Abstract
Pluripotent stem cells are capable of differentiating into all cell types of the body and therefore hold tremendous promise for regenerative medicine. Despite their widespread use in laboratories across the world, a detailed understanding of the molecular mechanisms that regulate the pluripotent state is currently lacking. Mouse embryonic (mESC) and epiblast (mEpiSC) stem cells are two closely related classes of pluripotent stem cells, derived from distinct embryonic tissues. Although both mESC and mEpiSC are pluripotent, these cell types show important differences in their properties suggesting distinct pluripotent ground states. To understand the molecular basis of pluripotency, we analyzed the nuclear proteomes of mESCs and mEpiSCs to identify protein networks that regulate their respective pluripotent states. Our study used label-free LC-MS/MS to identify and quantify 1597 proteins in embryonic and epiblast stem cell nuclei. Immunoblotting of a selected protein subset was used to confirm that key components of chromatin regulatory networks are differentially expressed in mESCs and mEpiSCs. Specifically, we identify differential expression of DNA methylation, ATP-dependent chromatin remodeling and nucleosome remodeling networks in mESC and mEpiSC nuclei. This study is the first comparative study of protein networks in cells representing the two distinct, pluripotent states, and points to the importance of DNA and chromatin modification processes in regulating pluripotency. In addition, by integrating our data with existing pluripotency networks, we provide detailed maps of protein networks that regulate pluripotency that will further both the fundamental understanding of pluripotency as well as efforts to reliably control the differentiation of these cells into functional cell fates.
Pluripotency, the ability to give rise to all cell types within the body, is a unique cellular property restricted to very early embryogenesis. Pluripotent stem cells have been derived from early embryos and represent a powerful system for understanding mammalian development as well as providing a scalable source of cells for regenerative medicine (1–7). Significant effort has been directed to understanding how the pluripotent state is specified in stem cells, and transcriptional regulatory mechanisms are now known to be central to maintenance of the pluripotent state. Embryonic stem cells are able to self-renew and yet remain in a poised state, ready to differentiate as required (8). In addition, pluripotent stem cells may be derived from different embryonic tissues, such as the epiblast (6), with distinct properties and responses to cellular signals suggesting that there are different pluripotent ground states.
Uncovering the molecular basis of stem cell pluripotency has been the principal goal of many 'omics studies. Protein expression analysis has been used to globally define proteins specific to embryonic stem cells (9, 10). In addition, targeted proteomics using affinity-purification methods has been used to define protein complexes important for maintenance of pluripotency (11, 12). Genome-wide profiling of DNA-binding sites and gene-expression analyses have been used to study stem cells and their differentiated derivatives and to uncover the molecular networks that regulate the pluripotent state (13). Meta-analyses of multiple data sets or aggregation of large-scale data has enabled the definition of pluripotency networks (14, 15). The latter study integrated gene-expression profiles from multiple different embryonic stem cell lines to construct a core pluripotency gene-network named PluriNet (15). A key conclusion from this and other studies is the existence of a defined network common to multiple different embryonic stem cells lines that differentiates these cells from nonpluripotent cells and functions to regulate the pluripotent state.
Processes that regulate chromatin assembly and conformation are major determinants of the required activation and repression of pluripotency gene-expression. Dynamic changes to chromatin occur as part of many developmental transitions in mammals, and also appear to regulate the maintenance of pluripotency. Processes such as DNA methylation and ATP-dependent chromatin-remodeling facilitate these developmental transitions through regulation and assembly of chromatin (16). The SWI/SNF complexes (also known as Brg associated factors, BAF), first described in yeast, have been shown to be important chromatin remodeling components in many other eukaryotes (16). Detailed maps of BAF complexes have shown that there are both core components and complex configurations that occur in specific cell types. In embryonic stem cells, the esBAF complex has been defined and shown to differ in its subunit composition from differentiated derivatives (17). The important role of BAF components such as Smarca4/Brg1 and Smarcc1/Baf155 in pluripotency has been shown by their role in direct repression of key pluripotency-regulating genes during differentiation of embryonic stem cells (18, 19).
Although many studies have focused on understanding the pluripotent state in mouse embryonic stem cells, it is now appreciated that distinct pluripotent ground states exist (mESC-like and mEpiSC-like) and it is important to decipher how pluripotency is regulated in these distinct states. These comparisons are especially pertinent because human embryonic stem cells share molecular properties and defining features with the epiblast state (6, 20, 21). Our study used label-free proteomics to quantitatively compare the nuclear proteomes of mouse embryonic stem cells (mESCs)1 and mouse epiblast stem cells (mEpiSCs). We show that proteomics approaches add significantly to the growing understanding of regulatory networks underlying pluripotency. We identify specific differences in expression of components of DNA methylation and chromatin remodeling protein networks, indicating that these processes differ between mESCs and mEpiSCs and may dictate the different pluripotent states of mESCs and mEpiSCs. This study provides the first global comparison of the mESC and mEpiSC proteomes, and will provide a foundation for manipulation and future functional studies of mESC and mEpiSC pluripotency.
EXPERIMENTAL PROCEDURES
Cell Culture, Cell Lysis, and Nuclear Extraction
mESCs and mEpiSCs were grown as described previously (6, 22). Whole colonies were lifted from the plates using 1.5 mg/ml (w/v) collagenase IV (Invitrogen). Colonies were pelleted at 200g for 5 min and washed twice with ice-cold PBS buffer (137 mm NaCl, 2.7 mm KCl, 100 mm Na2HPO4, and 2 mm KH2PO4) and either processed immediately or frozen at −80 °C until use. Cell lysis buffer I (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, 1% Triton-100, Protease inhibitor mixture) was added to the cell pellets to suspend the cells. The cell suspension was kept on ice for 30 min before low speed centrifugation at 500 g for 5 min in 4 °C. The supernatant (cytoplasmic fraction) and pellet (nuclei) were collected separately. Next, cell lysis buffer II (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, 1% Triton-100, 4% SDS, Protease inhibitor mixture) was added to the pelleted nuclei followed by sonication for 10 s twice. Nuclear fraction was then collected after centrifugation at 20,000 × g for 30 min. Protein concentration was measured by Bradford (BioRad, Hercules, CA; Cat.500-0006) quantification method using 2 mg/ml BSA (Pierce, Rockford, IL; Cat.23209) as standard.
One Dimensional SDS-PAGE Gel-based Approach and Liquid Chromatography-tandem MS (LC-MS/MS)
Equal amounts (20 μg) of protein from two cell types and two fractions were loaded on NuPAGE® Novex 4–12% Bis-Tris gel (Invitrogen, Cat.NP0321) and run at 150V for 1.5 hours. The gels were washed in distilled water twice for 5 min each and then put into fixation solution (40% acetic acid and 10% methanol) for 0.5 hour and washed twice in distilled water before adding Coomassie Blue G-250 staining buffer (BioRad, Cat.161-0787). After 1 hour, the gels were destained in ddH2O before taking images. Proteins were then separated into 10 fractions (1D gel fractionation) using 1D-SDS-PAGE prior to trypsin digestion and mass spectrometric analyses.
