Background: The pneumococcal HtrA protease represses competence selectively when the frequency of errors in protein synthesis is low.
Results: HtrA digests both the bacterial competence-stimulating peptide (CSP) and unfolded proteins.
Conclusion: The presence of proteins with exposed hydrophobic regions competitively reduces the degradation of CSP.
Significance: This suggests a mechanism by which HtrA functions as a sensor for biosynthetic errors.
Keywords: Bacterial Signal Transduction, Protein Misfolding, Quorum Sensing, Serine Protease, Streptococcus, Streptococcus pneumoniae, HtrA Protease, Competence-stimulating Peptide, Genetic Competence
Abstract
The HtrA protease of Streptococcus pneumoniae functions both in a general stress response role and as an error sensor that specifically represses genetic competence when the overall level of biosynthetic errors in cellular proteins is low. However, the mechanism through which HtrA inhibits development of competence has been unknown. We found that HtrA digested the pneumococcal competence-stimulating peptide (CSP) and constituted the primary extracytoplasmic CSP-degrading activity in cultures of S. pneumoniae. Mass spectrometry demonstrated that cleavage predominantly followed residue Phe-8 of the CSP-1 isoform of the peptide within its central hydrophobic patch, and in competition assays, both CSP-1 and CSP-2 interacted with HtrA with similar efficiencies. More generally, analysis of β-casein digestion and of digestion within HtrA itself revealed a preference for substrates with non-polar residues at the P1 site. Consistent with a specificity for exposed hydrophobic residues, competition from native BSA only weakly inhibited digestion of CSP, but denaturation converted BSA into a strong competitive inhibitor of such proteolysis. Together these findings support a model in which digestion of CSP by HtrA is reduced in the presence of other unfolded proteins that serve as alternative targets for degradation. Such competition may provide a mechanism by which HtrA functions in a quality control capacity to monitor the frequency of biosynthetic errors that result in protein misfolding.
Introduction
The regulated development of genetic competence in cultures of the respiratory tract pathogen S. pneumoniae (also known as the pneumococcus) is a coordinated multicellular activity generated by a microbial population. The ability to develop competence is critical for the virulence of this organism (1–4) and has promoted the development of antibiotic resistance (5, 6) that now restricts the options available for treating pneumococcal infections. A central component of this process (reviewed in Ref. 7) is the secretion and progressive accumulation in the extracellular environment of a peptide pheromone known as CSP. As the bacterial density rises, this peptide triggers competence by activating the transmembrane ComD sensor kinase and thereby initiating a well characterized signaling cascade that has been considered an example of bacterial quorum sensing.
In addition to its sensitivity to bacterial density, we have recently shown that pneumococcal competence is triggered when the frequency of biosynthetic errors during protein translation increases (8). Such responsiveness to the accuracy of protein synthesis supports the recent suggestion that competence in S. pneumoniae serves as a stress response (9) and is consistent with the observation that the competence regulon includes genes for cellular proteases and chaperones in addition to those genes responsible for transformation (10). Although the underlying mechanism has not been defined, this error-dependent pattern of regulation is generated in part through the activity of the bacterial HtrA protease, which acts selectively to repress the development of competence when the frequency of translational errors is low (8).
Pneumococcal HtrA belongs to the broadly distributed family of extracytoplasmic HtrA proteases (reviewed in Ref. 11) that are characterized by a serine protease domain followed by one or more PDZ domains. The substrate specificity of HtrA proteases ranges from that of DegS in Escherichia coli, which is highly selective for digestion of RseA as a key step in initiating the envelope-stress response (12), to that of proteases such as DegP in E. coli (13) and DegQ in Legionella (14) and homologues in Synechocystis (15), which are broadly active against unfolded proteins in which hydrophobic regions are exposed to solvent. In addition to such endogenous substrates, HtrA proteases have recently been characterized from the pathogens Helicobacter pylori (16) and Campylobacter jejuni (17) that cleave host proteins to promote bacterial migration across epithelial barriers. A shared feature among the HtrA proteases appears to be their ability to form oligomers starting at the level of trimers (14, 15, 18–25), which in some cases are then able to assemble and disassemble dynamically to produce large structures consisting of 24 or more monomers. Assembly of such multimeric structures is promoted by binding of peptides or denatured proteins to a PDZ domain of the protein in a process that also allosterically activates the proteolytic capacity of the enzyme (20–23, 26). Notably, members of the HtrA family, in addition to serving as proteases, commonly function as chaperones as well (14, 27, 28). The repression of competence by pneumococcal HtrA has been shown to depend specifically on its protease activity (8, 29), although the target through which it exerts this regulatory effect has not previously been determined.
We sought to test whether HtrA blocks competence by directly digesting CSP. The possibility of activity against CSP appeared to be consistent with the observation that repression of competence due to excess HtrA could be overcome by a point mutation activating the ComD sensor kinase (29), which indicated that repression by HtrA occurs early in the competence signaling pathway. The structure of CSP, characterized by a central amphipathic α-helix (30), also appeared to present a suitable target for an HtrA protease that we hypothesized would favor digestion at sites near non-polar residues in a manner similar to DegP. From a structural perspective, we considered that CSP might effectively mimic the exposed hydrophobic region of a misfolded protein and thereby give rise to a situation in which digestion of the signaling peptide was in competition with digestion of generic, misfolded proteins. Such competition would present a model to explain the loss of competence repression by HtrA that is observed when frequent translational errors generate proteins with intrinsic folding defects.
Here we present data confirming the digestion of CSP by recombinant HtrA as well as by the native protease in cultures of Streptococcus pneumoniae. Furthermore, we show that pneumococcal HtrA preferentially cleaves substrates in association with sites of hydrophobic residues and that unfolded proteins serve as effective competitors to reduce the degradation of CSP.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant HtrA Protease
Regions corresponding to the complete coding sequence of pneumococcal htrA with the exception of the predicted signal peptide were amplified from the genomes of strains R895 (31) and P1373 (29), encoding wild-type htrA and the htrA S234A catalytic site variant, respectively, using primers 5′-GGGAATTCCATATGACTCAACTAACTCAAAAAAGTAGTGTAAAC-3′ and 5′-GTACCGCTCGAGTTAAGATTCTAAATCACCTGAACTCTTG-3′. These fragments were digested with NdeI and XhoI and cloned into vector pET-28a (Novagen, Madison, WI). The resulting plasmids were transformed into a derivative of E. coli strain BL21(DE3)pLysS that had been modified to have a deletion of ompA (provided by the laboratory of Jeffrey Weiser). During the work, this expression host was further modified to delete degP by P1vir phage transduction of DNA from strain JW0157 (32) of the Keio collection followed by excision of the kanamycin resistance cassette by expression of the FLP recombinase.
Cultures for induction of rHtrA expression were grown in Luria-Bertani broth containing 1% glucose, 30 μg/ml kanamycin, and 34 μg/ml chloramphenicol at 37 °C to an OD620 nm of ∼0.35. The temperature was reduced shortly before the addition of 2 mm isopropyl-β-d-thiogalactopyranoside and maintained at 30 °C during a 1.5-h period of induction. Cells from 400 ml of culture were resuspended in 10 ml of a buffer containing 5 mm imidazole, 500 mm NaCl, 20 mm Tris-Cl, pH 7.9, 1× BugBuster detergent solution (Novagen, Madison, WI), 150 units/ml DNase I, and 0.5 mg/ml RNase A. After incubation for 20 min at room temperature with gentle shaking, the lysate was cleared by centrifugation at 15,000 × g for 20 min followed by passage through a 0.22-μm filter. The sample was purified by affinity chromatography using His-Bind resin (Novagen) and elution in a buffer containing 500 mm NaCl and 20 mm Tris-Cl, pH 7.9, with a linear gradient of imidazole increasing from 5 mm to 1 m. These purification steps following lysis were performed at 4 °C to limit autoproteolysis. Fractions enriched for rHtrA as determined by SDS-PAGE were further purified by size exclusion chromatography (SEC)2 using an AKTA FPLC system (GE Healthcare) and a Superdex 10/300 GL column (GE Healthcare) equilibrated with buffer containing 500 mm NaCl and 20 mm Tris-Cl, pH 7.9. Samples were injected in volumes of 0.5 ml and run at a flow rate of 0.5 ml/min in the same buffer. Fractions enriched for rHtrA were pooled and exchanged into a buffer of PBS using Amicon ultracentrifugal filters (nominal molecular weight limit 10,000; Millipore, Bedford, MA) and concentrated to ∼1–2 mg/ml as determined using a MicroBCA kit (Pierce). The same SEC conditions were used for analytical SEC, except the injection volume was reduced to 50 μl for rHtrA samples that had been previously purified as well as for a molecular size standard (Bio-Rad gel filtration standard).
