Background: The mechanism whereby CCT is auto-inhibited by its membrane-induced amphipathic helix (m-AH) is unknown.
Results: m-AH regions sharing an amphipathic 22-mer element can be interchanged between CCTs with retention of catalytic silencing and activation by lipids.
Conclusion: The 22-mer element is the principal auto-inhibitory motif.
Significance: Multi-tasking and conformationally malleable motifs are widely used to regulate protein function; the CCT m-AH is a novel example of this.
Keywords: α-Synuclein, Enzyme Inactivation, Intrinsically Disordered Proteins, Phosphatidylcholine, Protein Chimeras, Amphipathic α Helix, Auto-inhibition, Lipid Vesicle
Abstract
CTP:phosphocholine cytidylyltransferase (CCT), an amphitropic enzyme that regulates phosphatidylcholine synthesis, is composed of a catalytic head domain and a regulatory tail. The tail region has dual functions as a regulator of membrane binding/enzyme activation and as an inhibitor of catalysis in the unbound form of the enzyme, suggesting conformational plasticity. These functions are well conserved in CCTs across diverse phyla, although the sequences of the tail regions are not. CCT regulatory tails of diverse origins are composed of a long membrane lipid-inducible amphipathic helix (m-AH) followed by a highly disordered segment, reminiscent of the Parkinson disease-linked protein, α-synuclein, which we show shares a novel sequence motif with vertebrate CCTs. To unravel features required for silencing, we created chimeric enzymes by fusing the catalytic domain of rat CCTα to the regulatory tail of CCTs from Drosophila, Caenorhabditis elegans, or Saccharomyces cerevisiae or to α-synuclein. Only the tail domains of the two invertebrate CCTs were competent for both suppression of catalytic activity and for activation by lipid vesicles. Thus, both silencing and activating functions of the m-AH can tolerate significant changes in length and sequence. We identified a highly amphipathic 22-residue segment in the m-AH with features conserved among animal CCTs but not yeast CCT or α-synuclein. Deletion of this segment from rat CCT increased the lipid-independent Vmax by 10-fold, equivalent to the effect of deleting the entire tail, and severely weakened membrane binding affinity. However, membrane binding was required for additional increases in catalytic efficiency. Thus, full activation of CCT may require not only loss of a silencing conformation in the m-AH but a gain of an activating conformation, promoted by membrane binding.
Introduction
Membrane-binding amphipathic helices (m-AH)6 occur in proteins with a wide spectrum of functions, from membrane traffic control in eukaryotes (1, 2) to cell division control in bacteria (3). m-AH motifs are well adapted for a function in reversible membrane binding as they have intrinsic conformational plasticity and typically weak membrane affinity (4). In addition to this role, m-AH motifs are involved in curvature induction and sensing (5, 6). A less well recognized function is serving as a regulator of catalysis. Two diverse examples of catalytic silencing by an m-AH motif are the Escherichia coli pyruvate oxidase and eukaryotic CTP:phosphocholine cytidylyltransferase (CCT). Pyruvate oxidase binding to membranes, stimulated by increases in pyruvate concentration, facilitates the transfer of electrons between flavins and ubiquinone, the latter being located in the membrane (7, 8). CCT binding to PC-deficient membranes constitutes a way to sense membrane lipid composition and accelerate the supply of CDP-choline for the synthesis of PC (9, 10). For both enzymes, the membrane-bound forms are associated with catalytic activation, where the m-AH helical conformation is stabilized by membrane binding. In the catalytically silenced soluble forms, the m-AH is mostly nonhelical (7, 8, 11). For pyruvate oxidase, high resolution crystal structures revealed that the m-AH C-terminal peptide forms an ordered lid over the active site, restricting access of pyruvate and blocking electron transfer between thiamine diphosphate and FAD (7). For CCT, the mechanism of silencing is unknown. Although there is a high resolution structure of the catalytic dimer (12), a structure of a CCT containing the m-AH segment has not been solved. In fact, it is not known whether the regulatory AH segment makes contact with the catalytic domain as part of the silencing mechanism. Because silencing is a feature of CCT m-AH segments across phyla that have divergent M sequences, it is possible that the silencing mechanism does not involve an ordered contact between the two domains.
In the absence of lipids, the full-length rat CCTα is a very slow enzyme with a kcat of 0.2–0.3 s−1 (13, 14) and, according to one report, a Km value for its substrate CTP (∼ 25 mm) that is 100 times higher than the average cellular concentration of CTP (15). Binding to lipids increases the kcat value to ∼15 s−1 and lowers the CTP Km value to ≤1 mm (13, 14). This would suggest that flux through the CDP-choline pathway is highly dependent on membrane translocation of CCT, confirmed by many studies in cells showing a correlation between CCT membrane localization and PC synthesis rates (10). The CCT m-AH, responsible for maintaining its activity status, resides in a domain referred to as domain M, for membrane binding. We use the terms domain M and m-AH interchangeably. Although its boundaries are not certain, it includes at least 69 residues in rat CCTα, from Phe-234 to Met-302 (16). Domain M is followed by an intrinsically disordered, proline-rich segment (region P) that houses up to 16 phosphoserine sites in rat CCTα (17–19). In isolation, complete CCT tails (domain M + region P) from rat and Caenorhabditis elegans are predominantly unstructured, based on CD analysis, with a low α-helical content corresponding to <10 residues, but acquire helical content upon membrane binding (16). Synthetic domain M peptides from rat (11, 20–22) or the protist, Plasmodium falciparum (23), show a similar disorder to helix transition induced by membrane binding. Binding of the full-length rat CCTα to membranes was accompanied by an increase in helical content corresponding to ∼60 residues at the expense of unordered β-strand and turn conformations (11).
The evidence for domain M function in silencing is that truncation prior to domain M, but not after, results in a large increase in constitutive activity, defined as enzyme activity in the absence of lipid. This is seen for mammalian CCTα (12, 13, 24) and a CCT from C. elegans (25). On the other hand, Yang et al. (15) found that rat CCTα truncated at the boundary between the catalytic and M domain or truncated one-third of the way into domain M resulted in a very slow enzyme, leading them to suggest that rather than acting as a silencer of catalysis, domain M in its membrane-bound state functions as a positive regulator of activity. Although these models are not mutually exclusive, the conflicting results from very similar truncation variants need resolution.
Although several reports have tried to define the subregions and features of domain M required for membrane binding and lipid activation (15, 18, 21, 22, 24–28), the features important for the silencing function have not been systematically explored. Both functions of the M domain operate in CCTs even though the sequence diversity of this region is high between phyla. Vertebrate CCTs are characterized by a set of tandem 11-mer motifs, whose similarity with the 11-mer motif in the m-AH segment of the Parkinson disease protein, α-synuclein, has been noted (16, 29). In α-synuclein, the role of the 11-mer has been described as a means to enhance membrane interactions by maintaining the amphipathic register of a very long segment as a 3–11 α-helix (29). However, the 11-mer motif is not an obvious feature of invertebrate or yeast CCTs that are lipid-regulated, which raises the following question. What, if any, is the function of this motif in CCTs?
In this study, we have focused on features of the M domain critical for silencing. We initially asked three questions: Is the predicted structure (or lack of it) in this region conserved? Is there strong binding between the catalytic and M domains in the lipid-free form? Can the M domain of a CCT lacking a repeated 11-mer motif substitute for the native M domain in mammalian CCT with respect to silencing or activation? These explorations led to the identification of a conserved segment within domain M that is a key contributor to its silencing and activation functions.
EXPERIMENTAL PROCEDURES
Bioinformatics
Disorder Predictions for CCTs
Rat CCTα was analyzed with all 17 servers available at the rosetta design group server. We selected programs within this set for evaluating CCTs from other species based on two criteria: (i) accuracy in correlating order/disorder with elements from the solved structure of the catalytic domain of rat CCTα (Protein Data Bank code 3HL4); and (ii) low or negligible weighting of structural data from protein data banks obtained from sequence blasts. We found that DISOclust, DISOPRED, DISpro, and POODLE-I performed this task the best. The analysis of the physicochemical properties of the m-AH segments of M domains utilized the HeliQuest server (30).
Sequence Novelty of CCTs and Synuclein 11-mer Motifs
10 CCTα and -β (40 11-mers) and 11 α-, β-, and γ-synucleins (61 11-mers) from four classes of vertebrates were used to generate a hidden Markov sequence profile (31) using version 2.3.2 of the HMMER software. The input sequences and the output for the profile sequence are shown in the supplemental Table S1. This profile was used to search an ENSEMBL base (version 48) of vertebrate and invertebrate eukaryotic proteomes listed in the supplemental Table S1. The constraints imposed on the search were ≥2 11-mers with at least 1 11-mer matching the hidden Markov sequence profile. The retrieved sequences with e-values ≥2.0 were converted to FASTA format and aligned with ClustalW (32). To assemble a phylogenetic tree, we used PHYLIP (33) and TreeView programs (34). Caenorhabditis briggsae Dur-1 was arbitrarily assigned as the progenitor.