Standard in-gel tryptic digestion was performed according to published method (23). The combined elution fractions were lyophilized in SpeedVac Concentrator (Thermo Electron Corporation, Milford, MA), resuspended in 100 μl 0.1% formic acid and further cleaned up by reverse phase chromatography using C18 column (Harvard, Southborough, MA). The final volume was reduced to 10 μl by vacuum centrifugation and adding 0.1% formic acid.
Tryptic peptides were separated by on-line reverse phase nanoscale capillary liquid chromatography (nano-LC, Dionex Ultimate 3000 series HPLC system) coupled to electrospray injection (ESI) tandem mass spectrometer (MS-MS) with octopole collision cell (Thermo Fisher Scientific LTQ Orbitrap XL). Loaded peptides were trapped in a C18 trap column and eluted on nano-LC with 90-min gradients ranging from 6 to 66% Acetonitrile in 0.1% formic acid with a flow rate 300 nl/min. Data dependent acquisition was performed on the LTQ-Orbitrap using Xcalibur software (version 2.0.6, Thermo Scientific) in the positive ion mode with a full scan MS1 at resolution of 60,000 in the m/z range 325.0 to 1800.0 followed by collision-induced dissociation (CID) fragmentation of the top five precursor ions using 35% normalized collision energy. Appropriate dynamic exclusion settings were based on a nominal LC peak width of 45 s. Adequate LC re-equilibration time was factored into the LC-MS/MS method.
In-solution Based Approach and LC-MS/MS
Nuclear extracts (three replicates for each cell type) were added with cold acetone for protein precipitation overnight such that 40 μg of total protein was isolated from each sample. Solubilization of all the proteins was performed with the addition of 4% SDS in Tris 50 mm buffer, pH 8.0. Processing of the solubilized samples for in-solution digest was done using the FASP approach (24). Briefly, a 3K cutoff filter (Millipore Inc, Billerica, MA) was used as a proteomic reactor for detergent cleaning, cysteine reduction and alkylation prior to tryptic digestion. The digests prepared above were analyzed by LC-MS/MS system using a Waters nano acquity UPLC systems (Waters Inc, MA) that was interfaced to a LTQ Velos-Orbitrap mass spectrometer (Thermo-Finnigan, Bremen, Germany). The platform was operated in the nano-LC mode using the standard nano-ESI API stack fitted with a picotip emitter (uncoated fitting, 10 μm spray orifice, New Objective, Inc., Woburn, MA). The solvent flowrate through the column was maintained at 300 nL/min using the split-free acquity system. The protein digests (5 μl) were injected into a reversed-phase symmetry C18 trapping column (0.18 × 20 mm, 5 μm particle size, Waters Inc.) equilibrated with 0.1% formic acid/2% acetonitrile (v/v) and washed for 5 min with the equilibration solvent at a flow rate of 15 μl/min, using the sample trap mode of UPLC. After the washing step, the trapping column was switched in-line with a reversed-phase bridged ethyl hybrid (BEH) C18 nanoacquity UPLC column (0.075 × 250 mm, Waters Inc.) and the peptides were chromatographed using a linear gradient of acetonitrile from 5% to 50% in aqueous 0.1% formic acid over a period of 210 min at the above-mentioned flow rate such that the eluate was directly introduced to the mass spectrometer. A 100% acetonitrile elution step was subsequently performed for 15 min prior to resetting the analytical column to the initial equilibration conditions for 15 more minutes at the end of the chromatographic run, accounting for a total of 240 min/h of LC-MS/MS time. The mass spectrometer was operated in a data-dependent MS to MS/MS switching mode, with the 12 most intense ions in each MS scan subjected to MS/MS analysis. The full scan was performed at 60000 resolution in the Orbitrap detector and the MS/MS fragmentation scans were performed in the Velos dual ion trap detector (IT) CID mode such that the total scan cycle frequency was ∼1.5 s. The threshold intensity for the MS/MS trigger was always set at 1000 and the fragmentation was carried out using the CID mode using a normalized collision energy (NCE) of 35. The data was entirely collected in the profile mode for the full scan and centroid mode for the MS/MS scans. Dynamic exclusion function for previously selected precursor ions was enabled during the analysis such that the following parameters were applied: repeat count of 2, repeat duration of 45 s, exclusion duration of 60 s and exclusion size list of 450. Xcalibur software (version 2.0.7), Thermo-Finnigan Inc., San Jose, CA) was used for instrument control, data acquisition, and data processing.
Chromatographic reproducibility and full scan MS (also termed MS 1) intensity robustness are critical to the success of this label-free method. To monitor this efficiently and exhaustively, spiked external peptides from yeast enolase digest (400 fmoles on column-load) was used to keep a track of the chromatographic performance of the LC-MS/MS system.
Data Processing
Raw MS data were processed using Mascot search engine (version 2.2.0; Matrix Science, London, UK). The raw data were searched against Mouse International Protein Index (IPI) database (released on August 10, 2009 and containing 56,733 protein sequences) with decoy database search enabled. The searches were executed with fixed modification carbamidomethyl cysteine and variable modification oxidized methionine. Peptide tolerance and MS/MS tolerance were set at 15 ppm and 0.8 Da, respectively. In addition, peptide charges of +2 and +3 were selected and maximal missed cleavage of one was allowed. Scaffold (Proteome Software Inc., Portland, OR, USA; version 3.00.04) was used for spectral count analysis and to validate LC-MS/MS-based peptide and protein identifications (25). Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (26). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides (27). With these stringent parameters of Peptide Prophet and Protein Prophet, the false discovery rate was zero (27). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Data Analysis
For the gel-based fractionation, spectral counts and protein identifications from each gel slice were merged within each lane using Scaffold. Data normalization of spectral counts across experiments was performed using the Scaffold software. Pseudocounts were added to each spectral count value to account for missing values (28). Log2 fold-change values were computed for mESC versus mEpiSC and p values computed using Student's t test. The normalized gene expression data of mouse mESC and mEpiSC in three biological replicates using Agilent whole-genome microarrays was obtained from a previous study (6). From this data, the mean log2 fold change and t test were used to calculate statistical significance p value of each probe for a given gene. For those genes with multiple probes, the value of the probe with the most significant p value was used to represent the corresponding gene. The p values from the Student's T-tests were corrected using multiple hypothesis testing, and q-values were calculated using the bootstrap method (29). The set of 299 PluriNet genes were downloaded from http://www.stemcellmatrix.org (15).
