Abstract
Insulin-like growth factor 2 (IGF2) and the transformation related protein 53 (Trp53) are potent regulators of cell growth and metabolism in development and cancer. In vitro evidence suggests several mechanistic pathway interactions. Here, we tested whether loss of function of p53 leads to IGF2 ligand pathway dependency in vivo. Developmental lethality occurred in p53 homozygote null mice that lacked the paternal expressed allele of imprinted Igf2. Further lethality due to post-natal lung haemorrhage occurred in female progeny with Igf2 paternal null allele only if derived from double heterozygote null fathers, and was associated with a specific gene expression signature. Conditional deletion of Igf2fl/fl attenuated the rapid tumour onset promoted by homozygous deletion of p53fl/fl. Accelerated carcinoma and sarcoma tumour formation in p53+/− females with bi-allelic Igf2 expression was associated with reductions in p53 loss of heterozygosity and apoptosis. Igf2 genetic dependency of the p53 null phenotype during development and tumour formation suggests that targeting the IGF2 pathway may be useful in the prevention and treatment of human tumours with a disrupted Trp53 pathway.
Keywords: embryogenesis, Igf2, p53, tumour predisposition, X-linked
→ See accompanying article http://dx.doi.org/10.1002/emmm.201201509
INTRODUCTION
The tumour suppressor gene p53, expressed throughout development and adult life, is frequently disrupted in human cancers (Olivier et al, 2002). DNA damage, oncogene activation, telomere erosion, nucleotide depletion and hypoxia, can activate p53 cellular responses including cell-cycle arrest, apoptosis, senescence and DNA repair (Menendez et al, 2009; Riley et al, 2008; Vousden & Prives, 2009). Degradation of transformation related protein (Trp53) is mediated primarily by the human double minute 2 (MDM2/4) ubiquitin E3 ligase which is itself a transcriptional target of Trp53 (Marine & Lozano, 2010). Individuals with Li-Fraumeni syndrome (LFS) develop early onset cancers, and in 30–50% of cases, the tumours are attributed to germ-line heterozygote mutation of p53 with associated loss of heterozygosity (LOH) of the wild-type (WT) allele (Malkin, 2011; Srivastava et al, 1990). Modifiers alter the age of tumour onset in LFS. A single nucleotide polymorphism (SNP) of the MDM2 (SNP 309) gene promoter regulates the relative expression level of this negative regulator of p53 (Bond et al, 2004). SNP 309 associated gain of function of MDM2/4 expression renders p53 functionally null, and reduces the age of tumour onset in LFS by 7–15 years (Bond et al, 2004). A modifier in p53 itself (Trp53 codon 72) also reduces the age of tumour onset, but is more modest at 2–3 years (Fang et al, 2010). Screening of LFS patients for early onset tumours significantly impacts overall survival, such that identification of pathway modifiers of lethal tumour phenotypes have implications for both screening and prevention therapies (Villani et al, 2011).
Mice with homozygous disruption of p53 (p53−/−) are viable except for an exencephalic phenotype in females (Armstrong et al, 1995; Clarke et al, 1993; Donehower et al, 1992). Homozygote animals die within 6 months, due mainly to thymic lymphoma. p53 heterozygosity markedly reduces lymphoma, sarcoma and epithelial tumour latency and mirrors the phenotype of LFS (Donehower et al, 1992; Jacks et al, 1994). Increased p53 expression and conditional restoration of p53 function resulted in reduced tumour frequency, senescence and tumour regression (Martins et al, 2006; Ventura et al, 2007; Xue et al, 2007). p53 is frequently mutated at specific ‘hotspot’ bases in tumours. Mutations can lead to gain of function, as p53R172H/+ and p53R270H/+ can increase in carcinomas and osteosarcoma in the former, and carcinoma metastasis in the latter (Lang et al, 2004; Olive et al, 2004). Modelling of Mdm2 SNP alleles (SNP 309G of MDM2) in the mouse has validated effects on tumour latency (Post et al, 2010). Stabilization of mutant p53 is also required in tumours, and is frequently associated with aggressive tumour phenotypes that can be induced by similar modifiers to those that regulate WT p53 (Suh et al, 2011). Thus modelling potential pathway modifiers of p53 loss and gain of function are important intermediary steps in the validation of clinically important pathway targets for tumour treatment.
Evidence in vitro suggests convergence between IGF-PI3K-AKT-mTOR and ARF-MDM2-p53 pathways, mainly through promotion of IGF-PI3K-AKT-mTOR pathway activation following loss of p53 function. Interactions include; phosphorylation of MDM2 by Akt (Feng et al, 2004; Mayo & Donner, 2001), transcription of the ligand IGF2 (Lee et al, 2000; Zhang et al, 1996, 1998) and regulation of the insulin-like growth factor receptor 1 (IGF1R; Ungewitter & Scrable, 2010; Werner et al, 1996). Insulin-like growth factor binding proteins (IGFBP-2, -3 and -7) are negative regulators of IGF bio-availability and are also p53 target genes (Buckbinder et al, 1995; Grimberg et al, 2006; Suzuki et al, 2010). Function of p53 may be mediated by Phlda3, a negative regulator of Akt (Brady et al, 2011), and via activation of PTEN (Stambolic et al, 2001), TSC2, AMP1 (Feng et al, 2007), sestrins 1 and 2 (Budanov & Karin, 2008) or REDD1 (Ellisen et al, 2002).
IGF2 mRNA over-expression increases ligand supply in common human cancers mainly due to loss of imprinting (LOI, bi-allelic expression; Foulstone et al, 2005; Ito et al, 2008). Binding of insulin-like growth factor 2 (IGF2) to IGF1R and isoform A of the insulin receptor (IR-A) transmits downstream signalling via IRS1 and IRS2 to both the PI3K-AKT-mTOR and RAS-RAF-MEK-ERK pathways (Chalhoub & Baker, 2009; Ulanet et al, 2010). Supply of IGF2 is also controlled via its sequestration by the mannose 6-phosphate/insulin-like growth factor receptor 2 (Igf2r; Foulstone et al, 2005; Ghosh et al, 2003; Zaccheo et al, 2006). Igf2 expression is imprinted (maternal allele silenced) in the mouse, leading to paternal allele expression that is also highly regulated during embryonic and post-natal development (Baker et al, 1993). Targeted disruption of the paternal (p) allele of Igf2 (Igf2+m/−p) regulates whole organism growth (60% of the size of WT littermates; Burns & Hassan, 2001; DeChiara et al, 1990, 1991). Maternal (m) disruption of the imprinting control region (ICR) and the H19 gene (H19Δ13kb/+p) subsequently referred to as H19−m/+p, results in bi-allelic Igf2 expression and an overgrowth phenotype of 128% of the size of WT littermates (Leighton et al, 1995). Importantly, the H19−m/+p growth phenotype was rescued by Igf2+m/−p, suggesting that the predominate growth effects were mediated by Igf2, rather than as a consequence of the disruption of either the maternal allele locus, H19 non-coding RNA or the associated miR-675 (Gabory et al, 2009; Veronese et al, 2010; Yoshimizu et al, 2008). Igf2 can be reactivated in mouse tumours by LOI, where it promotes tumour progression (Christofori et al, 1994, 1995). DNA hypomethylation also results in increased Igf2 expression and tumour promotion (Gaudet et al, 2003; Linhart et al, 2007). Here we provide in vivo evidence of significant Igf2 dependency of the p53 null phenotype during development and tumour formation.
RESULTS
Igf2-dependent developmental lethality of p53 null mice
Mice with varying allelic doses of Igf2 and p53 were combined. Igf2+m/−p are deficient in Igf2 expression because of disruption of the expressed paternal allele associated with silencing (imprinting) of the maternal allele. H19−m/+p over-express Igf2 because of disruption of imprinting of the maternal allele ICR leading to bi-allelic expression (p, paternal allele; m, maternal allele; +, intact allele; −, disrupted allele). These lines were first crossed with heterozygote p53+/− mice. The double heterozygote (e.g. Igf2+m/−p, p53+/− or H19−m/+p, p53+/−) progeny were subsequently inter-crossed to generate combined genotypes including double homozygote alleles. As there are known strain-dependent phenotypic effects in p53 null mice (Harvey et al, 1993), we utilized both the 129S2/J (129) and C57BL/6J (B6) genetic backgrounds backcrossed by >20 generations. We were unable to backcross Igf2+m/−p onto a B6 background, and so 129/B6 F2 hybrids were generated (see Supporting Information Table S1).
The initial observation was that litter sizes from the 129 double heterozygote inter-cross (Igf2+m/−p, p53+/− × Igf2+m/−p, p53+/−) appeared significantly smaller, suggesting there was an overall loss of progeny (Fig 1A). This was only observed when breeding from double heterozygote mothers (Igf2+m/−p, p53+/−, Supporting Information Fig S1A and B) and appeared independent of the parental Igf2 dose (Supporting Information Fig S2). The majority of implantation sites in double heterozygote (Igf2+m/−p, p53+/−) mothers appeared abnormal (Supporting Information Fig S1C). Haemorrhage was observed within the maternal decidua at implantation sites in double heterozygote (Igf2+m/−p, p53+/−) mothers, and was less frequently observed in either p53+/− or Igf2+m/−p heterozygote mothers (Supporting Information Fig S1C and D).
