Abstract
Plant growth adjustment during water deficit is a crucial adaptive response. The rapid fine-tuned control achieved at the post-translational level is believed to be of considerable importance for regulating early changes in plant growth reprogramming. Aiming at a better understanding of early responses to contrasting plant water statuses, we carried out a survey of the protein phosphorylation events in the growing zone of maize leaves upon a range of water regimes. In this study, the impact of mild and severe water deficits were evaluated in comparison with constant optimal watering and with recovery periods lasting 5, 10, 20, 30, 45, and 60 min. Using four biological replicates per treatment and a robust quantitative phosphoproteomic methodology based on stable-isotope labeling, we identified 3664 unique phosphorylation sites on 2496 proteins. The abundance of nearly 1250 phosphorylated peptides was reproducibly quantified and profiled with high confidence among treatments. A total of 138 phosphopeptides displayed highly significant changes according to water regimes and enabled to identify specific patterns of response to changing plant water statuses. Further quantification of protein amounts emphasized that most phosphorylation changes did not reflect protein abundance variation. During water deficit and recovery, extensive changes in phosphorylation status occurred in critical regulators directly or indirectly involved in plant growth and development. These included proteins influencing epigenetic control, gene expression, cell cycle-dependent processes and phytohormone-mediated responses. Some of the changes depended on stress intensity whereas others depended on rehydration duration, including rapid recoveries that occurred as early as 5 or 10 mins after rewatering. By combining a physiological approach and a quantitative phosphoproteomic analysis, this work provides new insights into the in vivo early phosphorylation events triggered by rapid changes in plant water status, and their possible involvement in plant growth-related processes.
The maintenance of crop productivity upon reduced water resources will be a challenging issue to ensure food production under future climate conditions. Water deficit induces a wide range of responses at the whole plant, cellular and molecular levels (1). Among adjustments triggered by water deficit, leaf growth is one of the earliest and most sensitive processes that occurs independently of photosynthetic rates and plant carbon status (2, 3), which tends to show that growth reduction is a crucial adaptive response (4). A reduced leaf growth limits water losses by transpiration via a decreased leaf area but also limits the potential light interception. This reduces biomass accumulation and further, leads to substantial yield losses. Thus, leaf growth reduction has both a positive effect on plant stress avoidance and a negative effect on final crop yields. The optimization balance between the control of water losses and the potential carbon assimilation can differ according to climatic scenarios and increasing knowledge on its determinism may allow future progress in the maintenance of crop production on changing plant water statuses. In maize, leaf growth is restricted to the base of the leaf in a 6 to 8 cm-long fragment, named the growing zone (5, 6). This growing zone includes tissues in which both cell division and cell expansion happen, partly overlapping in time and space (6). Cells are produced near the leaf base in the meristematic region, pushing forward the expanding cells into the distal regions of the leaf growing zone. Upon water deficit, leaf growth reprogramming is achieved by modulating both cell division and cell expansion (7–9). Previous analyses have already reported that altered plant growth occurs concomitantly with changes in cyclin-dependent kinase activity, in cell wall expansion genes (such as expansin genes), in turgor or in cell wall properties (5, 10–14). In a recent paper, pioneer efforts in identifying the earliest molecular events controlling cell division upon osmotic stress have evidenced that ethylene signaling acts upstream on the cell cycle arrest and on cyclin-dependent kinase A activity, independently to a transcriptional control (15). However, the molecular mechanisms involved in the rapid plant growth adjustment in water stress responses is still fragmentary.
Phosphorylation of proteins is of considerable importance for regulating plant growth (16). As a rapid and transient post-translational modification, protein phosphorylation achieves a fine-tuned regulation of protein function in a wide array of cellular processes during development or in response to environmental cues, from signaling cascades to gene expression (17). Therefore, early protein phosphorylation and dephosphorylation events could play a pivotal role in the rapid growth adjustment occurring in plants facing water limitation. Studying phosphoproteome dynamics has already proven to be a useful strategy to decipher sucrose-induced responses in cell cultures (18) or ABA1-dependent changes in planta (19) for instance, but early responses to changing plant water status at the level of a growing tissue have not yet been well documented. To our knowledge, data dealing with water deficit-induced changes in protein phosphorylation have been mainly obtained from long-term stressed plants (20, 21), and no time-course analysis of rapid in vivo changes has been explored.
To investigate relations between early phosphorylation events and growth, the present exploratory study was carried out in planta on the phosphoproteome of the leaf growing zone of maize plants submitted to contrasting water statuses. Quantitative changes in phosphorylation status were assessed in two scenarios of water stress, an early and mild water deficit as well as a more severe one (MWD and SWD, respectively). Because water deficit is usually fluctuating in the fields with dehydration and rewatering events, we also analyzed a sequence of rehydration that causes rapid recovery of leaf elongation rate in maize. In maize, the full recovery of leaf growth occurs 40 min after rehydration, which is even faster than the recovery of leaf water potential (22). In addition, rewatering is instantaneous and enables to accurately define the response time to the changes in plant water status.
EXPERIMENTAL PROCEDURES
Plant Growth and Monitoring of Dehydration and Rehydration Conditions
Seeds of the maize (Zea mays L.) inbred line B73 were grown in 1-L pots filled with a soil:compost (2:3, v/v) mixture. Plants were grown in a growth cabinet at a light intensity of 450 μmol/m2/s (16 h photoperiod, 25 °C day and 18 °C night), with 50% air relative humidity. The design involved four randomized complete blocks (one biological replicate of each treatment per block), each surrounded with two bordering rows of supplementary plants. During the first hour of light, pots were weighed and watered to maintain a soil humidity of 60% (optimal irrigation) and the length of the fifth leaf was measured from pot edge to tip. Experimental treatment began when the fifth leaf reached a length of 30 cm (day 0), about 3 weeks after sowing. Control plants were maintained at a constant soil water status and two water deficits were imposed to other plants by withholding water, namely mild (MWD) and severe water deficit (SWD). MWD plants were sampled on day 1 whereas SWD plants were collected when the growth of the sixth leaf reached less than 1 cm/d, on days 4–5. A batch of SWD plants were rewatered to soil retention capacity and sampled 5, 10, 20, 30, 45, and 60 min after rewatering (RH5, RH10, RH20, RH30, RH45, and RH60, respectively). Control plants of the same ontogenic stage as stressed plants, were sampled on day 1 (CTRL1) and on days 4–5 when the fourth leaf ceased to grow (CTRL2). Five-centimeter-long fragments (cm 1 to 6 from leaf insertion point) of the leaf growing zone were harvested between the sixth and the seventh hour of light. Four biological replicates were collected for each treatment. Samples were immediately frozen in liquid nitrogen and stored at −80 °C. For each biological replicate, midday leaf water potential (Ψmd, MPa) was measured with a Scholander-type pressure chamber (PMS-670; PMS Instrument) on the fourth leaf (the last ligulated leaf) at the time of sampling.
Protein Extraction and In-solution Digestion
Leaf fragments were finely ground in a ball crusher (TissuLyser, Qiagen) cooled with liquid nitrogen. For each collected sample, a denaturing protein extraction using a trichloroacetic acid/acetone procedure was achieved as described in Méchin et al. (23), except that 40 μl of resolubilization solution [6 m urea, 2 m thiourea, 2% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (m/v), 30 mm Tris-HCl pH 7,8] was used to resuspend 1 mg of pellet. Protein content was determined using the 2-D Quant-kit (GE Healthcare), with bovine serum albumin (BSA) as standard. For each biological replicate, 1.5 mg of proteins were collected and reduced with DTT at a final concentration of 10 mm. Samples were subsequently alkylated with iodoacetamide at a final concentration of 40 mm. Both steps were performed during 45 min. The samples were diluted to < 2 m urea by adding 50 mm ammonium bicarbonate. Protein digestion (Trypsin Gold; Promega, Madison, WI) was performed at an enzyme/substrate ratio of 1:20 (w:w) by overnight incubation at 37 °C, and stopped by adding 1% formic acid (v/v).
Stable Isotope Dimethyl Labeling by Reductive Amination of Tryptic Peptides
Tryptic peptides were dried in vacuo and resuspended in 1 ml of 5% formic acid (v/v). Stable isotope dimethyl labeling was performed according to the on-column procedure described by Boersema et al. (24) using formaldehyde-H2 (light labeling) or formaldehyde-D2 (heavy labeling). Each sample was loaded on a separate SepPak C18 cartridge column (3 ml; Waters) and washed with 0.6% acetic acid (v/v). SepPak columns were flushed ten times with 1 ml of the respective labeling reagent (50 mm sodium phosphate buffer pH 7.5, 30 mm NaBHCN and 0.2% CH2O or CD2O (v/v) for light or heavy labeling, respectively). Labeled samples were eluted with 500 μl of 0.6% acetic acid (v/v) and 80% acetonitrile (ACN) (v/v). Biological replicates were labeled with the light isotope whereas an internal standard was labeled with the heavy isotope. The internal standard was designed by pooling equivalent amounts of peptides from each treatment. All heavy dimethyl-labeled peptides were homogenized before being mixed with the light dimethyl-labeled peptides in a 1:1 abundance ratio.
Peptide Fractionation Using Strong Cation Exchange Chromatography (SCX)
Prior to SCX, the dimethyl-labeled peptides were dried by vacuum centrifugation and subsequently reconstituted in 500 μl of solvent A (30% ACN (v/v), 5% formic acid (v/v), pH 2.5). SCX was performed at 200 μl/min using Zorbax BioSCX-Series II columns (0.8-mm inner diameter × 50-mm length; 3.5 μm particle size) and a Famos autosampler (LC Packings). After sample loading, the first 17 min were run isocratically at 100% solvent A, followed by an increasing pH gradient using solvent B (30% ACN (v/v), 5% formic acid (v/v), 540 mm ammonium formate, pH 4.7). SCX fractions were automatically collected using an on-line Probot system (LC Packings).