Protein Electrophoresis, Zymography, and Western Blotting
SDS-PAGE was performed under denaturing conditions using standard methods. For native gel electrophoresis, samples were first mixed 1:1 with a loading buffer of 20% glycerol, 100 mm Tris-Cl, pH 6.8, and 0.2% bromphenol blue before running on 4–15% acrylamide gradient gels with a running buffer of 25 mm Tris base and 0.19 m glycine. NativeMark (Invitrogen) was used as a molecular weight marker. Zymography was performed as described previously (29). Western blotting for HtrA was essentially as described (29) and for pneumolysin employed a 1:1000 dilution of an anti-pneumolysin monoclonal antibody (ab71810; Abcam, Cambridge, MA) and secondary detection with an anti-mouse immunoglobulin alkaline phosphatase conjugate (1:10,000; Sigma-Aldrich).
FRET Assays of CSP Proteolysis by rHtrA
A CSP-1 FRET reporter peptide was synthesized by Cambridge Research Biochemicals (Billingham, UK) with incorporation of a QSY-7 quencher (Invitrogen) and a Cys(Alexa488) fluorophore (Invitrogen) at the N terminus and C terminus of CSP-1, respectively. Purity greater than 95% was verified by HPLC and mass spectrometry. Samples of rHtrA or the S234A variant protein (10 μg) were incubated in 100 μl of buffer containing a 1 μm concentration of the CSP-1 FRET peptide together with 387 mm NaCl, 1.4 mm KCl, 5 mm Na2HPO4, 1 mm KH2PO4, 1.5 mm MgCl2, 1.5 mm CaCl2, and 83 mm Tris-Cl, pH 7.4. β-Casein (C6905, >98% purity; Sigma-Aldrich) was included at 10 μg/ml as indicated. Fluorescence was read (excitation 485 nm, emission 528 nm) every 2–5 min for samples incubated at 37 °C in 96-well plates using a Synergy2 plate reader (Bio-Tek, Winooski, VT).
Mass Spectrometry
To determine sites of CSP digestion, 1 μm unmodified CSP-1 (synthesized by GenScript, Piscataway, NJ) was incubated at 37 °C with 5 μg of either wild-type rHtrA or the S234A variant in 50 μl of the same buffer used for the FRET assay with these samples. To enhance the activity of rHtrA, 0.5 μg of β-casein was added, and a control reaction with only β-casein and CSP-1 was also analyzed. Following digestion, samples were flash frozen and stored at −80 °C until analysis by mass spectrometry. Aliquots were thawed, diluted 5-fold in 0.1% TFA, and immediately analyzed by LC-MS/MS using an LTQ-OrbiTrap XL mass spectrometer. The precursor ion series was examined for ions matching the predicted m/z for CSP-1 or fragments of the peptide. The MS2 spectra were searched against a sequence database comprising CSP, E. coli proteins (because rHtrA had been expressed in this background), and a list of common contaminants and other proteins of interest using the MASCOT search engine. The MS2 spectra were further analyzed using Scaffold (version 3.4.5; Proteome Software, Portland, OR) to verify identification of each peptide. MS2 total ion current values were summated for all of the spectra assigned to each peptide. Because the number of peptides that theoretically could be generated by digestion of β-casein and rHtrA would be much larger than those arising from CSP, degradation of these larger proteins was analyzed using Scaffold to identify MS2 spectra assigned to either of these proteins with a confidence probability of >80%.
CSP Digestion by Pneumococcal Culture Fractions
S. pneumoniae was grown in CAT medium (33) to an OD620 nm of 0.25. Cells were collected by centrifugation at 1875 × g for 20 min at 4 °C and resuspended in an equal volume of fresh medium. Supernatants were passed through a 0.22-μm filter before use. Selected samples of washed cells were disrupted by sonication at 10 watts for 6 bursts of 10 s each while chilled in ice water. For analysis of CSP digestion, 4 μl of culture samples were diluted in PBS to a volume of 200 μl in the presence of the CSP-1 FRET peptide at concentrations ranging from 10 to 160 nm. For competition assays, alternative substrates were added at the concentrations indicated. Denaturation of BSA was achieved by incubating 0.7 mm BSA with 20 mm dithiothreitol for 3 h at 40 °C immediately before each assay. Fluorescence was read for samples incubated at 37 °C as above.
Protoplast Formation and Proteinase K Digestion
Cultures were grown as above and concentrated 10-fold by centrifugation, washing, and resuspension in SMM buffer (0.5 m sucrose, 20 mm MgCl2, 20 mm MES). Following the addition of lysozyme (0.5 mg/ml; Epicenter Technologies, Madison, WI) and mutanolysin (0.01 mg/ml; Sigma-Aldrich), samples were incubated for 1 h at 37 °C, centrifuged at 3000 × g, and resuspended in an equal volume of fresh SMM buffer. Production of protoplasts was verified by testing osmotic fragility. Proteinase K (2 mg/ml; Roche Applied Science) was added to aliquots as indicated. After incubation at 37 °C for 1 h, Pefabloc SC (4 mg/ml; Roche Applied Science) was added to inhibit the protease during a 10-min incubation at room temperature. Protoplasts were then collected by centrifugation and resuspended in fresh SMM buffer containing Pefabloc before electrophoresis.
Generation of S. pneumoniae Mutants
An in-frame deletion of the pneumococcal comA gene (designated strain KSP209) was generated in the background of the R6 derivative strain R895 (31) using PCR ligation mutagenesis as described previously (34). Gene segments for ligation to the counterselectable Jauns cassette (35) were amplified flanking comA with the primers 5′-TTTTGTTTAGTGATTGGGGTAAG-3′ and 5′-ACGAGGATCCGAGAGCAGACCATTTTTTTGTTC-3′ for the 5′ region and 5′-AGCAGGGCCCAATACCAAGAAGGGGCAGAGGG-3′ and 5′-TAGCGAACAGAATCACCGAC-3′ for the 3′ region. A product encoding the in-frame deletion was constructed by overlap extension PCR using the primers 5′-TTTTGTTTAGTGATTGGGGTAAG-3′ and 5′-CTTCGACAATCTTGCCCTGTGGACTAGGTGCCATAAAAAGAGTC-3′ for the 5′ region and primers 5′-GACTCTTTTTATGGCACCTAGTCCACAGGGCAAGATTGTCGAAG-3′ and 5′-TAGCGAACAGAATCACCGAC-3′ for the 3′ region. A similar process was used previously in constructing strains KSP122 (htrA S234A) and KSP148 (htrA+) in the same background (8).
Competence Assays
A luciferase reporter consisting of an ssbB′-luc transcriptional fusion was used to measure activation of the competence system as described previously (8). Expression of ssbB is specifically induced during competence, and this reporter has been demonstrated to reflect competence for transformation (31, 36).
RNA Isolation and Quantitative RT-PCR
The comA deletion strain KSP209 was grown at 37 °C in C+YYB medium (8) supplemented with antibiotics at concentrations that previously had been shown to modulate development of competence in this medium (8). Samples were harvested in exponential growth phase (OD620 nm ∼0.2) and stored in RNAlater solution (Invitrogen). RNA was purified using the RNeasy Plus Mini kit (Qiagen, Valencia, CA) followed by DNase digestion and cDNA synthesis as described previously (37). Quantitative PCR for htrA employed primers 5′-AAAAGCCATCCAAACTGATACTGCTA-3′ and 5′-CCGATAACCTGCCCTTGAATATTGA-3′ and the probe 5′-6-carboxyfluorescein-CTCTGGCGGCCCACTG-nonfluorescent quencher-3′. Transcript levels for htrA were normalized to that of rpoA as an internal control after quantitative PCR as described previously (37).