Protein Functional Categorization and Network Visualization
For mESC and mEpiSC comparison, proteins with calculated log2 (mESC/mEpiSC) ratio obtained from label-free quantitation table (supplemental Table S2) were uploaded into the Ingenuity Pathways Analysis tool to identify biological processes, molecular networks, and functional pathways. For PluriNet (15) and PluriNetWork (30) overlap analyses, mESC and mEpiSC proteomics data was combined with PluriNet and PluriNetWork (supplemental Table S3). The Mammalian Phenotype resource provided by the Mouse Genome Database (31) was used to map mESC and mEpiSC proteins to their corresponding genes and embryonic phenotypes.
Immunoblot Analyses and Quantification
Equal amount (20 μg) of proteins from different samples were loaded on precast 4–12% Bis-Tris gel (NP-0335, Invitrogen, Carlsbad, CA) and subjected to electrophoresis. Gels were either stained with Coomassie Brilliant Blue or transferred to nitrocellulose membrane (10402594, Whatman, Dassel, Germany). Immunoblotting was used to detect the protein with super signal ELISA Pico chemiluminescent substrate (37070, Pierce, Rockford, IL). Primary antibodies anti-DNMT1 (5119, Cell Signaling Technology, Inc., Danvers, MA), anti-DNMT3L (ab3493, Abcam, Cambridge, MA), anti-Brg1 (3508, Cell Signaling Technology, Inc., Danvers, MA), anti-BAF53 (sc-271226, Santa Cruz Biotechnology, Inc.), anti-BAF155 (sc-10756, Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-BAF60a (sc-135843, Santa Cruz Biotechnology, Inc.) and anti-α-tubulin (2144, Cell Signaling Technology, Inc.) as loading control were applied at 1:1000, 1:1000, 1:1000, 1:1000, 1:1000, 1:1000 and 1:2000, respectively and secondary antibody HRP-conjugated anti-Rabbit (NB730-H, Novus Biologicals, Littleton, CO) were added at 1:20000. Chemiluminescence detection using SuperSignal* ELISA Pico Chemiluminescent Substrate (PI-37070, Thermo Scientific, Rockford, IL) was applied to all immunoblots. The bands shown in blots were then quantified by ImageJ (http://rsbweb.nih.gov/ij/) (32, 33) and the mean values were plotted as well as the standard deviation for the two replicates of each protein.
RESULTS
Analysis of mESC and mEpiSC Data Sets
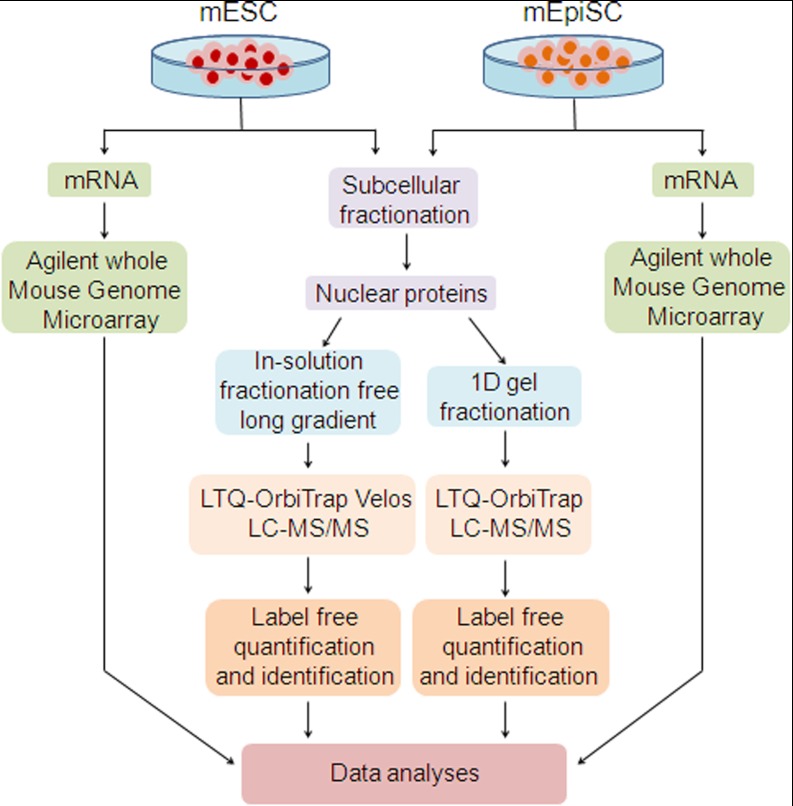
The goal of this study is to identify proteins and their associated networks that determine the distinct pluripotent states of mouse embryonic (mESC) and epiblast (mEpiSC) stem cells. An overview of the work flows used is shown in Fig. 1. Because pluripotency is largely regulated through control of gene-expression programs, we focused our proteomics experiments on analysis of the nuclear mESC and mEpiSC proteomes. In preliminary experiments, we analyzed both nuclear and cytoplasmic fractions of mESC and mEpiSCs and observed significant enrichment of known nuclear proteins and significant overlap with two previously described networks of pluripotency genes, PluriNet (15) and PluriNetWork (30) (Fig. 3). We also ascertained that spectral counts from nuclear and cytosolic fractions were similar for “housekeeping” proteins for mESCs and mEpiSCs showing that subcellular fractionation was similarly effective for each cell type (supplemental Table S7). In our extended studies we analyzed nuclear proteomes for both cell types (Fig. 1) and subsequent data analyses focused on these nuclear proteomes. To maximize coverage of the mESC and mEpiSC nuclear proteomes, two separate analytical strategies were applied. In the first, proteins were separated by 10-fold 1D-SDS-PAGE fractionation (hereafter referred to as the in-gel fractionation experiment) (supplemental Fig. S2). In the second study, an extended fractionation-free 4 h long chromatographic gradient was used (hereafter referred to as the in-solution experiment). In each study, 3 replicate mESC and 3 replicate mEpiSC samples were analyzed (a total of 6 × 10 = 60 MS/MS runs in the 1D SDS experiment and 6 MS/MS runs in the in-solution fractionation free long gradient experiment). These studies were performed using the same biological samples (3 mESC and 3 mEpiSC samples) after subcellular fractionation to enrich for nuclear associated proteins. In addition, we also previously acquired gene-expression microarray data (6) from these two cell types, enabling analysis of comparative gene-expression profiles from mESCs and mEpiSCs.
Fig. 1.
Experimental workflow. Sample preparation and comparative microarray and proteomics analysis of mouse embryonic stem cells (mESC) and mouse epiblast stem cells (mEpiSC) by LC-MS/MS.