Selective Enrichment of Phosphopeptides Using Immobilized Metal Ion Affinity Chromatography (IMAC)
SCX fractions were dried and resuspended in 300 μl of solvent C (250 mm acetic acid, 30% ACN (v/v)). Peptides were gently mixed with 80 μl of Phos-Select iron affinity gel (Sigma-Aldrich) and incubated for 1.5 h using a tube rotator, as described by Nühse et al. (25). The mixture was transferred into SigmaPrep spin columns (Sigma-Aldrich) and the flow-through fractions containing the nonphosphorylated peptides were collected. Iron affinity gel with bound phosphopeptides was rinsed twice with 200 μl of solvent C, then once with 200 μl of double distilled water. The elution of bound phosphopeptides was achieved with 100 μl of solvent D (400 mm NH4OH, 30% ACN) by centrifugation at 8200 g. Flow-through fractions and eluted phosphopeptides were dried and kept at −20 °C until LC-MS/MS analysis.
LC-MS/MS Analysis
On-line liquid chromatography was performed on a NanoLC-Ultra system (Eksigent). A 4 μl sample was loaded at 7.5 μl/min on a precolumn cartridge (stationary phase: C18 PepMap 100, particles of 5 μm; column: 100 μm i.d., 1 cm length; Dionex) and desalted with 0.1% formic acid in water. After 3 min, the precolumn cartridge was connected to the separating PepMap C18 column (stationary phase: C18 PepMap 100, particles of 3 μm; column: 75 μm i.d., 150 mm length; Dionex). Buffers were 0.1% formic acid in water (E) and 0.1% formic acid in ACN (F). Peptide separation was achieved using a linear gradient from 5 to 30% F at 300 nl/min.
Eluted peptides were analyzed with a LTQ XL ion trap (Thermo Electron) using a nano-electrospray interface. Ionization (1.5 kV ionization potential) was performed with liquid junction and a noncoated capillary probe (10 μm i.d.; New Objective). For phosphopeptide analysis, peptide ions were analyzed using Xcalibur 2.07 with the following data-dependent acquisition steps: (1) a full MS scan was recorded in enhanced scan rate mode for higher resolution and mass accuracy (mass-to-charge ratio (m/z) 300 to 1400, profile mode), (2) an MS/MS spectrum was acquired on the two highest-intensity species detected in the preceding MS scan (qz = 0.25, activation time = 30 ms, and collision energy = 35%; profile mode), and (3) an MS3 scan was triggered when a neutral loss of 98 or 80 Da was detected in the preceding MS/MS scan. Dynamic exclusion was set to 30 s. Non-phosphopeptides were analyzed using Xcalibur 2.07 with the following data-dependent acquisition steps: (1) a full MS scan (mass-to-charge ratio (m/z) 300 to 1400, profile mode) and (2) MS/MS (qz = 0.25, activation time = 30 ms, and collision energy = 35%; profile mode). Steps 2 was repeated for the three major ions detected in step 1. Dynamic exclusion was set to 30 s.
Identification of Peptides and Phosphorylation Sites
Database searches were performed using X!Tandem (http://www.thegpm.org/TANDEM/; 2010.01.01.4). Enzymatic cleavage was described to be because of trypsin digestion with one possible miscleavage. Cys carboxyamidomethylation and, light and heavy dimethylation of peptide N termini and lysine residues were set as static modifications whereas Met oxidation and phosphorylation of tyrosine, serine and threonine residues were set as variable modifications. Precursor mass and fragment mass tolerance were 2.0 and 0.5 Da, respectively. Identifications were performed using the MaizeSequence genome database (Release 4a.53, 53764 entries, October 2009, http://maizesequence.org) supplemented by the maize UniProt database (Release 15.11, 43694 entries, December 2009, http://www.uniprot.org) and by a contaminant database (trypsin, keratins, etc). Identified proteins were filtered and grouped using the X! Tandem pipeline v3.1.2, a bioinformatic tool developed by the PAPPSO platform (http://pappso.inra.fr/bioinfo/xtandempipeline/). Data filtering was achieved according to a peptide E value smaller than 0.01. The false discovery rate (FDR) at the peptide level was assessed from searches against a decoy database (using the reversed amino acid sequence for each protein). All peptide sequences and phosphorylation sites showing a significant water regime effect were confirmed by manual inspection of the raw data to verify the peptide sequence and the phosphorylation site assignment.
Identified MaizeSequence sequences and UniProt maize sequences matching unknown proteins were searched against the UniProtKB Plant database using the BLAST tool (http://www.uniprot.org/). Proteins were classified into functional groups according to the UniProtKB database, the DBGET system (http://www.genome.ad.jp/dbget/) or based on information retrieved from already published works.
Quantification of Peptides and Phosphorylation Sites
Relative quantification of nonphosphorylated peptides and phosphopeptides was performed using the MassChroQ software (26) by extracting ion chromatograms (XICs) of all identified peptides within a 0.3 Th window and by integrating the area of the XIC peak at their corresponding retention time. LC-MS/MS data alignment was performed using the “MS/MS method” that uses common MS/MS identifications as landmarks to evaluate retention time deviations along the chromatographic profile. Alignments were performed within groups of LC-MS/MS runs originating from similar SCX fractions. The following parameters were used: ms2 tendency halfwindow = 10 and ms2 smoothing halfwindow = 15. For each peptide detected in the heavy and light form in a single LC-MS/MS run, a light to heavy ratio was computed. To compensate for possible global variations between LC-MS/MS runs, a normalization step was performed by dividing ratios by their median value in each LC-MS/MS run. Only nonphosphopeptides were used for quantitation of protein amounts. Subsequent statistical analyses were performed on log2-transformed normalized data.
Data Availability
Raw MS output files, identified phosphopeptides and their corresponding spectra were uploaded to the ProticDB database (27). The data can be directly visualized or downloaded from http://moulon.inra.fr/protic/phospho. Tandem mass spectral data are also available via the Proteomics Identifications database (PRIDE) at http://www.ebi.ac.uk/pride/under the accession numbers 24901 to 26498. A detailed list of all peptides identified in all LC-MS/MS runs is given in supplemental Table S1, representative spectra of each phosphopeptide displaying significant water regime effect are shown in supplemental Fig. S1, and relative quantification data are available in supplemental Table S2.
Statistical Analysis
Results were analyzed with the R programming language v2.11.1 (R Development Core Team, 2010). Significance of water regime-dependent changes in phosphopeptide abundance was tested using a one-way ANOVA with the following model: Yik = μ + Ti + εik; in which Yik refers to individual values, μ is the general mean, Ti is the effect of treatment i (i = 10) and εik is the residual (k = 4). The R package QVALUE (version 1.1) was used to calculate q-values from the resulting p values assessed from the ANOVA. A q-value (28) lower than 0.05 was defined in order to control the positive false-discovery rate at a 5% level. Using such requirements, a p value < 0.01 was considered to be significant. A Self Organizing Tree Algorithm (SOTA) clustering was performed on phosphopeptides showing significant changes using the Multi Experiment Viewer (MEV) software (http://www.tm4.org/mev/). Clustering was performed from Z score transformed values, with the Pearson correlation as dissimilarity criteria, 10 cycles and a p value of 0.05 for determining the variability division criteria. The significance of recovery was assessed from the linear regression of phosphopeptide abundance on the duration of rehydration, including SWD to RH60. The relationship between protein phosphorylation changes and midday leaf water potential was evaluated by rank correlations (Kendall's tau coefficient, τ) performed by computing the average values observed in CTRL2 and in each time point of the rehydration kinetics, ranging from SWD to RH60. Changes in protein abundance were evaluated in response to SWD and RH60, in comparison to CTRL2 and SWD, respectively. Both effects were tested using a one-way ANOVA with a model similar as above.