Molecular Biology and Biochemical Methods
Preparation of CCT Constructs
The preparation of full-length His-tagged rat CCTα was described in Xie et al. (35). The preparation of the CCT head domain (CCT-236) encompassing residues 1–236 was described by Taneva et al. (11). Chimeric CCTs contained His tags linked to residues 1–236 of rat CCTα, which was fused to the tail domains of Drosophila melanogaster (residues 239–381), C. elegans (residues 226–347), Saccharomyces cerevisiae (residues 263–424), or the complete sequence (140 residues) of human α-synuclein. Chimeras were created by engineering complementary primers for PCR at the 3′ end of the rat CCT head domain and the 5′ end of the non-rat CCT tail domains, both containing a SapI type IIs restriction site (36). After amplification, digestion of the PCR products with SapI and ligation generated in-frame fusion of head and tail domains. Rat CCTα Δ272–293 (CCT-Δ22) was created by QuikChange mutagenesis using WT His6-rat CCTα as template and PCR primers complementary to the sequence flanking the 66 nucleotides to be excised. All constructs were checked for accuracy by sequencing. Constructs were expressed in COS cells, using 48 h of transfection time and purified by nickel-agarose affinity chromatography as described (14). His tags were not cleaved for our activity analyses; we and others (37) have shown that the tag does not influence enzyme activity, with or without lipids. The rat CCTα tail domain constructs were prepared as described (16). The unphosphorylated version has WT sequence from residues 237 to 367. The phosphomimic version is the same segment but with all 16 serines between residues 314 and 367 substituted with glutamate.
Pulldown Assay for Analysis of CCT Head-Tail Interactions in Trans
Purified rat CCTα head domain (His-CCT236) was pre-spun for 15 min at 13,000 rpm to remove insoluble particulates. His-CCT236 (0.3 nmol) was mixed with 0.3 nmol of rat CCTα tail segments (M + P or M + P − PM) in 40 μl of 30 mm phosphate, pH 8.15, 0.15 m NaCl, 40 mm imidazole, 2 mm DTT, where M is membrane-binding; P is phosphorylation region; PM is phosphomimic. The protein samples also contributed 15 μm octyl glucoside and 40 μm Triton X-100. This mix was added to pre-washed nickel-agarose beads and incubated for 20 min at 20 °C, end-over-end. After centrifugation at 2000 × g for 1 min, the supernatant was transferred to another tube. The beads were washed with ⅓ volume of mixing buffer and centrifuged as above. Proportional fractions of the combined supernatants and pellet were examined by SDS-PAGE. The Coomassie-stained protein bands were quantified using ImageQuant.
CCT Activity Assay
CCT activity was performed as described (14). The standard assays contained 0–25 pmol of enzyme, 10 mm CTP, 1.5 mm [14C]phosphocholine, and up to 7.5 μm Triton X-100, contributed by the CCT chimeric enzymes. Incubations were for 15 min in the presence of variable concentrations of lipids as follows: sonicated vesicles composed of egg PC/oleic acid (1:1) or egg PC/egg PG (1:1 or 4:1) (38, 39) or 100-nm extruded vesicles composed of DOPC/DOPE (2:3) (39). For the analysis of the [CTP] dependence, the concentration was varied from 0 to 30 mm CTP, and because for some CCT variants the dependence was nonhyperbolic, the CTP Km value is reported as S½.
CCT Membrane Partitioning in Cells
COS-1 cells were transiently transfected with pAX plasmids containing WT rat CCTα, CCT-236, or CCTΔ22. Expression of these CCTs was ∼50–100-fold above endogenous levels, so that the contribution from endogenous CCT was negligible. Membrane partitioning was measured as described (40) by fractionating the cells into lysate, 100,000 × g, supernatant, and 100,000 × g pellet. The latter was subfractionated into Triton X-100 soluble (membrane) and Triton-insoluble fractions (40). Minor modifications included harvesting the cells at 36 h post-transfection and lysing in 1.2 ml of hypotonic buffer (40). The total units of CCT in each subcellular fraction was assayed as described above in the presence of 1 mm sonicated egg PG vesicles, which fully activates the CCT. Triton X-100, which was used to solubilize the membrane fraction, was included in all samples in the enzyme assay at a concentration of 0.8 mm. An assay of the units of enzyme activity in each cell fraction using fully activating conditions is an accurate and quantitatively rigorous method for assessing CCT distribution between fractions (40).
Rates of PC Synthesis
COS cells on 6-cm dishes were transfected with 2 μg/dish of the pAX-CCT plasmids or with empty plasmid for 36 h. The growth medium was removed, and the cells were washed twice with PBS and incubated at 37 °C with 2 ml of DMEM containing 5 μCi/dish [3H]choline. At the end of this 30-min pulse, the medium was removed, and cells were washed twice with PBS, and fresh unlabeled DMEM containing 1 mg/ml BSA was added. The [3H]choline uptake was quenched with methanol/water (5:4); the cells were harvested at intervals thereafter, and [3H]PC was measured as described (41). To assess the relative expression levels of the various transfected CCTs for this experiment, parallel dishes with the same cell seeding density and transfection treatment were harvested, and lysates were prepared for Western blot. We loaded equivalent volumes with equivalent total lysate protein onto 10% SDS-PAGE, and CCT was detected by reaction with an antibody against the catalytic domain, at residues 164–176 (35, 40). Signal was captured using chemiluminescence and a Fujicam 1 LAS-4000 Image analyzer and quantified by ImageQuant.
RESULTS
Functional and Structural Features of Diverse CCT Tails
Table 1 provides a summary of the lipid regulation of CCTs from unicellular to mammalian sources. As an enzyme responsible for maintaining PC homeostasis, CCT is activated by membranes enriched in phospholipids other than PC. There are two major classes of lipids that have activating function, anionic lipids and type II lipids. Anionic lipids promote electrostatic attraction of the m-AH region of CCT, followed by insertion of the hydrophobic face of the nascent α-helix. Type II lipids destabilize bilayers due to their molecular shape and create negative curvature strain when incorporated into bilayers. CCT relieves this strain when its m-AH inserts into the outer leaflet of the vesicle (42, 43). All CCTs studied show a strong activating response to anionic lipid vesicles; for example, the activity of purified rat CCTα can be stimulated up to 100-fold by anionic phospholipid vesicles. The response to type II lipids is variable among species and to pure PC vesicles it is weak or negligible.
TABLE 1.
Comparison of the lipid regulation of diverse CCTs
ND means not determined.
| Species | Maximum activation | Response to anionic lipids | Response to type II lipids | Response to PC | Ref. |
|---|---|---|---|---|---|
| Mammalian | 50–100-Fold | Strong | Medium | Weak | 38, 43, 53 |
| Drosophila | ∼30-Fold | Strong | Medium | Weak | 37 |
| C. elegans | ∼25-Fold | Strong | Strong | Very weak | 25, 28 |
| Yeast | >30-Fold | Strong | Very weak (∼PC) | Very weak | 55, 56 |
| Castor bean | 100-Fold | Strong | ND | None | 57 |
| P. falciparum | ∼10-Fold | Medium | Very weak | None | 58 |
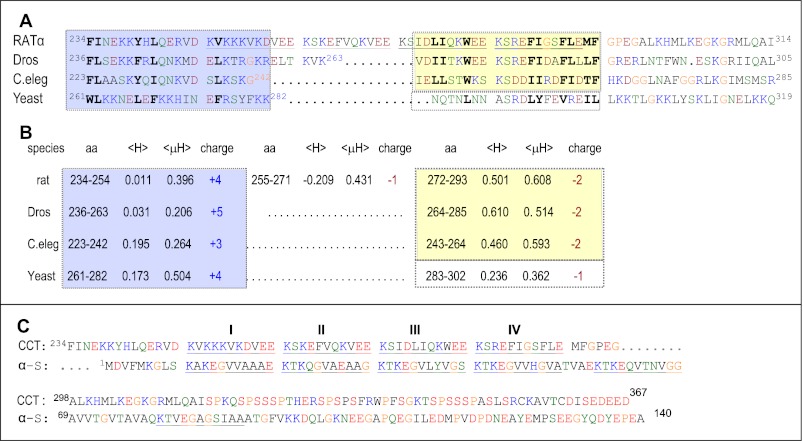
Fig. 1A aligns CCTs from S. cerevisiae, C. elegans, Drosophila, and rat. Direct evidence for the M region as the membrane-binding domain has been presented for CCTs from rat (44, 45), C. elegans (16, 25, 28), and from the plasmodium P. falciparum (23). Despite similar regulation by lipids the sequence of the M region is not well conserved. CCTs from the phylum chordata, exemplified by rat, have four degenerate tandem 11-mer repeats (underlined in Fig. 1). CCTs from insects, nematodes, and yeasts have interruptions of various lengths in this region, and no tandem 11-mers are obvious. CCTs from protists, such as P. falciparum, and from flowering plants, such as Arabidopsis thaliana, also lack the 11-mer repeat motif (data not shown). Almost all CCT sequences have preserved a 5-residue motif near the C terminus of domain M corresponding to 289FLEMF293 in the rat CCT (see Fig. 1A). Beyond this segment there is only weak sequence match across kingdoms, but there is a common pattern of spacing of hydrophobic and polar residues. The low sequence conservation across all eukaryotes raises the question as to whether the diverse CCTs share a common mechanism for membrane binding and for silencing catalysis, and whether the physicochemical features of the m-AH are at the root of the mechanism for both processes.
FIGURE 1.