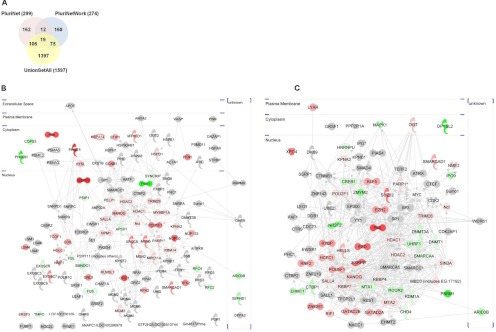
Fig. 3.
Intersecting proteomics studies with existing pluripotency networks. A, Overall overlap of all identified proteins in union set of proteins identified from gel-free and gel-based proteomics studies (“union set all” - 2 peptides; Scaffold probability ≥ 99%) with PluriNet and PluriNetWork. B, The intersection between proteomics data and PluriNet (analyzed using Ingenuity Pathways Analysis) - 125 proteins. C, The intersection between proteomics data and PluriNetwork - 94 proteins. Each node in (B) and (C) represents a protein identified in mESC or mEpiSC samples (red shaded nodes indicate mESC>mEpiSC protein expression with p < 0.1 (Student's t test), green shaded indicate mEpiSC>mESC protein expression with p < 0.1 (Student's t test) and gray shaded nodes indicate identified protein in either mESC or mEpiSC but without statistically significant quantitative difference.
Global comparison of the in-solution and in-gel fractionation experiments showed substantial overlap of the proteins identified. In total, the in-solution experiment identified 1462 proteins and the in-gel fractionation experiment identified 955 proteins according to the peptide and protein selection criteria used (≥ two peptides/protein, protein probability ≥ 0.99). Proteins identified in the in-gel fractionation experiment were largely a subset of those identified using the in-solution approach (85.1% of in-gel proteins were also identified in the in-solution experiment) (supplemental Fig. S3). To identify quantitative differences in protein expression between mESC and mEpiSC, spectral count analyses (Scaffold; Proteome Software Inc.) were performed. mESC/mEpiSC ratios, p values and q-values (to account for multiple testing) were then computed for each protein in each data set (in-solution or in-gel). supplemental Tables S1 and S2 provide complete sets of identified proteins and spectral counts analyses respectively.
We then assessed the concordance between the in-gel and in-solution experiments by analyzing the agreement of expression trends (mESC/mEpiSC) for proteins found in both. The trends between in-gel and in-solution experiments were highly concordant when proteins with p < 0.05 or p < 0.1 were compared (98.04% of proteins show same expression change trend; supplemental Fig. S1C). In addition, we compared the in-solution and in-gel experiments to the microarray data sets and for those features in common, found similar concordance (supplemental Fig. S4). Two data sets were initially created for subsequent analyses. In the first, we considered all proteins with p < 0.05 (in in-gel or in-solution experiments; maximal q-value = 0.08) (466 total proteins) and in the second, all proteins with p < 0.1 (in in-gel or in-solution experiments; maximal q-value = 0.16) (646 total proteins). These two data sets showed similar concordance in expression trends between proteins identified in in-solution and in-gel experiments. In addition, we noted that the extra proteins included by considering proteins with p < 0.1 largely fell in to similar biological process and pathways as proteins with p < 0.05. For subsequent global analyses therefore, a union set of proteins was created consisting of all proteins in the in-solution or in-gel experiments with expression differences significant at p < 0.1 in one or both experiments. This union data set consists of 646 proteins, 336 showing increased expression in mESC and 310 showing increased expression in mEpiSC (supplemental Table S5). For the focused analyses of protein networks in Figs. 3, 4 and 5, we primarily focused on proteins with p < 0.05 and included selected proteins with p < 0.1 where necessary.
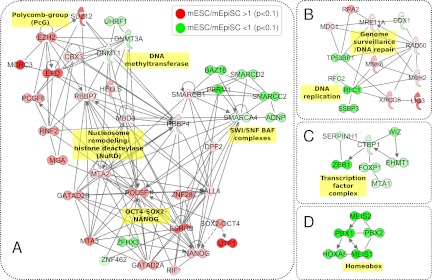
Fig. 4.
Selected mESC- and mEpiSC-associated protein networks. Ingenuity Pathway Analysis (IPA) reveals differentially expressed subnetworks including (A) Polycomb-group (PcG), DNA methyltransferase, Nucleosome remodeling/histone deacetylase (NuRD), OCT4-SOX2-NANOG and SWI/SNF BAF complexes, B, Protein networks functioning in DNA replication and Genome surveillance/DNA repair, (C) Transcription factor complexes, and (D) Homeobox protein complex. All proteins shown were identified as being either more abundant in mESC (red nodes) or more abundant in mEpiSC (green nodes) at p < 0.1 (Student's t test).
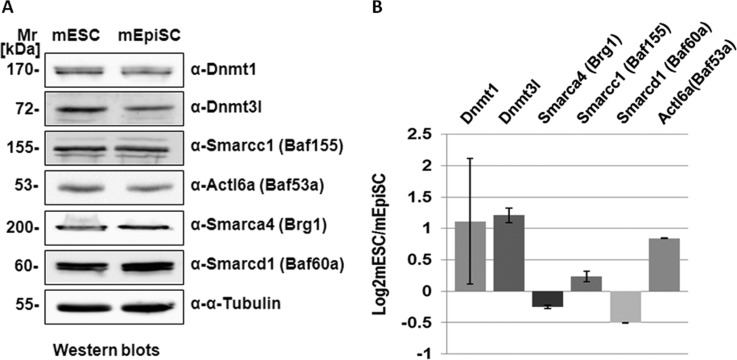
Fig. 5.
Immunoblot analyses of proteins with significantly different mESC/mEpiSC expression. A, Immunoblot analyses of selected DNA methyltransferase and Swi/Snf-related Baf complex components. Nuclear protein extracts (20 μg) from mESC and mEpiSC were loaded on 1D SDS-PAGE gel and transferred to membrane followed by immunoblotting using native antibodies. Mr refers to standard protein marker and α-tubulin was used as a loading control. B, Immunoblot quantification of two replicates of each protein verified. Bands for same protein in two replicated immunoblots were measured for intensity and the mean log ratio of mESC/mEpiSC fold change (the height of column) and the standard deviation (the error bar) between two blots were plotted.