RESULTS
Purification and Characterization of Recombinant Pneumococcal HtrA
We expressed and purified pneumococcal HtrA as a His6-tagged recombinant protein (rHtrA) by affinity chromatography followed by SEC (Fig. 1A). A variant of the protein containing an S234A mutation at the catalytic site was also prepared as a control. During the characterization of initial preparations of the protease, mass spectrometry revealed that small quantities of the homologous E. coli protein DegP were retained in the purified sample, possibly due to formation of heteromultimers. To circumvent this issue, the expression host was therefore modified to have a deletion of degP. Purification of rHtrA yielded a protein that was largely intact following SEC (Fig. 1A, lanes 1 and 3). The final step of purification, however, required concentration of the protein and exchange from a high salt buffer to PBS, without which process proteolytic activity was not seen. This step was accompanied by partial degradation of the intact protein, which appeared to be the result of autoproteolysis, considering that the change was not observed in the S234A catalytic site variant (Fig. 1A). Such autodigestion has been noted for other proteases in the HtrA family, including DegP (38). Although we have not directly compared the stability of our protease with that of DegP, self-cleavage may be enhanced in the case of pneumococcal HtrA due to the lack of cysteine residues and a disulfide bridge that has been shown to be important for stabilizing DegP against autodigestion (38).
FIGURE 1.
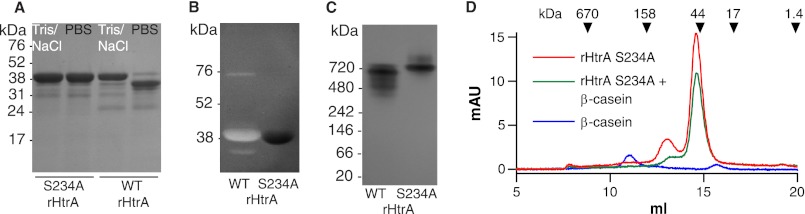
Characterization of wild-type (WT) rHtrA and the S234A catalytic site variant. A, SDS-PAGE of samples in Tris/NaCl buffer following the preparative SEC step and after exchange into PBS. B, β-casein zymography. C, native gel electrophoresis. D, analytical SEC in Tris/NaCl buffer in the presence or absence of β-casein. Elution volumes for standards are shown with arrows. mAU indicates milli-absorbance units.
Activity of the rHtrA protease was demonstrated by zymography using β-casein as a substrate (Fig. 1B). Importantly, no activity was observed in this assay for the S234A variant. The primary band of digestion observed for rHtrA was at the size expected for the intact protease, but two less intense bands were also noted to migrate with sizes both larger and smaller than the main band. The lower band is likely to represent a fragment of the protease generated by autodigestion but that retains activity. The origin of the faint upper band, which migrates in the range of 75 kDa, is uncertain, but mass spectrometry identified rHtrA in gel slices cut from each of these three bands on the zymogram. The absence of these bands in the S234A variant supports the conclusion that they result from the recombinant protease, albeit in different forms.
Because other HtrA proteases have been described to form multimers in a process that may be promoted by the presence of substrate proteins, we examined the recombinant pneumococcal protease by native gel electrophoresis. Under these conditions, the protease ran with an estimated molecular mass near 700 kDa (Fig. 1C), in the range expected for an 18-mer based on rHtrA monomers of 40 kDa. Consistent with the partial digestion of rHtrA monomers seen with denaturing electrophoresis, the active protease demonstrated multiple bands and a reduction in size relative to the S234A mutant on the native gel. No change in the migration of rHtrA was observed when the sample was incubated with β-casein prior to native electrophoresis (data not shown).
We also examined the purified protein by analytical SEC but unexpectedly did not observe the form of the protein that had been seen by native electrophoresis near 700 kDa. The inactive S234A variant was used for these multimerization assays to prevent self-digestion. In the same high salt buffer that had been used for SEC during the purification process, the protein eluted with a principal peak near the size expected for the monomer (Fig. 1D). A smaller peak was also observed in the range of 95 kDa that may represent either a trimer or the detergent-stable form of the protease corresponding to the faint upper band seen with zymography. A very small amount of material was also evident eluting near the exclusion limit of the column (1300 kDa). The addition of β-casein to the sample reduced the intensity of both peaks that had been seen with rHtrA alone but without the formation of larger species. Using PBS rather than the high salt buffer, the predominant peak on SEC remained that of the monomer without the production of larger forms (not shown). Despite injection of equivalent quantities of protein, the intensity of the monomer peak under these conditions was substantially reduced relative to assays in high salt buffer. As discussed below, these findings in conjunction with clear evidence of oligomerization by native electrophoresis suggest that pneumococcal rHtrA may form multimers that are unable to elute from the size exclusion column.
Digestion of CSP by Recombinant HtrA
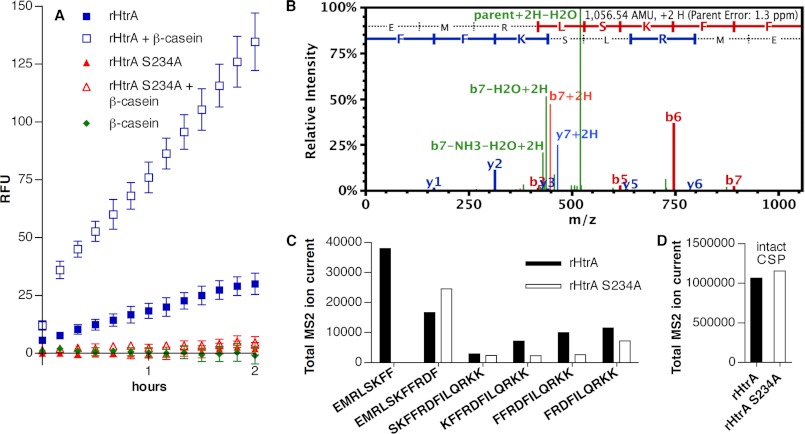
We tested the ability of rHtrA to digest CSP in FRET assays by incubating the protein with a synthetic version of the CSP-1 signaling peptide modified to have an Alexa 488 fluorophore at the C terminus and a quencher at the N terminus. Fluorescence resulting from digestion of the peptide was observed with the active form of the protease but not with the S234A catalytic site variant (Fig. 2A). Because other HtrA proteases, such as DegP, demonstrate allosteric activation in the presence of substrate proteins (20–23, 26), we evaluated the impact of adding β-casein to the reaction. The rate of proteolysis was significantly increased by the addition of β-casein. In a control reaction, β-casein alone did not affect the CSP FRET peptide.
FIGURE 2.
A, fluorescence generated by incubation of labeled CSP-1 FRET peptide with rHtrA or the catalytic site variant (rHtrA S234A) in the presence or absence of β-casein, expressed as relative fluorescence units (RFU). Symbols represent means ± S.D. (error bars) (n = 3). B, MS2 fragmentation spectrum from peptide EMRLSKFF produced by digestion of CSP-1 with rHtrA. C, total MS2 ion current from fragments of CSP-1 detected after incubation of intact peptide with rHtrA or rHtrA S234A. D, total MS2 ion current from intact CSP-1 detected in the same samples shown in C.
In order to determine the site at which HtrA cleaves the signaling peptide, samples of rHtrA as well as the inactive S234A variant were incubated with CSP-1, in the presence of β-casein to enhance activity of the protease, and then analyzed by mass spectrometry. This process demonstrated the generation of a peptide fragment (EMRLSKFF) in the rHtrA sample (corresponding to the first 8 residues from the N-terminal portion of CSP-1) that was not found in the rHtrA S234A sample (Fig. 2, B and C). This fragment was not seen in a control sample containing only β-casein and CSP-1, consistent with the FRET data indicating that the β-casein preparation did not itself contain protease activity. This finding demonstrates the ability of HtrA to digest CSP-1 following residue Phe-8 in the central α-helix of the peptide. Although production of the complementary C-terminal fragment RDFILQRKK would be expected to accompany this cleavage event, we were unable to detect this highly charged peptide in either the rHtrA sample or the S234A control. It is uncertain whether the absence of this peptide reflects a secondary degradation event that efficiently eliminates this fragment or biophysical properties of the peptide that impair its detection in this system.
Several other fragments of CSP-1 were also observed but were detected in both the rHtrA digest and the S234A control. For each of these other peptides, the MS2 total ion current signal was less intense than for the EMRLSKFF fragment (Fig. 2C). Such fragments present in the S234A control sample may represent either products of spontaneous hydrolysis of CSP-1 or impurities present in the chemical synthesis of the original peptide. The increased signal intensity noted for several of these minor fragments in the active protease sample may indicate the potential for less efficient digestion of CSP-1 by HtrA at multiple sites. Although these differences were seen for several peptide fragments, it is important to note that the rHtrA and S234A samples were well balanced with regard to the signal measured for the intact peptide (Fig. 2D).