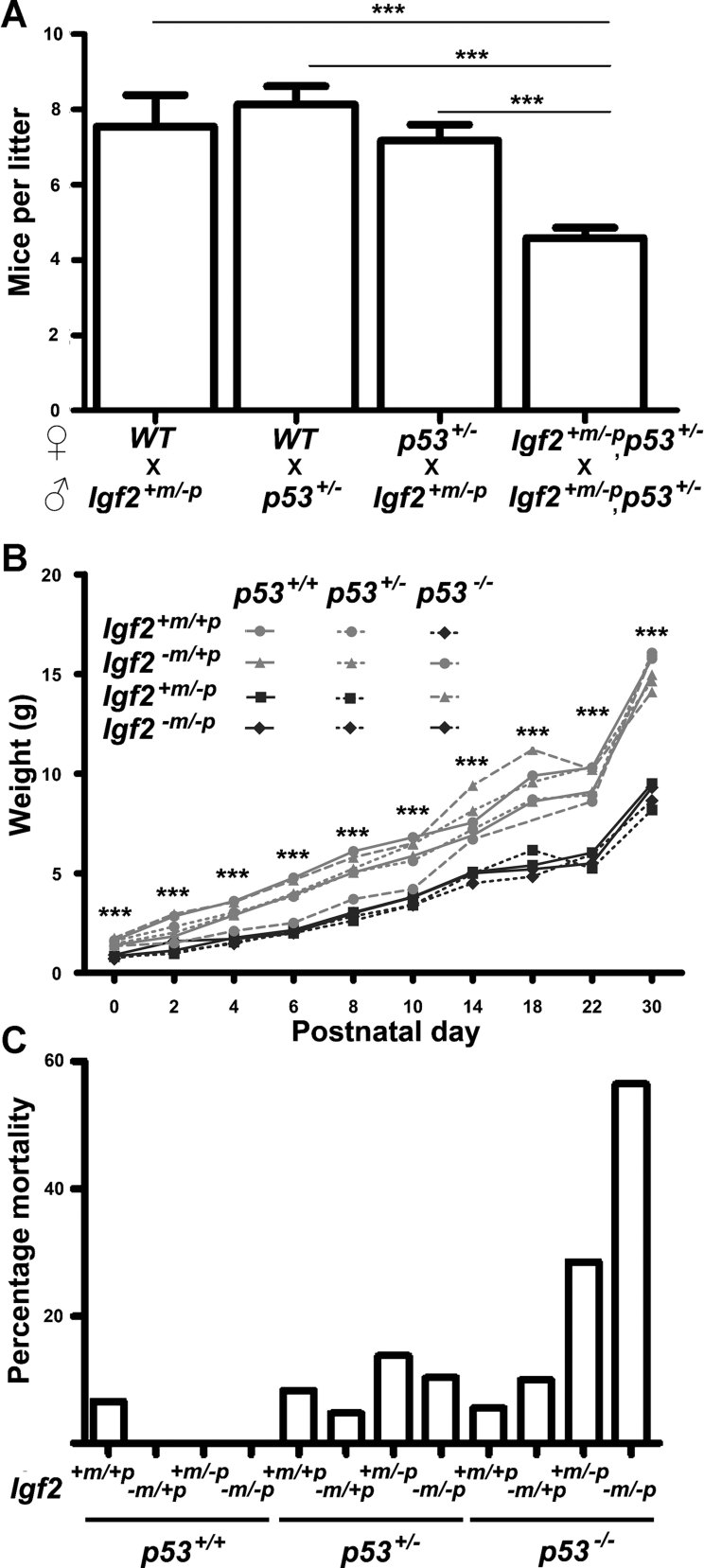
Figure 1. Combination of Igf2 and p53 null alleles results in developmental lethality independent of embryo growth.
- Mean litter size at P0 was significantly reduced from Igf2+m/−p, p53+/− inter-cross (***p < 0.0001, n = 67 litters) relative to Igf2+m/−p mated with WT (n = 11 litters), p53+/− mated with WT (n = 15 litters) and Igf2+m/−p mated with p53+/− (n = 36 litters; one-way ANOVA, Tukey's multiple comparison).
- Post-natal growth of progeny from a double heterozygote inter-cross (129, Igf2+m/−p, p53+/−). Mean weights of WT and Igf2−m/+p mice were significantly greater (***p < 0.0001, one-way ANOVA, Tukey's multiple comparison) than Igf2+m/−p and Igf2−m/−p mice regardless of allelic dosage of p53.
- Increased post-natal mortality by p30 in progeny null for both Igf2 and p53 (Igf2+m/−p, p53−/−, 2 of 7 and Igf2−m/−p, p53−/−, 5 of 9) relative to WT mice (1 of 15, p = 0.037, Fisher's exact test).
At birth, either H19−m/+p, Igf2+m/−p or p53+/− alleles exhibited the expected Mendelian segregation (129, B6 and F1/F2, Supporting Information Tables S2 and S3). The exception, however, were the results from a 129 double heterozygote null inter-cross (e.g. Igf2+m/−p, p53+/− × Igf2+m/−p × p53+/−; Table 1). Evaluation of 297 mice from 34 litters led to three further observations.
Table 1.
Abnormal Mendelian segregation of progeny from a Igf2+m/−p, p53+/− inter-cross in 129J
| Strain | WT | Igf2+/− | Igf2−/− | p53+/− | Igf2+/−, p53+/− | Igf2−/−, p53+/− | p53−/− | Igf2+/−, p53−/− | Igf2−/−, p53−/− | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | ||||||||||
| χ2-Value | ||||||||||
| 129 | 15 | 17 | 7 | 24 | 71 | 29 | 18 | 17 | 9 | 207 |
| (12.94) | (25.88) | (12.94) | (25.88) | (51.75) | (25.88) | (12.94) | (25.88) | (12.94) | p < 0.02 | |
| 0.329 | 3.044 | 2.725 | 0.136 | 7.161 | 0.377 | 1.981 | 3.044 | 1.198 | 19.995 | |
| 129B6F2 | 7 | 22 | 5 | 19 | 33 | 12 | 8 | 13 | 7 | 126 |
| (7.88) | (15.75) | (7.88) | (15.75) | (31.5) | (15.75) | (7.88) | (15.75) | (7.88) | p = NS | |
| 0.097 | 2.480 | 1.049 | 0.671 | 0.071 | 0.893 | 0.002 | 0.480 | 0.097 | 5.841 |
Progeny did not segregate according to a normal Mendelian distribution (p < 0.02, χ2-test). In contrast, progeny from the 129B6F1 Igf2+/−, p53+/− inter-cross had a normal Mendelian distribution. Observed (upper), expected (middle, in brackets) and χ2 (lower).
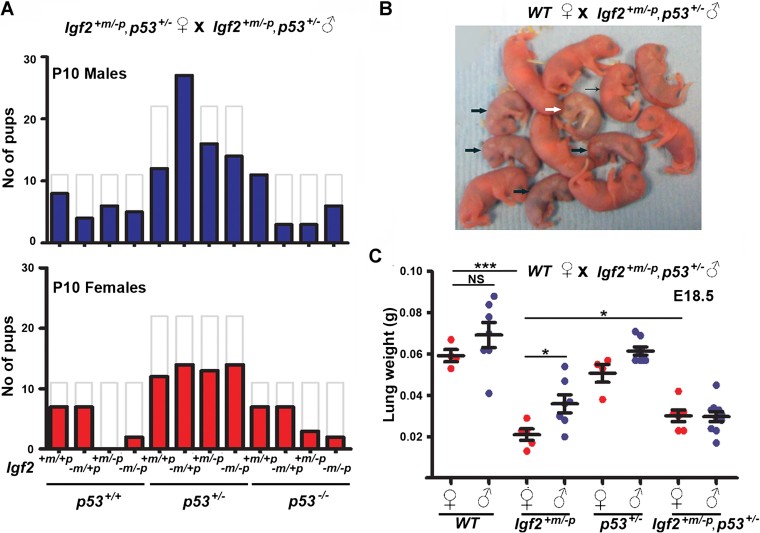
First, genotypes associated with loss of function of the paternal expressed Igf2 allele were 60% smaller from birth (Fig 1B). Second, pre-natal and post-natal lethality occurred in homozygote null p53 combined with disruption of the paternal expressed allele of Igf2 (Igf2+m/−p, p53−/− and Igf2−m/−p, p53−/− mice). The lethality was observed by birth (16 of 44 expected), but also continued during the post-natal period and was associated with no obvious developmental abnormalities (Fig 1C). Third, there was specific absence of Igf2+m/−p female progeny. Post-natal (P3.5) loss of Igf2+m/−p in both sexes was expected based on steroid responsive delayed lung development (Table 1; Silva et al, 2006). However, a deficit specific to females occurred in our case (Fig 2A and Table 2). If double heterozygote (Igf2+m/−p, p53+/−) males were crossed with either WT or p53+/− females (Table 2), we again failed to obtain any live Igf2+m/−p females. Thus, combined loss of alleles for both p53 and Igf2 in the paternal germ-line appeared incompatible with specific survival of subsequent Igf2+m/−p, p53+/+ female progeny. In contrast, double heterozygote (Igf2+m/−p, p53+/−) female littermates survived beyond post-natal day 10, indicating the lethality could be rescued by loss of an allele of p53. Morphological appearances of embryos appeared normal between E9.5 and E18.5, even though Igf2+m/−p embryos were smaller (Supporting Information Fig S3). Following delivery by caesarean section at E19.5, Igf2+m/−p females became increasingly cyanotic (Fig 2B). Analysis of organ growth revealed that the mean wet weights of lungs from Igf2+m/−p females were significantly reduced compared either to WT, Igf2+m/−p males or double heterozygote females (Igf2+m/−p, p53+/−, Fig 2C and Supporting Information Fig S3B and C).
