Abstract
The attachment of N-acetylglucosamine to serine or threonine residues (O-GlcNAc) is a post-translational modification on nuclear and cytoplasmic proteins with emerging roles in numerous cellular processes, such as signal transduction, transcription, and translation. It is further presumed that O-GlcNAc can exhibit a site-specific, dynamic and possibly functional interplay with phosphorylation. O-GlcNAc proteins are commonly identified by tandem mass spectrometry following some form of biochemical enrichment. In the present study, we assessed if, and to which extent, O-GlcNAc-modified proteins can be discovered from existing large-scale proteome data sets. To this end, we conceived a straightforward O-GlcNAc identification strategy based on our recently developed Oscore software that automatically analyzes tandem mass spectra for the presence and intensity of O-GlcNAc diagnostic fragment ions. Using the Oscore, we discovered hundreds of O-GlcNAc peptides not initially identified in these studies, and most of which have not been described before. Merely re-searching this data extended the number of known O-GlcNAc proteins by almost 100 suggesting that this modification exists even more widely than previously anticipated and the modification is often sufficiently abundant to be detected without enrichment. However, a comparison of O-GlcNAc and phospho-identifications from the very same data indicates that the O-GlcNAc modification is considerably less abundant than phosphorylation. The discovery of numerous doubly modified peptides (i.e. peptides with one or multiple O-GlcNAc or phosphate moieties), suggests that O-GlcNAc and phosphorylation are not necessarily mutually exclusive, but can occur simultaneously at adjacent sites.
The modification of proteins with N-acetylglucosamine (O-GlcNAc)1 is an emerging dynamic post-translational modification of serine or threonine residues of proteins. O-GlcNAc is found on a wide range of proteins involved in virtually all cellular processes as well as various human diseases (1, 2) including cancer (3). In addition, O-GlcNAc can interplay with phosphorylation, which, for instance, modulates the stability and activity of p53 (4). Despite its biological importance, the analysis of O-GlcNAc-modified proteins remains highly challenging. In fact, of the ∼800 reported O-GlcNAc proteins, direct and unambiguous evidence for the site of O-glycosylation is available for less than 25% of these (5).
The identification of O-GlcNAc proteins is typically achieved by combining selective enrichment and liquid chromatography tandem mass spectrometry (LC-MS/MS). Albeit powerful, the identification of modified peptides and sites is hindered by the substoichiometric occupancy of O-GlcNAc sites (2) and the lability of the O-glycosidic bond in the gas phase (6). In mass spectrometry-based proteomics, peptides are usually sequenced via collision-induced dissociation (CID). However, under typical CID conditions, the concurrent O-GlcNAc peptide and site identification is difficult, because peptides readily lose the GlcNAc moiety, and spectra are dominated by neutral loss species along with the GlcNAc oxonium ion and fragments thereof (7). Peptide sequence identification is often still possible from fragments that lost the O-GlcNAc moiety, but site information is irretrievably lost upon dissociation of the O-glycosidic bond. In contrast, the fragmentation of peptides with electron capture dissociation (ECD) or electron transfer dissociation (ETD) typically preserves PTM sites and allows the direct and simultaneous identification of O-GlcNAc peptide sequences and sites (8, 9) but these techniques also have shortcomings notably concerning sensitivity on most current commercial platforms.
Although not ideal for O-GlcNAc site localization, the initial detection of O-GlcNAc peptides is strongly facilitated in CID-type experiments (10, 11) because diagnostic GlcNAc losses along with the GlcNAc oxonium ion and its fragments define a characteristic pattern, which identifies O-GlcNAc peptides even in very complex proteomics samples (9). The availability of high resolution and high mass accuracy instruments further improves the selectivity of these diagnostic fragment ions (12, 13).
We have recently developed a bioinformatics tool, termed Oscore that automatically assesses tandem MS spectra for the presence and intensity of O-GlcNAc diagnostic fragment ions and, in turn, allows ranking spectra according their probability of representing an O-GlcNAc peptide (12). On a test data set of 750 O-GlcNAc spectra and 11,300 spectra from unmodified peptides, the Oscore was able to discriminate O-GlcNAc spectra from spectra of unmodified peptides with 95% sensitivity and >99% specificity and outperformed alternative approaches such as the simple filtering for diagnostic ions. In the present study, we show that the Oscore can be applied to existing large-scale proteomic data to discover hundreds of O-GlcNAc peptides not initially identified in these studies. Merely re-searching this data extended the number of known O-GlcNAc proteins by almost 100 suggesting that this modification exists even more widely than previously anticipated and is often abundant enough to be detected without specific biochemical enrichment.