Cleavage Site Profiles for HtrA
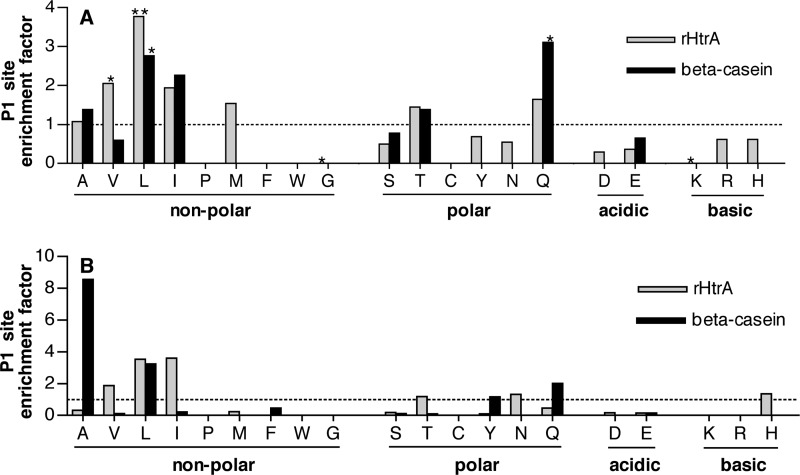
The target site selectivity of HtrA was investigated at a more general level by examining the pattern of peptide fragments resulting from digestion of β-casein and of rHtrA autodigestion within the same samples that were analyzed by mass spectrometry to determine the site of CSP cleavage. This process revealed a pattern of fragments indicating 66 breakage sites in rHtrA and 22 sites in β-casein. Of these, 62 and 18 sites, respectively, were not found in the rHtrA S234A sample and were considered to represent sites of HtrA proteolysis for subsequent analysis. No breakage sites were found exclusively in the inactive rHtrA S234A sample. We focused our analysis on residues found at the P1 site of the substrate (immediately on the N-terminal side of the hydrolyzed bond) because interactions between these residues and the S1 binding pocket of the enzyme are principal determinants of the specificity of serine proteases in the chymotrypsin family (39), of which HtrA is a member. The distribution of amino acid residues at the P1 site was analyzed by calculating the frequency of each residue among the P1 sites and then normalized to the frequency of that residue in the overall protein sequence. The resulting enrichment score was greater than 1 for residues that were overrepresented at the P1 site and less than 1 for residues that were underrepresented.
For both β-casein and rHtrA, a preference existed for non-polar, aliphatic residues at the P1 site (Fig. 3A), although enrichment for glutamine was also noted in the case of β-casein. With both substrates, a bias against charged P1 site residues was further evident. Because the efficiency of digestion among these sites would be expected to vary, we also repeated the analysis using weighted values for each P1 site based on the strength of the MS2 total ion current measured for the individual spectra associated with each peptide fragment. This analysis also indicated a preference for aliphatic residues at the P1 site (Fig. 3B). Interestingly, neither β-casein nor rHtrA showed substantial digestion following phenylalanine such as had been seen with CSP-1, although the frequency of this residue in the proteins was low (1 and 4% for rHtrA and β-casein, respectively) and may have limited the availability of sites having phenylalanine displayed in an appropriate context. This observation, however, raises the possibility that the selectivity of pneumococcal HtrA may have been modified during its evolution in order to accommodate activity against CSP while retaining general activity for proteolysis near aliphatic residues.
FIGURE 3.
A, P1 residues associated with sites of HtrA cleavage of either rHtrA (62 sites) or β-casein (18 sites) based on mapping of fragments identified by mass spectrometry. The frequency of each residue among the P1 sites was divided by the overall frequency of that residue in the target protein to generate an enrichment factor score. *, p < 0.05; **, p < 0.001 by Fisher's exact test comparing the frequency of a residue at the P1 site versus the frequency of that residue within the entire protein. The power of this test was limited for residues that were rare both in the P1 sample and within the proteins overall. Power was also greater for rHtrA sites than for β-casein sites because the sample of cut sites was larger in the former case. Residues for which bars are not shown were not found at the P1 site. Note that whereas all residues are represented in β-casein, residues Trp (W) and Cys (C) are not present in rHtrA. B, P1 residues associated with sites of HtrA cleavage as in A, except that frequencies were weighted by MS2 total ion current signal intensity of the spectra attributed to each fragment. Conversion of frequencies to weighted continuous values prevented the application of statistical tests based on discrete variables.
Activity of HtrA in Pneumococcal Cultures
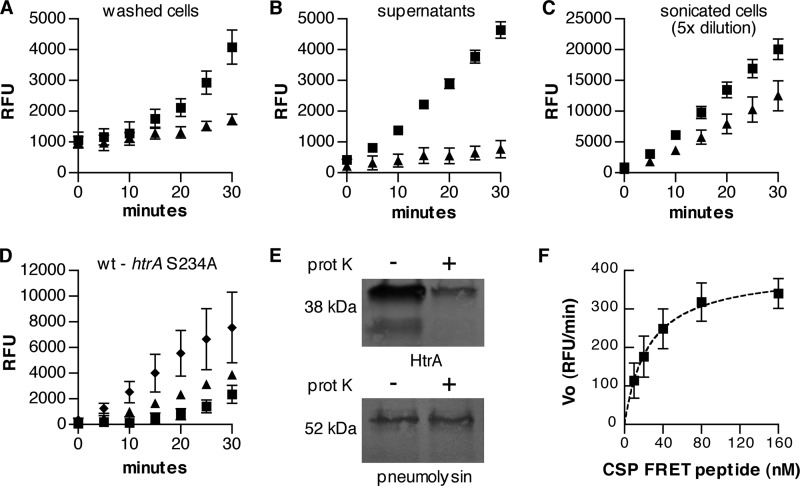
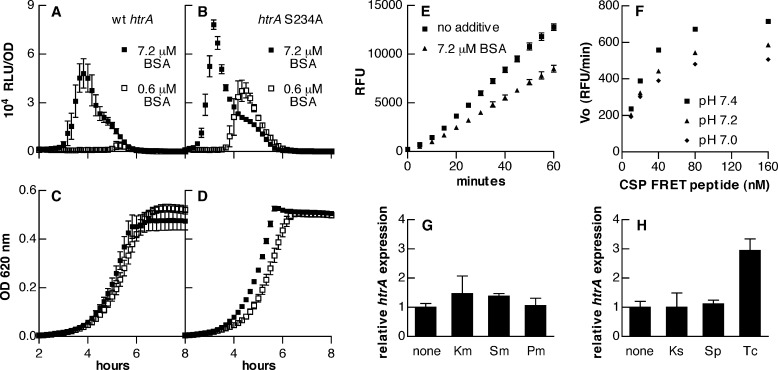
We next used the FRET method to measure the ability of native HtrA to digest CSP in the setting of growing pneumococcal cultures. In these assays, the activity of an htrA wild-type strain was compared with that of an htrA S234A catalytic site mutant measured in parallel. Importantly, the bacterial densities in these samples were not significantly different between the wild-type and mutant strains when measured in terms of either cfu/ml or the average number of bacteria comprising an individual chain of pneumococci (data not shown). For both washed cells and culture supernatants prepared from equal volumes of the same cultures, wild-type samples digested CSP more rapidly than did the htrA S234A mutant (Fig. 4, A and B). Notably, the rate of CSP degradation in the mutant was minimal, suggesting that HtrA is the principal extracytoplasmic protease active against this peptide.
FIGURE 4.
A–C, fluorescence generated by incubation of labeled CSP-1 FRET peptide with fractions consisting of washed cells (A), supernatants (B), or sonicated cells (C). Samples were derived from equal volumes of the same S. pneumoniae cultures (but then diluted 5-fold in the case of sonicated cells) expressing either wild-type htrA (strain KSP148; squares) or htrA S234A (strain KSP122; triangles). D, the difference in fluorescence signal between wild-type and htrA S234A cultures for washed cells (squares), supernatants (triangles), or sonicated cells (diamonds; 5-fold diluted). E, Western blots for HtrA and pneumolysin from protoplasts incubated with or without proteinase K. F, initial reaction velocity (Vo, expressed in relative fluorescence units (RFU)/min) for digestion of varying concentrations of the CSP-1 FRET peptide with HtrA in culture supernatants. The best fit of the data to the Michealis-Menten equation is shown with a dashed line. Symbols represent means ± S.D. (error bars), n = 3 (A–D) or n = 4 (F).