Differing Functional Properties of the mESC and mEpiSC Nuclear Proteomes
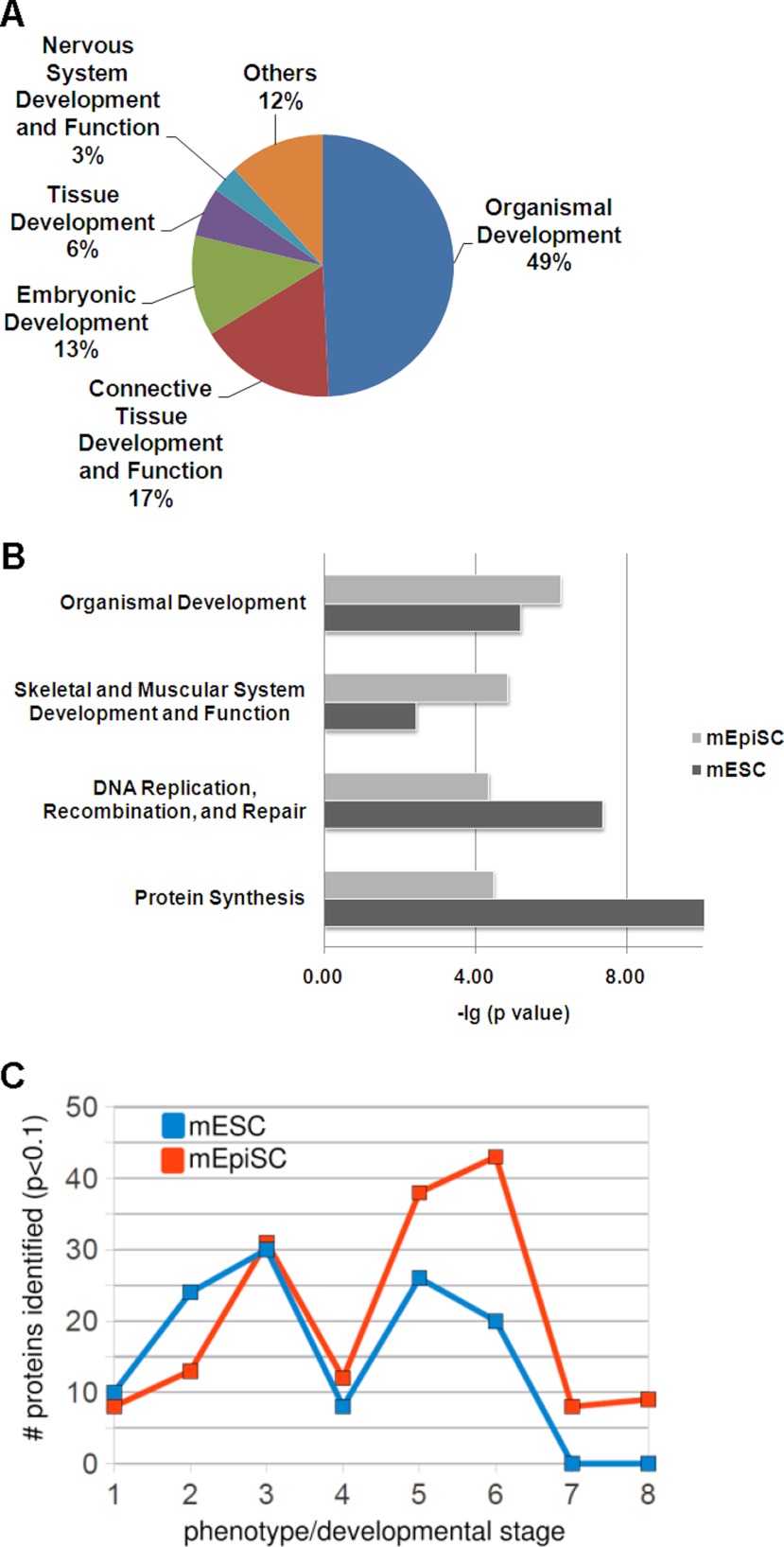
In analyzing functional annotations of the sets of mESC and mEpiSC proteins, we noted very significant representation of proteins involved in developmental processes. As shown in Fig. 2A, the most significant functional categories pertain to developmental processes such as embryonic and tissue development.
Fig. 2.
Biological trends in mESC and mEpiSC nuclear proteomes. A, Top functional categories represented in combined mESC and mEpiSC data sets (using all identified proteins) showing significant representation of proteins involved in embryogenesis and development. B, Proteins with significantly increased expression (p < 0.1; Student's t test) in either mESC and mEpiSC show significant differences in representation of several functional categories. C, Analysis of mutant phenotypes of genes corresponding to mESC and mEpiSC proteins (proteins differentially expressed in mESCs or mEpiSCs with p < 0.1; Student's t test). mESC proteins represent genes with embryonic lethal phenotypes at earlier developmental stages than mEpiSC proteins. Phenotypes are arranged according to developmental stage: (1) Prenatal lethality, (2) embryonic lethality before implantation (E0-E4.5), (3) embryonic lethality before somite formation (E4.5-E8), (4) embryonic lethality before turning of embryo (E8-E9), (5) embryonic lethality during organogenesis (E19-E14), (6) postnatal lethality, (7) absent somites, (8) open neural tube.
To compare active cellular processes in mESCs and mEpiSCs, the sets of proteins either enriched (p < 0.1) in mESC cells (336 proteins) or enriched (p < 0.1) in mEpiSC (310 proteins) were analyzed. The most significant functional categories for each of these two sets of proteins are shown in Fig. 2B. Notably, mESCs are enriched in proteins encoding fundamental cellular processes such as DNA replication, recombination and repair (p value = 4.46E-8). A greater enrichment of proteins involved in embryonic development is found in the mESC than in mEpiSC samples (p values equal to 6.69E-06 and 3.66E-04 respectively). Of the 56 proteins from mEpiSC included in this category, several pertain directly to epiblast related functions such as survival of embryonic stem cells (DNMT1), hatching of blastocyst (SMARCA4) and the formation or morphology of the visceral endoderm (MYH10). These global analyses show that proteins with pertinent biological functions are identified in the proteomics screens and identify functional categories that significantly differ between mESCs and mEpiSCs.
To further understand the differences in developmental functions represented by mESC and mEpiSC proteins, we analyzed mouse phenotypes of corresponding mutations. All mESC and mEpiSC proteins from the union set were mapped to embryonic phenotypes using the Mammalian Phenotype resource (31). Embryonic lethal phenotypes and their associated numbers of genes represented in the mESC and mEpiSC sets are shown in Fig. 2C. The overall numbers of proteins that could be assigned to a phenotype using the Mouse Genome Informatics database (31) were approximately similar for mESC and mEpiSC proteins (140 and 147 respectively), representing ∼50% of the mESC and mEpiSC sets overall. However, the phenotypes differ markedly between mESC and mEpiSC. We focused on embryonic lethality phenotypes as shown in Fig. 1C, and found that mESC proteins were more highly associated with very early embryonic lethality (pre-implantation failure or lethality prior to somite formation) than mEpiSC proteins, which showed greater representation of genes with lethality occurring during organogenesis and post-natally. Thus, aside from the differences in functional categorizations (Fig. 1B), the mESC and mEpiSC proteins represent at least in part, proteins expressed as part of the ordered process of embryonic development. In addition, the association of significant proportions of the identified proteins with embryonic mutant phenotypes suggests that many of the identified proteins play key, nondispensable roles in development.