Figure 2. Lethality of female Igf2 +m/−p progeny derived from Igf2 +m/−p, p53 +/− fathers is associated with reduced lung growth.
- Male (n = 115) and female (n = 87) progeny at P10 from the 129 Igf2+m/−p, p53+/− inter-cross did not segregate according to the expected Mendelian distribution (see Table 1; p < 0.05) and were significantly different to expected numbers estimated from previous litter sizes (total expected numbers: males = 175, females = 175 based on mean litter size from earlier 129 breeding, p < 0.0001, χ2-test). Solid bars, observed numbers, grey unfilled bars, expected numbers (see also Table 2).
- Litter of 13 pups delivered by caesarean section at E19.5 from a WT female mated with Igf2+m/−p, p53+/− male. One Igf2+m/−p female (white arrow) became increasingly cyanotic and died 1hr post-delivery. The remaining four Igf2+m/−p females (thick black arrows) were also cyanotic compared to their littermates (thin black arrow points to a representative Igf2+m/−p male).
- Lung weights from E18.5 Igf2+m/−p female progeny derived from Igf2+m/−p, p53+/− fathers were significantly lower than those from Igf2+m/−p male progeny (*p = 0.025), WT female progeny (***p < 0.0001) and Igf2+m/−p, p53+/− female progeny (*p = 0.049, two-tailed t-test, NS = not significant).
Table 2.
Complete perinatal lethality of 129J Igf2+m/−p female progeny was only observed from females mated with 129J Igf2+m/−p, p53+/− males.
| Cross | Time-point | WT | Igf2+m/−p | p53+/− | Igf2+m/−p, p53+/− | p53−/− | Igf2+m/−p, p53−/− | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | ||||||||||||||
| χ2-Value | ||||||||||||||
| ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | |||
| WT ♀ x | P0 | 74 | 82 | 54 | 59 | – | – | – | – | – | – | – | – | 269 |
| Igf2+m/−p ♂ | (67) | (67) | (67) | (67) | p = NS | |||||||||
| 0.67 | 3.23 | 2.61 | 1.01 | 7.54 | ||||||||||
| P3.5 | 66 | 77 | 32 | 35 | – | – | – | – | – | – | – | – | 210 | |
| (53) | (53) | (53) | (53) | p < 0.0001 | ||||||||||
| 0.35 | 11.4 | 8.00 | 5.83 | 28.74 | ||||||||||
| p53+/− ♀ x | P10 | 20 | 18 | 13 | 18 | 16 | 22 | 13 | 17 | – | – | – | – | 137 |
| Igf2+m/−p ♂ | (17) | (17) | (17) | (17) | (17) | (17) | (17) | (17) | p = NS | |||||
| 0.48 | 0.04 | 0.99 | 0.04 | 0.99 | 1.39 | 0.07 | 0.00 | 4.02 | ||||||
| WT ♀ x | E18.5 | 7 | 3 | 7 | 4 | 6 | 1 | 6 | 3 | – | – | – | – | 37 |
| Igf2+m/−p, p53+/− ♂ | (5) | (5) | (5) | (5) | (5) | (5) | (5) | (5) | p = NS | |||||
| 1.22 | 0.57 | 1.22 | 0.08 | 0.48 | 2.84 | 0.48 | 0.57 | 7.32 | ||||||
| P0 | 9 | 9 | 5 | 0 | 8 | 8 | 9 | 2 | 50 p < 0.05 | |||||
| (6.3) | (6.3) | (6.3) | (6.3) | (6.3) | (6.3) | (6.3) | (6.3) | 14.00 | ||||||
| 1.21 | 1.21 | 0.25 | 6.25 | 0.49 | 0.49 | 1.21 | 2.89 | |||||||
| P10 | 6 | 12 | 4 | 0 | 6 | 13 | 6 | 5 | – | – | – | – | 52 | |
| (6.5) | (6.5) | (6.5) | (6.5) | (6.5) | (6.5) | (6.5) | (6.5) | p < 0.01 | ||||||
| 0.04 | 4.65 | 0.96 | 6.5 | 0.04 | 6.5 | 0.04 | 0.37 | 19.08 | ||||||
| p53+/− ♀ x | E9.5 | 6 | 7 | 4 | 4 | 10 | 15 | 9 | 11 | 2 | 3 | 4 | 5 | 80 |
| Igf2+m/−p, p53+/− ♂ | (5) | (5) | (5) | (5) | (10) | (10) | (10) | (10) | (5) | (5) | (5) | (5) | p = NS | |
| 0.20 | 0.80 | 0.20 | 0.20 | 0.00 | 2.50 | 0.10 | 0.10 | 1.80 | 0.80 | 0.20 | 0.00 | 4.40 | ||
| E18.5 | 3 | 2 | 1 | 3 | 5 | 7 | 5 | 4 | 3 | 2 | 2 | 3 | 40 | |
| (2.5) | (2.5) | (2.5) | (2.5) | (5) | (5) | (5) | (5) | (2.5) | (2.5) | (2.5) | (2.5) | p = NS | ||
| 0.10 | 0.10 | 0.90 | 0.10 | 0.00 | 0.80 | 0.00 | 0.20 | 0.10 | 0.10 | 0.10 | 0.10 | 1.40 | ||
| P10 | 10 | 8 | 7 | 0 | 15 | 12 | 10 | 10 | 2 | 3 | 1 | 1 | 75 | |
| (4.7) | (4.7) | (4.7) | (4.7) | (9.4) | (9.4) | (9.4) | (9.4) | (4.7) | (4.7) | (4.7) | (4.7) | p = NS † | ||
| 6.02 | 2.34 | 1.14 | 4.7 | 3.38 | 0.74 | 0.04 | 0.04 | 1.54 | 0.61 | 2.90 | 2.90 | 12.487 | ||
Significant deviation from the normal expected Mendelian ratio of Igf2+m/−p progeny from WT mothers at P3.5 in both females and males was detected, suggesting an effect of the paternal allele on survival. The Igf2+m/−p genotyped progeny derived from Igf2+/−, p53+/− males were absent at P10 compared to E18.5, although the significance was not detected in the segregation of genotypes in progeny derived from p53+/− females (†).
Igf2+m/−p female specific lethality was associated with a gene expression signature and lung haemorrhage
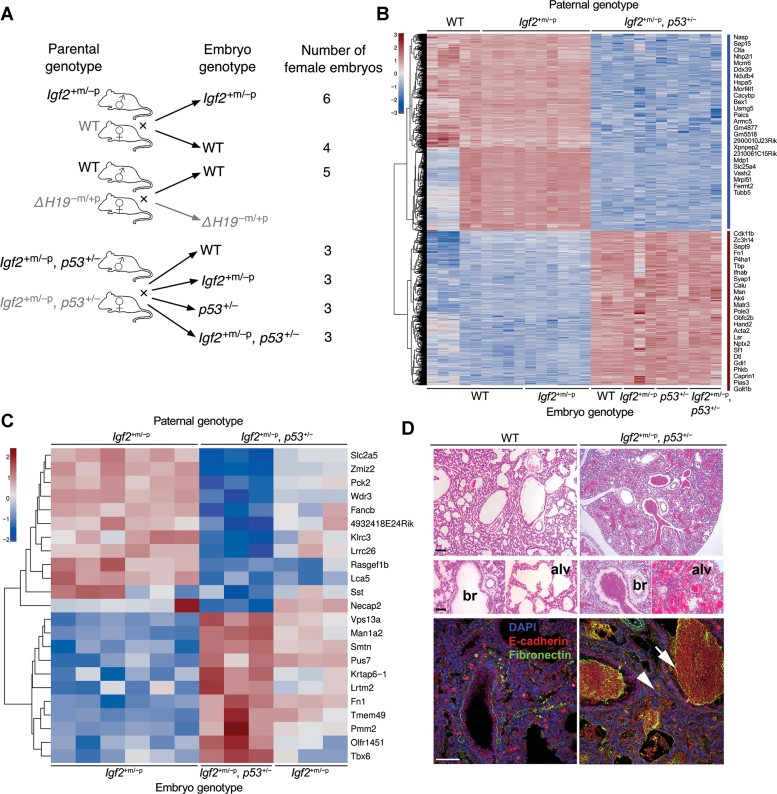
We next investigated effects of paternal genotype on gene expression. Whole female embryos derived from WT, Igf2+m/−p or double heterozygote (Igf2+m/−p, p53+/−) fathers were obtained at E9.5 (Fig 3A). This time was chosen because it is known that WT and Igf2+m/−p embryos are of similar size at E9.5, and we wished to eliminate confounding effects from later differences in growth (Burns & Hassan, 2001). Illumina RNA expression profiles of all female progeny at E9.5 were then compared (Fig 3B). A specific gene expression signature was observed in progeny that were derived from double heterozygote fathers (Igf2+m/−p, p53+/−; Fig 3B). Greater than two-fold differences in gene expression were observed for 1461 genes (Fig 3B lists the most differentially expressed genes) with the top 50 differentially expressed genes further annotated (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-1001; note all microarray data and tables of genes described are available from Arrayexpress). Importantly, no effects were consistent with co-segregation of the gene expression signature with the p53 null allele, suggesting the signature was a direct consequence of the combined paternal genotype rather than a segregating modifier allele. By filtering the initial signature of 1461 genes according to chromosomal location, we obtained 53 X-linked genes. These genes were distributed along the entire length of the X-chromosome and failed to show any differential expression patterns (data not shown).