EXPERIMENTAL PROCEDURES
Publically Available Data
Publically available raw mass spectrometric data from published proteome-wide studies of 11 different cell lines (14), HeLa cells (15), as well as data from published proteome-wide and phospho-proteome studies of hES and iPS cells (16) were downloaded from respective repositories (see also supplemental Table S1).
Data Analysis
The mass spectrometric data were processed essentially as described (12). Briefly, peak picking and processing was performed using Mascot Distiller 2.4.2.0 (Matrix Science, London, UK) in which merging of tandem MS spectra from the same precursor as well as isotope fitting of fragments below m/z 205 was disabled. The resulting peak list files were processed by the Oscore perl script, which calculates the Oscore for every peptide precursor for which the tandem MS spectrum contains at least one diagnostic O-GlcNAc feature within a tolerance of 10 ppm. The peak list files were searched with Mascot 2.3.0 against the UniProtKB complete human (download date 26.10.2010, 110,550 sequences) combined with sequences of common contaminants. In case of the phospho-proteome dataset of hES and iPS cells (16), the spectra were searched against a subset database generated with Scaffold 3.3.1 (Proteome Software, Portland, OR) including only protein identifications from the respective full proteome data set (11,288 sequences). Carbamidomethylation of cysteine residues, oxidation of methionine, and HexNAc modification of serine, threonine and asparagine residues were taken into account as variable modifications. Where applicable, phosphorylation of serine, threonine and tyrosine residues was set as variable modification. Likewise, 4-plex or 8-plex iTRAQ was set as fixed modification at the peptide amino terminus and lysine side chain for data sources using these peptide tags. According to the proteases employed in the original studies, enzyme specificity was set to trypsin (lysine, arginine), LysC (lysine), or GluC (aspartic acid, glutamic acid) allowing for up to two missed cleavage sites. The modification definition for HexNAc is described in detail in supplemental Fig. S1. The target-decoy option of Mascot was enabled and peptide mass tolerance was set to 10 ppm and fragment mass tolerance to 0.02 Da. Search results were imported into Scaffold 3.3.1. Proteins were required to have at least 99% protein probability and 80% peptide probability (supplemental Table S2). Candidate O-GlcNAc spectra were filtered against false-positive O-GlcNAc peptide-spectrum-matches (PSMs) to retain only O-GlcNAc PSMs with Oscores smaller than 2.3. Candidate O-GlcNAc PSMs were inspected and validated manually (see supplemental Spectra).
A list of known human and murine O-GlcNAc proteins and sites was compiled from recent publications (13, 17–19) as well as from the databases dbOGAP (5) and PhosphositePlus (20). Information on phosphorylated and ubiquitinylated proteins was retrieved from the PhosphositePlus database. Reported N-linked glycosylation sites were extracted from UniProtKB, and subcellular localization information from Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA).
The Oscore script is available from www.wzw.tum.de/proteomics/content/research/software/; and the peaklist files for all processed data can be downloaded from ProteomeCommons.org Tranche using the following hash key:
ChunHqKHVaLCoocgKoyBjphK1QntOh6ehU0MzuLgwf+FZHjEfAntIyzzY38Rv051iVNoNFNJQHibLYJl4dDRotCm1UAAAAAAAAEpg==(passphrase: sa3sh7mgcf6eolskt57p).
RESULTS AND DISCUSSION
Oscore-based O-GlcNAc Protein Identification Strategy
We recently developed the Oscore as a means to assess the probability of a tandem MS spectrum to represent an O-GlcNAc modified peptide (12). The high specificity of the score is further increased by the high mass accuracy provided by modern mass spectrometers. We therefore reasoned that it may be possible to identify O-GlcNAc modified peptides from large-scale proteomic data and, if so, to assess the overall abundance of the modification. To this end, we downloaded a number of published data sets from public data repositories (supplemental Table S1), which were all acquired on dual pressure linear ion trap Orbitrap hybrid mass spectrometers using HCD fragmentation (21). The first data set comprises the label-free comparison of 11 commonly used cell lines (14); the second data set comprises a comprehensive characterization of the HeLa cancer cell line proteome employing multiple protease digestion (15), and the third data set represents an iTRAQ-based quantitative comparison of the proteome and the phospho-proteome of four human embryonic stem (hES) cell lines and four induced pluripotent stem (iPS) cell lines (16). Together, these data sets constitute 13,897,945 tandem MS spectra.