A, sequence alignment of the M region of CCT. The boundaries of M were chosen based on empirical evidence for insertion of Phe-234 of rat CCTα into lipid vesicles (37) and phosphorylation of rat CCTα beginning at residues Ser-315 (17, 19). Rat, α isoform, P19836; Dros (D. melanogaster) CCT-2, B4QMG1; C. elegans, Q3HKC4; yeast (S. cerevisiae), P13259. The alignment was done manually. There is discrepancy in translation products for the C. elegans CCT in the databanks due to three potential start sites. We use the start site associated with the clone characterized by Friesen et al. (25). The blue-shaded box highlights the positively charged N-terminal segment, and the yellow-shaded box highlights the regulatory 22-mer segment containing 10–11 conserved hydrophobic amino acids. The four 11-mer repeats in rat CCTα are underlined. Previous literature refers to three 11-mer repeats in rat CCTs, based on a frame starting with 256VEEKS … (15, 21, 22, 27, 59). Amino acids are color-coded as follows: red, acidic and phosphoserine; blue, basic; green, hydrophilic; black, hydrophobic; orange, glycine, proline. B, physicochemical features of subsections of domain M. The sequences were analyzed with HeliQuest (30), where the amino acid (aa) is the sequence range for the analysis; 〈H〉 is the mean hydrophobicity, and 〈μH〉 is the mean helical hydrophobic moment calculated using the Eisenberg formula (60), based on the hydrophobicity scale of Fauchere and Pliska (61). The output of the analysis as 3–11 helices is shown; analysis as canonical α-helices yielded only slight variation in 〈μH〉 values. C, sequence alignment of rat CCTα (P19836) with human α-synuclein (α-S; P37840.1). The 11-mer repeats are underlined, and the color coding is as for A.
Fig. 1B compares the physicochemical properties of segments within the M domain. In animal CCTs, the N terminus of domain M has a high positive charge density, low hydrophobicity (〈H〉), and moderate amphipathy (〈μH〉). The C terminus has net negative charge, a higher density of hydrophobic residues, and a very high amphipathy. We shall later address the critical role of this 22-mer segment highlighted in Fig. 1B with a yellow box (see below). Interestingly, the yeast M domain does not fit this pattern but has a rather low hydrophobicity distributed throughout and amphipathic character concentrated at the N terminus.
We evaluated secondary structure predictions for the four CCT tail sequences shown in Fig. 1 and found a degree of variability between species and between predictive programs. For example, PsiPred predicts extensive helical configuration for all four CCTs in the M region, but PROFsec predicts very little helical content for C. elegans CCT and a lack of secondary structure for residues 256–272 for rat CCT (data not shown). Although CD analyses show adoption of helical structure for the CCT tails of rat and C. elegans when membrane-bound (14), the structure of the M domain in the absence of lipids is less clear. A subtractive CD approach suggested that the M domain of full-length rat CCTα is mainly unstructured in the absence of lipids (11).
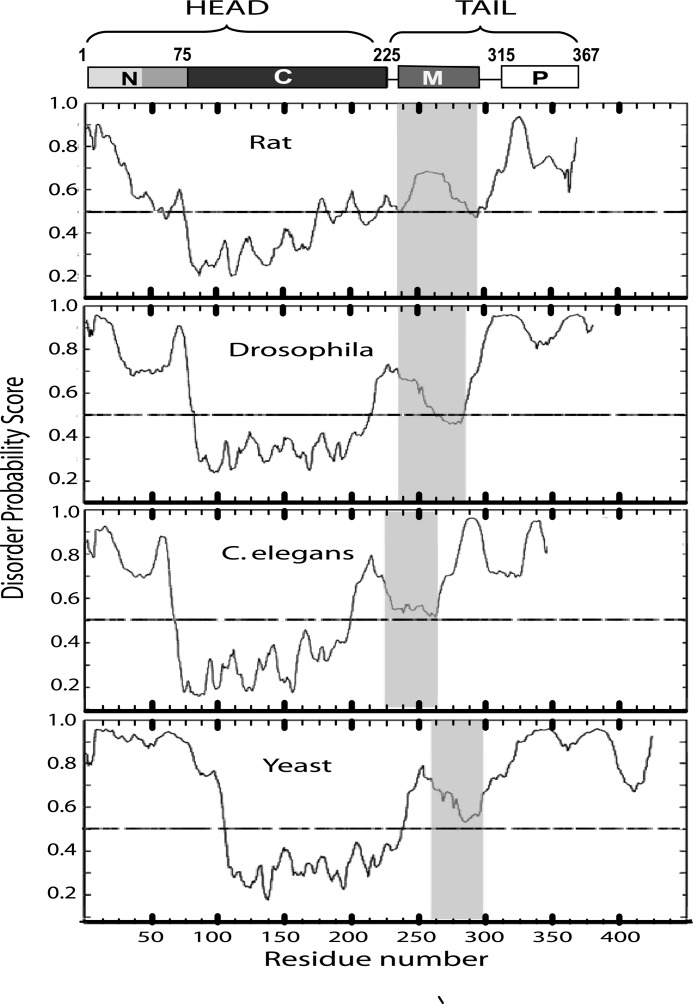
We then compared the four CCT sequences using a set of order/disorder predictive programs available on line. Fig. 2 shows the output from DISOclust, a program that relies heavily on sequences associated with high thermal factors and missing electron density in the Protein Data Bank of solved structures (46). The M domains of all four CCTs are relatively disordered and show a biphasic pattern as follows: the N-terminal portion scores high for disorder and the C terminus nears the threshold for an ordered structure. This pattern was also observed using DISOPRED, DISprot, POODLE, RONN, PONDR, Met Predict, and several others. Some of these programs incorporate machine training with known ordered and disordered sequences for their predictions. In summary, these analyses suggest that in the absence of lipids the M domains in the regulatory tails of CCTs are likely to experience conformational plasticity with a predisposition toward helix formation.
FIGURE 2.
Disorder prediction of complete CCT protein sequences using DISOClust (46). The diagram at the top shows the domain structure of a CCT monomer with rat CCTα numbering. All CCTs have disordered N and C termini, highly ordered catalytic (C) domains, and biphasic membrane-binding (M) domains. The gray-shaded box in each panel corresponds to the M domain. The sequence identifiers are provided in the legend to Fig. 1. N, N-terminal region; P, phosphorylation region.
Novel Sequence Match between α-Synuclein and CCT
The regulatory tail domain of CCT resembles α-synuclein in that it contains a long lipid-inducible amphipathic helix followed by a disordered acidic segment. An alignment of the rat CCTα tail and human α-synuclein (Fig. 1B) highlights the similarities. α-Synuclein has seven 11-mer repeats, the first five of which have a conserved KTKEGVφXφφX motif, where φ denotes a hydrophobic residue. Vertebrate CCTα and β forms have four 11-mer repeats with a consensus resembling synuclein, KSKEXVφXφEE. We probed whether the sequence similarity between the CCTs and synucleins is unique to these proteins by generating a profile sequence and searching a large sequence database for matches. 100 CCT and synuclein 11-mer sequences from four classes of vertebrates were used to generate a Hidden Markov sequence profile (see supplemental material). Given that in both proteins this repeated motif makes up a continuous α-helix when membrane-bound (47–49), we searched a set of 56 eukaryotic proteomes spanning S. cerevisiae to Homo sapiens (see supplemental material) for matches to ≥2 contiguous 11-mer repeats with at least one match to our profile sequence. 134 sequence matches were retrieved, and of these, only one authentic protein sequence was not derived from CCT or synuclein (Dur-1, a dehydrin; see supplemental Fig. S1). If this motif were common in these proteomes, the search would have identified many other proteins. Thus, this repeated 11-mer sequence motif is quite unique to CCT and synucleins. When the retrieved sequences were assembled into a phylogenetic tree, the CCTs and synucleins segregated into separate branches (supplemental Fig. 1). This suggested a separate evolutionary history for the two proteins.
To further evaluate this conclusion, we prepared separate sequence profiles of CCT and synuclein as queries for probing the same database and with the same criteria for a match. The profile built from synuclein sequences identified 69 matches to α-, β-, or γ-synucleins, and several dehydrin/LEA proteins from the nematode C. briggsae. No CCT sequences were identified from this search. The profile built from CCT sequences identified 64 matches to CCTs from invertebrates and vertebrates. No synuclein sequences were identified from this search. These data add further support for the hypothesis that the CCTs and synucleins evolved separately.