We also observed many known markers of embryonic stem cells in our proteomics data sets. Table I shows these markers with their respective fold-differences between mESC and mEpiSCs. Notably, most of the known markers show increased expression in mESC samples. We observed 4 distinct peptides corresponding to Nanog protein (uniquely in mESC samples). Although the Nanog protein is a known Oct4 binding partner, Nanog is not consistently detected using Oct4 immuno-purification (12, 34, 58), possibly because of difficulty in digesting the Nanog protein to tryptic peptides (34). Thus proteomics profiling approaches, such as our own, are complementary to focused immuno-purification of pluripotency associated proteins.
Table I. Selected known embryonic stem cell markers observed in proteomics data sets.
*Log2 (mESC/mEpiSC) values (positive values indicate higher expression in mESCs and negative values indicate higher expression in mEpiSCs).
| Gene symbol | Protein name | mESC/mEpiSC protein fold change* | p value (T-test) | References |
|---|---|---|---|---|
| Oct4/Pou5f1 | POU class 5 homeobox 1 | 1.59 | 0.0005 | 51 |
| Sox2 | SRY (sex determining region Y)-box 2 | 2.00 | NA | 51 |
| Nanog | Nanog homeobox | 0.96 | 0.098 | 51 |
| Sall4 | Sal-like 4 | 0.57 | 0.008 | 52 |
| Dppa4 | Developmental pluripotency associated 4 | 1.56 | 0.002 | 53 |
| Utf1 | Undifferentiated embryonic cell transcription factor 1 | 3.53 | 0.002 | 54 |
| Trim28 | tripartite motif containing 28 | 0.66 | 0.009 | 51, 55, 56 |
| Dnmt3l | DNA (cytosine-5-)-methyltransferase 3-like | 4.39 | NA | 45, 57 |
| Dnmt3a | DNA (cytosine-5-)-methyltransferase 3 alpha | −1.00 | 0.02 | 57 |
Integrated Pluripotency Networks
To understand more fully what our proteomics data reveal about pluripotency, we integrated our data with two previously defined pluripotency networks. A previous study used gene-expression profiling to provide a large-scale classification of diverse stem cell lines (15). By integrating these transcriptional profiles with protein-protein interaction networks, a core pluripotency network, named PluriNet (15), was defined. A second study, using an alternative approach of curating literature, created a pluripotency network, PluriNetwork, focused on molecules and interactions underlying pluripotency mechanisms in mouse stem cells (30). We first compared the complete set of proteins identified in our study to those proteins represented in PluriNet and PluriNetwork. As shown in Fig. 3A, there are significant overlaps of our identified proteins with PluriNet and PluriNetwork, showing that the mESC and mEpiSC nuclear proteomes are highly enriched for proteins with roles in pluripotency. We detected 42% of the total proteins in PluriNet and 35% of the total proteins represented in PluriNetwork. In addition, we noted that there is relatively little overlap between PluriNet and PluriNetwork; indeed only 31 genes are represented in both PluriNet and PluriNetwork (∼10% of each network). This likely reflects the different methods used to derive PluriNet (unbiased clustering of gene-expression profiles and integration with high-throughput protein-protein interaction data) and PluriNetwork (curation of pluripotency-related publications). It also suggests that each resource most likely represents an incomplete network of pluripotency related genes/proteins. This finding prompted us to further integrate our data with these pluripotency networks to see whether the networks might be extended using our proteomics data.
Proteomics data was integrated with PluriNet and PluriNetwork as shown in Figs. 3B and 3C respectively. In each case, all proteins present in both our proteomics data and PluriNet (Fig. 3B) or our proteomics data and PluriNetwork (Fig. 3C) were analyzed using Ingenuity Pathways Analysis. Proteins shown in gray were detected in our study but not significantly different between mESC and mEpiSC, whereas proteins in red are those proteins significantly (p < 0.1) more abundant in mESC than mEpiSC and proteins in green those proteins significantly more abundant in mEpiSC than mESC proteomes. Considering only proteins with statistically significant differences between mESC or mEpiSC (p < 0.1) in the union data set, we observed a greater overlap between mESC and PluriNet (44 proteins) than between mEpiSC and PluriNet (15 proteins) (Fig. 3B). Similarly, we observed a greater overlap between mESC and PluriNetWork (38 proteins) than between mEpiSC and PluriNetWork (19 proteins) (Fig. 3C). The greater representation of proteins enriched in mESC as compared with mEpiSC suggests that existing pluripotency networks represent the mESC pluripotency state better than the mEpiSC pluripotency state. In addition, there is greater representation of embryonic stem cell samples in PluriNet than epiblast cell samples (15).
We also noted differences in the properties of the PluriNet/proteomics intersecting network (Fig. 3B) and the PluriNetwork/proteomics intersecting network (Fig. 3C). The PluriNet network (Fig. 3B) is noticeably less dense in terms of connectivity than the PluriNetwork (Fig. 3C), presumably attributable to the different sources of information for PluriNet and PluriNetwork. In addition, proteins represented in Figs. 3B and 3C differ in their functional categories. Table II lists the most significant functional categories. Interestingly, although some of these functional categories are the same, the degree of enrichment of the PluriNetwork associated categories is greater than that for PluriNet. For example, Embryonic development (p value = 3.41E-09 for PluriNetwork versus p value = 5.53E-05 for PluriNet) and Tissue development (p value = 1.85E-08 for PluriNetwork versus p value = 5.53E-05 for PluriNet).
Table II. Enriched functional categories and canonical pathways in mESC and mEpiSC intersecting proteins with PluriNet or PluriNetWork.