Our previous fractionation experiments with S. pneumoniae showed that the largest amount of HtrA protein was associated with the cell membrane upon fractionation and that only smaller quantities were present in the supernatant (29). In contrast, these FRET assays indicated that the activity of washed cells was no greater than that of supernatants. Because there was a delay in the development of HtrA-specific fluorescence signal in assays using washed cells (Fig. 4A, compare wild-type versus htrA S234A samples during the first half of the assay), it was uncertain whether the increased signal measured during the later part of the assay reflected surface-associated HtrA or enzyme that had been shed from the cells. Sonication of these cells, however, dramatically increased digestion of CSP such that a 5-fold dilution of the samples was required to avoid saturation of the assay through rapid consumption of the substrate (Fig. 4C). Sonicated samples from both wild-type and htrA S234A cultures demonstrated enhanced digestion of CSP, suggesting the release of other proteases from within the bacteria. Importantly, however, the component of the fluorescence signal attributable to HtrA, which was defined by the difference between the wild-type and mutant samples, also increased more than 10-fold upon sonication (Fig. 4D, in which the sonicated samples were diluted 5-fold).
Proteases in the HtrA family have been uniformly found in the periplasmic space of Gram-negative bacteria or as extracellular proteins in Gram-positive organisms. Considering the impact of sonication, however, we sought to verify the orientation of pneumococcal HtrA relative to the membrane. Exposing protoplasts to proteinase K, we demonstrated that HtrA was indeed susceptible to enzymatic degradation as expected for a protein associated with the exterior aspect of the cell membrane (Fig. 4E). In contrast, proteinase K failed to digest pneumolysin, which is localized to the cytoplasm in this strain (29). These findings indicate that the enhanced HtrA activity generated by sonication cannot simply result from the enzyme being released from an intracellular store. Instead, these observations suggest that HtrA is likely to exist on the surface of S. pneumoniae as part of a complex that in the absence of sonication limits the access of exogenous peptides to the protease.
Kinetics and Inhibition of HtrA Activity
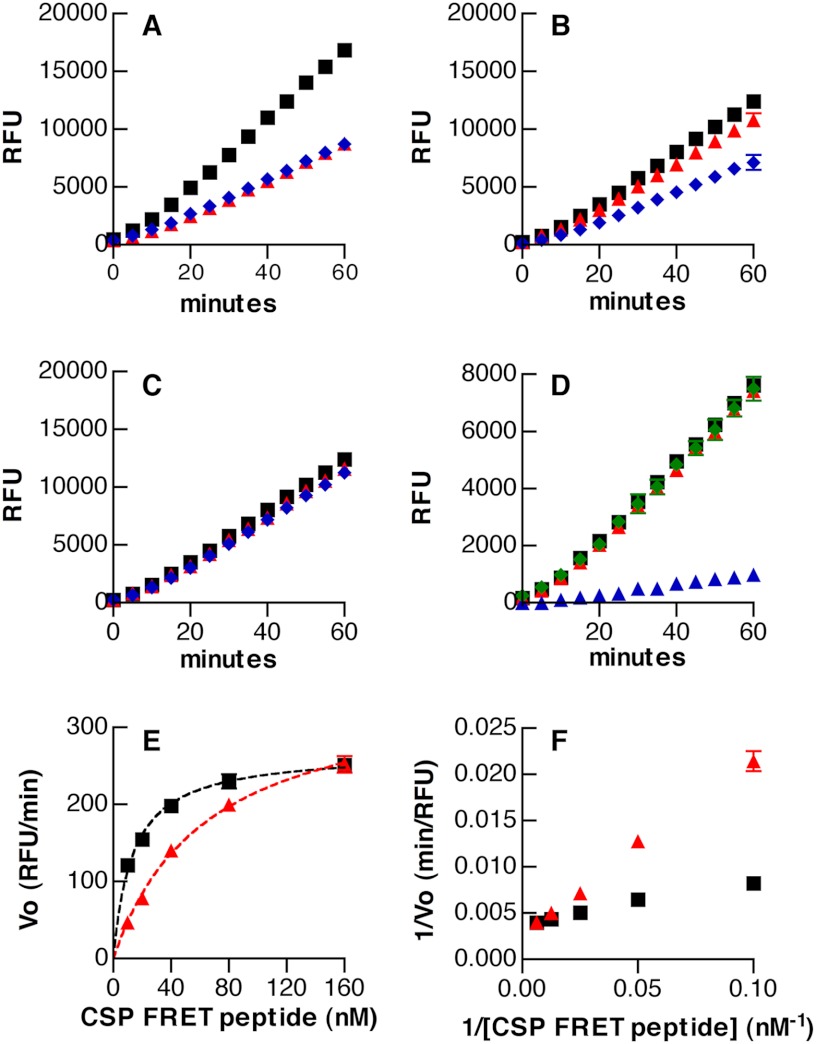
Because assays using pneumococcal supernatants demonstrated linear increases in FRET signal over time (Fig. 4B), we used these samples to investigate the kinetics of CSP digestion by native HtrA. Fixed dilutions of supernatant were incubated with concentrations of the CSP FRET peptide ranging from 10 to 160 nm. HtrA activity measured from the difference between fluorescence curves for wild-type and htrA S234A mutant samples fit Michealis-Menten kinetics well, with an apparent Km value of 24.7 ± 6.0 nm (Fig. 4F). To test the ability of HtrA to interact with the unlabeled, native form of CSP-1 as well as with the other prevalent variant of the peptide (CSP-2), we performed FRET assays with and without these peptides. As anticipated, the presence of unlabeled peptide as an alternative substrate for HtrA reduced the fluorescence signal from degradation of the labeled peptide (Fig. 5A). In this assay, both CSP-1 and CSP-2 appeared to be recognized by HtrA with equivalent efficiencies.
FIGURE 5.
A–D, fluorescence from digestion of labeled CSP-1 FRET peptide (20 nm) in the presence (colored symbols) or absence (black squares) of alternative substrates. Alternative substrates were tested as follows. A, unlabeled CSP-1 (200 nm; red triangles) or CSP-2 (200 nm; blue diamonds); B, β-casein (200 nm (red triangles) and 1000 nm (blue diamonds)); C, BSA (200 nm (red triangles) and 1000 nm (blue diamonds)); D, BSA denatured with DTT (200 nm; blue triangles) compared with native BSA (200 nm; red triangles) or DTT alone (29 μm; green diamonds). Fluorescence values represent differences in relative fluorescence units between samples containing wild-type versus htrA S234A supernatants. E, initial reaction velocities versus substrate concentration for digestion of the CSP-1 FRET peptide by HtrA in culture supernatants. Samples with 50 nm denatured BSA (red triangles) are compared with reactions without additive (black squares). Best fits to the Michaelis-Menten equation are shown with dashed lines. F, Lineweaver-Burk transformation of kinetic data from E. Symbols represent means ± S.D., n = 4 (A–D) or n = 3 (E and F).
Because β-casein had been able to serve as a target for HtrA in zymography but had also enhanced activity of the recombinant protein in FRET assays, we tested the impact of this protein on the ability of HtrA to digest CSP in the context of pneumococcal supernatants. In contrast to the increased digestion triggered by β-casein when using recombinant HtrA, β-casein at concentrations of 200 or 1000 nm reduced the ability of HtrA derived from culture supernatants to degrade CSP (Fig. 5B). Lowering the concentration of β-casein to 20 nm resulted in minimal inhibition of the reaction (data not shown) but still did not enhance digestion in the way that had been seen with the recombinant protease. The inability of β-casein to enhance the activity of HtrA in the setting of bacterial cultures may be the result of exposure to other proteins or peptides, whether present in the growth medium or produced by S. pneumoniae itself, that may activate the protease on an ongoing basis.