RESULTS
Plant Growth and Physiological Response to Changes in Plant Water Status
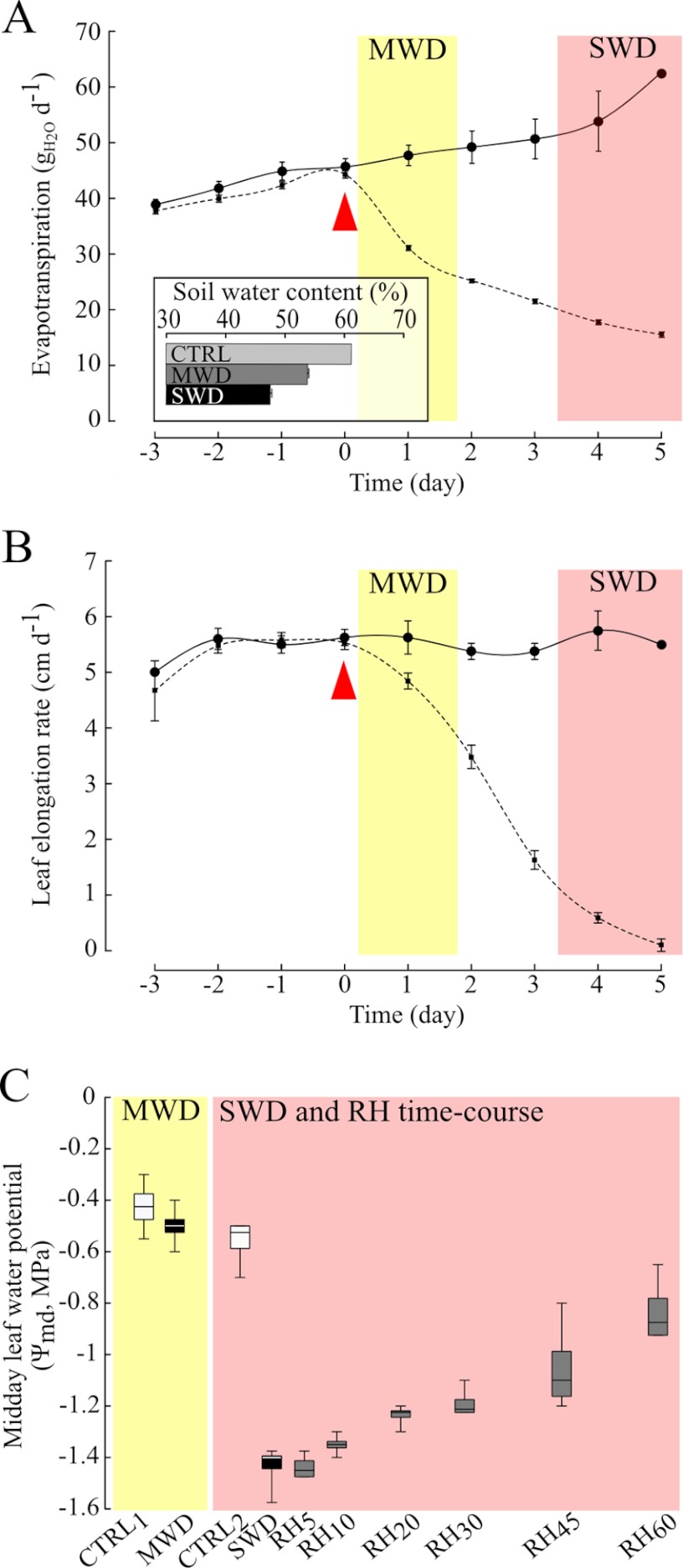
In our experimental design, plants were submitted to a range of water regimes including a mild and a severe water deficit (MWD and SWD), and rehydration periods lasting 5, 10, 20, 30, 45, and 60 min starting from SWD (RH5, RH10, RH20, RH30, RH45, and RH60, respectively). Each water regime led to homogeneous phenotypes. Under optimal irrigation, the evapotranspiration increased along with plant growth, whereas it declined relative to control plants, by 35% during MWD and by about 70% during SWD (Fig. 1A). Compared with control conditions, water deficit led to a 12% and a 22% decrease of soil water content in MWD and SWD, respectively (Fig. 1A inset). The growth decrease of the 5th leaf was initiated during MWD and a 90% growth drop was observed as a result of SWD (Fig. 1B). Although MWD induced changes in evapotranspiration, in soil water status and in plant growth, it was not sufficient to affect midday leaf water potential (Ψmd, Fig. 1C), which suggested that plants controlled water losses via stomatal regulation (29). On the opposite, SWD resulted in an extensive decrease in Ψmd by nearly 1 MPa. Rehydration events induced a significant partial recovery of Ψmd as early as 20 min after rewatering (Fig. 1C), emphasizing that water transport had recovered in plants at that time.
Fig. 1.
Evapotranspiration, leaf growth, and plant water status characterization. A, the evapotranspiration (gH2O/d) is shown for control (black line, n = 8) and stressed (broken line, n = 32) plants. Mean values ± S.E. are shown. The red arrow indicates the beginning of water shortage in stressed plants. The inset shows the soil water content assessed in control (CTRL, n = 8), mild water deficit (MWD, n = 4), and severe water deficit (SWD, n = 28) treatments. B, the elongation rate of the fifth leaf (cm/d) is depicted for control (black line, n = 8) and stressed (broken line, n = 32) plants. Mean values ± S.E. are shown. C, the midday leaf water potential of plants (Ψmd, MPa) was measured concomitantly to the samplings of the leaf growing zone. Each box represents the quartile below (Q1) and above (Q3) the median value. Vertical bars show minimum and maximum values. Box plots are shown for mild water deficit (MWD), severe water deficit (SWD), and for the rehydration time course (RH5 to RH60), in comparison to control plants (CTRL1 and CTRL2) at the same ontogenic development.
Assessing the Dynamics of Protein Phosphorylation Status in the Growing Zone of Maize Leaves
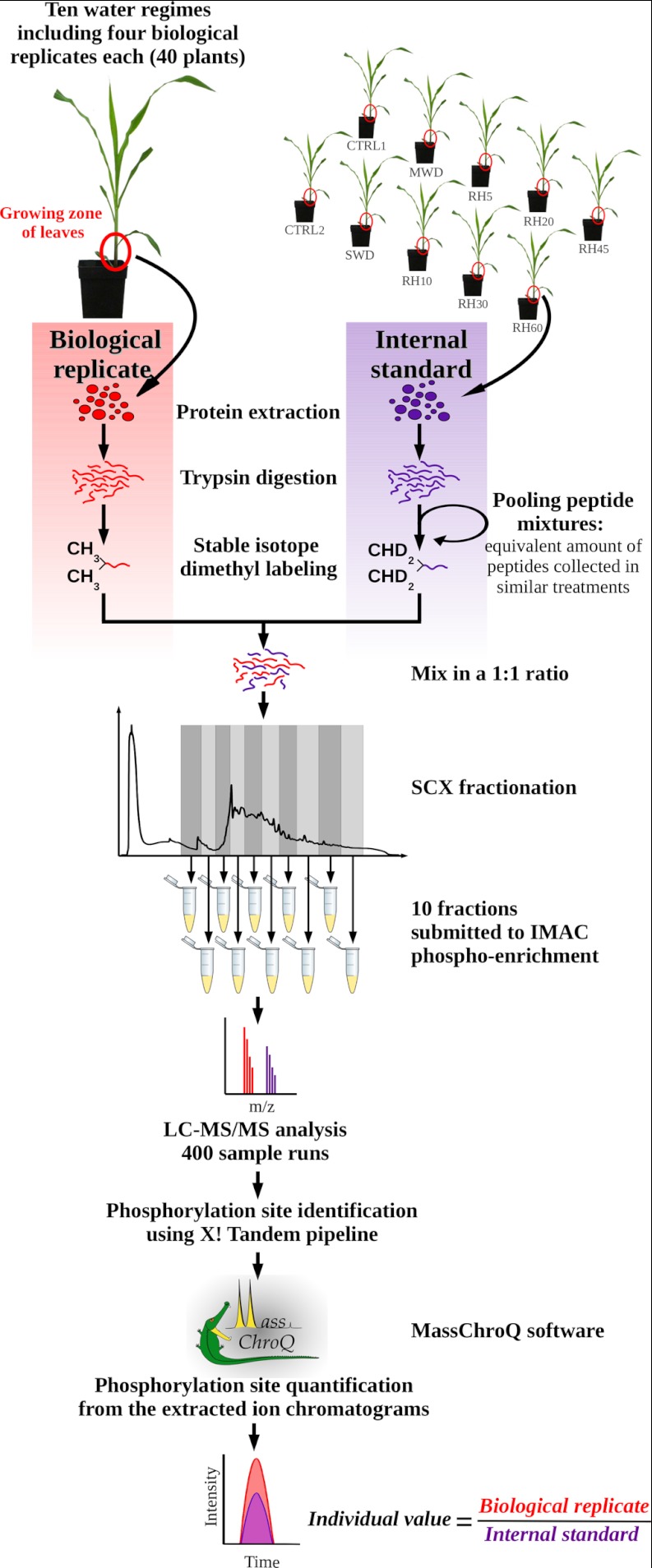
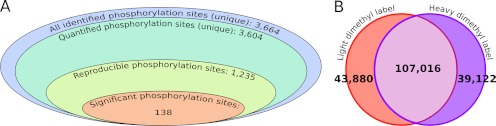
To assess a comprehensive scheme of the protein phosphorylation dynamics in the growing zone of maize leaves, we designed a quantitative phosphoproteomic approach based on stable isotope incorporation at the peptide level using dimethyl labeling. Stable isotope dimethyl labeling has already proven to be a useful strategy for assessing reciprocal measurements between treatments (30) in a single MS analysis using the mass difference of the dimethyl labels. In this study, an alternative labeling strategy was designed: the same internal standard was systematically mixed with all analyzed samples at a 1:1 ratio. The internal standard was designed by pooling an equivalent amount of peptides from all the analyzed samples (Fig. 2). To control for any isotope effect, the internal standard peptide mixture was labeled with the same heavy isotope composition whereas all analyzed samples were labeled with the light isotope composition (Fig. 2). Thus, samples from all treatments were analyzed exactly the same way, and the use of the internal standard maximized the number of peptides for which quantification was possible on both the heavy and the light isotope labels. After mixing the internal standard with each biological replicate, peptide mixtures were submitted to an SCX prefractionation, yielding ten individual eluates (Fig. 2). Each eluate was submitted to affinity chromatography over IMAC-(Fe3+) for phosphopeptide enrichment. As a whole, 400 LC-MS/MS runs were carried out (ten per sample), which resulted in the identification of 80,217 phosphopeptides at a false discovery rate of < 0.5% (supplemental Table S1). They accounted for 3,664 unique phosphorylation sites (Fig. 3A) belonging to 2,496 different phosphoproteins. Phosphopeptide abundances were then compared through their extracted ion chromatograms (XIC). Here, 3604 nonredundant phosphorylation sites were quantified, accounting for 297,034 individual quantitative values. Considering all the quantified phosphorylation sites, nearly 60% were observed in both light and heavy isotope forms within each LC-MS/MS run (Fig. 3B). According to our strategy, phosphorylation site quantification was achieved by computing their light to heavy ratio in each LC-MS/MS run, which required that both isotope envelopes had been extracted from the total ion current. Before any normalization, the median value of the raw ratios was nearly 1 (supplemental Fig. S2). After normalization, quantitative changes of phosphopeptides were assessed from ratios that were quantified in at least 2/3 of all biological replicates (Fig. 3A). This stringent criterion offered a high confidence for approving the phosphorylation site identification and relative quantification. Furthermore, most of the reported phosphorylation sites were identified from MS/MS spectra recorded in more than one biological replicate and in more than one experimental condition (supplemental Table S1).