| Union Set (mESC and mEpiSC) overlap with PluriNet |
Union Set (mESC and mEpiSC) overlap with PluriNetWork |
||
|---|---|---|---|
| Functional categories | p value | Functional categories | p value |
| Connective Tissue Development and Function | 1.96E-05 | Organismal development | 9.48E-20 |
| Organismal Survival | 2.20E-05 | Embryonic Development | 3.41E-09 |
| Embryonic Development | 5.53E-05 | Tissue development | 1.85E-08 |
| Nervous System Development and Function | 5.53E-05 | Organ development | 3.96E-08 |
| Tissue Development | 5.53E-05 | Hematological System Development and Function | 1.68E-07 |
| Canonical pathways | p value | Canonical pathways | p value |
|---|---|---|---|
| Mismatch Repair in Eukaryotes | 2.52E-15 | DNA Methylation and Transcriptional Repression Signaling | 3.8E-21 |
| Cell Cycle Control of Chromosomal Replication | 2.89E-15 | Role of Oct4 in Mammalian Embryonic Stem Cell Pluripotency | 2.26E-09 |
| Hereditary Breast Cancer Signaling | 8.75E-11 | Chronic Myeloid Leukemia Signaling | 4.18E-07 |
| Role of CHK Proteins in Cell Cycle Checkpoint Control | 4.01E-09 | Role of NANOG in Mammalian Embryonic Stem Cell Pluripotency | 1.15E-06 |
| Role of BRCA1 in DNA Damage Response | 8.21E-09 | Huntington's Disease Signaling | 1.67E-06 |
Integrating the proteomics data with PluriNet and PluriNetWork also suggested that known pluripotency networks might be extended. The relatively small overlap between PluriNet and PluriNetWork (31 proteins representing ∼10% of the total from each network) and the different properties of the networks as noted above suggest that the underlying pluripotency network is incomplete. By considering PluriNet, PluriNetWork and the proteomics data set, we observed several protein complexes that are incompletely represented in the known pluripotency networks. For example, although PluriNet includes components of the chromatin remodeling BAF (Brg1 associated factor) complexes, such as SMARCD1 and SMARCAD1, and PluriNetWork includes SMARCA4 and SMARCAD1, our data suggest that additional BAF complex components SMARCB1, SMARCD2, and SMARCC2 are also intrinsic to the pluripotency network. Similarly, a tightly clustered homeobox subnetwork of PBX1, PBX2, MEIS1, MEIS2, and HOXA5 (Fig. 4) suggests that these may also be included in the core pluripotency network along with PBX1. In summary, our data add many key proteins to the clustered functional protein complexes in PluriNet and PluriNetWork.
Nuclear Proteomics Differentiates mESC and mEpiSC Protein Networks
The principal objective of this study is to identify proteins and networks that determine the pluripotent ground states of mESCs and mEpiSCs. We therefore analyzed the complete set of proteins in the union set (p < 0.1) using Ingenuity Pathways Analysis to identify subnetworks connecting proteins with significant differential expression in either the mESC or mEpiSC proteome. We noted in particular, that networks regulating gene-expression through chromatin modification and DNA methylation were highly represented with significant differential expression in the mESC or mEpiSC proteomes. Fig. 4 shows several selected protein networks showing coherent differential expression in mESC and mEpiSC proteomes. In Fig. 4A, several DNA and chromatin-modification associated protein networks showed significant enrichment in either mESCs or mEpiSCs. The Oct4-Nanog subnetwork and associated proteins is more highly expressed in the mESC proteome, along with Polycomb complex components. In contrast, several DNA methyltransferases and multiple components of SWI/SNF nucleosome remodeling complexes (also known as BRG associated factors, BAF) were found to be enriched in the mEpiSC proteome. Other notable subnetworks are shown in Figs. 4B–4D. C-terminal binding protein 1 (CTBP1) is a transcriptional co-repressor with known roles in transcriptional repression during development and in oncogenesis (35). As shown in Fig. 3C, several known interacting partners of CTBP1 show significant increased expression in the mEpiSC proteome. ZEB1 binds to CTBP1 and functions in concert with CTBP1 as a transcriptional repressor and regulator of the epithelial to mesenchymal transition (EMT) (36). The zinc-finger protein, WIZ, was shown to physically link the EHMT1 histone methyltransferase to the CTBP repressor machinery in mESCs (37). Thus, coherent increased expression of this complex suggests increased activity in mEpiSCs. Fig. 4D shows a subnetwork of interacting homeobox transcription factors also significantly enriched in mEpiSCs. PBX, MEIS, and HOX classes of homeobox are known to form trimeric complexes (38). PBX and MEIS homeoproteins act as cofactors for HOX domain transcription factors and play important roles during early mammalian development (39).
We were particularly intrigued by the differential expression of DNA methyltransferases, the polycomb complexes and the SWI/SNF-related Baf components that we observed between mESC and mEpiSC proteomes. To further qualify these observations, we cross-referenced our proteomics data set to our previous microarray-based comparison of mESC and mEpiSC gene-expression (Gene Expression Omnibus accession GSE26814; supplemental Table S4). By integrating the gene-expression microarray and proteomics data at the gene level, we observed that the global trends between the data sets show significant similarity (supplemental Fig. S4). We analyzed in more detail subsets of genes represented in our protein networks. Supplemental Table S4 contains mESC/mEpiSC ratios from the stem cell proteomics and gene-expression data sets for genes represented in the subnetworks in Fig. 4.
We first analyzed the expression of several SWI/SNF-related Baf complex components. Two Baf complex components, Smarcd1 and Smarca4 were observed to be more abundant in mEpiSCs than in mESCs at the protein and gene-expression levels. To further validate the findings from proteomics and gene-expression microarray data sets, several proteins were selected for immunoblot analysis. Fig. 5 shows immunoblot analysis (Fig. 5A) and quantification of expression (Fig. 5B) of selected components from the chromatin and DNA modification subnetwork in Fig. 4A. Proteins were selected based upon their observed differences in expression according to the spectral count analyses as well as the availability of a suitable antibody. Two independent immunoblots were performed for each protein and the results used to quantify and measure ratios of expression (Fig. 5B). We were also interested to further explore differences in expression of DNA methyltransferases. In particular, in our preliminary proteomics comparisons of mESCs and mEpiSCs, we observed dramatically increased expression of Dnmt3l (DNA methyltransferase-3 like) protein in mESCs as compared with mEpiSCs (supplemental Table S6). Since we could not confirm this finding with subsequent experiments (peptides corresponding to Dnmt3l protein were not observed in our union data set), we used anti-Dnmt3l antibody in immunoblots to compare expression in mESCs and mEpiSCs. As shown in Fig. 5, Dnmt3l protein shows markedly increased abundance in mESCs as compared with mEpiSCs. In contrast, a binding partner of Dnmt3l, Dnmt1 was more abundant in mEpiSC proteomes than in mESCs (Fig. 4A). Thus, these results combined with the proteomics data show that specific perturbations of DNA and chromatin modification networks are detectable using nuclear proteomics of mESCs and mEpiSCs and that these perturbations may determine the pluripotency ground states of mESCs and mEpiSCs.
DISCUSSION
Understanding the molecular basis of pluripotency in stem cells is imperative to being able to manipulate stem cells for applications in regenerative medicine. In addition, it is clear that there exist multiple types of stem cell with potentially different therapeutic applications. The mechanisms that maintain pluripotency in different classes of embryonic stem cells, exemplified by mESCs and mEpiSCs, may differ and it is important to understand these differences for subsequent efforts to manipulate these different cell types. In this study, we have undertaken an in depth comparison of the nuclear proteomes of mESCs and mEpiSCs. Our proteomics data sets are highly enriched in proteins important during embryogenesis and overlap and extend known networks of pluripotency associated proteins, providing a framework for future manipulation of the pluripotency state. In addition, our analysis of mutation phenotypes associated with mESCs or mEpiSCs proteins showed that there is an association of proteins expressed in each cell with known mutations at different stages of embryogenesis (Fig. 2C). mESC proteins are more likely to be correspond to genes that when mutated show embryonic lethality prior to somite formation, whereas mEpiSC proteins correspond to genes associated to lethality during organogenesis and post-natal lethality. This is significant because it shows that cultured epiblast stem cells recapitulate developmentally dependent patterns of expression, thus reinforcing their value as a model for early mammalian development. Finally, qualitative and quantitative comparisons of the mESC and mEpiSC proteomes reveal key differences in particular in the expression of protein networks associated with DNA and chromatin-modification, pointing to the importance of these processes in specifying mESC and mEpiSC pluripotency states.