Figure 3. Paternal p53 genotype alters embryonic gene expression and Igf2+m/−p female specific lethality.
- Schema of embryonic RNA expression analysis. Three parental genotype pairs were crossed to yield a number of female embryos of specific genotype as illustrated.
- Comparison of gene expression in E9.5 female embryos derived from Igf2+m/−p or WT fathers (WT and Igf2+m/−p) with the expression in embryos from Igf2+m/−p, p53+/− fathers (WT, Igf2+m/−p, p53+/− and Igf2+m/−p, p53+/−). 1461 genes were significantly different in expression by SAM analysis and varied more than twofold, with most differentially expressed genes displayed in the heatmap.
- Comparison of gene expression in Igf2+m/−p female embryos derived from WT fathers with those Igf2+m/−p and Igf2+m/−p, p53+/−embryos from Igf2+m/−p, p53+/− fathers. ‘Rescue’ signature of 23 genes shown in the heatmap.
- H & E-stained lungs from E19.5 Igf2+m/−p female embryos (low power above, bar 1 mm, high power 100 µm) were filled with blood in both bronchioles (br) alveolar air-spaces (alv) when compared to WT littermates. Confocal images showed there were no gross differences in lung structure labelled with E-cadherin, but fibronectin was predominantly present in blood within the airspaces (white arrow), and subjectively increased in small airway and alveolar regions (white arrow head). Bar = 100 µm.
We evaluated whether the gene lethality expression profiles represented a hierarchy of transcriptional regulation. PScan (Zambelli et al, 2009) determined transcription factors whose binding sites were statistically over-represented in the 50 most differentially expressed genes, including those located on the X-chromosome. A single X-linked transcription factor, Zfx, was a regulator of the gene sets, although there were no differences in Zfx mRNA levels (not shown; Cellot & Sauvageau, 2007; Luoh et al, 1997). We also identified Igf2-dependent genes in all females, with respect to the paternal genotype. A total of 41 Igf2-dependent genes, including Igf2 itself, were obtained from this further analysis, with some apparent fold change differences dependent on paternal p53+/− status, suggesting that Igf2 regulated genes may also be differentially modified by p53. Using PScan (Zambelli et al, 2009) to locate putative p53 binding sites in the promoters of the 50 most differentially expressed genes between progeny that were derived from double heterozygote fathers (Igf2+m/−p, p53+/−) and controls, we detected 48/50 potential p53 regulated genes. These were located in autosomes, without overlap with Igf2 regulated genes.
We next focused on the p53-dependent rescue of lethality in Igf2+m/−p females. We reasoned that genes responsible for lethality would be differentially expressed in Igf2+m/−p female progeny of double heterozygote fathers (Igf2+m/−p, p53+/−), relative to the Igf2+m/−p, p53+/− female littermates that survive. The expression levels of the lethality rescue genes would be expected to be closer to those in Igf2+m/−p embryos from Igf2+m/−p or WT fathers, i.e. that expression was normalized. The genes identified as significantly different in Igf2+m/−p females compared to Igf2+m/−p and Igf2+m/−p, p53+/− embryos female littermates were then compared (Fig 3C). We obtained a signature that most likely represented the gene changes associated with p53+/− rescue. The E9.5 ‘lethality (rescue) signature’ (Fig 3C) contained 23 genes including those encoding transcription factors (Zmiz2, Tbx6), endocrine regulators (Sst), endocytosis (Necap2), metabolic enzymes (Pck2, Man1a, Pus7) and the extracellular matrix protein, fibronectin (Fn1).
We noted that the relative expression levels of Fn1 were high compared to the other genes (Fig 3C). Previous work had identified Fn1 gain of function as a modifier of lung branching morphogenesis, associated with gain of cell–matrix adhesion relative to the reciprocal repression of E-cadherin cell–cell adherence (Sakai et al, 2003). Lung airspaces of the Igf2+m/−p, p53+/+ females were filled with blood suggesting that haemorrhage was the likely cause of death (Fig 3D). Laser scanning confocal microscopy (LSCM) detected no obvious gross structural alterations in vessels and airways, neither in E-cadherin, Ki67 nor cleaved caspase-3 labelling (Fig 3D and not shown). Localization of fibronectin showed increased labelling in blood present in both large and small airways, presumably related to its known role in coagulation. However, an increase in fibronectin appeared in regions of the small airways and alveoli in Igf2+m/−p, p53+/+ females compared to female and male WT and Igf2+m/−p, p53+/− littermates (Fig 3D and not shown).
Accelerated tumour formation in p53 heterozygote females combined with loss of imprinting of Igf2 (H19−m/+p)
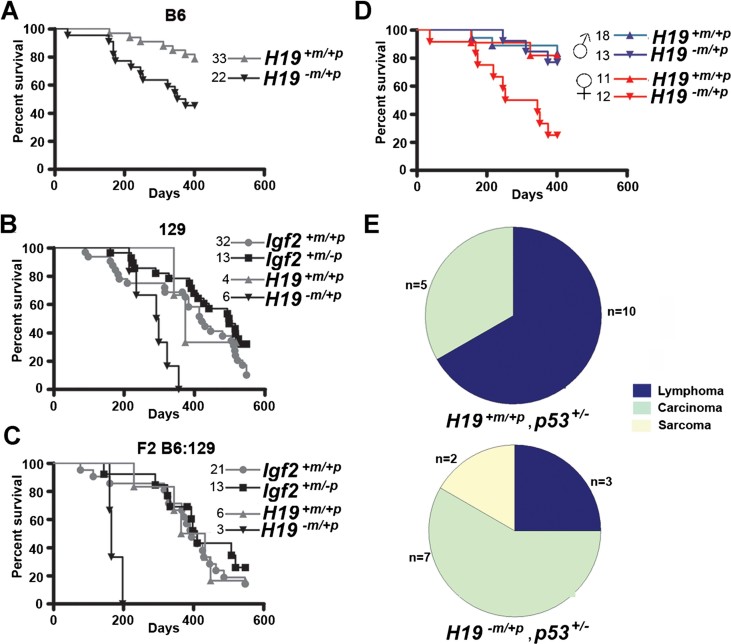
We next investigated dependencies of the p53 and Igf2 pathways during tumour development. Neither allelic loss nor gain of Igf2 expression in WT (p53+/+) and homozygote null (p53−/−) mice had any effect on tumour latency and overall longevity to 18 months, even though the IGF and p53 pathways were both implicated in aging and longevity in the mouse (Donehower, 2002, 2009; Supporting Information Fig S4). Heterozygosity of p53 (p53+/−) combined with bi-allelic expression of Igf2 (in this case associated with the 13 kb deletion of the H19 gene and ICR H19Δ13kb), resulted in accelerated tumour formation in B6, 129 or F2 backgrounds (Fig 4A–C). Interestingly, the effect on tumour progression also appeared unusually specific to females rather than males (Fig 4D). Control p53+/− females and males had similar tumour latencies (Fig 4D) indicating that the acceleration in tumour formation in females was primarily due to the consequences of Δ13 kb deletion on the Igf2-H19 locus. The consequences of H19−m/+p include the expected bi-allelic expression of Igf2, loss of function of the H19 ncRNA and associated miR-675, and the potential up-regulation of miR-483 in the Igf2 locus (Gabory et al, 2009).
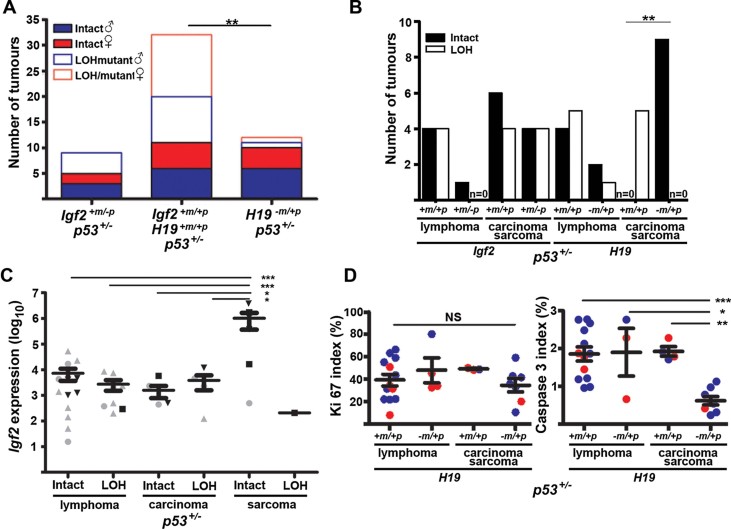
Figure 4. Bi-allelic Igf2 expression accelerates tumour formation in p53+/− female mice, irrespective of strain.