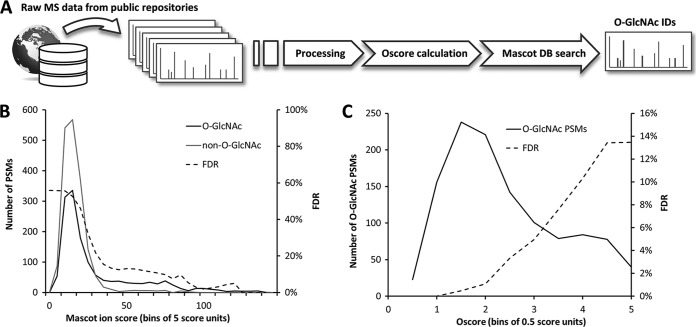
We conceived a straightforward strategy for data re-analysis, which combines standard Mascot database searching and Oscoring of tandem mass spectra for the assessment of potential O-GlcNAc spectra (Fig. 1A). Both algorithms exploit complementary properties of tandem MS spectra. Although the Mascot ion score reflects peptide sequence information, the Oscore assesses tandem MS spectra solely based on the presence of O-GlcNAc diagnostic fragment ions (supplemental Fig. S2). Given the particular fragmentation behavior of O-GlcNAc peptides, the Mascot ion score alone is not able to discriminate accurately between O-GlcNAc and non-O-GlcNAc spectra (Fig. 1B). However, when O-GlcNAc PSMs assigned by Mascot are re-assessed according to their Oscore, it is easily possible to discriminate between O-GlcNAc and non-O-GlcNAc spectra. Low Oscores represent strong O-GlcNAc spectra, high Oscores represent weak or unlikely O-GlcNAc spectra and no Oscore represent the absence of typical O-GlcNAc features. The Oscore-based ranking of O-GlcNAc PSMs then allows filtering the data at the desired target-decoy FDR while maintaining adequate sensitivity (Fig. 1C).
Fig. 1.
O-GlcNAc protein identification strategy. A, Raw LC-MS/MS data is downloaded from public data repositories, tandem mass spectra are processed into peak lists. These are examined for candidate O-GlcNAc information using the Oscore and identified by database searching using Mascot. B, Mascot ion score distribution of candidate O-GlcNAc PSMs. For FDR estimation, PSMs which were assigned to O-GlcNAc-modified sequences, but did not contain O-GlcNAc diagnostic features, were considered as false-positive hits and are indicated as “non-O-GlcNAc” PSMs. C, Oscore distribution of candidate O-GlcNAc PSMs. The FDR is calculated on target and decoy PSMs and spectra with Oscores of <2.3 correspond to an FDR of 2.5%.