Catalytic Domain of Rat CCTα Does Not Interact with Its Tail in Trans
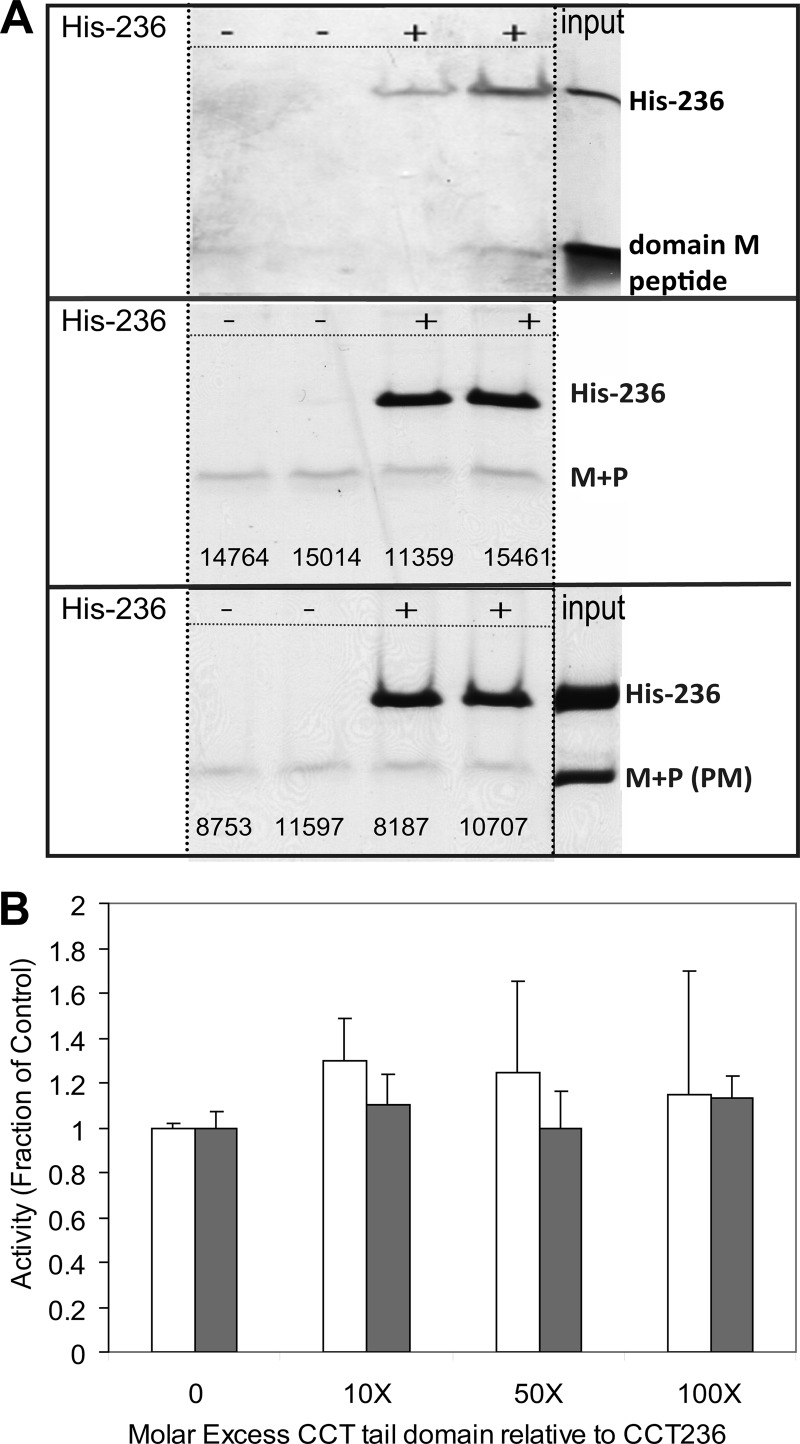
To probe for specific features involved in silencing, we attempted to establish a simple assay for probing the interaction between the catalytic and regulatory domains. We tested whether the head and tail domains could form a stable complex in trans. We immobilized the head domain via a His tag to a nickel resin and probed for capture of the untagged tail domain. Fig. 3A shows that neither a fragment corresponding to the m-AH segment nor the full tail domain bound to the resin in a manner specifically requiring the CCT head domain. This was true for both an unphosphorylated version of the tail and a phosphomimic (PM) version. Because a successful interaction of this type is predicated on a strong long lived interaction, we then probed for softer interactions by assessing whether the tail domain could inhibit the activity of the constitutively active head domain. Fig. 3B shows that even a 100 molar excess of tail segment relative to head domain did not diminish the activity of the head, as assayed in the absence of lipids. These results are compatible with a model of regulation involving only a weak interaction between the tail and head domain that requires connection through the polypeptide backbone for silencing function.
FIGURE 3.
Probing interactions between catalytic head and regulatory tail in trans. A, rat CCTα catalytic domain does not capture its tail domain in trans. We measured the capture of a domain M peptide (top panel), M + P tail fragment (middle), or M + P phosphomimic (PM) fragment (bottom) onto nickel-agarose beads without (−) or with (+) His-tagged catalytic fragment (His-236). Samples were subjected to SDS-PAGE using 12% Tricine gels. The pellet fractions containing the nickel beads are shown. The numbers represent the density of the tail bands determined by ImageQuant. B, rat CCTα catalytic domain is not inhibited by its tail domain in trans. Catalytic fragment (CCT-236) was mixed with the indicated molar excess of tail fragment for 5 min prior to activity analysis. M + P, open bar; M + P(PM), filled bar. Data are the average of two independent determinations and are normalized to the activity obtained without tail fragment.
M Domains of Invertebrate CCTs Can Substitute for the Native M Domain in a Mammalian CCT
Given the lack of head-tail complex formation in trans, we turned to analysis of chimeric proteins to explore head-tail interactions. We asked specifically if the tail regions of CCTs from Drosophila, C. elegans, or yeast could substitute for the tail of the rat CCT in its silencing function and in mediating membrane binding and activation of the rat catalytic domain. Because these CCT tails lack the 11-mer repeat featured in vertebrate CCT tails, we could probe their regulatory role. The catalytic domains of CCTs from Drosophila, C. elegans, or S. cerevisiae share 73, 64, or 63% percent identity (91, 89, or 85.5% similarity) with the catalytic domain of rat CCTα (residues 75–233). The chimeras were fusions of the first 236 residues of rat CCTα with the tail domains of these representative CCTs beginning at the residue corresponding to residue 237 of the rat CCT. CCT-236 is an independently folded catalytic domain, whose structure has been solved (12). We have shown that the tail region, starting from residue 237 (rat) or 226 (C. elegans) to the C terminus, can fold independently of the catalytic domain when presented to lipid vesicles (16). If the foreign tail can substitute, then we would observe very low activity in the absence of lipid and full activity (∼50-fold stimulation) in the presence of saturating lipid. If the tail cannot substitute, then we expected to observe constitutive (lipid-independent) activity, indicating that the foreign tail domain is missing key elements for silencing.
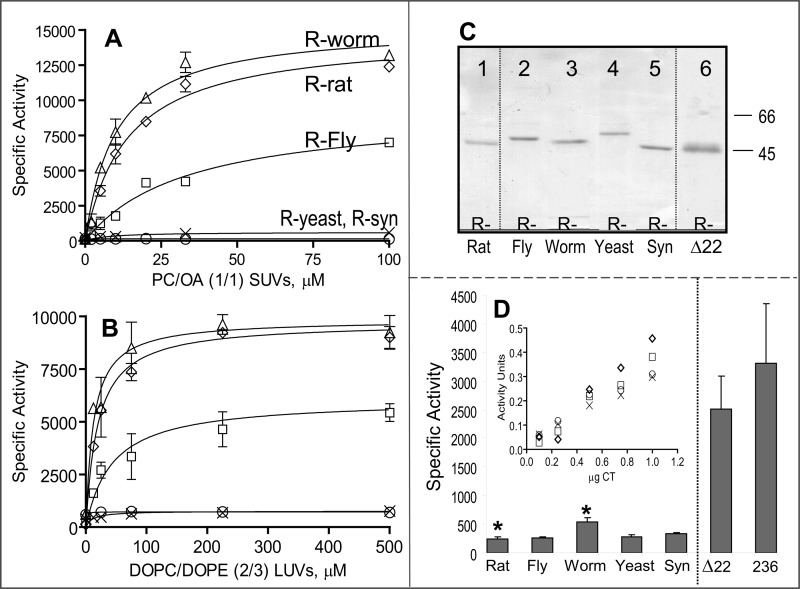
The chimeric CCTs were expressed and purified from the soluble fraction of transfected COS cells (Fig. 4C). We measured the activity of the hybrid CCTs as a function of lipid concentration using two lipid vesicle systems that have been widely used to activate CCTs from diverse sources (13, 15, 37, 38, 43). PC/oleic acid is an example of an anionic lipid vesicle, and dioleoyl phosphatidylcholine/dioleoyl phosphatidylethanolamine (DOPC/DOPE) is an example of a vesicle with type II lipids. Despite the deletion of 9 or 18 residues from the AH region of the tail, the rat-fly or rat-worm chimeras showed activation profiles that were very similar to that of unmodified rat CCTα (Fig. 4, A and B). Although the profiles were virtually identical for the rat CCT control and the rat-worm hybrid, the maximal specific activity of the rat-fly hybrid was reduced by a factor of 2, and its response to lipid was weaker by ∼2-fold compared with the rat control and rat-worm chimera. In the absence of lipid, the three animal CCTs had low but detectable activities (Fig. 4D). The specific activity of the rat-fly hybrid was not significantly different from the rat CCT control, but the rat-worm hybrid was ∼2-fold higher. These data suggest that the rat CCT catalytic domain can be regulated very well by shorter M domains that lack the four 11-mers.
FIGURE 4.