β-Casein is distinctive in having a relatively open conformation that does not adopt a well ordered tertiary structure (40). This relaxed structure together with the amphipathic character of β-casein (40) is likely to expose hydrophobic regions for proteolysis by an enzyme, such as HtrA, that shows selectivity for cleavage near non-polar residues. From this perspective, assays using β-casein simulate the capacity of a denatured protein to serve as a substrate for HtrA. To evaluate the potential of a well folded protein to serve as a substrate, we tested the ability of BSA to interact with HtrA. BSA was unable to inhibit the digestion of the FRET peptide by HtrA at concentrations of up to 1000 nm (Fig. 5C), at which level β-casein had been an effective inhibitor. Denaturing the protein by reducing its intramolecular disulfide bonds with dithiothreitol (DTT), however, converted BSA into a potent inhibitor of CSP digestion (Fig. 5D). As expected, this inhibition by denatured BSA was competitive in nature, as demonstrated by reduced efficacy as the concentration of the FRET CSP peptide was increased and convergence of the Lineweaver-Burk plots (Fig. 5, E and F).
Because the presence of BSA or serum in the growth medium has been required for development of pneumococcal competence in vitro, we tested for evidence of interaction between this component of the medium and HtrA. Notably, our standard competence medium contains 7.2 μm BSA, a concentration high enough that a reduction of HtrA activity could be seen against the FRET CSP peptide even in the absence of DTT (Fig. 6E). We speculate that this inhibition may result from occasional proteins with the stock of BSA that may be damaged or misfolded. Without any supplemental BSA, cultures failed to grow (data not shown). Reducing the concentration of BSA from 7.2 to 0.6 μm, however, resulted in loss of competence with an htrA wild-type strain but not in an htrA S234A catalytic site mutant (Fig. 6, A–D).
FIGURE 6.
A and B, luciferase activity of the ssbB′-luc reporter for competence in strains KSP148 (wild-type htrA) (A) and KSP122 (htrA S234A) (B) grown in media containing either 7.2 μm or 0.6 μm BSA. Symbols represent means ± S.E. for 12 replicate cultures. C and D, optical density (OD at 620 nm) for the same cultures shown in A and B, respectively. E, fluorescence from digestion of the CSP-1 FRET peptide (20 nm) in the presence (triangles) or absence (squares) of 7.2 μm BSA. Fluorescence values represent differences in relative fluorescence units between samples containing wild-type versus htrA S234A supernatants and are shown as means ± S.D., n = 4. F, initial reaction velocities versus substrate concentration for digestion of the CSP-1 FRET peptide are shown for identical aliquots of supernatant diluted into PBS that had been adjusted to the pH values indicated. G, relative expression of htrA as measured by quantitative RT-PCR in cultures of strain KSP209 grown in the presence of antibiotics that promote development of competence (kanamycin (Km), 6 μg/ml; streptomycin (Sm), 3 μg/ml; paromomycin (Pm), 3 μg/ml) compared with that of untreated samples. H, relative expression of htrA in KSP209 treated with antibiotics that repress development of competence (kasugamycin (Ks), 6 μg/ml; spectinomycin (Sp), 6 μg/ml; tetracycline (Tc), 60 ng/ml). Bars, means ± S.D. (error bars), n = 3. Differences between treated and untreated samples are significant only for tetracycline (p < 0.01 by analysis of variance with Dunnett's multiple comparison test).
Effects on HtrA of Other Factors That Modulate Competence
Because the development of competence is impaired by small decreases in pH of the growth medium, we examined the effect of pH on the activity of HtrA from pneumococcal supernatants. The observation that the HtrA family protease DegQ from E. coli displays increased activity at lower pH (41) raised the prospect that modulation of HtrA activity might explain the pH sensitivity of the competence system. In contrast to the report for DegQ, however, we found that activity of pneumococcal HtrA did not increase at lower pH but rather declined modestly as the pH was reduced from 7.4 to 7.0 (Fig. 6F). Direct modulation of HtrA activity by changes in pH therefore would not appear to be responsible for the effect of pH on competence.
We have shown previously that competence is induced by antibiotics that increase the frequency of translational errors and is blocked by antibiotics that reduce such errors (8). These agents would be expected to affect the level of protein folding stress but might also trigger changes in the level of HtrA in response. Although we had shown previously that the antibiotics streptomycin, kanamycin, and paromomycin did not appear to influence the quantity of HtrA detected by Western blotting, we sought to evaluate the possibility of error-responsive htrA regulation more quantitatively using RT-PCR. In order to separate possible effects of antibiotics on htrA from the known induction of htrA that is a delayed part of the competence response (10), these assays were conducted in the background of a strain with a deletion of comA. This strain is unable to secrete CSP and does not develop spontaneous competence. We found that antibiotics that stimulate ribosomal errors and induce competence (Fig. 6G) did not significantly affect the expression of htrA. Of the antibiotics that increase translational accuracy and repress competence (Fig. 6H), kasugamycin and spectinomycin did not change htrA expression. Although tetracycline appeared to increase htrA expression (Fig. 6H), a change limited to this antibiotic does not itself explain the repression of competence exerted by this entire group of antibiotics. Together these observations suggest that the dominant interaction modulating the digestion of CSP by HtrA is competition from misfolded proteins rather than changes in expression of the enzyme or alterations in its intrinsic activity.
DISCUSSION
In this work, we showed that the HtrA protease of S. pneumoniae was able to digest the peptide pheromone CSP, which regulates development of genetic competence. This protease furthermore accounted for the large majority of the CSP-degrading activity present in extracytoplasmic fractions of pneumococcal cultures. The apparent Km for native HtrA catalyzing this reaction in the context of culture supernatants was ∼25 nm, which is close to the concentration of CSP required to initiate competence signaling (30, 42). Specifically, maximal transformation was reported to be induced by CSP-1 at concentrations of at least 13 nm (42), whereas half-maximal activation of competence signaling by CSP-1 and CSP-2 was seen at 4.4 and 14 nm, respectively (30). The similarity of these values suggests that HtrA should be effective in degrading CSP produced by S. pneumoniae during growth.
Digestion of CSP by HtrA thereby appears to provide a mechanism to explain the earlier observations that competence was enhanced by either deletion of htrA or a point mutation inactivating the catalytic site of the enzyme (8, 29). HtrA appears to prevent the onset of competence under specific conditions discussed below, such as when biosynthetic errors during protein production are rare (8) or when the extracellular protein concentration is low (this work). Increased expression of HtrA through activation of the CiaRH two-component signaling system also inhibits development of competence (29). In contrast to the ability of HtrA to block competence initiation in these settings, the role of the protease in terminating competence is less clear. Although htrA expression is induced ∼8-fold as part of a delayed response following the activation of competence (10), mutants in which htrA has been inactivated or deleted terminate competence in a manner similar to what is seen in the wild type and do not extend the response into the stationary phase of growth (Fig. 6, A and B) (8, 29). Because the onset of competence is somewhat earlier in strains without active HtrA (Fig. 6) (8, 29), the net effect is a modest prolongation of the competence response. Although HtrA may contribute to shutting off competence signaling by digesting CSP, these observations indicate that other factors, which remain to be defined, are sufficient to terminate the response. Considering that positive feedback strongly amplifies the production of CSP once competence has begun, the reduced efficacy of HtrA in turning competence off as opposed to blocking its initiation may be the result of higher pheromone levels at the later stage. Finally, it is important to note that although HtrA expression is regulated by at least two signaling systems (10, 43), changes in HtrA levels are not responsible for the modulation of competence by this protease that allows it to serve as a sensor for the accuracy of protein biosynthesis (Fig. 6, G and H) (8).