- Survival of B6 H19−m/+p, p53+/− mice was significantly reduced compared to p53+/− mice (p = 0.0065, log-rank test). Due to accelerated tumour latency the humane end point was brought forward to 400 days.
- Survival of 129 H19−m/+p, p53+/− mice was significantly reduced compared to Igf2+/−p, p53+/− (p = 0.0001), Igf2+m/+p, p53+/− (p = 0.008) and H19+m/+p, p53+/− mice (p = 0.036, log-rank test).
- Survival of F2 hybrid H19−m/+p, p53+/− mice was significantly reduced compared to Igf2+m/−p, p53+/− (p = 0.0008), Igf2+m/+p, p53+/− (p = 0.0017) or H19+m/+p, p53+/− mice (p = 0.0018, log-rank test).
- Tumour latency in B6 H19−m/+p, p53+/− female mice was significantly reduced compared to H19−m/+p, p53+/− males and p53+/− females and male mice (p < 0.0007, log-rank test).
- B6 H19−m/+p, p53+/− mice had significantly more solid tumours (carcinomas and sarcomas) and fewer lymphomas than B6 H19+m/+p, p53+/− mice (p = 0.037, Fisher's exact test).
A statistically significant alteration in tumour spectrum occurred in p53+/− mice with H19−m/+p, with more tumours classified as carcinoma and sarcoma compared to p53+/− mice (Fig 4E, Supporting Information Fig S5). Inactivation of the remaining WT p53 allele by LOH is normally required for tumour formation. We next determined the frequency of p53 LOH, mutation and the presence of p53 protein by IHC in tumours and correlated this with latency and tumour spectrum in H19−m/+p, p53+/− (Fig 5A; Supporting Information Table S4). Overall, 70% (21/30) of tumours from p53+/− littermates had either LOH or mutation of p53 compared to 16.7% (2/12) of tumours in H19−m/+p, p53+/− (Fig 5A). In carcinomas and sarcomas, and to a lesser extent in lymphoma, p53 LOH frequency appeared independent of loss of the paternal Igf2 allele (Fig 5B). In contrast, the untargeted (intact) p53 allele was retained in sarcoma and carcinomas associated with bi-allelic expression of Igf2 (Fig 5B), although this was independent of sex (not shown). High Igf2 expression determined by quantitative PCR was particularly associated with an intact p53 allele in sarcomas that arose in H19−m/+p, p53+/− mice, but was without consistent differences in Mdm2 localization (Supporting Information Figs S6 and S7). p53 LOH was detected with equal frequency to intact alleles in all tumours with lower Igf2 expression (Fig 5C, Supporting Information Fig S7). For the tumours that arose in H19−m/+p, p53+/−, we detected 1/10 with mutations in the p53 DNA-binding domain (Supporting Information Table S4). Our data suggested that bi-allelic expression of Igf2 and the associated disruption of the H19 locus may have reduced the selection pressure for inactivation of the remaining WT allele of p53 mainly in carcinoma and sarcoma.
Figure 5. p53 mutation, LOH and apoptosis in tumours from p53 +/− mice with bi-allelic Igf2 expression (H19 −m/+p).
- Proportion of p53+/− tumours with LOH/mutation was significantly reduced in tumours from mice bi-allelic for Igf2 (H19−m/+p), compared to mice null for Igf2+m/−p and WT (**p < 0.002, Fisher's exact test).
- 100% of sarcomas and carcinomas from H19−m/+p, p53+/− retain an intact WT p53 allele (n = 9), compared to 100% of LOH in the littermate p53+/− control mice (n = 5; **p = 0.0005, Fisher's exact test)
- Sarcomas with intact WT p53 (n = 6) had significantly higher Igf2 mRNA expression than lymphomas and carcinomas, irrespective of p53 allelic status (p < *0.05–***0.01, one-way ANOVA, Tukey's multiple comparison). Sample annotation is as for genotypes in Fig 4A and B.
- Solid tumours (n = 8) from H19−m/+p, p53+/− mice had significantly fewer apoptotic cells, assessed by staining for cleaved caspase-3 (right) than solid tumours from H19+m/+p, p53+/− mice (n = 4, *p = < 0.05, **p = < 0.01, ***p < 0.001, one-way ANOVA, Tukey's multiple comparison). Blue, male; Red, female.
To determine whether proliferation and apoptosis were modified in the accelerated tumour formation from bi-allelic H19−m/+p, p53+/− females, we performed immuno-labelling for Ki67 proliferative marker and cleaved caspase-3 apoptotic marker. In contrast to Ki67 labelling, cleaved caspase-3 labelling was significantly reduced in solid tumours (carcinoma and sarcoma) from p53+/− mice when combined with bi-allelic Igf2 expression compared to tumours from mice with mono-allelic Igf2 expression (Fig 5D). The correlation of cleaved caspase 3 labelling with Igf2 expression approached significance (Supporting Information Fig S8). The cause of the sex-specific acceleration of tumour development in females still remains obscure. The effect may be either paternal or X-linked, as oestrogen receptor (ER-α) immuno-labelling appeared independent of sex, tumour histology or genotype (Supporting Information Fig S9).
Igf2 dependency of the rapid tumour onset in p53 null using combined conditional alleles
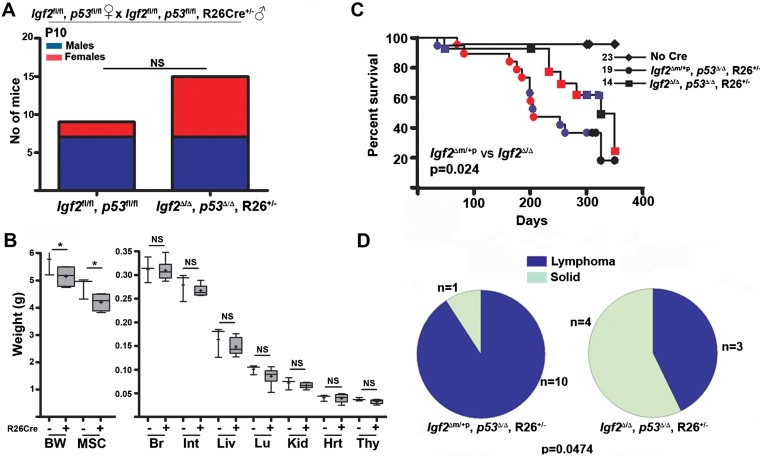
We next utilized a new conditional allele for Igf2 with loxp sites that flanked either side of exons 4–6 (Sandovici et al, in preparation), a conditional allele previously characterized for p53, and combined these with a conditional tamoxifen inducible Cre (ROSA26Cre; see Supporting Information Table S5). Igf2fl/fl, p53fl/fl females were successfully bred with Igf2fl/+, p53fl/+, R26CreER+/− males on a C57BL6-FVBN background (Fig 6A). Injection of tamoxifen (1 mg) to mothers at E10.5 generated deletion alleles and the expected segregation of progeny (37 males and 28 females, Supporting Information Fig S10A). Non-quantitative PCR suggested that recombination was incomplete as most tissues retained the loxp flanked allele (Supporting Information Fig S10B). We were unable to either conditionally modify both alleles prior to embryonic day 8, or to generate germ-line transmission of deleted alleles. Continued breeding of males with p53 loxp flanked alleles resulted in 73 mice from 5.6 litters with normal transmission of non-recombined alleles. Following recombination in utero, males with deleted alleles (p53Δ/Δ or p53+/Δ) generated only seven progeny from 3 litters, with no subsequent transmission of the deleted (recombined) p53 allele. By P30 the mean total body weights (BW) and that of the eviscerated carcass of Igf2Δ/Δ, p53Δ/Δ, R26CreER+/− progeny was significantly less than that of Igf2fl/fl, p53fl/fl littermates, indicating that at least in these tissues Igf2-dependent functional growth effects could be detected (Fig 6B). Compared to other organs, the growth findings imply that Igf2-dependent total body and musculoskeletal growth may extend beyond E10.5 (Burns & Hassan, 2001).
Figure 6. Decreased tumour latency and altered tumour spectrum following homozygous conditional deletion of both Igf2 and p53.
- Progeny from the homozygous conditional cross of Igf2fl/fl, p53fl/fl females × Igf2fl/fl, p53fl/fl, R26+/− males segregated according to the expected Mendelian distribution at P10, regardless of sex.
- Following maternal IP injection of tamoxifen at E10.5, BW and musculoskeletal weights (MS) of carcasses, were significantly less for Igf2Δ/Δ, p53Δ/Δ, R26Cre+/− mice (n = 6) compared to sham injected controls Igf2fl/fl, p53fl/fl, R26Cre+/− (n = 3, *p = 0.05, unpaired t-test). No significant differences in the weight of viscera were detected; Br, brain, Int, small intestine; Liv, liver; Lu, lungs; Kid, kidney; Hrt, heart; Thy, thymus. Plot shows 95% of the range (whiskers), interquartile range (box), median (horizontal crossbar) and mean (cross).
- Mice with homozygous conditional deletion of Igf2 and p53 (Igf2Δ/Δ, p53Δ/Δ, R26+/−) developed tumours later and survived for longer (median survival = 325 days) than mice with an intact paternal allele of Igf2 (Igf2Δm/+p, p53Δ/Δ, R26+/−, median survival = 206 days, p = 0.024, log-rank test, blue, males; red, females).