Fig. 2.
Experimental work flow used for analyzing the phosphoproteome of the growing zone of maize leaves. Each biological replicate was individually analyzed relative to an internal standard. The internal standard was designed by pooling equivalent amounts of digested peptides from all analyzed plants. Peptide samples were then submitted to prefractionation prior to phosphopeptide enrichment. Phosphorylated peptides were analyzed using LC-MS/MS and relative quantification was achieved by XIC extraction.
Fig. 3.
Results of large-scale phosphorylation site identification and quantification. A, overview of the number of phosphorylation sites throughout the work flow of the quantitative analysis. B, Venn diagram depicting the number of quantifications of all phosphorylated peptides in light- and heavy-dimethyl labeled forms.
Impact of Contrasting Plant Water Statuses on Protein Phosphorylation in the Growing Zone of Maize Leaves
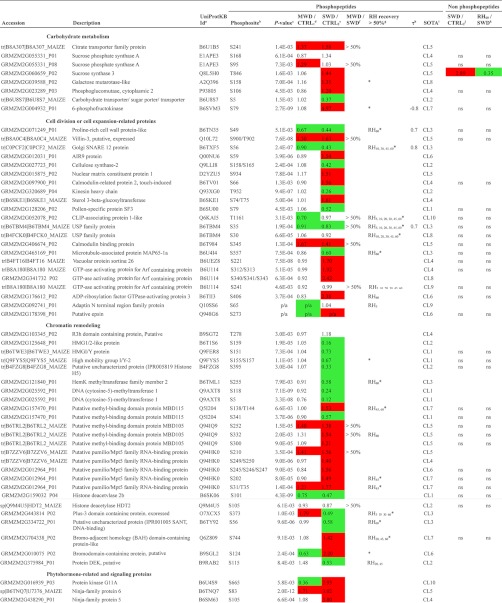
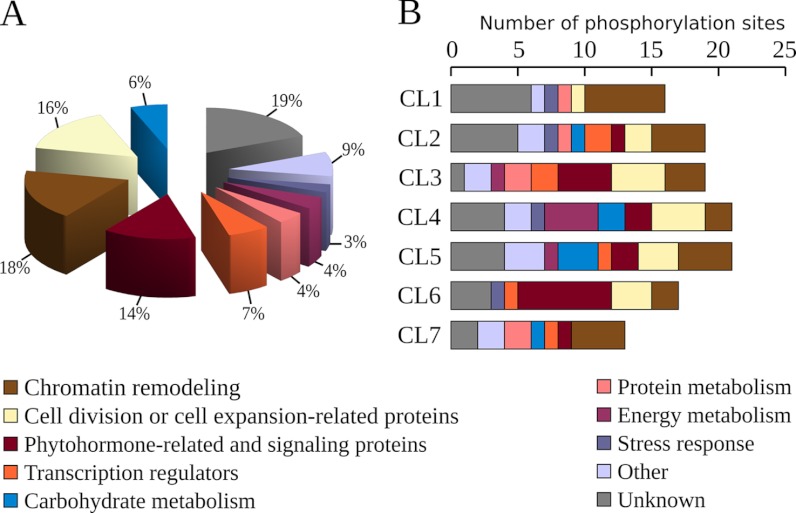
Among 1235 reproducible phosphopeptides (Fig. 3A), 132 displayed highly significant quantitative changes (p < 0.01) in response to water regimes and 6 showed unambiguous qualitative changes (Table I, supplemental Table S3). Qualitative changes concerned three phosphorylation sites that were not detected upon MWD only, another one that was not detected upon SWD only, and two others that were reproducibly detected only during SWD and the rehydration time-course. Overall, quantitative and qualitative changes concerned 124 unique proteins mainly involved in chromatin remodeling (18%), in cell division or cell expansion-related processes (16%), in phytohormone-responses or signaling (14%), in transcription regulation (7%), and in carbohydrate metabolism (6%) (Fig. 4A). Other processes were related to protein metabolism (4%), energy metabolism (4%), stress response (3%), and other miscellaneous processes (9%). Proteins whose function could not be ascertained (“unknown proteins”) accounted for 19% of the significant changes.
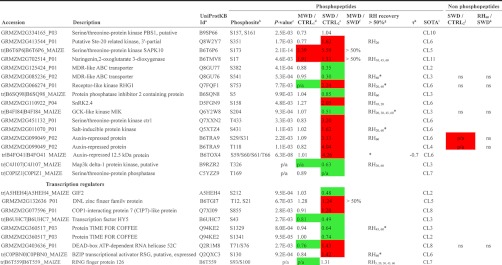
Table I. Partial list of phosphorylation sites significantly affected by changing plant water statuses.
a Refers to UniProtKB Identifier retrieved from sequence similarity searches using the UniProt BLAST tool (http://www.uniprot.org/).
b Multiple sites of phosphorylation are separated by a coma while ambiguous sites are separated by /. All phosphorylation sites reported here were confirmed by manual inspection of the raw data to verify the phosphorylation site assignment obtained using X! Tandem.
c Refers to the p values obtained from the one-way ANOVA, “p/a” refers to qualitative changes.
d Refers to the fold change computed between the light to heavy ratio measured during MWD and the light to heavy ratio measured during CTRL1. Decreasing trends in phosphopeptide abundance are highlighted in green while increasing trends in phosphopeptide abundance are highlighted in red.
e Refers to the fold change computed between the light to heavy ratio measured during SWD and the light to heavy ratio measured during CTRL2. Decreasing trends in phosphopeptide abundance are highlighted in green whereas increasing trends in phosphopeptide abundance are highlighted in red.
f “> 50%” indicates that the ratio between quantitative changes relative to control plants detected on mild water deficit (MWD) and those detected on sever water deficit (SWD) reached more than 0.5.
g Refers to time points of the rehydration kinetics which displayed a >50% recovery compared to the variation of phosphopeptide abundances detected during SWD. “*” indicates when the linear regression of phosphopeptide abundances vs. duration of the rewatering events, was significant (p value < 0.05).
h Kendall τ coefficient refers to the correlation between relative abundance of phosphorylation site and midday leaf water potential. Only significant Kendall τ coefficients are showed (p value < 0.02). Plots are shown in supplemental Fig. S3.
i Refers to the cluster number shown in Fig. 5 and in supplemental Fig. S2.
j Refers to the fold change computed between the light to heavy ratio measured during SWD and the light to heavy ratio measured during CTRL2 at the protein level. Information is provided for each reliably quantified protein, “ns” indicates that no significant change was detected. SWD-induced decreases in protein abundance are highlighted in green while SWD-induced increases in protein abundance are highlighted in red.
k Refers to the fold change computed between the light to heavy ratio measured during RH60 and the light to heavy ratio measured during SWD at the protein level. Information is provided for each reliably quantified protein, “ns” indicates that no significant change was detected. RH60-induced decreases in protein abundance are highlighted in green while RH60-induced increases in protein abundance are highlighted in red.
Fig. 4.
Functional cataloging of proteins displaying changing phosphorylation status in the growing zone of maize leaves upon contrasting water regimes. A, proteins were functionally categorized using the UNIPROT database, the DBGET system or from information retrieved from already published works. B, the number of phosphorylation sites associated with the seven main SOTA clusters are shown according to their functional classification. The cluster identifications (CL1, CL2, CL3, CL4, CL5, CL6, and CL7) refer to Fig. 5 and Table I.