Lipid-dependent and -independent activity of CCT chimeras. CCT was activated by egg PC/oleic acid (1:1)-sonicated vesicles (A) or DOPC/DOPE (2:3) 100 nm extruded vesicles (B). R-worm refers to the rat catalytic domain fused to the regulatory tail of the CCT from C. elegans, etc. Data are means ± S.E. of four independent determinations; in many cases, the error bar is within the symbol. Symbols in B are defined in A. C, purity of CCT chimeras. ∼2 μg of protein purified by nickel-agarose chromatography was separated on 10% SDS-PAGE and stained with Coomassie Brilliant Blue-R. Δ22 refers to rat CCTΔ272–293. D, activity of all CCT constructs in the absence of lipid. Data are means ± average deviations of 4–7 independent determinations. *, p = 0.037. Inset, micrograms of pure CCT in the assay are plotted versus activity units (nanomoles of CDP-choline formed/min) for the yeast, synuclein, and Drosophila chimeras as well as the rat control CCT. Symbols are as in A. SUV, small unilamellar vesicle; LUV, large unilamellar vesicle.
The chimeric enzyme incorporating a tail region from yeast had low but detectable activity in the absence of lipids (Fig. 4D and inset), but in contrast to the rat-fly or rat-worm chimeras, its activity was increased only ∼2-fold by either lipid system and required an ∼5-fold higher lipid concentration for this weak effect (see supplemental Fig. S2). The low activity for the rat-yeast construct even with saturating lipid concentrations suggested that the yeast tail compromises the rat CCT catalytic domain in some way. We also tested α-synuclein as a surrogate regulatory region for rat CCT. The CCT-synuclein chimera displayed very low but detectable activity in the absence of lipid, but unlike the other constructs, the activity was not affected at all by lipids (Fig. 4, A and B), including PC/PG (1:1) small unilamellar vesicles (data not shown). A sedimentation analysis revealed that the lack of activation by lipids was not because of an inability of this construct to bind to the anionic lipid vesicles (supplemental Fig. S3). These data suggest that the catalytic function of rat CCT is not compatible with simply any amphipathic motif that can convert to an m-AH upon membrane binding.
Conserved 22-Residue Segment in Domain M Contributes to Both Silencing and Membrane Binding/Activation of Metazoan CCTs
The chimeric enzyme analysis suggested that M domains from other metazoans, but not from yeast, can be interchanged. Metazoan CCTs contain a 22-mer segment highlighted in Fig. 1 (yellow box) that, although having only 27% identity, has a common pattern of spacing of hydrophobic and polar residues. This sequence element is not apparent in the CCT sequence from yeast (Fig. 1) or other fungi and is more cryptic in CCTs from plants and protists. The supplemental Fig. S4 provides a more extensive multiple sequence alignment and shows that this segment in both animal and plant CCTs features both high hydrophobicity and amphipathy. The 22-mer has a register and a pattern that is distinct from the tandem 11-mer repeats that are shared by synucleins and mammalian CCT M domains.
We deleted this segment from the rat CCTα (CCT-Δ22) and assessed the effect on the lipid-independent and lipid-dependent activity, in comparison with wild-type (WT) rat CCTα and a truncation mutant (CCT-236) missing the entire regulatory tail. Although there is consensus that activation of WT CCT by lipid vesicles involves a large increase in kcat values and no effect on the Km values for phosphocholine, there are discrepant reports for effects on the Km values for CTP (13, 15). We found, in fair agreement with Yang et al. (15), that WT CCT activation involves a 10-fold decrease in the CTP Km value (Table 2). Our assays of CCTα-236 found its activity to be relatively lipid-insensitive, but with a Vmax value less than half that of the WT enzyme and a CTP S½ value intermediate between that of silenced and activated WT CCT (Table 2), in agreement with previous analyses of tail-less CCTs (12, 13). Thus, deletion of the CCT tail generates an enzyme with compromised catalytic function. For the CCT-Δ22 variant, the lipid-independent activity increased by a factor of ∼10-fold (Fig. 4D; Table 2), to a level that approximates that of CCT-236, missing the entire regulatory tail. CCT-Δ22 also had a very high S½ for CTP of 9 mm in the absence of lipid. These results suggested that the 22-residue segment is a key element involved in catalytic silencing of rat CCT, but the results also revealed that neither deletion of the 22-mer segment nor the entire regulatory tail was sufficient to fully activate CCT. Thus, domain M in its membrane-bound state must somehow exert a positive effect on catalysis.
TABLE 2.
Kinetic constants for rat CCTs
CCT activity was assayed in the presence of 0–30 mm CTP and other standard conditions of the assay. The lipid vesicles were saturating concentrations (0.1–0.5 mm) of egg PC/PG (1:1). For the CCT-Δ22 and 236 variants, the dependence on [CTP] was sigmoidal, and these plots were evaluated in GraphPad Prism using the sigmoidal-variable slope to obtain values for S½. The analyses for WT-CCT utilized a Michaelis equation. Data are the average ± S.E. of at least two independent determinations.
| Lipid | CCT- WT |
CCT-Δ22 |
CCT-236 |
|||
|---|---|---|---|---|---|---|
| − | + | − | + | − | + | |
| Vmaxa | 240 ± 35 | 13,200 ± 990 | 3500 ± 200 | 13,000 ± 400 | 3350 ± 250 | 5550 ± 650 |
| S½·CTPb | 10 ± 3 | 0.9 ± 0.2 | 9.2 ± 1.4 | 2.3 ± 0.2 | 5.6 ± 0.9 | 7.6 ± 0.7 |
a Units are in nanomoles of CDP-choline/min/mg CCT.
b Units are in mm.
We examined the effect of the 22-mer deletion on the response to strong and weakly activating lipid vesicles in comparison with WT rat-CCTα and the CCT-236 variant. CCT-236 activity was increased slightly (≤50%) by each of the lipid vesicle systems (Fig. 5, A–C). The mechanism is unknown, since it lacks an m-AH. Strongly anionic vesicles (egg PC/PG (1:1) stimulated WT CCT activity up to 50-fold, with an EC50 of ∼1 μm (Fig. 5A). CCT-Δ22 was activated ∼4-fold by the strongly anionic vesicles, and its affinity was lower than that of WT, with an EC50 of ∼ 6 μm. Both the Vmax and CTP S½ parameters were affected by lipid vesicles, much like WT-CCT (Table 2). To further probe the consequences of the 22-mer deletion on the CCT membrane affinity, we measured the response to vesicles to which WT CCT binds more weakly, PC/PG (4:1) and PC/PE (2:3) vesicles. Membrane binding in these systems would be much more reliant on hydrophobic rather than electrostatic interactions. The CCT-Δ22 response was dramatically muted toward these vesicles (Fig. 5, B and C). In fact, the weak increase in activity by these vesicles was equivalent to that of CCT-236. Thus, the 22-residue element not only participates in silencing in the absence of lipid but is a major driver of the hydrophobic component of the membrane interaction that leads to enzymatic activation. It has a net negative charge (−2) that would tend to antagonize binding to anionic lipid surfaces, in contrast to the positively charged N-terminal segment, which would be the major driver of the electrostatic binding component (22).
FIGURE 5.
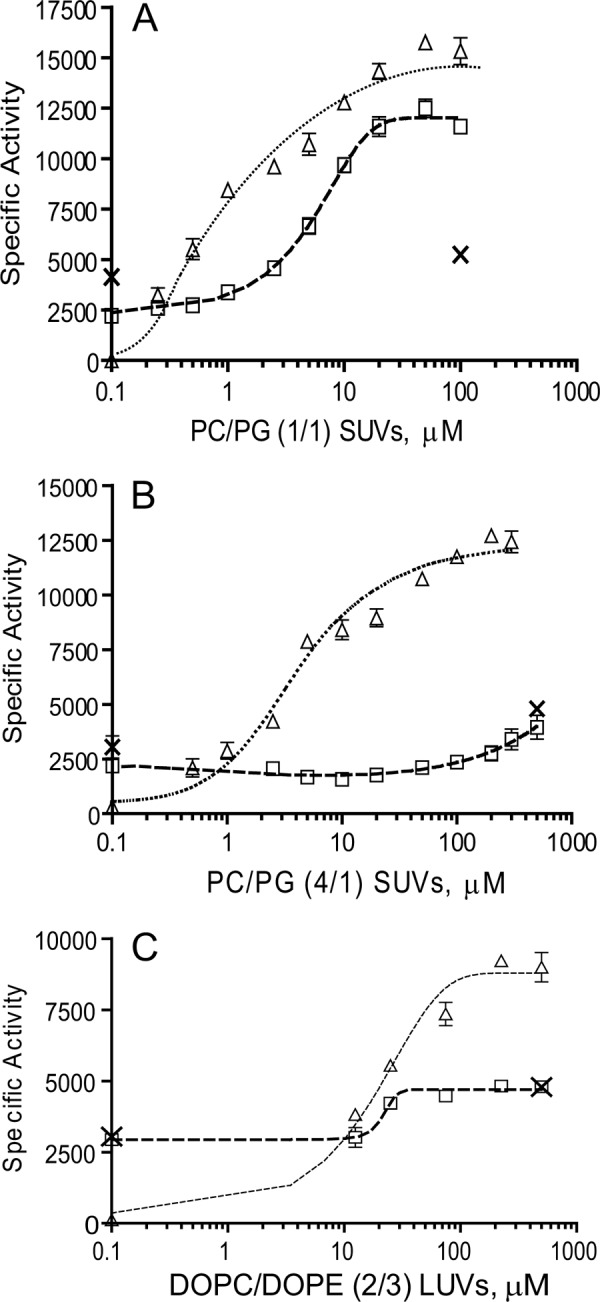
Deletion of 22-mer segment decreases the response to activating lipids. Activities were measured in the presence of the indicated concentrations of egg PC/egg PG (1:1) sonicated vesicles (A), egg PC/egg PG (4:1) sonicated vesicles (B), or DOPC/DOPE (2:3) 100 nm extruded vesicles (C). Data are means ± S.E. of two independent determinations. △, WT rat CCTα; □, rat CCT-Δ22; ×, rat CCTα-236; SUV, small unilamellar vesicle; LUV, large unilamellar vesicle.