O-GlcNAc Sites From HCD Spectra
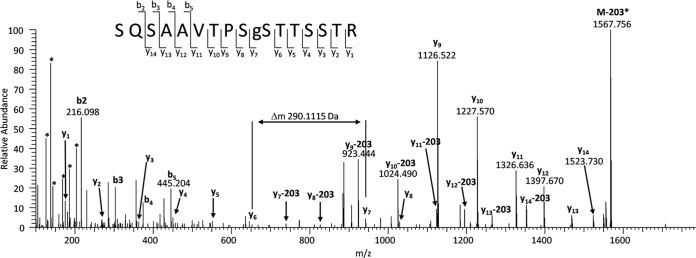
The Oscore-based re-analysis of three comprehensive cell line proteome data sets resulted in the identification of 158 O-GlcNAc peptides containing 194 sites from 628 spectra (Table I). Manual interpretation of the best PSM for every peptide allowed the unambiguous localization of 26 O-linked GlcNAc and 12 N-linked GlcNAc sites (see below). The localization of 13 sites could be narrowed down to three or less residues, and the localization of 140 sites remained ambiguous. An example O-GlcNAc HCD spectrum is depicted in Fig. 2 (see supplemental Spectra for all annotated spectra). The high mass accuracy and the large dynamic range of HCD spectra facilitate not only the identification of the SQSAAVTPSgSTTSSTR peptide from ADRM1, but also support the detection of the PTM via diagnostic fragments and allows the unambiguous localization of the O-GlcNAc site even in the presence of nine alternative sites. Although it has been possible to identify numerous O-GlcNAc sites from HCD spectra, the low stability of the O-glycosidic bond during CID conditions render the localization of O-GlcNAc sites very difficult. Clearly, the fragmentation method of choice for an accurate O-GlcNAC site localization is ETD, which retains the O-GlcNAc site during fragmentation and enables direct site localization. However, stretches of serine and threonine residues around the actual O-GlcNAc site further impede site localization. Only five out of 158 peptides have only a single possible O-GlcNAc site (Ser, Thr), and the average number of potential sites per peptide is 5.6. This is consistent with published O-GlcNAc transferase consensus motifs (5, 17, 19). Interestingly, nonmodified peptides contain only 1.5 possible O-GlcNAc acceptor sites and phospho-peptides (see below) harbor 3.3 possible O-GlcNAc sites, suggesting that O-GlcNAc is more likely to occur on serine/threonine-rich peptides.
Table I. O-GlcNAc protein and peptide identifications from published large-scale proteome studies.
| Project | MS/MS | PSM | Peptides | Sites | Proteins |
|---|---|---|---|---|---|
| Geiger et al. | 5,985,620 | 454 | 104 | 125 | 76 |
| Nagaraj et al. | 4,829,525 | 75 | 36 | 38 | 29 |
| Phanstiel et al. | 1,766,566 | 99 | 41 | 50 | 32 |
| Total | 12,581,711 | 628 | 158a | 194a | 114a |
| Phanstiel et al. (phospho data set) | 1,316,234 | 107 | 28 | 34 | 22 |
| Total + phospho | 13,897,945 | 735 | 174a | 204a | 124a |
a nonredundant peptides, sites, and proteins.
Fig. 2.
Example HCD spectrum of a novel O-GlcNAc site corresponding to the sequence SQSAAVTPSgSTTSSTR of the proteasomal ubiquitin receptor ADRM1. The large dynamic range of the HCD spectrum and the high mass accuracy allows determining the peptide sequence and the localization of the O-GlcNAc site despite the presence of nine alternative modification sites. Diagnostic O-GlcNAc features (i.e. reporter ions and the GlcNAc oxonium loss) are depicted as well (*).
Among the 158 GlcNAc peptides are 12 peptides for which the GlcNAc modification could be localized to N-linked asparagine residues within an NX[ST] consensus motif. In addition, 20 peptides for which the site of modification could not be reliably deduced from tandem mass spectra, harbor N-linked glycosylation sites reported in UniProt (also see supplemental Table S4). Although single N-linked GlcNAc residues are not generally expected to be present on proteins, our result is in accordance with previous findings (18). A possible explanation raised by Chalkley et al. is that these N-linked HexNAc peptides are artifacts formed upon cell lysis by the activity of the cytosolic endo-β-N-acetylglucosaminidase. The enzyme cleaves the β-1,4-glycosidic bond in the N,N′-diactylchitobiose core of high mannose glycopeptides and glycoproteins leaving an N-linked GlcNAc residue. However, these N-GlcNAc peptides, as well as peptides from O-glycans, may also arise from in-source fragmentation of the glycan structure in the high pressure region at the front end of the mass spectrometer.