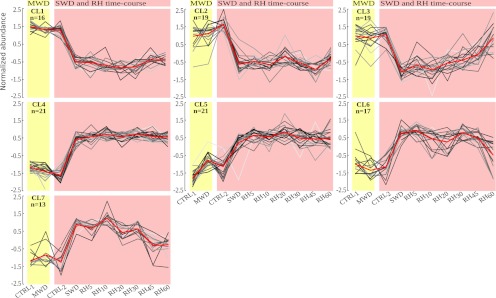
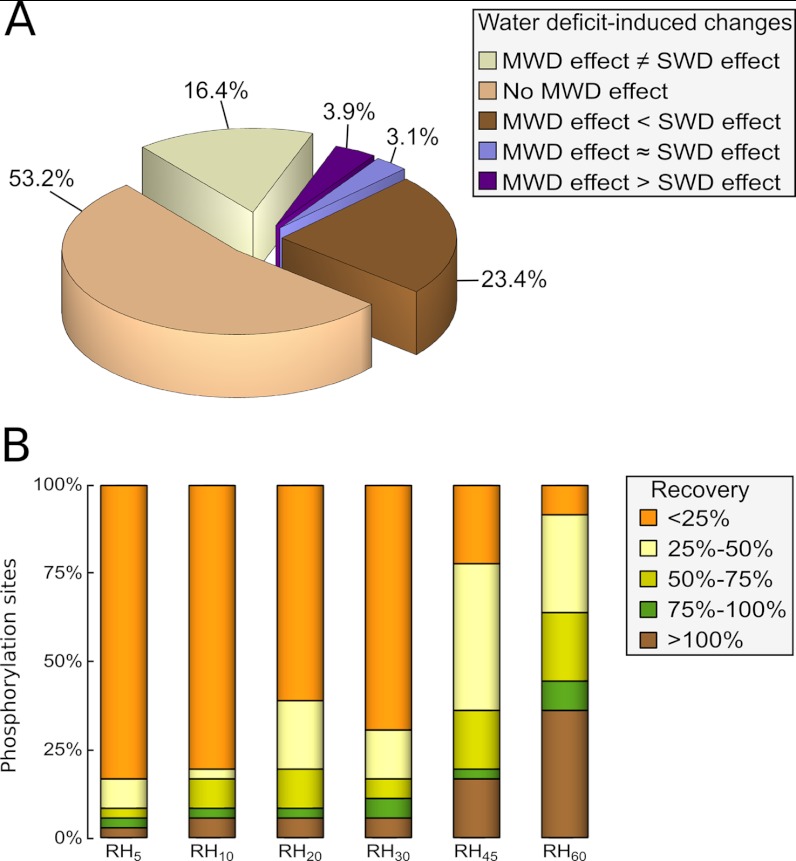
To uncover patterns in the changes in protein phosphorylation during dehydration and rehydration, a self organizing tree algorithm (SOTA) was used on phosphorylated peptides showing significant abundance variations (Fig. 5 and supplemental Fig. S3). Seven main clusters with distinct phosphorylation dynamics, gathered more than 91% of the whole quantitative kinetic profiles (Fig. 5). Regardless of the whole cluster patterns, the largest variations of phosphorylation status were observed in response to SWD, either through decreasing or increasing abundances. Among SWD-induced changes, more than the half were not detected during MWD or were not exceeding ± 25% of the SWD variation, and about 16% displayed opposite changes (Fig. 6A). However, part of the SWD-induced changes was already observed upon MWD and most of them varied with a smaller magnitude, as evidenced in CL5 for instance (Fig. 5). The abundance of 30 phosphorylated peptides displayed a variation in response to MWD ranging from 25% to 75% of that detected during SWD, whereas comparable changes (75% to 125% of that detected during SWD) accounted for four phosphorylation sites (Fig. 6A). Changes in abundance of higher magnitude during MWD compared with SWD, referred to five phosphorylated peptides. Furthermore, although CL1 or CL4 reflected persistent SWD-induced changes even after a 60 min-long rewatering, CL3, CL6, and CL7 depict that part of the SWD-induced changes partially or fully recovered along with the rehydration time-course (Fig. 5). These three clusters included 60% of the observed changes belonging to phytohormone-related and signaling proteins, 45% of those evidenced on transcription regulators, one third of those related to chromatin-related or cell cycle-related proteins, and less than 12% of those involved in the other identified processes (Fig. 4B). In addition, CL2, CL6, and CL7 clusters showed that part of the changes displayed sudden and reversible variations within the rehydration time course. These transitory changes were mainly observed for protein kinases or phosphatases in the range of 10 to 30 min after rewatering. As a whole, nearly 40% of the 132 phosphopeptides showing a significant quantitative change, displayed at least a half-recovery to their control level at one of the time points of the rehydration kinetic. To assess the occurrence of a recovery, a linear regression was computed from phosphopeptide abundances versus duration of the rewatering events. Although the expected response was not necessarily linear, this enables to detect protein phosphorylation statuses which recovered at least partially to the status of the control plants. A significant linear regression (p < 0.05) was detected for 36 phosphopeptides (Fig. 6B) among which ten showed a highly significant Kendall correlation (p < 0.02) of their abundance along with the Ψmd changes (Table I, supplemental Table S3 and supplemental Fig. S4). In all significant linear regressions except one, the change in abundance over the rehydration time-course was consistent with a recovery. Interestingly, more than a half-recovery was observed for three and six phosphorylation sites as early as 5 and 10 min rewatering respectively, whereas 23 were observed after 60 min rehydration, including 14 that displayed more than a 90% recovery (Fig. 6B).
Fig. 5.
Self organizing tree algorithm (SOTA clustering) of the phosphorylation sites significantly affected by plant water status. The seven main clusters are shown (the four additional clusters are displayed in supplemental Fig. S3). Individual profiles are depicted by gray lines (Z score) for mild water deficit (MWD), severe water deficit (SWD), and for the rehydration time course (RH5 to RH60), in comparison to control plants (CTRL1 and CTRL2) at the similar ontogenic development (for a detailed view of individual profiles, see supplemental Fig. S6). The average profile is marked in red. The cluster identification and the number of profiles included in each cluster are indicated in the left upper corner.
Fig. 6.
Contrasts between mild versus severe water deficit responses, and magnitude of the recoveries in protein phosphorylation status during the rehydration time course. A, the distribution of phosphorylated peptides are shown according to the consistency/discrepancy of their changes in abundance detected upon mild water deficit (MWD) and compared with severe water deficit (SWD). Consistency/discrepancy criterion was assessed by computing the ratio between the quantitative changes relative to control plants detected upon MWD and those detected upon SWD. “MWD effect ≠ SWD effect”: MWD induced antagonizing changes compared with those observed upon SWD; “No MWD effect”: MWD did not induce any changes in protein phosphorylation status (MWD-induced changes were less than 25% of those detected upon SWD); “MWD effect < SWD effect”: MWD-induced changes were of smaller magnitude compared with SWD-induced changes (ranging from 25% to 75% of those detected upon SWD); “MWD effect ≈ SWD effect”: changes of comparable magnitude were observed regardless of the water deficit intensity (MWD-induced changes reached 75% to 125% of those detected upon SWD); “MWD effect > SWD effect”: MWD induced changes of larger magnitude compared with SWD-induced changes (more than 125% of those observed upon SWD). B, for each rehydration time point (RH5, RH10, RH20, RH30, RH45, and RH60), the magnitude of the recovery in protein phosphorylation status is indicated for the 36 phosphorylation sites displaying a significant linear regression between phosphopeptide abundance and the duration of the rehydration.
Furthermore, the abundance of proteins displaying water regime-responsive phosphopeptides was explored. To quantify protein abundance, we investigated their nonphosphorylated peptides (NP) found in the IMAC flow-through fractions (FT). Since SCX prefractionation intrinsically delays NP elution (data not shown), only the seven last FT per biological replicate (among the 10 available ones) were analyzed. NP was traced in SWD plants in comparison to CTRL2, and in RH60 relative to SWD i.e. a total of 84 LC-MS/MS runs. Among the 124 unique proteins showing phosphopeptide quantitative variations, 83 were identified with at least one specific NP. Reproducible quantitative values were reliably obtained for 49 proteins (Table I and supplemental Table S3). This result emphasizes that phosphopeptide enrichment obviously enabled the identification of very low-abundance phosphorylated proteins. This substantially prevents joint quantification of both NP and phosphopeptides for many proteins because of contrasting dynamic range. Relative to CTRL2, SWD induced significant changes in NP abundances for only six proteins, including five covariations between NP and phosphopeptide abundance changes (Table I, supplemental Table S3 and Supplemental Fig. S5). Only one protein, a sucrose synthase 3, showed a significant quantitative variation between SWD and RH60 (Table I and supplemental Fig. S5), whereas this change was not observed in the phosphopeptide abundance. Overall, the abundance variations of phosphopeptides and nonphosphopeptides were consistent across CTRL2, SWD and RH60 for only two proteins (a dehydrin COR410 and an auxin-repressed protein). Along with the early phosphoproteomic variations observed during the first minutes of rehydration, these results support the hypothesis that most of the detected phosphorylation changes was related to variations of the phosphorylation status rather than to variations of protein abundance. Likewise, several proteins displayed two or more phosphorylation sites that behaved differently, either changing/unchanging or belonging to different clusters (e.g. S312/S313 and S241 in a GTP-ase activating protein for Arf containing protein, S177 and S166 in a putative uncharacterized protein; Table I and supplemental Table S3). Contrasting phosphorylation dynamics occurring on different sites within the same protein cannot be explained by a variation of protein abundance as well. In addition, such independent variations at different sites of the same protein emphasize that different phosphorylation events have specific roles (31).
DISCUSSION
In this study, we report the first analysis of the phosphorylation dynamics in the growing zone of leaves that resolves an in vivo time course of dehydration and rehydration. Quantitative changes of protein phosphorylation status were evaluated during an early and mild water deficit (MWD), and a severe water deficit (SWD) both impacting plant growth in comparison with constant optimal watering at the same ontogenic stage (CTRL1 and CTRL2, respectively). Phosphoproteome changes were also analyzed in comparison with SWD, over a time course of recovery periods lasting 5, 10, 20, 30, 45, and 60 min (RH5, RH10, RH20, RH30, RH45, and RH60, respectively) that induced partial recovery of the midday leaf water potential. As leaf elongation rate recovers faster than leaf water potential (22), reactivation of plant growth processes was likely to occur within the duration of the studied rehydration kinetics.
Phosphoproteomics of the Growing Zone of Leaves Evidences In Vivo Covariations Between Protein Phosphorylation Events and Plant Water Status
In the growing zone of maize leaves, the length of the zone with cell division is close to 70 mm in the sixth leaf and lengthens in younger leaves (10). Within this range, cell division rate is maximum in the first 20 mm and decreases along with the distance to the leaf insertion point, overlapping in space with expanding cells. This means that our analysis, carried out on a 5 cm-long leaf fragment, provides a picture of phosphorylation events occurring in tissues where both cell division and expansion happen. Upon water deficit, cell division rate is affected all along the growing zone, and cell expansion is altered approximately to the same extent (32). Thus, phosphopeptide variations observed between controls and WD samples should not be primarily related to the variation of the ratio between dividing and expanding cells. The variation of this ratio is even less expected to occur during the short period of rehydration.
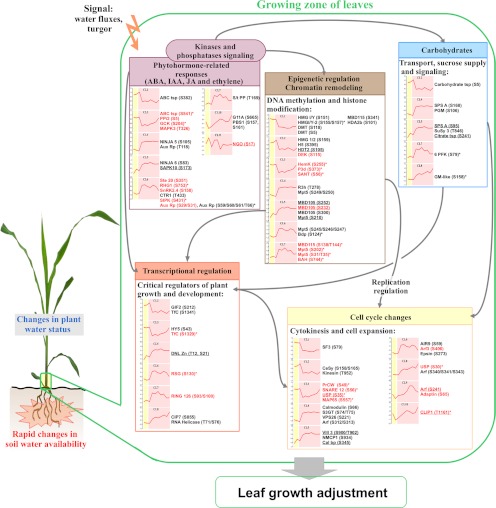
Using quantitative phosphoproteomics, we identified 138 phosphorylation sites that were altered upon changing plant water statuses in the leaf growing zone. Interestingly, 75% of the changes referring to proteins of known functions, belonged to five main processes directly or indirectly involved in the regulation of plant development, either through epigenetic and transcriptional regulations, cell cycle-related changes, hormone-mediated responses or carbohydrate metabolism adjustments (Fig. 7).