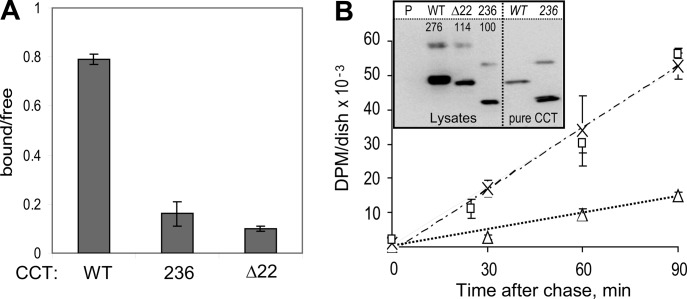
The effects of the 22-mer deletion on silencing and membrane binding were also observed in cells. We expressed WT rat CCTα, CCTΔ22, or CCT-236 in COS cells. We fractionated the cells in parallel. The WT CCT distribution expressed as the ratio of membrane-bound to soluble form was 0.8 (Fig. 6A). This ratio fell to only 0.16 for CCT-236 and 0.10 for CCTΔ22, indicative of a membrane-binding defect for both. These data show that the 22-mer sequence confers cell membrane affinity for CCT. To assess its silencing role in cells, we measured the effects of these three CCT constructs on rates of PC synthesis. Expression of WT CCT resulted in a 3-fold acceleration of PC synthesis, identical to a previous report (50). The PC synthesis rate was increased an additional ∼4-fold in the cells transfected with the constitutively active CCT-236, compared with WT-CCT, in agreement with previous reports showing acceleration of PC synthesis in CCT-236-transfected cells (13, 24, 51). This effect was duplicated by transfection with CCTΔ22. This suggests the 22-mer is the key auto-inhibitory (AI) motif within domain M required for silencing CCT in cells, and in this way it contributes to the control of PC synthesis.
FIGURE 6.
Deletion of the 22-mer reduces CCT cell membrane affinity and accelerates PC synthesis. A, membrane partitioning in cells. COS cells expressing rat CCTα-WT, CCT-236, or CCT-Δ22 were fractionated, and the total activity units in each fraction were determined. The graph shows the ratio of units in the membrane fraction versus soluble fraction. The data are means ± range of two independent experiments. B, PC synthesis was monitored by a pulse-chase protocol for cells transfected with CCT: △, WT rat CCTα; □, rat CCT-Δ22; ×, rat CCTα-236. Data are means ± range of two independent determinations. The disintegration/min values in the lipid fraction from cells transfected with empty vector were determined in parallel, and these values were subtracted as base line. The disintegration/min values were normalized to the CCT expression levels, determined via immunoblot of equivalent cell lysate volumes using an antibody against the catalytic domain (inset). Lysates analyzed were from cells transfected with empty vector (P) or the indicated CCTs. Pure CCT standards are in the right lanes. The numbers on the blot are relative signal volumes. The upper band in each lane represents the CCT dimer, and the lower band represents the CCT monomer. Western analysis was repeated using different volumes of lysate, yielding similar results.
DISCUSSION
In this study, we examined the degree of variation in the design features of M domains from diverse species of CCT that have lipid-dependent activity to discover the limits of variation that are tolerated for effective silencing and activating functions. In the process, a loosely conserved regulatory element emerged, a 22-mer segment that we propose houses the important information for silencing and participates strongly in the activating response to lipids.
Domain M Is Conformationally Pliable
M domains from diverse CCTs are predicted to be disordered, with the greatest degree of disorder corresponding to the polar, net-positive N-terminal region. The similar disorder predictions for all lipid-regulated CCTs suggest this is a common feature that may be important for silencing and to facilitate membrane binding. The dynamic nature of the M domain in its soluble form also fits with our finding that rat CCTα M domain or complete tail domain does not interact tightly with the catalytic domain when supplied in trans. It will be important to determine how the malleability of domain M mediates its regulatory functions. Such a study is underway in which we monitor changes in anisotropy of the M domain of variants with regulatory defects.
Examples of induced folding of regulatory domains by binding to ligands such as DNA, peptides, or hormones are numerous (52). CCT is exceptional in that the ligand influencing the folding landscape is a membrane. Disorder to order transitions upon membrane vesicle binding in vitro have been documented for the M domains of rat (11, 16, 21, 22), C. elegans (16), and P. falciparum (23). In cells, CCT conversion between soluble and membrane-bound forms is triggered by small changes in membrane lipid content that enhance the charge density and/or the hydrophobicity of the bilayer surface (9, 10, 38, 39, 42, 43, 53, 54). We propose that both electrostatics and hydrophobicity govern a competition between the bilayer surface and a site on the catalytic domain for binding segments of domain M, especially the 22-mer AI segment, a process that requires domain M plasticity. The site of the silencing interaction between domain M and the catalytic domain is the subject of a future publication.7
Domain M Function Is Tolerant of Sequence and Length Variation
The Rat CCTα catalytic domain can be effectively regulated by M domains from two invertebrates that are shorter and are missing the mid-section of domain M. Thus, the specific features of the vertebrate M domain (four tandem 11-mer repeats and a length exceeding 60 residues) have not evolved to solve special regulatory needs of the vertebrate catalytic domain. The domain M mid-section appears to be dispensable. The repeating 11-mer motif that is associated with the longer M domains may have evolved as the solution to maintaining the register of the hydrophobic face of a 3–11 helix and thus optimize membrane binding, as has been proposed for α-synuclein (29). The M domain of yeast can effectively perform silencing and activating roles on its cognate catalytic domain, but can it regulate the rat catalytic domain? At first glance, it is tempting to conclude that the yeast and synuclein constructs are competent for silencing. But the failure of these constructs to become active when presented with high concentrations of lipid vesicles could be a reflection of an incompatibility of these tails with the rat catalytic domain. Thus, we are cautious in drawing conclusions on the silencing functions from these constructs because the low enzyme activity might be due to a structural change in the active site. Nevertheless, it remains a possibility for future study that the yeast CCT M domain is competent for silencing and that the poor response to lipids reflects an inability to acquire a critical conformation required for activation, a conformation that may be facilitated by the 22-mer element conserved only in the animal CCTs.
Core 22-mer Segment Is the Key Contributor to Regulation of Activity
Our analyses highlight the regulatory role of a 22-mer segment at the C-terminal region of domain M of the CCTs from animal sources. M domains containing this 22-mer segment were effective silencers of the rat catalytic domain, and when deleted the rat CCT M domain was ineffective in its silencing function and weakened in its membrane affinity. This behavior was observed in cells as well as in vitro, in that CCT-Δ22 phenocopied CCT-236 missing the entire tail region with respect to reduced cell membrane partitioning and enhanced rate of PC synthesis. Although the 22-mer element is required for silencing of rat CCTα, our data do not eliminate a supporting role for the N-terminal segment of domain M in silencing.
Although deletion of a 22-residue segment might have global structural effects, we argue that this is unlikely for deletion of the AI motif. (i) The catalytic and regulatory domains are discrete folding units, as revealed by the fact that they can be expressed independently and function in vitro independently of each other (12, 16). (ii) The deletion results in a gain-of-function, as witnessed by in vitro enzyme assays and by measuring the rates of PC synthesis in cells expressing CCT-Δ22, and thus the catalytic domain is not impaired by this deletion. (iii) Order/disorder predictions show only localized effects due to the deletion, i.e. no effect on the catalytic domain. (iv) The portion of domain M that remains after deletion of the AI segment is intrinsically disordered, as is region P (see Fig. 2). There is no structure in these regions to perturb by deletion of the AI motif, which appears to be the only structured segment in the entire regulatory tail.
The 22-mer segment corresponds to the predicted peak of order in the M region in all three CCTs and has the strongest hydrophobic and amphipathic character. In the CCT-soluble form, it may exist as a preformed structural element on a flexible tether that engages a site in the catalytic domain to mediate silencing. This sequence is not in-frame with the 11-mer motifs that match to α-synuclein. Friesen et al. (25) had previously identified a 21-mer (essentially the same motif shifted by three residues) required for lipid activation in C. elegans. Their analysis relied on a series of truncations that successively eliminated portions of the tail region. We manually aligned the sequences capturing conserved sequence patterns among diverse CCTs; this highlighted the 22-mer, which we have now shown is required for auto-inhibition, at least in the rat CCT. Previous alignments and mutagenesis relied too heavily on the 11-mer motifs repeated in the vertebrate CCT sequences but missing or interrupted in other animal CCTs (26, 27).