Identified O-GlcNAc Proteins
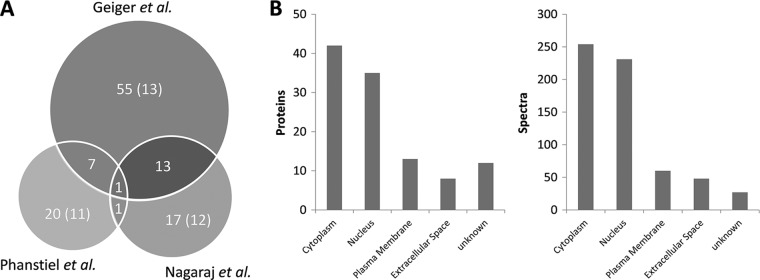
After processing more than 12 million tandem mass spectra, 628 O-GlcNAc spectra corresponding to 158 peptides and 114 candidate O-GlcNAc proteins were identified (supplemental Tables S3–S5). The three re-examined studies contribute common and exclusive protein identifications (Fig. 3A). The highest number of modified proteins originates from the 11 cell line proteomes profiled by Geiger et al. (14). Within that study, the number of identified spectra and proteins varies significantly between cell lines (supplemental Fig. S3) and may reflect cell-type specific differences of protein expression and O-GlcNAcylation. Interestingly, the analysis of the HeLa deep proteome published by Nagaraj et al. (15) also contributed a significant number of exclusive and novel O-GlcNAc proteins, even though the HeLa cell line was also part of the panel analyzed by Geiger et al. (14). A closer inspection of the data revealed that 16 out of the 18 exclusive protein identifications originate from GluC (7 proteins) or LysC digests (nine proteins), underscoring the usefulness of multiple protease digestion for proteomics in general and O-GlcNAc and PTM studies in particular. Interestingly, the only O-GlcNAc protein identified in all studies is the Host cell factor 1, a protein known to be highly O-GlcNAcylated.
Fig. 3.
Number of O-GlcNAc proteins identified in different studies from various cell lines. A, Number of O-GlcNAc proteins and known proteins (in parentheses) from three different data sets. B, Subcellular localization of candidate O-GlcNAc proteins and spectra.
We note that for ten proteins, the GlcNAc site was assigned to an asparagine residue (N-GlcNAc). Moreover, although O-GlcNAc has been reported for proteins of almost all cellular compartments as well as on extracellular proteins (22), we cannot rule out the possibility that several of the identified ER- and Golgi-resident proteins are early synthesis products of O-GalNAc-type glycans. The subcellular localization of candidate O-GlcNAc proteins is depicted in Fig. 3B. For 47 of the identified proteins, the O-GlcNAc modification has been previously reported, while 57 represent novel O-GlcNAc proteins. In addition, for nine of the known O-GlcNAc proteins, we report direct evidence for the modification for the first time. Collectively, this data shows that O-GlcNAc modified peptides can be identified from large-scale proteomic data, which makes a point in favor sharing proteomic data with the scientific community.
O-GlcNAc is Less Abundant Than Phosphorylation
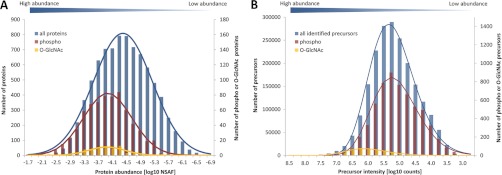
The modified and unmodified peptides identified in the present re-analysis of proteomic data enabled us to perform a crude estimation of the frequency and abundance of these modifications on the most abundant modified proteins. From the Geiger et al. data (11 cell lines), we identified 2,023,960 tandem mass spectra, 6124 of which correspond to phosphorylated peptides and 454 matched to O-GlcNAc peptides. Hence, the frequency of phospho-spectra is 1 in 334 and the frequency of O-GlcNAc spectra is 1 in 4500 indicating that O-GlcNAc is numerically ∼13-fold less frequent than phosphorylation. We are aware that this estimation rests upon the assumption that O-GlcNAcylated peptides are, by and large, identified at the same rate as phosphopeptides from HCD data, which may not necessarily be the case (although probably approximately true). We also expressed the protein abundance for all 11 cell lines as the logarithmic normalized spectral abundance factor (23) (NSAF, Fig. 4A). As expected, the detected modified proteins are mostly among the medium to high abundant proteins. Interestingly, but somewhat unexpectedly, the NSAF distributions of O-GlcNAc- and phospho-proteins are quite similar. This clearly indicates that the observed O-GlcNAc- and phospho-proteins are, by and large, equally abundant, but that the O-GlcNAc modification is less frequent. Alternatively, we also used the distribution of peptide precursor intensities (Fig. 4B) as a proxy for the abundance of the detected (modified) peptides.
Fig. 4.