Fig. 7.
Overview of the main processes involving proteins whose phosphorylation status was rapidly altered in response to changing plant water statuses in the growing zone of maize leaves. The five main processes identified in this study are shown. Graphs refer to the SOTA clusters shown in Fig. 5, only the average profile is depicted in red. Individual profiles are shown in supplemental Fig. S6. Proteins displaying at least one phosphopeptide belonging to any SOTA clusters are indicated. Underlined proteins show that mild water deficit (MWD) induced at least half of the change in phosphopeptide abundance that was detected during severe water deficit (SWD). Proteins depicted in red show that 5, 10, 20, 30, 45, or 60 min rehydration (RH5, RH10, RH20, RH30, RH45, or RH60, respectively) enabled at least a half recovery and * indicates when the linear regression of phosphopeptide abundances versus duration of the rewatering events, was significant (p value < 0.05). Arrows refer to functional links between processes. Abbreviations: ABA, abscisic acid; IAA, auxin; JA, jasmonic acid; ABC tsp, MDR-like ABC transporter; PPi2, protein phosphatase inhibitor 2; GCK, GCK-like kinase MIK; MAPK3, Map3k delta-1 protein kinase; NINJA, novel Interactor of JAZ-family proteins; Aux Rp, auxin-repressed protein; SAPK10, serine/threonine-protein kinase SAPK10; Ste 20, Ste-20 related kinase; RHG1, receptor-like kinase RHG1; SnRK, sucrose nonfermenting related kinase; CTR1, serine/threonine-protein kinase ctr1; SIPK, salt-inducible protein kinase; S/t PP, Serine/threonine-protein phosphatase; G11A, protein kinase G11A; PBS1, Serine/threonine-protein kinase PBS1; NGO, naringenin,2-oxoglutarate 3-dioxygenase; HMG, high mobility group protein; DMT, DNA (cytosine-5)-methyltransferase 1; H5, histone H5; HDT2, histone deacetylase HDT2; DEK, protein DEK; MBD105/115, methyl-binding domain protein 105/115; HDA2b, Histone deacetylase 2b; HemK, methyltransferase family member 2; P3d, plus-3 domain containing protein; SANT, SANT DNA-binding protein; R3h, R3h domain containing protein; Mpt5, pumilio/Mpt5 family RNA-binding protein; Bdp, Bromodomain-containing protein; BAH, bromo-adjacent homology (BAH) domain-containing protein; Carbohydrate tsp, carbohydrate transporter/sugar porter/transporter; SPS A, sucrose phosphate synthase A; PGM, phosphoglucomutase; SuSy 3, sucrose synthase 3; Citrate tsp, citrate transporter family protein; 6 PFK, 6-phosphofructokinase; GM-like, galactose mutarotase-like; GIF2, GRF-interacting factor GIF2; TfC, protein TIME FOR COFFEE; HY5, LONG HYPOCOTYL 5 bZIP transcription factor; DNL Zn, DNL zinc finger family protein; RSG, bZIP transcriptional activator RSG; RING 126, RING finger protein 126; CIP7, COP1-interacting protein 7; SF3, pollen-specific protein SF3; AIR9, microtubule-associated protein AIR9; Arf, ADP-ribosylation factor GTPase-activating protein; CeSy, cellulose synthase-2; USP, universal stress protein; PrCW, proline-rich cell wall protein; SNARE 12, golgi SNARE 12 protein; MAP65, microtubule-associated protein MAP65–1a; S3GT, sterol 3-beta-glucosyltransferase; VPS26, vacuolar protein sorting 26; CLIP1, CLIP-associating protein 1; Vill 3, villin-3; NMCP1, nuclear matrix constituent protein 1; Cal bp, calmodulin binding protein.
In this study, MWD exemplified the first stage of the growth decrease when leaf water potential was not altered yet. Therefore, the similar changes in phosphorylation status observed in both MWD and SWD could correspond to early responses which were maintained over the establishment of water deficit, whereas the changes of lower magnitude suggest that part of the variations could depend on the duration and/or on the intensity of water shortage. On the contrary, the changes of higher magnitude or opposite to those detected during SWD, reveal specific responses of the early stage of water stress response. Our findings also emphasize rapid events in protein phosphorylation occurring in vivo within minutes, during early changes in plant water status. The picture of these rapid phosphorylation events, that were basically observed during the rehydration kinetics, evidenced that the earliest phosphorylation events mainly concerned cell cycle-related proteins. This emphasizes that the regulation of the cell cycle-related processes are among the primary targets of the phosphorylation and dephosphorylation machinery. Early changes in the phosphorylation status of cell cycle-related proteins could sustain the rapid recovery of leaf growth occurring within minutes in maize (22).
Transitory phosphorylation/dephosphorylation events in the range of 10 to 30 min after rewatering were mainly observed for kinases and phosphatases, including hormone-related proteins (Fig. 7). This could be a consequence of the recovery of water fluxes and turgor in the leaf, which was likely to occur concomitantly with the recovery of leaf water potential. These phosphorylation events detected on kinase and phosphatase proteins could act within signaling cascades, in an upstream control of further changes detected later during the rehydration time-course. On the contrary, no change was detected in kinases or phosphatases within 5 min rehydration although signaling events are rather expected at an early stage upon changing environments, and despite the identification, as early as 5 min after watering, of changing phosphorylation statuses of proteins involved in other functional processes. Inherently to exploratory phosphoproteomics approaches, some fine-tuned events were probably not detected in this study.
Fully or partially reversible events were also widely observed in the phosphorylation status of proteins related to epigenetic and transcriptional regulation (Fig. 7), which is consistent with the fact that the cell cycle-related changes require extensive epigenetic and transcriptional regulations for achieving cell cycle transitions, as already reported in growing organs of Arabidopsis thaliana plants (33, 34). In contrast, the phosphorylation events occurring in carbohydrate metabolism-related proteins, appeared rather as sustainable changes over the whole rehydration time course (Fig. 7).
As a whole, the dependence on stress intensity, the significant correlations with plant water status as well as the rapid recoveries observed during rehydration, collectively suggest that a substantial part of the observed phosphorylation dynamics are of functional relevance in the response of maize leaves to contrasting water regimes.
Earliest Changes in Plant Water Status Involve Cytokinesis- and Cell Expansion-related Proteins
The cell cycle is likely to encounter substantial changes in response to adverse environmental conditions (8–10). In many plant organs, including leaves, roots and seeds, water deficit rapidly decreases cell division rate (4, 7, 35). In this study, 19 changes in phosphorylation status involved proteins acting in cell cycle-related processes (Fig. 7). These proteins are likely involved in a general control of the cell cycle (i.e. USP family proteins, calmodulin binding protein and calmodulin), in the phragmoplast and the nuclear envelope assembly (i.e. vacuolar protein sorting 26, nuclear matrix constituent protein 1, pollen-specific protein, villin-3, MAP65–1a microtubule-associated protein, CLIP-associating protein 1, and kinesin), and in the cell plate initiation and maturation (i.e. Arf-GTPase, epsin, adaptin, Golgi-SNARE 12 protein, and AIR9). Although previous studies mainly reported the impact of water deficit in delaying the G1-to-S transition as a result of a reduced cyclin-dependent kinase activity (5, 36), here we identified quantitative changes of phosphoproteins involved in the mitosis phase and in cytokinesis.
Another evidence of changes in growth-related processes was the identification of phosphoproteins involved in cell wall biogenesis and remodeling, because both processes of cytokinesis and cell expansion require the addition of new cell wall components (37, 38). Here, we identified three altered phosphosites belonging to a cellulose synthase, a proline-rich cell wall protein (two proteins involved in cell wall building), and a sterol glycosyltransferase providing sterolglycosides which can act as precursors for cellulose biosynthesis. Another change was observed in a sucrose synthase which could be involved in the synthesis of cell wall components by providing UDP-glucose directly to the cellulose synthases and/or callose synthases (39).
Recovery of Leaf Water Potential Mainly Matches With Hormone-mediated Responses and Cell Signaling Events
Plant hormones are major regulators of growth and development, and they rapidly affect many aspects of plant biology during responses to biotic and abiotic stresses (40, 41). For instance, abscisic acid, jasmonate, and ethylene have already been reported as critical regulators of the early plant growth adjustments during drought or osmotic stress (14, 15). Here, we identified ten sites with significant changes in phosphorylation status belonging to proteins involved in either ABA-, ethylene-, auxin- or jasmonate-related responses. Our study evidenced single sites in two kinase proteins, SAPK10 and SnRK2.4, both belonging to the SnRK2 family proteins which play a pivotal role in regulating sucrose metabolism, several ion channels and ABA-induced gene expression (42, 43). Interestingly, the phosphorylation status of SnRK2.4 transiently recovered to the control level in the range of 10 to 20 min after rewatering. A rapid phosphorylation increase has already been observed in SnRK2 family proteins following 30-min ABA treatment (19). A slight and transitory recovery was also identified during 10 to 20 min rewatering for a phosphopeptide of Ctr1, a Raf-like protein kinase acting as the main negative regulator of the ethylene signal transduction chain. This is an interesting finding because ethylene has recently been reported as an early upstream regulator of the cell cycle machinery (15). Regarding auxin-related proteins, two drought-induced phosphorylation changes recovered in two auxin-repressed proteins (one recovered after 60 min rewatering whereas the other one was correlated with the recovery of plant water status). Finally, two increasing phosphorylated forms were detected in NINJA-family proteins 5 and 6 (Novel Interactor of JAZ-family proteins). NINJA is a member of the ABI five binding proteins that interact with the Jasmonate ZIM-domain proteins (JAZ proteins) for repressing jasmonate response genes (44).