- Igf2Δ/Δ, p53Δ/Δ, R26+/− mice (right) developed more solid tumours than Igf2Δm/+p, p53Δ/Δ, R26+/− mice (left, 4 and 1, respectively), and fewer lymphomas (3 and 10, respectively, p = 0.0474, Fisher's exact test).
Tamoxifen treatment of homozygous conditional null Igf2Δ/Δ, p53Δ/Δ mice with ROSA 26 driven Cre (R26CreER+/−), avoided the combined lethality observed using germ-line transmission to create Igf2−m/−p, p53−/−. Moreover, bi-allelic disruption of the Igf2 allele eliminated the potential for tumour specific re-expression of the imprinted Igf2 (silenced) maternal allele (LOI). Mice with conditional deletion of both alleles of Igf2 and p53 developed a longer tumour latency (n = 14, three lymphomas, four solid tumours, two unknown causes, no metastasis, median survival = 325 days) compared to mice with conditional deletion of p53 but retention of the paternal Igf2 allele (n = 19, 10 lymphomas, 1 solid tumour, 5 unknown causes, median survival = 206 days, Fig 6C). Importantly, the time-dependent onset of tumours mimicked that of the p53−/− germ-line modification (Supporting Information Fig S4). There was a significant shift in tumour spectrum of Igf2Δm/+p, p53Δ/Δ, R26+/− mice relative to Igf2Δ/Δ, p53Δ/Δ, R26+/− mice with a reduction in lymphoma development relative to carcinoma and sarcoma (Fig 6D). These results confirm that allelic dose of Igf2 significantly reduced the onset of homozygote p53 null tumour phenotype for all types of tumours.
DISCUSSION
Single cell type in vitro models tend to lack tissue specific auto-regulatory feedback loops, and so, although informative, significance for in vivo function may be limited (Feng & Levine, 2010). Here, we exploited the imprinting regulation of Igf2, a developmental and tumour growth factor, to both increase and decrease the allelic dosage of Igf2 and ligand supply. An assumption was that genetic modifications altered Igf2 expression alone. As in most mouse models, disruption of genomic regions can generate direct and indirect effects, with altered expression of microRNAs as an example. Loss of miR-675 in the H19ncRNA can promote tumour formation, as can an increase in miR-483 within the Igf2 region (Gabory et al, 2009; Veronese et al, 2010; Yoshimizu et al, 2008). A second assumption is that alteration of IGF2 ligand supply leads to physiological activation of the pathway. The proportionate growth effects of the Igf2 models support this assumption when compared to un-physiological expression derived from introduced transgenes. The resulting increase in supply of IGF2 would bind cell surface IGF1R and IR-A, to activate downstream signalling to all intact pathway components, and specific feedback mechanisms (Ulanet et al, 2010).
Igf2-dependent developmental lethality and an Igf2-dependent modification of tumour formation occurred in p53 null mice. When Igf2+m/−p has been previously combined with genetic models of tumour susceptibility, including several developmental genes, there has been little evidence for combined lethality with this allele, suggesting that the synergistic lethality phenotype we now describe with p53 is novel (Christofori et al, 1994; Hassan & Howell, 2000; Ho et al, 2009; Wu et al, 2006). Thus, our observation that homozygote null p53 mice fail to survive to adulthood in the context of disruption of the paternal allele of Igf2 suggests Igf2 is required to rescue a p53 loss of function-dependent lethality.
The specific lethality in Igf2+m/−p female progeny was genetically attributed to double heterozygote fathers. The paternal X-chromosome appears a prerequisite for lethality of the Igf2+m/−p females because of the survival of male Igf2+m/−p progeny. Female Igf2+m/−p, p53+/− progeny also survived, albeit at lower than expected numbers, indicating that loss of one allele of p53 from the genotype of the offspring was sufficient for a partial rescue of the female specific lethality. We speculate that potential rescue mechanisms might include interactions of p53 with a range of epigenetic and X-specific targets, e.g. DNA methyltransferases (Dnmt; Park et al, 2005; Peterson et al, 2003) and Xist expression (Panning & Jaenisch, 1996). In Igf2−m/−p mice, the canalicular phase of lung maturation during late gestation appeared delayed, leading to lungs having poorly organized alveoli with thick septae and reduced airspaces (Silva et al, 2006). Reduced exposure to maternal corticosteroids may have been responsible for the delayed lung development since progeny from Igf2−m/−p mothers treated with this glucocorticoid had lung architecture similar to that of WT controls (Silva et al, 2006). None of these effects were sex or female specific as in our case. Fibronectin (Fn1) is the major component of the extracellular matrix in the lung and excess supply promotes epithelial branching morphogenesis (Sakai et al, 2003). The ten-fold increase in expression of Fn1 in Igf2+m/−p, p53+/+ female genotyped embryos destined to die, with lower levels in surviving Igf2+m/−p, p53+/− littermates, appeared significant, yet we could not reliably infer the phenotype was Fn1 dependent. Expression of Man1a2 gene, which encodes golgi α1,2-mannosidase IB, was also elevated in Igf2+m/−p, p53+/+ female progeny. Homozygous knock-out of Man1a2 produced a surprisingly severe lung phenotype that resulted in neonatal lethality (Tremblay et al, 2007). Importantly, Man1a2−/− mice died hours after birth from respiratory distress and alveolar haemorrhage, mirroring our phenotype even though in our case Man1a2 appeared up-regulated (Tremblay et al, 2007).
Deletion of the H19 locus (Leighton et al, 1995) is an imperfect model of LOI for Igf2. Importantly, we cannot exclude the possibility that some of the effects that we observe may be due to removal of H19 non-coding RNA, as been demonstrated in the small intestine (Yoshimizu et al, 2008). H19 long non-coding RNA has putative roles as a tumour suppressor (Yoshimizu et al, 2008) and regulator of embryonic growth (Gabory et al, 2009), mainly through loss of microRNA, miR-675, which is hosted by H19 within its first exon (Cai & Cullen, 2007; Mineno et al, 2006). Thus the consequences of the 13 kb deletion in the H19−m/+p model we utilized may also have had Igf2-independent gain of function effects on the p53 pathway. However, our previous analysis of miRNA profiles from these Igf2 models revealed numerous other miRNA changes, despite Igf2 specific phenotypic effects in the context of Pten heterozygosity (Church et al, 2011).
The tumour-promoting effect of H19−m/+p appeared independent of the direct loss of the WT p53 allele (LOH) in carcinoma and sarcoma tumours. We cannot however explain the female dependency of this phenotype, except to propose that it may depend on either the X chromosome, a strain modifier or via early hormonal changes (Harvey et al, 1993; Jacks et al, 1994). It has been observed that anti-tumour effects can occur following re-introduction of a WT p53 allele in p53 null tumours (Martins et al, 2006; Ventura et al, 2007; Xue et al, 2007). Our data suggest that counteracting the inhibitory effects of the IGF2 pathway on the endogenous WT p53 allele, may also lead to re-activation of the endogenous WT p53 allele, and promote tumour suppression. In tumours with intact p53 alleles and Igf2 over-expression, inhibiting the IGF2 signalling pathway may offer a therapeutic strategy. Conditional homozygous deletion of both Igf2 and p53 alleles slowed the development of tumours compared with the conditional deletion of p53 alleles alone. Thus, p53 loss of function in both development and tumour formation appears dependent on Igf2. The Igf2 dependency of tumours that arise in the context of other models associated with p53 loss of function implicates Igf2 as a significant factor that regulates the progression of tumours through regulation of apoptosis (Christofori et al, 1994, 1995; Lopez & Hanahan, 2002), more recently observed in a mouse model of high grade chondrosarcoma (Ho et al, 2009). In the chondrosarcoma model, a p53-dependent reduction of IGFBP levels may have accounted for a secondary increase in IGF2 ligand bio-availability, and hence dependence on Igf2. The dependency of tumour growth on the IGF pathway is further supported by observations in Li-Fraumeni associated tumours, as adrenal carcinoma and osteosarcoma have both been associated with dependency on IGF2, and are the basis of interventional clinical trials of agents that inhibit the activation of the IGF1 receptor (Avnet et al, 2009; Barlaskar et al, 2009). These data suggest that regulation of the IGF2 ligand in tumour susceptibility syndromes associated with p53 (e.g. Li-Fraumeni, p53 and MDM2 associated SNPs) may be a potential strategy for tumour prevention. Finally, we cannot extend these observations to either gain of function mutants of p53, where partial, complete and altered pathway interactions may be evident (Hinkal et al, 2009). These issues require further genetic investigation during development and tumour formation.
MATERIALS AND METHODS
Animals and genotyping
Animal work was approved by University of Oxford ethics committee and UK Home Office. Mice [Mus musculus C57BL/6J (B6) and 129S2/J (129)] were maintained and genotyped as previously described (Hassan & Howell, 2000; Supporting Information Table S5, primers available on request). The new Igf2 conditional allele has loxp sites flanking exons 4–6 and was maintained (>10 generations) on a C57Bl6 background (Sandovici et al in preparation). Seminal plugs at 9.00 am and conception was taken ±12 h. Weighed embryos were fixed in either 4% neutral buffered formalin (NBF) or RNAlater (Qiagen, Crawley, UK). DNA from E9.5 embryo yolk sacs was extracted by TRI reagent (Applied Biosystems, Warrington, UK). To distinguish Igf2+m/−p from Igf2−m/+p, embryos with reduced CR length and low Igf2 expression with qPCR were designated as being Igf2+m/−p. Recombination of conditional alleles by R26CreER+/− was with Tamoxifen (1 mg, 5% v/v ethanol and 95% v/v corn-oil, Sigma, Poole, UK) injected I.P. at E10.5. Humane end-point for tumour burden was set at 18 months. Animals were checked daily for palpable tumours and evidence of systemic decline (weight loss, lack of grooming, reduced activity and piloerection).
