 53.Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmülling T, Heyl A. Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J Proteome Res. 2008;7:3649–60. doi: 10.1021/pr0703831. [DOI] [PubMed] [Google Scholar]
- 54.Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;32:227–54. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- 55.Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–71. doi: 10.1016/S0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 56.Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–8. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- 57.Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci U S A. 1998;95:5401–6. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou X, Liu Q, Xie F, Wen CK. RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiol. 2007;145:75–86. doi: 10.1104/pp.107.104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong CH, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 2008;53:275–86. doi: 10.1111/j.1365-313X.2007.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci U S A. 1998;95:7825–9. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang ZG, Zhou HL, Chen T, Gong Y, Cao WH, Wang YJ, et al. Evidence for serine/threonine and histidine kinase activity in the tobacco ethylene receptor protein NTHK2. Plant Physiol. 2004;136:2971–81. doi: 10.1104/pp.103.034686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, Hall AE, O’Malley R, Bleecker AB. Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci U S A. 2003;100:352–7. doi: 10.1073/pnas.0237085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hall BP, Shakeel SN, Amir M, Ul Haq N, Qu X, Schaller GE. Histidine kinase activity of the ethylene receptor ETR1 facilitates the ethylene response in Arabidopsis. Plant Physiol. 2012;159:682–95. doi: 10.1104/pp.112.196790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lei G, Shen M, Li ZG, Zhang B, Duan KX, Wang N, et al. EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 2011;34:1678–92. doi: 10.1111/j.1365-3040.2011.02363.x. [DOI] [PubMed] [Google Scholar]
- 65.Singh K, Singla-Pareek SL, Pareek A. Dissecting out the crosstalk between salinity and hormones in roots of Arabidopsis. OMICS. 2011;15:913–24. doi: 10.1089/omi.2011.0098. [DOI] [PubMed] [Google Scholar]
- 66.Karan R, Singla-Pareek SL, Pareek A. Histidine kinase and response regulator genes as they relate to salinity tolerance in rice. Funct Integr Genomics. 2009;9:411–7. doi: 10.1007/s10142-009-0119-x. [DOI] [PubMed] [Google Scholar]