Furthermore, we evidenced phosphorylation events in seven additional protein kinases and two phosphatases, named G11A, PBS1, Ste-20 related kinase, Map3k delta-1, GCK-like kinase MIK, salt-inducible protein kinase (SIPK), receptor-like kinase RHG1, protein phosphatase inhibitor 2 containing protein, and a serine/threonine-protein phosphatase, although little information is available for most of them. GCK MIK is believed to play a role in plant growth and development. Belonging to the GCK subgroup of MAP4Ks, it interacts with the maize atypical receptor kinase (MARK), a receptor expressed during embryogenesis and in the meristems of maize (45). Noteworthy, phosphorylation changes in the Ste-20 related kinase, in the Map3k delta-1 protein kinase, in SIPK and in RHG1 displayed transient recoveries after 20 min rewatering.
Changes in Plant Water Status Induce Reversible Modifications Preparing Epigenetic and Transcriptional Regulations
Up to 25% of the phosphorylation sites displaying significant changes in response to water regimes belonged to proteins involved in an upstream control of gene expression, mainly influencing chromatin structure and transcriptional regulation. Here, significant changes were observed in 25 phosphorylation sites belonging to proteins directly or indirectly involved in histone modification and DNA methylation. In response to environmental cues, gene expression and plant development are influenced by extensive chromatin remodeling, mainly achieved through histone post-translational modifications or DNA methylation patterns (46). Likewise, specific chromatin modifications occurring at both histone and DNA levels are also a prerequisite to sustain the transcriptional regulation involved in the cell cycle progression (33, 47). In this study, decreased phosphorylated forms in response to dehydration were detected in 12 phosphorylation sites belonging to proteins acting on chromatin structure or in DNA methylation (Fig. 7). Partial or full recoveries were observed for five of them, including a small chromatin-associated proteins (HMG I/Y-2), a DEK protein (a nuclear chromatin-associated phosphoprotein (48), a SANT domain- (a histone-interaction module (49)), a plus-3 domain-containing proteins, and a HemK protein (a class of methyl transferase proteins). Furthermore, increased phosphorylation upon SWD was evidenced in 11 phosphorylation sites belonging to seven proteins. Partial or full recoveries were observed for six of them, including two sites of a Pumilio/MPT5 proteins and single sites belonging to a MBD105 proteins, a MBD115 protein, a bromodomain- and a BAH domain- containing proteins. BAH and bromodomains are structural motifs commonly found in chromatin-associated proteins that recognize methylated and acetylated lysine residues, respectively. Their phosphorylation changes were concomitant to those observed on DNA methylation or histone deacetylation-related proteins.
Along with these nuclear architectural events changing the overall DNA accessibility, significant modifications of five critical regulators of plant growth and development were evidenced, including single sites in a GRF-interacting factor GIF2, in a bZIP transcriptional activator RSG (for repression of shoot growth), in a COP1-interacting protein 7 (CIP7), in a LONG HYPOCOTYL 5 bZIP transcription factor (HY5) and two sites in a TIME FOR COFFEE protein (TIC). GIF genes act in organ size control through a possible role in cell proliferation by positively regulating cell-cycle gene expression (50, 51) and RSG is involved in cell elongation by controlling the endogenous amounts of gibberellins (52). The function of RSG is known to be negatively regulated by 14–3-3 proteins that bind to RSG depending on its phosphorylation status, thereby sequestering RSG in the cytoplasm and making it unable to regulate its targets in the nucleus (53). These results suggest that the above processes may trigger further changes in cell division and expansion under water deficit. Likewise, TIC is a nuclear regulator of the circadian clock and could control various aspects of hormone-related processes involved in plant growth and development (54). CIP7 and HY5 are pivotal regulators of plant photomorphogenic development and both are interactors of COP1 (55). Being required for hypocotyl inhibition in all light conditions, HY5 is linked to a wide range of processes involving auxin, jasmonate, abscissic acid, ethylene and gibberellins pathways (56) which is in agreement with the hormone-mediated responses that have already been suggested above. CIP7 is a negative regulator of COP1 whereas this latter is a direct repressor of HY5 by inducing its proteasome-mediated degradation, resulting in the reduced expression of photomorphogenic genes (56, 57). Previous experiments suggested a role of HY5 phosphorylation in changing COP1-HY5 interaction.
Phosphorylation Changes Related to Carbohydrate Metabolism Refer to Long-term Adjustments
Phosphorylation changes occurring in proteins involved in carbohydrate metabolism accounted for a minor part of the whole detected variations, although sugars are major metabolites involved in stress responses. Glucose and especially sucrose can act as signaling molecules influencing the expression of a wide spectrum of genes and as the most suitable source of carbon and energy for many aspects of plant growth and development (58). In response to contrasting water regimes, two phosphopeptides of a sucrose phosphate synthase (SPS) and single phosphopeptides in a sucrose synthase (SuSy), in a phosphoglucomutase (PGM), in a phosphofructokinase (PFK), in a carbohydrate transporter and in a citrate transporter displayed significant changes in relative abundance. SPS phosphorylation occurs on multiple seryl residues in vivo and involves SnRK protein kinases at least (43, 59), which is in agreement with the rapid changes in the two SnRK2 proteins, described above. Changes in SPS activity during osmotic stress have already been observed as a consequence of phosphorylation events that differently occur on different sites (59). Here, except for the site identified in the carbohydrate transporter, detected phosphorylation sites were overall increased during SWD, with a partial recovery during rehydration for PFK only. Our results suggest long-term changes in the sucrose supply through phosphorylation events in both SPS and SuSy, but also in PGM and PFK which provide substrates for sucrose synthesis.
In conclusion, by combining physiological characterizations with phosphoproteome analysis, we reported here the first phosphoproteome dynamics occurring in vivo in the growing zone of maize leaves upon rapid changes in plant water status. Severe water deficit deeply modified the phosphorylation status of proteins, and part of these modifications was observed early, i.e. at a time when leaf water potential and plant growth were only slightly affected. During rehydration, that enables leaf growth resumption, this work enabled to identify transitory phosphorylation events as well as rapid in vivo changes occurring within five to 10 min after watering, independently to protein abundance variations. This exploratory approach emphasizes a comprehensive picture of the molecular plasticity of the phosphoproteome of leaf growing tissues upon dehydration/rehydration, and reveals a number early protein targets of the phosphorylation/dephosphorylation process potentially involved in an upstream control of the rapid leaf growth adjustment. This provides first insights into the in vivo dynamic behavior of individual protein phosphorylation sites upon changing plant water statuses. These findings are meant to guide future research on early plant water stress responses and on the understanding of processes involved in the rapid growth adjustment occurring upon fluctuating plant water status.
Supplementary Material
Acknowledgments
We thank Edlira Nano for her excellent technical assistance in the use of the MassChroQ software and to Olivier Langella for making LC-MS/MS data available in the PROTICdb database. We gratefully acknowledge Delphine Pflieger for her careful reading and editing of this manuscript. We also acknowledge Attila Csordas, from the PRIDE staff, for his excellent technical assistance in MS data submission.
Footnotes
* This research work was supported by a grant from the Agence National de la Recherche (DROMADAiR project, 08-GENM-003).
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S3.
1 The abbreviations used are:
- ABA
- abscisic acid
- MWD
- mild water deficit
- SWD
- severe water deficit
- RH5
- RH10, RH20, RH30, RH45, and RH60, rewatering duration of 5, 10, 20, 30, 45 and 60 min, respectively
- SCX
- strong cation exchange chromatography
- FDR
- false discovery rate
- XIC
- extracted ion chromatogram
- SOTA
- self organizing tree algorithm
- CL
- SOTA cluster.