Mutagenesis of four conserved hydrophobic residues to polar ones in the M domain of C. elegans CCT had little effect on silencing, but when substituted en bloc lipid-independent activity increased >10-fold (28). When the conserved Phe-260 was substituted with charged residues, there was an ∼30-fold increase in constitutive activity. These data strongly support a role for hydrophobic residues in silencing. The same mutations were reported to lower lipid-dependent activity, suggesting that both silencing and lipid interactions rely on multiple hydrophobic residues in the M domain, with some more important than others (28). We are currently evaluating the single and cooperative contributions of multiple hydrophobic residues in rat CCT to the silencing of its catalytic function.
Membrane binding resulting in catalytic activation is not relegated to just the 22-mer segment. The entire 70-residue stretch forms a helical domain when membrane-bound (11, 16, 47). This is true for the M domain of rat CCT and also of the shorter M domain of C. elegans (16). Helix stabilization and membrane insertion are linked (22). We propose a role for the 22-mer motif in stabilizing the amphipathic α-helical structure of the entire M domain. Without the 22-mer segment, binding is more dependent on the electrostatic interactions in the N-terminal segment and thus is responsive only to highly charged vesicles. On the other hand, it should be stressed that the 22-mer segment is not required for full activation of the enzyme, provided the enzyme is supplied with sufficient lipid to force binding of the N-terminal portion of domain M.
Domain M Has Both Negative and Positive Contributions to Activity Regulation
If the M domain acts purely as a silencer, then deletion would result in full constitutive activity. If the M domain acts strictly as an activator, its deletion should yield a dead enzyme. We obtained something in between, suggesting that domain M works in both ways. A comparison of the behavior of the rat CCTα WT, Δ22 deletion, and CCT-236 illustrates two aspects of the activation process as follows: (i) dissociation of a domain M auto-inhibitory segment from the catalytic domain, and (ii) induction of a domain M conformation that facilitates activation. The first aspect is construed from the effects of deletion of the entire tail or of the core 22-mer. This increases Vmax ∼10-fold compared with the WT enzyme in the absence of lipid activators but only marginally reduces the CTP S½ value. This effect falls short of achieving the full catalytic potential of the enzyme. Full activation requires a second aspect, deduced from the additional effects of membrane binding of any CCT with sufficient lipid-inducible m-AH character. The membrane stabilization of the AH structure results in an overall ∼50-fold increase in Vmax and an ∼10-fold reduction in the S½ for CTP. These quantitative effects on the kinetics of the enzyme cannot be achieved merely by deletion or debilitation of domain M by mutation. Domain M functions as more than just a silencer. The specific acquisition of a helical structure for domain M by membrane insertion may be required (via an indirect route) to form a fully functional active site. Future work will strive to bring to light why this is so.
Acknowledgments
We thank Joseph Lee for preparation of the CCT tail constructs; Dr. Ismael Vergara Correa for assistance with the bioinformatic analysis; Dr. Rama Mallampalli for the gift of the antibody against domain C, and Dr. Jaeyong Lee for helpful discussions.
This work was supported by a grant from the Canadian Institutes for Health Research (to R. B. C.) and a Natural Science and Engineering Research Council Discovery Grant (to N. C.).

This article contains supplemental Figs. S1–S4, Table S1, and additional references.
H. K.-H. Huang, S. G. Taneva, J. Lee, L. Silva, D. Shriemer, and R. B. Cornell, submitted for publication.
- m-AH
- membrane-inducible amphipathic helix
- CCT
- CTP:phosphocholine cytidylyltransferase
- PC
- egg phosphatidylcholine
- PG
- egg phosphatidylglycerol
- DOPE
- 1,2-dioleoyl phosphatidylethanolamine
- OA
- oleic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- AH
- amphipathic helix
- DOPC
- dioleoyl phosphatidylcholine.
REFERENCES
- 1. Behnia R., Munro S. (2005) Organelle identity and the signposts for membrane traffic. Nature 438, 597–604 [DOI] [PubMed] [Google Scholar]
- 2. Frost A. (2011) Membrane trafficking. Decoding vesicle identity with contrasting chemistries. Curr. Biol. 21, R811–813 [DOI] [PubMed] [Google Scholar]
- 3. Shih Y. L., Huang K. F., Lai H. M., Liao J. H., Lee C. S., Chang C. M., Mak H. M., Hsieh C. W., Lin C. C. (2011) The N-terminal amphipathic helix of the topological specificity factor MinE is associated with shaping membrane curvature. PLoS ONE 6, e21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cornell R. B., Taneva S. G. (2006) Amphipathic helices as mediators of the membrane interaction of amphitropic proteins and as modulators of bilayer physical properties. Curr. Protein Pept. Sci. 7, 539–552 [DOI] [PubMed] [Google Scholar]
- 5. Campelo F., McMahon H. T., Kozlov M. M. (2008) The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J. 95, 2325–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drin G., Antonny B. (2010) Amphipathic helices and membrane curvature. FEBS Lett. 584, 1840–1847 [DOI] [PubMed] [Google Scholar]
- 7. Neumann P., Weidner A., Pech A., Stubbs M. T., Tittmann K. (2008) Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 17390–17395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weidner A., Neumann P., Pech A., Stubbs M. T., Tittmann K. (2009) New insights into the membrane-binding and activation mechanism of pyruvate oxidase from Escherichia coli. J. Mol. Catal. B Enzym. 61, 88–92 [Google Scholar]
- 9. Cornell R. B. (1998) How cytidylyltransferase uses an amphipathic helix to sense membrane phospholipid composition. Biochem. Soc. Trans. 26, 539–544 [DOI] [PubMed] [Google Scholar]
- 10. Cornell R. B., Northwood I. C. (2000) Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem. Sci. 25, 441–447 [DOI] [PubMed] [Google Scholar]
- 11. Taneva S., Johnson J. E., Cornell R. B. (2003) Lipid-induced conformational switch in the membrane binding domain of CTP:phosphocholine cytidylyltransferase. A circular dichroism study. Biochemistry 42, 11768–11776 [DOI] [PubMed] [Google Scholar]
- 12. Lee J., Johnson J., Ding Z., Paetzel M., Cornell R. B. (2009) Crystal structure of a mammalian CTP:phosphocholine cytidylyltransferase catalytic domain reveals novel active site residues within a highly conserved nucleotidyltransferase fold. J. Biol. Chem. 284, 33535–33548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friesen J. A., Campbell H. A., Kent C. (1999) Enzymatic and cellular characterization of a catalytic fragment of CTP:phosphocholine cytidylyltransferase α. J. Biol. Chem. 274, 13384–13389 [DOI] [PubMed] [Google Scholar]
- 14. Taneva S., Dennis M. K., Ding Z., Smith J. L., Cornell R. B. (2008) Contribution of each membrane binding domain of the CTP:phosphocholine cytidylyltransferase-α dimer to its activation, membrane binding, and membrane cross-bridging. J. Biol. Chem. 283, 28137–28148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang W., Boggs K. P., Jackowski S. (1995) The association of lipid activators with the amphipathic helical domain of CTP:phosphocholine cytidylyltransferase accelerates catalysis by increasing the affinity of the enzyme for CTP. J. Biol. Chem. 270, 23951–23957 [DOI] [PubMed] [Google Scholar]
- 16. Taneva S. G., Lee J. M., Cornell R. B. (2012) The amphipathic helix of an enzyme that regulates phosphatidylcholine synthesis remodels membranes into highly curved nanotubules. Biochim. Biophys. Acta 1818, 1173–1186 [DOI] [PubMed] [Google Scholar]
- 17. Bogan M. J., Agnes G. R., Pio F., Cornell R. B. (2005) Interdomain and membrane interactions of CTP:phosphocholine cytidylyltransferase revealed via limited proteolysis and mass spectrometry. J. Biol. Chem. 280, 19613–19624 [DOI] [PubMed] [Google Scholar]
- 18. Cornell R. B., Kalmar G. B., Kay R. J., Johnson M. A., Sanghera J. S., Pelech S. L. (1995) Functions of the C-terminal domain of CTP:phosphocholine cytidylyltransferase. Effects of C-terminal deletions on enzyme activity, intracellular localization, and phosphorylation potential. Biochem. J. 310, 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. MacDonald J. I., Kent C. (1994) Identification of phosphorylation sites in rat liver CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 269, 10529–10537 [PubMed] [Google Scholar]
- 20. Johnson J. E., Cornell R. B. (1994) Membrane-binding amphipathic α-helical peptide derived from CTP:phosphocholine cytidylyltransferase. Biochemistry 33, 4327–4335 [DOI] [PubMed] [Google Scholar]
- 21. Johnson J. E., Rao N. M., Hui S. W., Cornell R. B. (1998) Conformation and lipid binding properties of four peptides derived from the membrane-binding domain of CTP:phosphocholine cytidylyltransferase. Biochemistry 37, 9509–9519 [DOI] [PubMed] [Google Scholar]
- 22. Johnson J. E., Xie M., Singh L. M., Edge R., Cornell R. B. (2003) Both acidic and basic amino acids in an amphitropic enzyme, CTP:phosphocholine cytidylyltransferase, dictate its selectivity for anionic membranes. J. Biol. Chem. 278, 514–522 [DOI] [PubMed] [Google Scholar]