Whereas the role of pneumococcal HtrA in modulating cell-to-cell communication appears to be unique, other general characteristics of the HtrA protease in S. pneumoniae reflect conserved properties similar to those described for related members of the HtrA family. Among these features are its ability to form multimers and the enhancement of protease activity by exposure to molecules, such as β-casein, that serve as substrates for the enzyme. The ability of β-casein to stimulate the activity of recombinant pneumococcal HtrA against CSP was evident (Fig. 2), but its impact on assembly of higher order multimers, similar to the substrate-induced multimerization shown for other HtrA proteases, was less clear. On native gel electrophoresis, our recombinant protein appeared to have a size of ∼700 kDa, which, given the propensity of other HtrA proteases to combine subunits consisting of trimers, suggests assembly into 18-mers. Remarkably, however, peaks corresponding to multimers in this size range were not evident by SEC although the system used had an exclusion limit of 1300 kDa. Although the high salt buffer (0.5 m NaCl) employed during SEC may have limited intermolecular interactions, performing SEC using PBS substantially reduced the apparent amount of the eluted monomer without generating new peaks corresponding to larger forms. Similarly, we noted that the addition of β-casein to the HtrA sample reduced the intensity of the peak eluting at the size of the monomer without producing additional peaks. These observations suggest that both reduction in the salt concentration and the addition of β-casein may affect the state of HtrA so as to promote its retention on the column. Whether these changes reflect increased affinity of the protein for the resin or formation of aggregates that are too large or collectively bind too avidly to elute from the column is uncertain. The transition from the high salt buffer to PBS during purification of rHtrA, however, was also associated with increased protease activity (Fig. 1A) such as accompanies formation of larger DegP multimers. We therefore favor the interpretation that pneumococcal HtrA may be able to form oligomers that are larger even than what was seen on native gel electrophoresis and that exposure to substrate molecules, such as β-casein, may promote this transition. Notably, multimers of DegP have been described that contain up to 30 monomer subunits (19). The precise nature, however, of HtrA multimers in S. pneumoniae remains a subject for investigation.
The increased rate of CSP proteolysis by purified rHtrA in the presence of β-casein suggests an allosteric activation that would be consistent with the recent report that the PDZ domain of pneumococcal HtrA contains a peptide-binding groove able to recognize several peptides having hydrophobic residues at the C terminus (44). Binding of similar peptides to the PDZ domains of other HtrA proteases has been shown to be responsible for substrate-induced activation of these enzymes (12). Contrasting with such behavior of the purified protease, our work shows that, in the context of supernatants from growing bacterial cultures, pneumococcal HtrA cannot be further activated by the addition of an exogenous substrate protein. In this setting, protein components of the complex growth medium and potentially also from the organism itself may maintain HtrA in a continual state of activation. Under such circumstances, the predominant effect of the accumulation of additional substrates for HtrA is likely to be that of competition as discussed below.
In addition to its activity against CSP, pneumococcal HtrA displayed the ability to digest a range of other proteins. Based on sites of digestion within β-casein and within HtrA itself, such proteolysis appeared to occur preferentially on the C-terminal side of non-polar residues. Combined with the finding that denaturation is required to convert BSA into an effective competitive inhibitor of HtrA, this pattern of digestion suggested an overall selectivity for exposed hydrophobic regions such as result from improper protein folding. This activity against unfolded proteins resembles what has been characterized for DegP but, when combined with activity against CSP, generates the potential for cell signaling that is responsive to protein folding stress. The ability of HtrA to digest denatured proteins is also consistent with previous reports that pneumococcal htrA mutants have a reduced ability to tolerate thermal or oxidative stresses (45) and show a greater reduction in growth rate with subinhibitory concentrations of the antibiotic streptomycin, which causes ribosomal errors (8).
The predominant cut site for HtrA within CSP-1 was on the C-terminal side of residue Phe-8 within the hydrophobic patch of the central α-helix of the peptide. The conservation of hydrophobic residues at this site among CSP variants (30, 46) is likely to account for the ability of HtrA to recognize both CSP-1 and CSP-2. From the perspective of the protease, this region of CSP therefore seems likely to resemble an exposed hydrophobic segment of a misfolded protein. This similarity places the degradation of CSP in direct competition with the degradation of misfolded proteins. Competition between CSP and other substrates for HtrA proteolysis would then have the effect of making the pneumococcal competence pathway sensitive to the level of misfolded proteins on the exterior of the cell. These observations therefore raise the possibility that CSP functions, rather than as a dedicated quorum-sensing pheromone for measuring bacterial density, as part of a mechanism for the bacterium to monitor the folding status of its extracytoplasmic proteome.
Misfolded proteins that interact with HtrA may be derived from a combination of secondary denaturation of mature proteins as a result of environmental stress and primary biosynthetic errors that result in production of proteins with intrinsic folding defects. Consistent with the ability of HtrA to sense proteins in the environment, a pneumococcal strain with wild-type htrA showed repression of competence when the concentration of BSA in the growth medium was reduced, and this repression could be relieved by either inactivating htrA or increasing the amount of BSA. Our in vitro data predict that denatured proteins should be more effective than well folded proteins in promoting competence by acting as efficient alternative substrates for HtrA and competitively reducing the digestion of CSP. Attempts to demonstrate this effect in the setting of pneumococcal cultures using exogenous denatured proteins, however, have not been successful and are likely to be complicated by the intrinsic tendency of denatured proteins to aggregate and to stick to other proteins and surfaces. For competence assays, denatured proteins would need to survive as soluble molecules in the presence of abundant proteins in the medium and on the bacterial cells themselves over several h of culture growth. Although neither heating nor DTT treatment enhanced the ability of BSA to promote competence, these findings may reflect limited stability of denatured proteins under culture conditions.
Because HtrA is at least approximately co-localized with the Sec apparatus for protein translocation (47), it appears possible that HtrA may preferentially monitor the folding status of recently secreted proteins. Consistent with this model for stimulation of competence by primary biosynthetic errors, we recently showed that competence can be induced by increasing the frequency of translational errors during protein synthesis and that repression of competence by HtrA is relieved when such errors are common (8). Development of competence in the setting of such folding stress would seem to be an adaptive response because the competence regulon encodes cellular proteases and chaperones, such as ClpL, GrpE, DnaJ, DnaK, GroEL, and GroES, as well as HtrA itself (10) that are critical for both refolding and digesting misfolded proteins. Beyond the immediate response to folding stress, it is furthermore interesting to speculate that genetic exchange resulting from transformation may function in repairing upstream genetic lesions that contribute to the burden of misfolded proteins.
In summary, we have demonstrated that pneumococcal HtrA digests the bacterial signaling peptide CSP in a manner that is competitively inhibited by the presence of misfolded proteins, which serve as alternative substrates for the protease. This finding contributes to an emerging view of competence in S. pneumoniae as a stress response that is triggered by errors in protein folding and points to a key regulatory role for the HtrA protease in this process.
Acknowledgments
We thank M. Goulian for providing strain JW0157 and the P1vir phage for deletion of degP and J. Bergelson and S. Dawid for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI075194 (to M. E. S.).
- SEC
- size exclusion chromatography
- CSP
- competence-stimulating peptide
- rHtrA
- recombinant HtrA.