The paper explained
PROBLEM:
The p53 gene product is a transcription factor that controls the cells' response to genome toxins, such as radiation, and is a frequently mutated target in cancer. Inherited mutation of the p53 gene can predispose humans to early onset cancers (Li-Fraumeni syndrome, LFS), and this effect can be reproduced in the mouse. When tumours form, the remaining wild-type (WT) allele is lost either through loss of genomic DNA in a process called ‘loss of heterozygosity (LOH)’, or by inhibition of p53. The pathways that modify the age of onset of LFS cancers are potential targets for screening and therapy, and such pathways may also be potential targets to restore function when p53 function is lost. Molecular interactions between p53 and a growth signalling pathway regulated by insulin-like growth factor (IGF) ligands have been observed in cell culture models. To date there has been no systematic analysis of the consequences of gain and loss of supply of the IGF2 ligand in the context of loss of p53 function in the whole organism.
RESULTS:
We addressed the problem by taking a comprehensive genetic approach in the mouse. We combined mice that lack supply of the IGF2 ligand (Igf2 null) and mice with increased supply of Igf2 (bi-allelic expressed alleles) with mice that were p53 heterozygote and homozygote. If both these genes were homozygote null in all tissues, then embryonic development failed, suggesting an important pathway interaction. Interestingly, if the breeding male was heterozygote for both genes, only females with loss of the male derived Igf2 allele developed a haemorrhagic lung syndrome, suggesting a more specific interaction. Importantly, the age of tumour onset was either reduced or increased if the supply of IGF2 was decreased and increased, respectively. When IGF2 supply was increased, we also observed that the WT p53 allele remained intact in an altered tumour spectrum towards carcinoma and sarcoma, suggesting the mechanism of suppression of p53 was independent of LOH in these tumours. Targeting IGF2 supply in this instance may reactivate the suppressed p53 and generate anti-tumour mechanisms.
IMPACT:
Screening patients with LFS can be a very effective method to detect early tumour formation and prevent development of lethal cancers. For some sporadic cancers, such as bone and soft tissue sarcoma that are frequently observed in LFS, defects in the p53 pathway have also been observed, suggesting that the sporadic development of p53-dependent cancers may in fact represent a spectrum of susceptibility ranging from germ-line mutation to pathway polymorphisms. To date, no therapeutic agents have been developed that modify the age of onset either in LFS tumours or in tumours (sarcoma) that arise in the context of p53 pathway modification rather than inherited gene mutation. Interruption of the IGF2 signaling pathway, for example by specific targeting of the ligand, may therefore be a useful mechanism to test the prevention of human tumours with a disrupted p53 pathway. Moreover, targeting IGF2 may be a treatment strategy in selected patients with tumours that arise in the context of p53 pathway disruption without LOH but with increased IGF2 pathway activity.
RNA extraction, reverse transcription and qPCR
Material stored at −80°C in RNAlater (Applied Biosystems) were homogenized with either an Ultra-Turrax T8 (Ika Labortechnik, Staufen, Germany) or by a 25-gauge needle. A total nucleic acid isolation kit (Applied Biosystems) was used to extract RNA from formalin-fixed paraffin embedded (FFPE) tissue. RNA was quantified by NanoDrop 1000 (Thermo Fisher Scientific, Loughborough, UK). RT-PCR (High-Capacity cDNA Reverse Transcription Kit, Applied Biosystems) prior to quantitative PCR normalized to the expression of β-actin (primer sequences available on request).
Immunohistochemistry and microscopy
Paraffin-embedded tissues were sectioned and stained with haematoxylin and eosin (H & E; Hassan & Howell, 2000). Antibodies and antigen retrieval methods are available on request. Sections were blocked (1 h, 1.5% v/v) with goat or rabbit serum, or overnight using a Mouse-on-Mouse kit (Vector Laboratories, Peterborough, UK), and incubated with primary antibody either at 4°C (overnight) or at room temperature (RT, 1 h). Secondary labelling was with either biotinylated (Vectastain elite avidin–biotin complex kit, Vector Laboratories) or alexa-fluor 594-conjugated secondary antibodies and counterstained with haematoxylin or 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Digital images were acquired using a Zeiss LSM 510 confocal microscope (Carl Zeiss, Welwyn Garden City, UK) and a Nuance MSI camera (CRi, Woburn, MA) and analysed using Nuance 2.4.10 software. The percentage of Ki67 or cleaved-caspase 3 positive cells was scored by counting at least 500–1000 cells from 4 to 5 fields per tumour.
Microarray gene expression profiling and bioinformatics
RNA from E9.5 embryos were hybridized to Illumina Mouse Whole Genome 6 v1.1 or v2 BeadChips (Illumina, San Diego, CA) by Cambridge Genomic Services (Cambridge, UK) or Wellcome Trust Centre for Human Genetics (Oxford, UK). Bioinformatics included R 2.12.0, (R Development Core Team, 2010) with microarray-specific packages obtained from the Bioconductor repository (Bioconductor 2.7, (Gentleman et al, 2004). Raw data were normalized with ‘lumi’ (Du et al, 2008) and Significance Analysis of Microarrays (Tusher et al, 2001) with ‘siggenes’. Mapping of Illumina identifiers to annotation terms was done with ‘biomaRt’ and data from the Ensembl BioMart database (European Bioinformatics Institute, Cambridge, UK).
LOH and mutation analysis
PCR for p53 LOH with primers are described in Supporting Information Table S5. PCR products were purified with a Zymoclean Gel DNA Recovery Kit (Cambridge Bioscience, Cambridge, UK) and sequenced by GeneService (Oxford, UK).
Author contributions
Experiments were conceived and developed by ABH and VLH; VLH performed all experimental breeding except Igf2 knockout backcrosses which were performed by CFG; VLH provided all of the experimental data, except array data analysis (DJB), histopathology diagnosis (FP), confocal imaging (CB) and some mouse genotyping (EC); IS and MC generated mice with Igf2 conditional alleles utilized by VLH; The manuscript and figures were written by VLH, DJB and ABH, and all authors provided detailed comments.
Acknowledgments
We thank Shuobo Zhang and Louise Falk for genotyping, Richard Stillion for assistance with histology, Andrew Wilkie, Gareth Bond, Elisabeth Bikoff, Elisabeth Robertson, Andrew Ward and David Church for valuable discussion. This work was supported by CR-UK Programme (ABH, C429), CRUK Studentship (VH/ABH) and the Department of Oncology, University of Oxford.
Supporting Information is available at EMBO Molecular Medicine online.
The authors declare that they have no conflict of interest.
For more information
P53 OMIM: http://omim.org/entry/191170
IARC p53 database: http://www-p53.iarc.fr/
IGF2 OMIM: http://omim.org/entry/147470
Imprinting: http://www.geneimprint.com/
Li-Fraumeni syndrome Orphanet: http://www.orpha.net/
Li-Fraumeni support: http://www.mdjunction.com/li-fraumeni-syndrome
LFS association: http://www.lfsassociation.org/our-story/lfs-board/
Constancia lab: http://www.mrl.ims.cam.ac.uk/staff/Constancia.php
Hassan lab: http://www.path.ox.ac.uk/dirsci/cellbiol/hassan
Supplementaary material
Detailed facts of importance to specialist readers are published as ”Supporting Information”. Such documents are peer-reviewed, but not copy-edited or typeset. They are made available as submitted by the authors.