Protein abundance data for the eleven cell lines analyzed by Geiger et al. A, Protein abundance distribution (expressed as logarithmic NSAF) of 76 O-GlcNAc- and 736 phospho-proteins. B, Protein abundance distribution (expressed as summed peptide precursor intensity) of 454 O-GlcNAc spectra, 6124 phospho-spectra and >2 million unmodified spectra. Note the secondary y axis for O-GlcNAc- and phospho-identifications.
The data shows that the distributions of phospho-peptides and ordinary peptides are very similar. In contrast, the distribution of O-GlcNAc peptides is massively skewed toward high intensity proteins indicating that many high abundance proteins are also O-GlcNAc modified and that the site occupancy of the detected peptides is likely significantly higher for O-GlcNAc peptides than for phospho-peptides. To test this hypothesis, we estimated the site occupancy of all identified O-GlcNAc and phospho-peptides via the summed precursor intensities for modified and unmodified peptides. By this method, we found an average site occupancy of 0.73 for phospho-peptides and of 0.90 for O-GlcNAc peptides. This difference in site occupancy is supported by the fact that the unmodified peptide counterpart could be identified for 46% of all phospho-peptides, but only for 26% of the O-GlcNAc peptides. We do realize that the above estimates are crude because the assumption that the detection efficiencies of modified and unmodified peptides by the employed methods are not grossly different may not be well justified. Still, we think the data suggests that the O-GlcNAc modification appears to be considerably less frequent than phosphorylation. At the same time, however, the average occupancy of the sites that we detected appears to be rather high indicating that many of the observed (i.e. abundant) O-GlcNAc proteins are stably modified under physiological conditions. This is consistent with recent in vitro data on human O-GlcNAc transferase suggesting that some substrates are constitutively modified (24).
Simultaneous O-GlcNAc/Phospho Occupancy of Proximal Sites
Given the potential interplay of O-GlcNAc and phosphorylation (25), we investigated whether O-GlcNAc peptide identifications are also possible from large-scale phospho-proteome data. To this end, we employed the Oscore-strategy to identify O-GlcNAc sites from the phospho-proteome of hES and iPS cells (16). Overall, we identified 107 spectra corresponding to 28 O-GlcNAc-modified peptides and 34 O-GlcNAc sites on 22 proteins (Table I and supplemental Tables S6–S8). Of these peptides, 67% were doubly modified with one or multiple O-GlcNAc and phosphate moieties. The identification of O-GlcNAc peptides, which are not phosphorylated, is not surprising given that only around 50% of all identified peptides from the phospho-proteome data harbor phosphorylation sites.
According to common notion, the cross-talk between O-GlcNAc and phosphorylation on identical or proximal sites is extensive and usually referred to as being either antagonistic or synergistic (1). Most of the reported cases in the literature show competitive occupancy by O-GlcNAc or phosphate of the same or neighboring residues, and it is argued that the reciprocal exclusion results from either the large size of an O-GlcNAc residue (with an Stokes radius four to fivefold larger than a phosphate moiety) or by the negative charge of the phosphate group or by conformational changes induced by either modification (26). The observation of 23 doubly modified peptides with a median length of 24 residues suggest that both modifications cannot only occur simultaneously on distal sites of the same protein, but that also proximal residues can be occupied by O-GlcNAc and phosphate simultaneously. A striking example is given by the peptide SEApSg(SS)PPVVTSSSHSR of the SOX2 transcription factor. Here, the tandem mass spectrum (supplemental Spectrum #208) localizes the phosphorylation at S4 and the O-GlcNAc modification at either S5 or S6, indicating that both modifications can, at the same time, occur even on (almost) adjacent sites.
Functional Roles of Novel Human O-GlcNAc Proteins
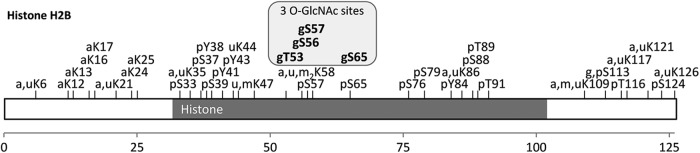
Numerous of the novel O-GlcNAc proteins (supplemental Table S9) highlight the emerging role of O-GlcNAc as part of the histone code and in the regulation of histone modifications (27, 1). Among the novel proteins identified, histone H2B is a particularly interesting case as we identified three O-GlcNAc sites that are in close proximity to (di-)methylation, ubiquitination, and phosphorylation sites (Fig. 5). O-GlcNAcylation of S113 has, very recently, been reported to facilitate monoubiquitination at K121. Interestingly, here, the O-GlcNAc moiety seems to act as primer for a histone H2B ubiquitin ligase, and monoubiquitination presumably results in transcriptional activation (28). Although the precise roles of the novel O-GlcNAc sites between T53 and S65 on H2B are unknown, one might speculate about further relationships of O-GlcNAc and ubiquitination.