- 23. Larvor M. P., Cerdan R., Gumila C., Maurin L., Seta P., Roustan C., Vial H. (2003) Characterization of the lipid-binding domain of the Plasmodium falciparum CTP:phosphocholine cytidylyltransferase through synthetic peptide studies. Biochem. J. 375, 653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y., Kent C. (1995) Identification of an inhibitory domain of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 270, 18948–18952 [DOI] [PubMed] [Google Scholar]
- 25. Friesen J. A., Liu M. F., Kent C. (2001) Cloning and characterization of a lipid-activated CTP:phosphocholine cytidylyltransferase from Caenorhabditis elegans. Identification of a 21-residue segment critical for lipid activation. Biochim. Biophys. Acta 1533, 86–98 [DOI] [PubMed] [Google Scholar]
- 26. Ridsdale R., Tseu I., Wang J., Post M. (2010) Functions of membrane binding domain of CTP:phosphocholine cytidylyltransferase in alveolar type II cells. Am. J. Respir. Cell Mol. Biol. 43, 74–87 [DOI] [PubMed] [Google Scholar]
- 27. Yang J., Wang J., Tseu I., Kuliszewski M., Lee W., Post M. (1997) Identification of an 11-residue portion of CTP-phosphocholine cytidylyltransferase that is required for enzyme-membrane interactions. Biochem. J. 325, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Braker J. D., Hodel K. J., Mullins D. R., Friesen J. A. (2009) Identification of hydrophobic amino acids required for lipid activation of C. elegans CTP:phosphocholine cytidylyltransferase. Arch. Biochem. Biophys. 492, 10–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bussell R., Jr., Eliezer D. (2003) A structural and functional role for 11-mer repeats in α-synuclein and other exchangeable lipid-binding proteins. J. Mol. Biol. 329, 763–778 [DOI] [PubMed] [Google Scholar]
- 30. Gautier R., Douguet D., Antonny B., Drin G. (2008) HELIQUEST. A web server to screen sequences with specific α-helical properties. Bioinformatics 24, 2101–2102 [DOI] [PubMed] [Google Scholar]
- 31. Bateman A., Birney E., Durbin R., Eddy S. R., Finn R. D., Sonnhammer E. L. (1999) Pfam 3.1. 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27, 260–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Higgins D. G., Thompson J. D., Gibson T. J. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266, 383–402 [DOI] [PubMed] [Google Scholar]
- 33. Felsenstein J. (1988) Phylogenies from molecular sequences. Inference and reliability. Annu. Rev. Genet. 22, 521–565 [DOI] [PubMed] [Google Scholar]
- 34. Page R. D. (1996) TreeView. An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 35. Xie M., Smith J. L., Ding Z., Zhang D., Cornell R. B. (2004) Membrane binding modulates the quaternary structure of CTP:phosphocholine cytidylyltransferase. J. Biol. Chem. 279, 28817–28825 [DOI] [PubMed] [Google Scholar]
- 36. Beck R., Burtscher H. (1994) Introduction of arbitrary sequences into genes by use of class IIs restriction enzymes. Nucleic Acids Res. 22, 886–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Helmink B. A., Friesen J. A. (2004) Characterization of a lipid-activated CTP:phosphocholine cytidylyltransferase from Drosophila melanogaster. Biochim. Biophys. Acta 1683, 78–88 [DOI] [PubMed] [Google Scholar]
- 38. Cornell R. B. (1991) Regulation of CTP:phosphocholine cytidylyltransferase by lipids. 2. Surface curvature, acyl chain length, and lipid-phase dependence for activation. Biochemistry 30, 5881–5888 [DOI] [PubMed] [Google Scholar]
- 39. Drobnies A. E., Davies S. M., Kraayenhof R., Epand R. F., Epand R. M., Cornell R. B. (2002) CTP:phosphocholine cytidylyltransferase and protein kinase C recognize different physical features of membranes. Differential responses to an oxidized phosphatidylcholine. Biochim. Biophys. Acta 1564, 82–90 [DOI] [PubMed] [Google Scholar]
- 40. Dennis M. K., Taneva S. G., Cornell R. B. (2011) The intrinsically disordered nuclear localization signal and phosphorylation segments distinguish the membrane affinity of two cytidylyltransferase isoforms. J. Biol. Chem. 286, 12349–12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ng M. N., Kitos T. E., Cornell R. B. (2004) Contribution of lipid second messengers to the regulation of phosphatidylcholine synthesis during cell cycle re-entry. Biochim. Biophys. Acta 1686, 85–99 [DOI] [PubMed] [Google Scholar]
- 42. Attard G. S., Templer R. H., Smith W. S., Hunt A. N., Jackowski S. (2000) Modulation of CTP:phosphocholine cytidylyltransferase by membrane curvature elastic stress. Proc. Natl. Acad. Sci. U.S.A. 97, 9032–9036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davies S. M., Epand R. M., Kraayenhof R., Cornell R. B. (2001) Regulation of CTP:phosphocholine cytidylyltransferase activity by the physical properties of lipid membranes. An important role for stored curvature strain energy. Biochemistry 40, 10522–10531 [DOI] [PubMed] [Google Scholar]
- 44. Craig L., Johnson J. E., Cornell R. B. (1994) Identification of the membrane-binding domain of rat liver CTP:phosphocholine cytidylyltransferase using chymotrypsin proteolysis. J. Biol. Chem. 269, 3311–3317 [PubMed] [Google Scholar]
- 45. Johnson J. E., Aebersold R., Cornell R. B. (1997) An amphipathic α-helix is the principal membrane-embedded region of CTP:phosphocholine cytidylyltransferase. Identification of the 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)diazirine photolabeled domain. Biochim. Biophys. Acta 1324, 273–284 [DOI] [PubMed] [Google Scholar]
- 46. McGuffin L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics 24, 1798–1804 [DOI] [PubMed] [Google Scholar]
- 47. Dunne S. J., Cornell R. B., Johnson J. E., Glover N. R., Tracey A. S. (1996) Structure of the membrane binding domain of CTP:phosphocholine cytidylyltransferase. Biochemistry 35, 11975–11984 [DOI] [PubMed] [Google Scholar]
- 48. Georgieva E. R., Ramlall T. F., Borbat P. P., Freed J. H., Eliezer D. (2010) The lipid-binding domain of wild-type and mutant α-synuclein. Compactness and interconversion between the broken and extended helix forms. J. Biol. Chem. 285, 28261–28274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lokappa S. B., Ulmer T. S. (2011) α-Synuclein populates both elongated and broken helix states on small unilamellar vesicles. J. Biol. Chem. 286, 21450–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walkey C. J., Kalmar G. B., Cornell R. B. (1994) Overexpression of rat liver CTP:phosphocholine cytidylyltransferase accelerates phosphatidylcholine synthesis and degradation. J. Biol. Chem. 269, 5742–5749 [PubMed] [Google Scholar]
- 51. Gehrig K., Morton C. C., Ridgway N. D. (2009) Nuclear export of the rate-limiting enzyme in phosphatidylcholine synthesis is mediated by its membrane binding domain. J. Lipid Res. 50, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wright P. E., Dyson H. J. (2009) Linking folding and binding. Curr. Opin. Struct. Biol. 19, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arnold R. S., Cornell R. B. (1996) Lipid regulation of CTP:phosphocholine cytidylyltransferase. Electrostatic, hydrophobic, and synergistic interactions of anionic phospholipids and diacylglycerol. Biochemistry 35, 9917–9924 [DOI] [PubMed] [Google Scholar]
- 54. Jamil H., Hatch G. M., Vance D. E. (1993) Evidence that binding of CTP:phosphocholine cytidylyltransferase to membranes in rat hepatocytes is modulated by the ratio of bilayer- to nonbilayer-forming lipids. Biochem. J. 291, 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Friesen J. A., Park Y. S., Kent C. (2001) Purification and kinetic characterization of CTP:phosphocholine cytidylyltransferase from Saccharomyces cerevisiae. Protein Expr. Purif. 21, 141–148 [DOI] [PubMed] [Google Scholar]
- 56. Johnson J. E., Kalmar G. B., Sohal P. S., Walkey C. J., Yamashita S., Cornell R. B. (1992) Comparison of the lipid regulation of yeast and rat CTP:phosphocholine cytidylyltransferase expressed in COS cells. Biochem. J. 285, 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang X., Moore T. S. (1990) Phosphatidylcholine biosynthesis in castor bean endosperm. Purification and properties of cytidine 5′-triphosphate:choline-phosphate cytidylyltransferase. Plant Physiol. 93, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yeo H. J., Larvor M. P., Ancelin M. L., Vial H. J. (1997) Plasmodium falciparum CTP:phosphocholine cytidylyltransferase expressed in Escherichia coli. Purification, characterization, and lipid regulation. Biochem. J. 324, 903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kalmar G. B., Kay R. J., Lachance A., Aebersold R., Cornell R. B. (1990) Cloning and expression of rat liver CTP:phosphocholine cytidylyltransferase. An amphipathic protein that controls phosphatidylcholine synthesis. Proc. Natl. Acad. Sci. U.S.A. 87, 6029–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Eisenberg D., Weiss R. M., Terwilliger T. C. (1982. The helical hydrophobic moment. A measure of the amphiphilicity of a helix. Nature 299, 271–374 [DOI] [PubMed] [Google Scholar]
- 61. Fauchere J., Pliska V. (1983) Hydrophobic parameters of amino acid side chains from the partitioning of N-acetylamino acid amides. Eur. J. Med. Chem. 8, 369–375 [Google Scholar]