References
- Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5:931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- Avnet S, Sciacca L, Salerno M, Gancitano G, Cassarino MF, Longhi A, Zakikhani M, Carboni JM, Gottardis M, Giunti A, et al. Insulin receptor isoform A and insulin-like growth factor II as additional treatment targets in human osteosarcoma. Cancer Res. 2009;69:2443–2452. doi: 10.1158/0008-5472.CAN-08-2645. [DOI] [PubMed] [Google Scholar]
- Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- Barlaskar FM, Spalding AC, Heaton JH, Kuick R, Kim AC, Thomas DG, Giordano TJ, Ben-Josef E, Hammer GD. Preclinical targeting of the type I insulin-like growth factor receptor in adrenocortical carcinoma. J Clin Endocrinol Metab. 2009;94:204–212. doi: 10.1210/jc.2008-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Broz DK, Basak S, Park EJ, McLaughlin ME, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JL, Hassan AB. Cell survival and proliferation are modified by insulin-like growth factor 2 between days 9 and 10 of mouse gestation. Development. 2001;128:3819–3830. doi: 10.1242/dev.128.19.3819. [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellot S, Sauvageau G. Zfx: at the crossroads of survival and self-renewal. Cell. 2007;129:239–241. doi: 10.1016/j.cell.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofori G, Naik P, Hanahan D. A second signal supplied by insulin-like growth factor II in oncogene-induced tumorigenesis. Nature. 1994;369:414–418. doi: 10.1038/369414a0. [DOI] [PubMed] [Google Scholar]
- Christofori G, Naik P, Hanahan D. Deregulation of both imprinted and expressed alleles of the insulin-like growth factor 2 gene during beta-cell tumorigenesis. Nat Genet. 1995;10:196–201. doi: 10.1038/ng0695-196. [DOI] [PubMed] [Google Scholar]
- Church DN, Phillips BR, Stuckey DJ, Barnes DJ, Buffa FM, Manek S, Clarke K, Harris AL, Carter EJ, Hassan AB. Igf2 ligand dependency of Pten(+/−) developmental and tumour phenotypes in the mouse. Oncogene. 2011 doi: 10.1038/onc.2011.526. DOI: 10.1038/onc.2011.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, Wyllie AH. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Efstratiadis A, Robertson EJ. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature. 1990;345:78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Donehower LA. Does p53 affect organismal aging. J Cell Physiol. 2002;192:23–33. doi: 10.1002/jcp.10104. [DOI] [PubMed] [Google Scholar]
- Donehower LA. Longevity regulation in flies: a role for p53. Aging (Albany NY) 2009;1:6–8. doi: 10.18632/aging.100010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- Fang S, Krahe R, Lozano G, Han Y, Chen W, Post SM, Zhang B, Wilson CD, Bachinski LL, Strong LC, et al. Effects of MDM2, MDM4 and TP53 codon 72 polymorphisms on cancer risk in a cohort study of carriers of TP53 germline mutations. PLoS ONE. 2010;5:e10813. doi: 10.1371/journal.pone.0010813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Kachnic L, Zhang J, Powell SN, Xia F. DNA damage induces p53-dependent BRCA1 nuclear export. J Biol Chem. 2004;279:28574–28584. doi: 10.1074/jbc.M404137200. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–33053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- Foulstone E, Prince S, Zaccheo O, Burns JL, Harper J, Jacobs C, Church D, Hassan AB. Insulin-like growth factor ligands, receptors, and binding proteins in cancer. J Pathol. 2005;205:145–153. doi: 10.1002/path.1712. [DOI] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- Grimberg A, Coleman CM, Shi Z, Burns TF, MacLachlan TK, Wang W, El-Deiry WS. Insulin-like growth factor factor binding protein-2 is a novel mediator of p53 inhibition of insulin-like growth factor signaling. Cancer Biol Ther. 2006;5:1408–1414. doi: 10.4161/cbt.5.10.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey M, McArthur MJ, Montgomery CA, Jr, Bradley A, Donehower LA. Genetic background alters the spectrum of tumors that develop in p53-deficient mice. FASEB J. 1993;7:938–943. doi: 10.1096/fasebj.7.10.8344491. [DOI] [PubMed] [Google Scholar]
- Hassan AB, Howell JA. Insulin-like growth factor II supply modifies growth of intestinal adenoma in Apc(Min/+) mice. Cancer Res. 2000;60:1070–1076. [PubMed] [Google Scholar]
- Hinkal G, Parikh N, Donehower LA. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS ONE. 2009;4:e6654. doi: 10.1371/journal.pone.0006654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Stojanovski A, Whetstone H, Wei QX, Mau E, Wunder JS, Alman B. Gli2 and p53 cooperate to regulate IGFBP-3- mediated chondrocyte apoptosis in the progression from benign to malignant cartilage tumors. Cancer Cell. 2009;16:126–1136. doi: 10.1016/j.ccr.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Ito Y, Koessler T, Ibrahim AE, Rai S, Vowler SL, Abu-Amero S, Silva AL, Maia AT, Huddleston JE, Uribe-Lewis S, et al. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17:2633–2643. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lee YI, Lee S, Das GC, Park US, Park SM. Activation of the insulin-like growth factor II transcription by aflatoxin B1 induced p53 mutant 249 is caused by activation of transcription complexes; implications for a gain-of-function during the formation of hepatocellular carcinoma. Oncogene. 2000;19:3717–3726. doi: 10.1038/sj.onc.1203694. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez T, Hanahan D. Elevated levels of IGF-1 receptor convey invasive and metastatic capability in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2002;1:339–353. doi: 10.1016/s1535-6108(02)00055-7. [DOI] [PubMed] [Google Scholar]
- Luoh SW, Bain PA, Polakiewicz RD, Goodheart ML, Gardner H, Jaenisch R, Page DC. Zfx mutation results in small animal size and reduced germ cell number in male and female mice. Development. 1997;124:2275–2284. doi: 10.1242/dev.124.11.2275. [DOI] [PubMed] [Google Scholar]
- Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2:475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- Park IY, Sohn BH, Choo JH, Joe CO, Seong JK, Lee YI, Chung JH. Deregulation of DNA methyltransferases and loss of parental methylation at the insulin-like growth factor II (Igf2)/H19 loci in p53 knockout mice prior to tumor development. J Cell Biochem. 2005;94:585–596. doi: 10.1002/jcb.20263. [DOI] [PubMed] [Google Scholar]
- Peterson EJ, Bogler O, Taylor SM. p53-mediated repression of DNA methyltransferase 1 expression by specific DNA binding. Cancer Res. 2003;63:6579–6582. [PubMed] [Google Scholar]
- Post SM, Quintas-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, Maccio DR, Bond GL, Johnson DG, Levine AJ, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18:220–230. doi: 10.1016/j.ccr.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- Silva D, Venihaki M, Guo WH, Lopez MF. Igf2 deficiency results in delayed lung development at the end of gestation. Endocrinology. 2006;147:5584–5591. doi: 10.1210/en.2006-0498. [DOI] [PubMed] [Google Scholar]
- Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- Suh YA, Post SM, Elizondo-Fraire AC, Maccio DR, Jackson JG, El-Naggar AK, Van Pelt CS, Terzian T, Lozano G. Multiple stress signals activate mutant p53 in vivo. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Igarashi S, Nojima M, Maruyama R, Yamamoto E, Kai M, Akashi H, Watanabe Y, Yamamoto H, Sasaki Y, et al. IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis. 2010;31:342–349. doi: 10.1093/carcin/bgp179. [DOI] [PubMed] [Google Scholar]
- Tremblay LO, Nagy Kovacs E, Daniels E, Wong NK, Sutton-Smith M, Morris HR, Dell A, Marcinkiewicz E, Seidah NG, McKerlie C, et al. Respiratory distress and neonatal lethality in mice lacking Golgi alpha1,2-mannosidase IB involved in N-glycan maturation. J Biol Chem. 2007;282:2558–2566. doi: 10.1074/jbc.M608661200. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanet DB, Ludwig DL, Kahn CR, Hanahan D. Insulin receptor functionally enhances multistage tumor progression and conveys intrinsic resistance to IGF-1R targeted therapy. Proc Natl Acad Sci USA. 2010;107:10791–10798. doi: 10.1073/pnas.0914076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewitter E, Scrable H. Delta40p53 controls the switch from pluripotency to differentiation by regulating IGF signaling in ESCs. Genes Dev. 2010;24:2408–2419. doi: 10.1101/gad.1987810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Veronese A, Lupini L, Consiglio J, Visone R, Ferracin M, Fornari F, Zanesi N, Alder H, D'Elia G, Gramantieri L, et al. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res. 2010;70:3140–3149. doi: 10.1158/0008-5472.CAN-09-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A, Tabori U, Schiffman J, Shlien A, Beyene J, Druker H, Novokmet A, Finlay J, Malkin D. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: a prospective observational study. Lancet Oncol. 2011;12:559–567. doi: 10.1016/S1470-2045(11)70119-X. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Werner H, Karnieli E, Rauscher FJ, LeRoith D. Wild-type and mutant p53 differentially regulate transcription of the insulin-like growth factor I receptor gene. Proc Natl Acad Sci USA. 1996;93:8318–8323. doi: 10.1073/pnas.93.16.8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Shete S, Amos CI, Strong LC. Joint effects of germ-line p53 mutation and sex on cancer risk in Li-Fraumeni syndrome. Cancer Res. 2006;66:8287–8292. doi: 10.1158/0008-5472.CAN-05-4247. [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci USA. 2008;105:12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccheo OJ, Prince SN, Miller DM, Williams C, Kemp CF, Brown J, Jones EY, Catto LE, Crump MP, Hassan AB. Kinetics of insulin-like growth factor II (IGF-II) interaction with domain 11 of the human IGF-II/mannose 6-phosphate receptor: function of CD and AB loop solvent-exposed residues. J Mol Biol. 2006;359:403–421. doi: 10.1016/j.jmb.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kashanchi F, Zhan Q, Zhan S, Brady JN, Fornace AJ, Seth P, Helman LJ. Regulation of insulin-like growth factor II P3 promotor by p53: a potential mechanism for tumorigenesis. Cancer Res. 1996;56:1367–1373. [PubMed] [Google Scholar]
- Zhang L, Zhan Q, Zhan S, Kashanchi F, Fornace A,J, Jr, Seth P, Helman LJ. p53 regulates human insulin-like growth factor II gene expression through active P4 promoter in rhabdomyosarcoma cells. DNA Cell Biol. 1998;17:125–131. doi: 10.1089/dna.1998.17.125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.