Fig. 5.
Graphical representation of post-translational modifications along the sequence of histone H2B (a, acetylation; g, O-GlcNAc; m, methylation; p, phosphorylation; u, ubiquitination).
Further noteworthy examples for O-GlcNAc modified proteins include the transcription factors SOX-2 and Sal-like protein 4 (SALL4) as well as STAT3, which have been discovered in the hES and iPS cell proteomes (16). Although SALL4 and SOX-2 have been previously reported to be O-GlcNAc-modified in mouse (19), no site has been determined yet for STAT3 (29). The STAT3 O-GlcNAc site could be localized between T714 and T721 (supplemental Spectrum #193). For SALL4, three novel O-GlcNAc sites have been found: one site between S480 and T501, one site at T608, S609, or S612; and one additional site between T608 and S628 (supplemental Spectra: #203, 149, and 156, respectively). All three proteins are involved in maintaining stem cell identity and governing stem cell-renewal (30, 31) by up-regulating pluripotency genes and down-regulating developmental genes. The discovery of novel O-GlcNAc-modified stem cell transcription factors is in line with the finding that O-GlcNAc transferase might regulate transcription during early development via the modification of proteins required to maintain the embryonic stem cell transcriptional repertoire (19).
CONCLUSIONS
We revisited >13 million tandem mass spectra from four large-scale human proteome and phosphoproteome data sets and identified several hundred O-GlcNAc modified peptides, most of which have not been reported before. This shows that at least some O-GlcNAc modified proteins are abundant enough so that they can be identified without biochemical enrichment. The current study also makes a point in favor of sharing data between laboratories because one can expect to be able to discover many hundreds more modified peptides from the vast quantities of published proteomic data. Interestingly, the number of O-GlcNAc peptides and sites reported in this work is larger than those of most other O-GlcNAc studies which all use some form of biochemical enrichment. This may indicate that the development of such enrichment methods is still in its infancy. The fact that the number and abundance of O-GlcNAc peptides we identify “in passing” as it were, is much smaller than those of phosphorylated peptides further highlights the need for the development of better biochemical tools.
Supplementary Material
Acknowledgments
We thank the originators of the mass spectrometry data used in this study for making this data available to the community.
Footnotes
* We gratefully acknowledge the Studienstiftung des deutschen Volkes e. V. for a PhD fellowship to HH, and the support of the Faculty Graduate Center Weihenstephan of TUM Graduate School at the Technische Universität München, Germany.
 This article contains supplemental Figs. S1 to S3, Spectra, and Tables S1 to S9.
This article contains supplemental Figs. S1 to S3, Spectra, and Tables S1 to S9.
1 The abbreviations used are:
- O-GlcNAc
- O-linked N-acetylglucosamine
- HCD
- higher collision energy dissociation
- HexNAc
- N-acetylgalactosamin, N-acetylglucosamin
- NSAF
- normalized spectral abundance factor
- PSM
- peptide-spectrum-match.