REFERENCES
- 1. Lau G. W., Haataja S., Lonetto M., Kensit S. E., Marra A., Bryant A. P., McDevitt D., Morrison D. A., Holden D. W. (2001) A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40, 555–571 [DOI] [PubMed] [Google Scholar]
- 2. Bartilson M., Marra A., Christine J., Asundi J. S., Schneider W. P., Hromockyj A. E. (2001) Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 39, 126–135 [DOI] [PubMed] [Google Scholar]
- 3. Hava D. L., Camilli A. (2002) Large scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 4. Oggioni M. R., Trappetti C., Kadioglu A., Cassone M., Iannelli F., Ricci S., Andrew P. W., Pozzi G. (2006) Switch from planktonic to sessile life. A major event in pneumococcal pathogenesis. Mol. Microbiol. 61, 1196–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sibold C., Henrichsen J., König A., Martin C., Chalkley L., Hakenbeck R. (1994) Mosaic pbpX genes of major clones of penicillin-resistant Streptococcus pneumoniae have evolved from pbpX genes of a penicillin-sensitive Streptococcus oralis. Mol. Microbiol. 12, 1013–1023 [DOI] [PubMed] [Google Scholar]
- 6. Croucher N. J., Harris S. R., Fraser C., Quail M. A., Burton J., van der Linden M., McGee L., von Gottberg A., Song J. H., Ko K. S., Pichon B., Baker S., Parry C. M., Lambertsen L. M., Shahinas D., Pillai D. R., Mitchell T. J., Dougan G., Tomasz A., Klugman K. P., Parkhill J., Hanage W. P., Bentley S. D. (2011) Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnsborg O., Håvarstein L. S. (2009) Regulation of natural genetic tranfsormation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol. Rev. 33, 627–642 [DOI] [PubMed] [Google Scholar]
- 8. Stevens K. E., Chang D., Zwack E. E., Sebert M. E. (2011) Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. mBio 2, e00071–00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Claverys J. P., Prudhomme M., Martin B. (2006) Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60, 451–475 [DOI] [PubMed] [Google Scholar]
- 10. Peterson S. N., Sung C. K., Cline R., Desai B. V., Snesrud E. C., Luo P., Walling J., Li H., Mintz M., Tsegaye G., Burr P. C., Do Y., Ahn S., Gilbert J., Fleischmann R. D., Morrison D. A. (2004) Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51, 1051–1070 [DOI] [PubMed] [Google Scholar]
- 11. Clausen T., Kaiser M., Huber R., Ehrmann M. (2011) HTRA proteases. Regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 12, 152–162 [DOI] [PubMed] [Google Scholar]
- 12. Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T. (2003) OMP peptide signals initiate the envelope stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113, 61–71 [DOI] [PubMed] [Google Scholar]
- 13. Kolmar H., Waller P. R., Sauer R. T. (1996) The DegP and DegQ periplasmic endoproteases of Escherichia coli. Specificity for cleavage sites and substrate conformation. J. Bacteriol. 178, 5925–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wrase R., Scott H., Hilgenfeld R., Hansen G. (2011) The Legionella HtrA homologue DegQ is a self-compartmentizing protease that forms large 12-meric assemblies. Proc. Natl. Acad. Sci. U.S.A. 108, 10490–10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huesgen P. F., Miranda H., Lam X., Perthold M., Schuhmann H., Adamska I., Funk C. (2011) Recombinant Deg/HtrA proteases from Synechocystis sp. PCC 6803 differ in substrate specificity, biochemical characteristics and mechanism. Biochem. J. 435, 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoy B., Löwer M., Weydig C., Carra G., Tegtmeyer N., Geppert T., Schröder P., Sewald N., Backert S., Schneider G., Wessler S. (2010) Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 11, 798–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boehm M., Hoy B., Rohde M., Tegtmeyer N., Bæk K. T., Oyarzabal O. A., Brøndsted L., Wessler S., Backert S. (2012) Rapid paracellular transmigration of Campylobacter jejuni across polarized epithelial cells without affecting TER. Role of proteolytic-active HtrA cleaving E-cadherin but not fibronectin. Gut Pathog. 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang J., Zhang X., Chen Y., Wu Y., Zhou Z. H., Chang Z., Sui S. F. (2008) Activation of DegP chaperone-protease via formation of large cage-like oligomers upon binding to substrate proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 11939–11944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S., Grant R. A., Sauer R. T. (2011) Covalent linkage of distinct substrate degrons controls assembly and disassembly of DegP proteolytic cages. Cell 145, 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huston W. M., Tyndall J. D., Lott W. B., Stansfield S. H., Timms P. (2011) Unique residues involved in activation of the multitasking protease/chaperone HtrA from Chlamydia trachomatis. PLoS One 6, e24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krojer T., Sawa J., Schäfer E., Saibil H. R., Ehrmann M., Clausen T. (2008) Structural basis for the regulated protease and chaperone function of DegP. Nature 453, 885–890 [DOI] [PubMed] [Google Scholar]
- 22. Merdanovic M., Mamant N., Meltzer M., Poepsel S., Auckenthaler A., Melgaard R., Hauske P., Nagel-Steger L., Clarke A. R., Kaiser M., Huber R., Ehrmann M. (2010) Determinants of structural and functional plasticity of a widely conserved protease chaperone complex. Nat. Struct. Mol. Biol. 17, 837–843 [DOI] [PubMed] [Google Scholar]
- 23. Kim S., Sauer R. T. (2012) Cage assembly of DegP protease is not required for substrate-dependent regulation of proteolytic activity or high temperature cell survival. Proc. Natl. Acad. Sci. U.S.A. 109, 7263–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shen Q. T., Bai X. C., Chang L. F., Wu Y., Wang H. W., Sui S. F. (2009) Bowl-shaped oligomeric structures on membranes as DegP's new functional forms in protein quality control. Proc. Natl. Acad. Sci. U.S.A. 106, 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baud C., Gutsche I., Willery E., de Paepe D., Drobecq H., Gilleron M., Locht C., Jamin M., Jacob-Dubuisson F. (2011) Membrane-associated DegP in Bordetella chaperones a repeat-rich secretory protein. Mol. Microbiol. 80, 1625–1636 [DOI] [PubMed] [Google Scholar]
- 26. Meltzer M., Hasenbein S., Hauske P., Kucz N., Merdanovic M., Grau S., Beil A., Jones D., Krojer T., Clausen T., Ehrmann M., Kaiser M. (2008) Allosteric activation of HtrA protease DegP by stress signals during bacterial protein quality control. Angew. Chem. Int. Ed. Engl. 47, 1332–1334 [DOI] [PubMed] [Google Scholar]
- 27. Malet H., Canellas F., Sawa J., Yan J., Thalassinos K., Ehrmann M., Clausen T., Saibil H. R. (2012) Newly folded substrates inside the molecular cage of the HtrA chaperone DegQ. Nat. Struct. Mol. Biol. 19, 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spiess C., Beil A., Ehrmann M. (1999) A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97, 339–347 [DOI] [PubMed] [Google Scholar]
- 29. Sebert M. E., Patel K. P., Plotnick M., Weiser J. N. (2005) Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J. Bacteriol. 187, 3969–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnsborg O., Kristiansen P. E., Blomqvist T., Håvarstein L. S. (2006) A hydrophobic patch in the competence-stimulating peptide, a pneumococcal competence pheromone, is essential for specificity and biological activity. J. Bacteriol. 188, 1744–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chastanet A., Prudhomme M., Claverys J. P., Msadek T. (2001) Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183, 7295–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants. The Keio collection. Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Porter R. D., Guild W. R. (1976) Characterization of some pneumococcal bacteriophages. J. Virol. 19, 659–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lau P. C., Sung C. K., Lee J. H., Morrison D. A., Cvitkovitch D. G. (2002) PCR ligation mutagenesis in transformable streptococci. Application and efficiency. J. Microbiol. Methods 49, 193–205 [DOI] [PubMed] [Google Scholar]
- 35. Sung C. K., Li H., Claverys J. P., Morrison D. A. (2001) An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67, 5190–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prudhomme M., Attaiech L., Sanchez G., Martin B., Claverys J. P. (2006) Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313, 89–92 [DOI] [PubMed] [Google Scholar]
- 37. Kowalko J. E., Sebert M. E. (2008) The Streptococcus pneumoniae competence regulatory system influences respiratory tract colonization. Infect. Immun. 76, 3131–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Skórko-Glonek J., Żurawa D., Tanfani F., Scirè A., Wawrzynów A., Narkiewicz J., Bertoli E., Lipińska B. (2003) The N-terminal region of HtrA heat shock protease from Escherichia coli is essential for stabilization of HtrA primary structure and maintaining of its oligomeric structure. Biochim. Biophys. Acta 1649, 171–182 [DOI] [PubMed] [Google Scholar]
- 39. Perona J. J., Craik C. S. (1995) Structural basis of substrate specificity in the serine proteases. Protein Sci. 4, 337–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Livney Y. D., Schwan A. L., Dalgleish D. G. (2004) A study of β-casein tertiary structure by intramolecular cross-linking and mass spectrometry. J. Dairy Sci. 87, 3638–3847 [DOI] [PubMed] [Google Scholar]
- 41. Sawa J., Malet H., Krojer T., Canellas F., Ehrmann M., Clausen T. (2011) Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope. J. Biol. Chem. 286, 30680–30690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Håvarstein L. S., Coomaraswamy G., Morrison D. A. (1995) An unmodified heptadecapeptide phermone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92, 11140–11144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sebert M. E., Palmer L. M., Rosenberg M., Weiser J. N. (2002) Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70, 4059–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fan K., Zhang J., Zhang X., Tu X. (2011) Solution structure of HtrA PDZ domain from Streptococcus pneumoniae and its interaction with YYF-COOH containing peptides. J. Struct. Biol. 176, 16–23 [DOI] [PubMed] [Google Scholar]
- 45. Ibrahim Y. M., Kerr A. R., McCluskey J., Mitchell T. J. (2004) Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72, 3584–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Whatmore A. M., Barcus V. A., Dowson C. G. (1999) Genetic diversity of the streptococcal competence (com) gene locus. J. Bacteriol. 181, 3144–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsui H. C., Keen S. K., Sham L. T., Wayne K. J., Winkler M. E. (2011) Dynamic distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. mBio 2, e00202–00211 [DOI] [PMC free article] [PubMed] [Google Scholar]