REFERENCES
- 1. Chaves M. M., Maroco J. P., Pereira J. S. (2003) Understanding plant responses to drought - from genes to the whole plant. Func. Plant Biol. 30, 239–264 [DOI] [PubMed] [Google Scholar]
- 2. Saab I. N., Sharp R. E. (1989) Non-hydraulic signals from maize roots in drying soil: inhibition of leaf elongation but not stomatal conductance. Planta. 179, 466–474 [DOI] [PubMed] [Google Scholar]
- 3. Westgate M. E., Boyer J. S. (1985) Osmotic adjustment and the inhibition of leaf, root, stem and silk growth at low water potentials in maize. Planta 164, 540–549 [DOI] [PubMed] [Google Scholar]
- 4. Tardieu F., Granier C. (2011) Water deficit and growth. Co-ordinating processes without an orchestrator? Curr. Opin. Plant Biol. 14, 283–289 [DOI] [PubMed] [Google Scholar]
- 5. Granier C., Tardieu F. (1999) Water deficit and spatial pattern of leaf development. Variability in responses can be simulated using a simple model of leaf development. Plant Physiol. 119, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ben-Haj-Salah H., Tardieu F. (1995) Temperature affects expansion rate of maize leaves without change in spatial distribution of cell length (Analysis of the coordination between cell division and cell expansion). Plant Physiol. 109, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skirycz A., Inzé D. (2010) More from less: plant growth under limited water. Curr. Opin. Biotech. 21, 197–203 [DOI] [PubMed] [Google Scholar]
- 8. Schuppler U., He P. H., John P. C., Munns R. (1998) Effect of water stress on cell division and cell-division-cycle 2-like cell-cycle kinase activity in wheat leaves. Plant Physiol. 117, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aguirrezabal L., Bouchier-Combaud S., Radziejwoski A., Dauzat M., Cookson S. J., Granier C. (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ. 29, 2216–2227 [DOI] [PubMed] [Google Scholar]
- 10. Granier C., Inzé D., Tardieu F. (2000) Spatial distribution of cell division rate can be deduced from that of p34(cdc2) kinase activity in maize leaves grown at contrasting temperatures and soil water conditions. Plant Physiol. 124, 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cosgrove D. J. (2005) Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 [DOI] [PubMed] [Google Scholar]
- 12. Vincent D., Lapierre C., Pollet B., Cornic G., Negroni L., Zivy M. (2005) Water deficits affect caffeate O-methyltransferase, lignification, and related enzymes in maize leaves. A proteomic investigation. Plant Physiol. 137, 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muller B., Bourdais G., Reidy B., Bencivenni C., Massonneau A., Condamine P., Rolland G., Conéjéro G., Rogowsky P., Tardieu F. (2007) Association of specific expansins with growth in maize leaves is maintained under environmental, genetic, and developmental sources of variation. Plant Physiol. 143, 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harb A., Krishnan A., Ambavaram M. M., Pereira A. (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 154, 1254–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Skirycz A., Claeys H., De Bodt S., Oikawa A., Shinoda S., Andriankaja M., Maleux K., Eloy N. B., Coppens F., Yoo S. D., Saito K., Inzé D. (2011) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell. 23, 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Novak B., Kapuy O., Domingo-Sananes M. R., Tyson J. J. (2010) Regulated protein kinases and phosphatases in cell cycle decisions. Curr. Opin. Cell Biol. 22, 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schulze W. X. (2010) Proteomics approaches to understand protein phosphorylation in pathway modulation. Curr. Opin. Plant Biol. 13, 279–286 [DOI] [PubMed] [Google Scholar]
- 18. Niittylä T., Fuglsang A. T., Palmgren M. G., Frommer W. B., Schulze W. X. (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics. 6, 1711–1726 [DOI] [PubMed] [Google Scholar]
- 19. Kline K. G., Barrett-Wilt G. A., Sussman M. R. (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ke Y., Han G., He H., Li J. (2009) Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem. Biophys. Res. Commun. 379, 133–138 [DOI] [PubMed] [Google Scholar]
- 21. Röhrig H., Schmidt J., Colby T., Bräutigam A., Hufnagel P., Bartels D. (2006) Desiccation of the resurrection plant Craterostigma plantagineum induces dynamic changes in protein phosphorylation. Plant Cell Environ. 29, 1606–1617 [DOI] [PubMed] [Google Scholar]
- 22. Parent B., Hachez C, Redondo E., Simonneau T., Chaumont F., Tardieu F. (2009) Drought and abscisic acid effects on aquaporin content translate into changes in hydraulic conductivity and leaf growth rate: a trans-scale approach. Plant Physiol. 149, 2000–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Méchin V., Damerval C., Zivy M. (2007) Plant Proteomics: Methods and Protocols, pp 1–8, Humana Press, Totowa, New Jersey, U.S.A [Google Scholar]
- 24. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. R. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 25. Nühse T. S., Bottrill A. R., Jones A. M., Peck S. C. (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Valot B., Langella O., Nano E., Zivy M. (2011) MassChroQ: A versatile tool for mass spectrometry quantification. Proteomics. 17, 3572–3577 [DOI] [PubMed] [Google Scholar]
- 27. Ferry-Dumazet H., Houel G., Montalent P., Moreau L., Langella O., Negroni L., Vincent D., Lalanne C., de Daruvar A., Plomion C., Zivy M., Joets J. (2005) PROTICdb: a web-based application to store, track, query, and compare plant proteome data. Proteomics. 5, 2069–2081 [DOI] [PubMed] [Google Scholar]
- 28. Storey J. D., Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sperry J. S., Hacke U. G., Oren R., Comstock J. P. (2002) Water deficits and hydraulic limits to leaf water supply. Plant Cell Environ. 25, 251–263 [DOI] [PubMed] [Google Scholar]
- 30. Boersema P. J., Foong L. Y., Ding V. M., Lemeer S., van Breukelen B., Philp R., Boekhorst J., Snel B., den Hertog J., Choo A. B., Heck A. J. R. (2010) In-depth qualitative and quantitative profiling of tyrosine phosphorylation using a combination of phosphopeptide immunoaffinity purification and stable isotope dimethyl labeling. Mol. Cell. Proteomics. 9, 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 32. Tardieu F., Reymond M., Hamard P., Granier C., Muller B. (2000) Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: a synthesis of the effects of soil water status, evaporative demand and temperature. J. Exp. Bot. 51, 1505–1514 [DOI] [PubMed] [Google Scholar]
- 33. Beemster G. T., De Veylder L., Vercruysse S., West G., Rombaut D., Van Hummelen P., Galichet A., Gruissem W., Inzé D., Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol. 138, 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Desvoyes B., Sanchez M. P., Ramirez-Parra E., Gutierrez C. (2010) Impact of nucleosome dynamics and histone modifications on cell proliferation during Arabidopsis development. Heredity. 105, 80–91 [DOI] [PubMed] [Google Scholar]
- 35. Stals H., Inzé D. (2001) When plant cells decide to divide. Trends Plant Sci. 6, 359–364 [DOI] [PubMed] [Google Scholar]
- 36. Inzé D., De Veylder L. (2006) Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105 [DOI] [PubMed] [Google Scholar]
- 37. Backues S. K., Konopka C. A., McMichael C. M., Bednarek S. Y. (2007) Bridging the divide between cytokinesis and cell expansion. Curr. Opin. Plant Biol. 10, 607–615 [DOI] [PubMed] [Google Scholar]
- 38. Jürgens G. (2005) Cytokinesis in higher plants. Annu. Rev. Plant Biol. 56, 281–299 [DOI] [PubMed] [Google Scholar]
- 39. Amor Y., Haigler C. H., Johnson S., Wainscott M., Delmer D. P. (1995) A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. U.S.A. 92, 9353–9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santner A., Calderon-Villalobos L. I., Estelle M. (2009) Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307 [DOI] [PubMed] [Google Scholar]
- 41. Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Christmann A., Moes D., Himmelbach A., Yang Y., Tang Y., Grill E. (2006) Integration of abscisic acid signalling into plant responses. Plant Biol. 8, 314–325 [DOI] [PubMed] [Google Scholar]
- 43. Zheng Z., Xiaoping X., Crosley R. A., Greenwalt S. A., Sun Y., Blakeslee B., Wang L., Ni W., Sopko M. S., Yao C., Yau K., Burton S., Zhuang M., McCaskill D. G., Gachotte D., Thompson M., Greene T. W. (2010) The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 153, 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pauwels L., Barbero G. F., Geerinck J., Tilleman S., Grunewald W., Pérez A. C., Chico J. M., Bossche R. V., Sewell J., Gil E., García-Casado G., Witters E., Inzé D., Long J. A., De Jaeger G., Solano R., Goossens A. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 464, 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Llompart B., Castells E., Río A., Roca R., Ferrando A., Stiefel V., Puigdomenech P., Casacuberta J. M. (2003) The direct activation of MIK, a germinal center kinase (GCK) -like kinase, by MARK, a maize atypical receptor kinase, suggests a new mechanism for signaling through kinase-dead receptors. J. Biol. Chem. 278, 48105–48111 [DOI] [PubMed] [Google Scholar]
- 46. Chinnusamy V., Zhu J. K. (2009) Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sanchez, Mde L., Caro E., Desvoyes B., Ramirez-Parra E., Gutierrez C. (2008) Chromatin dynamics during the plant cell cycle. Semin. Cell. Dev. Biol. 19, 537–546 [DOI] [PubMed] [Google Scholar]
- 48. Kappes F., Scholten I., Richter N., Gruss C. (2004) Functional domains of the ubiquitous chromatin protein DEK. Mol. Cell. Biol. 24, 6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boyer L., Latek R. (2004) The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell. 5, 1–6 [DOI] [PubMed] [Google Scholar]
- 50. Kim J. H., Choi D., Kende H. (2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104 [DOI] [PubMed] [Google Scholar]
- 51. Lee B. H., Ko J. H., Lee S., Lee Y., Pak J. H., Kim J. H. (2009) The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151, 655–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fukazawa J., Sakai T., Ishida S., Yamaguchi I., Kamiya Y., Takahashi Y. (2000) Repression of shoot growth, a bZIP transcriptional activator, regulates cell elongation by controlling the level of gibberellins. Plant Cell. 12, 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Igarashi D., Ishida S., Fukazawa J., Takahashi Y. (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell. 13, 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ding Z., Millar A. J., Davis A. M., Davis S. J. (2007) TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell. 19, 1522–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yi C., Deng X. W. (2005) COP1 – from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625 [DOI] [PubMed] [Google Scholar]
- 56. Nemhauser J. L. (2008) Dawning of a new era: photomorphogenesis as an integrated molecular network. Curr. Opin. Plant Biol. 11, 4–8 [DOI] [PubMed] [Google Scholar]
- 57. Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X. W. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 19, 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramon M., Rolland F., Sheen J. (2008) Sugar sensing and signaling. The Arabidopsis Book. 6, 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Toroser D., Huber S. C. (1997) Protein phosphorylation as a mechanism for osmotic-stress activation of sucrose-phosphate synthase in spinach leaves. Plant Physiol. 114, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw MS output files, identified phosphopeptides and their corresponding spectra were uploaded to the ProticDB database (27). The data can be directly visualized or downloaded from http://moulon.inra.fr/protic/phospho. Tandem mass spectral data are also available via the Proteomics Identifications database (PRIDE) at http://www.ebi.ac.uk/pride/under the accession numbers 24901 to 26498. A detailed list of all peptides identified in all LC-MS/MS runs is given in supplemental Table S1, representative spectra of each phosphopeptide displaying significant water regime effect are shown in supplemental Fig. S1, and relative quantification data are available in supplemental Table S2.