REFERENCES
- 1. Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hu P., Shimoji S., Hart G. W. (2010) Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 584, 2526–2538 [DOI] [PubMed] [Google Scholar]
- 3. Slawson C., Hart G. W. (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., Cho J. W. (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 5. Wang J., Torii M., Liu H., Hart G. W., Hu Z. Z. (2011) dbOGAP - an integrated bioinformatics resource for protein O-GlcNAcylation. BMC Bioinformatics 12, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huddleston M. J., Bean M. F., Carr S. A. (1993) Collisional fragmentation of glycopeptides by electrospray ionization LC/MS and LC/MS/MS: methods for selective detection of glycopeptides in protein digests. Anal. Chem. 65, 877–884 [DOI] [PubMed] [Google Scholar]
- 7. Chalkley R. J., Burlingame A. L. (2001) Identification of GlcNAcylation sites of peptides and alpha-crystallin using Q-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 12, 1106–1113 [DOI] [PubMed] [Google Scholar]
- 8. Mirgorodskaya E., Roepstorff P., Zubarev R. A. (1999) Localization of O-glycosylation sites in peptides by electron capture dissociation in a Fourier transform mass spectrometer. Anal. Chem. 71, 4431–4436 [DOI] [PubMed] [Google Scholar]
- 9. Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., Burlingame A. L. (2006) O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 10. Haynes P. A., Aebersold R. (2000) Simultaneous detection and identification of O-GlcNAc-modified glycoproteins using liquid chromatography-tandem mass spectrometry. Anal. Chem. 72, 5402–5410 [DOI] [PubMed] [Google Scholar]
- 11. Chalkley R. J., Burlingame A. L. (2003) Identification of novel sites of O-N-acetylglucosamine modification of serum response factor using quadrupole time-of-flight mass spectrometry. Mol. Cell. Proteomics 2, 182–190 [DOI] [PubMed] [Google Scholar]
- 12. Hahne H., Kuster B. (2011) A novel two-stage tandem mass spectrometry approach and scoring scheme for the identification of O-GlcNAc modified peptides. J. Am. Soc. Mass Spectrom. 22, 931–942 [DOI] [PubMed] [Google Scholar]
- 13. Zhao P., Viner R., Teo C. F., Boons G. J., Horn D., Wells L. (2011) Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. J. Proteome Res. 10, 4088–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geiger T., Wehner A., Schaab C., Cox J., Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phanstiel D. H., Brumbaugh J., Wenger C. D., Tian S., Probasco M. D., Bailey D. J., Swaney D. L., Tervo M. A., Bolin J. M., Ruotti V., Stewart R., Thomson J. A., Coon J. J. (2011) Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods 8, 821–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Z., Udeshi N. D., Slawson C., Compton P. D., Sakabe K., Cheung W. D., Shabanowitz J., Hunt D. F., Hart G. W. (2010) Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 3, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chalkley R. J., Thalhammer A., Schoepfer R., Burlingame A. L. (2009) Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Myers S. A., Panning B., Burlingame A. L. (2011) Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 9490–9495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hornbeck P. V., Kornhauser J. M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsuura A., Ito M., Sakaidani Y., Kondo T., Murakami K., Furukawa K., Nadano D., Matsuda T., Okajima T. (2008) O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. J. Biol. Chem. 283, 35486–35495 [DOI] [PubMed] [Google Scholar]
- 23. Zybailov B., Mosley A. L., Sardiu M. E., Coleman M. K., Florens L., Washburn M. P. (2006) Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 [DOI] [PubMed] [Google Scholar]
- 24. Shen D. L., Gloster T. M., Yuzwa S. A., Vocadlo D. J. (2012) Insights into O-GlcNAc processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J. Biol. Chem. 287, 15395–15408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hart G. W., Housley M. P., Slawson C. (2007) Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 26. Chen Y. X., Du J. T., Zhou L. X., Liu X. H., Zhao Y. F., Nakanishi H., Li Y. M. (2006) Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N terminus of murine estrogen receptor beta. Chem. Biol. 13, 937–944 [DOI] [PubMed] [Google Scholar]
- 27. Hanover J. A. (2010) Epigenetics gets sweeter: O-GlcNAc joins the “histone code”. Chem. Biol. 17, 1272–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fujiki R., Hashiba W., Sekine H., Yokoyama A., Chikanishi T., Ito S., Imai Y., Kim J., He H. H., Igarashi K., Kanno J., Ohtake F., Kitagawa H., Roeder R. G., Brown M., Kato S. (2011) GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480, 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whelan S. A., Lane M. D., Hart G. W. (2008) Regulation of the O-linked beta-N-acetylglucosamine transferase by insulin signaling. J. Biol. Chem. 283, 21411–21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang J., Tam W. L., Tong G. Q., Wu Q., Chan H. Y., Soh B. S., Lou Y., Yang J., Ma Y., Chai L., Ng H. H., Lufkin T., Robson P., Lim B. (2006) Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 8, 1114–1123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.