This study represents the first in-depth proteomics study of epiblast stem cell pluripotency, significant because of the apparent similarity between mEpiSC state and human embryonic stem cells. The quantitative and qualitative differences between the mESC and mEpiSC proteomes have the potential to shed significant light on the different pluripotency states and properties of these two stem cell types. We observed many proteins, biological processes and protein networks that were more abundant in mEpiSCs than mESCs (Fig. 4). In particular, we observed networks of transcription factors such as the Meis-Pbx-Hox complex (Fig. 4D) that suggest the epiblast stem cells are primed for the expression of downstream developmental programs. Increased expression of Ctbp and associated proteins (Fig. 4C) in the epiblast proteome and their role in transcriptional repression and the epithelial to mesenchymal transition, also suggest that whereas the epiblast stem cells maintain pluripotency, they are also primed for key developmental transitions.
Our results emphasize the importance of chromatin remodeling protein complexes and networks in regulating the pluripotent state. Together with other studies indicating the specific role of BAF complex components (11, 17), our results suggest that different pluripotent states may in part be determined by the balance (or reconfiguration) of BAF complex components that both operate to activate and repress gene-expression. Specifically, we identified several components of ATP-dependent chromatin remodeling complexes that exhibit differential expression between mESCs and mEpiSCs. Expression of Smarcd1/Baf60a and Smarca4/Brg1 were both significantly higher in mEpiSCs than mESCs. Interestingly, Smarcd1/Baf60a mediates the interaction between several signaling pathways and BAF complexes (40). In human cells, a direct interaction between p53 with SMARCD1/BAF60A (but not between p53 and SMARCA4/BRG1) has been reported (41). Uncoupling of the interaction between SMARCD1/BAF60A and p53 resulted in repression of p53-dependent cellular functions. SMARCD1/BAF60A also mediates the interaction between nuclear hormone receptors and BAF complexes (42), governing the efficacy of chromatin remodeling in response to glucocorticoid receptor hormone signaling. Increased expression of Smarcd1/Baf60a expression in mEpiSCs, as we have observed here, may suggest that mEpiSCs are primed to respond transcriptionally to developmental and other signaling pathways. Other SWI/SNF/BAF complex components that show preferential mEpiSC or mESC expression include Baz1b, Smarcc2, Arid1a, and Arid1b (mEpiSC) and Smarcad1 (mESC). Of particular interest, an embryonic stem cell chromatin remodeling complex (esBAF) was defined by comparing mouse embryonic fibroblasts, mouse embryonic stem cells and neuron progenitors (11). Although core BAF complex components were identified across these cell types, Smarcc2/Baf170 occurred most abundantly in immuno-precipitates from neuron progenitors cells and was not a component of the esBAF complex. Our observation that Smarcc2/Baf170 is more abundant in mEpiSCs than mESCs suggests that the epiblast stem cell state is more closely related to cells such as neuron progenitors, primed for differentiation into specific cell types. We also identified Arid1a and Arid1b as more abundantly expressed in mEpiSC than mESC nuclei. These two proteins, however, are considered components of the esBAF complex (11), suggesting that differential expression of BAF complex components between mESCs and mEpiSCs is not simply because of coordinated higher expression of esBAF complex components in mESCs. As previously noted, there is considerable diversity of SWI/SNF/BAF complexes, with both generic core components and functionally-specialized components (16).
Our data also show differential expression of DNA methyltransferases between mESCs and mEpiSCs. In particular, Dnmt3l (DNA-methyltransferase 3-like) protein and mRNA are substantially more abundant in mESCs than in mEpiSCs. A recent gene-expression microarray study of induced epiblast stem cells (iEpiSCs) showed a similarly high ratio of Dnmt3l in mESCs as compared with iEpiSCs (43), in keeping with results from other studies (44, 45). In addition, Dnmt3l expression drops as pluripotent cells differentiate (45, 57). Interestingly, Dnmt3l is also expressed at low to undetectable levels in human pluripotent cell-lines, in keeping with the similarity of human embryonic stem cells to mEpiSCs (6). In contrast to Dnmt3a and Dnmt3b, Dnmt3l is catalytically inactive, but promotes methylation of DNA through functional and physical interactions with Dnmt3a and Dnmt3b (46). By interacting with other Dnmt3 family proteins, Dnmt3l facilitates binding of Dnmt3s to DNA as well as increasing their catalytic activity (59). Dnmt3a and Dnmt3b have been shown to synergistically function in de novo methylation of DNA in embryonic stem cells (47), although DNA methylation per se is apparently not required for maintenance of pluripotent stem cells, because mESCs lacking Dnmt1, Dnmt3a, and Dnmt3b are able to proliferate and maintain undifferentiated characteristics (48). Our analysis suggests intricate communication between the processes of DNA and chromatin modification (Fig. 4). Previous work showed that DNA methyltransferases may physically associate with chromatin remodeling components. In mouse lymphosarcoma cells, Dnmt3a was shown to co-immunoprecipitate with Smarca4/Brg1 (49). Given the roles of DNA and chromatin modification networks in transcriptional regulation, future studies that identify the DNA target sites of these proteins in mESCs and mEpiSCs should be highly informative. Genome binding sites of core SWI/SNF components and their protein-protein interactions, have revealed a complex picture of diverse activities and both repression and activation of gene-expression by SWI/SNF complexes (50). The potentially different and significant patterns of DNA-binding of SWI/SNF components in mESCs and mEpiSCs and their significance remain to be discovered.
Integration of mESC and mEpiSC nuclear proteomics data with existing pluripotency networks, PluriNet and PluriNetwork, shows that protein expression profiling may significantly augment existing pluripotency networks. The pluripotency network serves as a framework for studies such as ours, but as evident from our study, expression differences between components of pluripotency networks may be key to understanding the differences between pluripotent stem cell types and their pluripotent ground states. In this study, we have identified specific protein expression differences between mESCs and mEpiSCs, in protein complexes with known roles in maintaining the pluripotent state. This study supports the notion that mESCs and mEpiSCs maintain distinct pluripotent ground states. Future work will focus on understanding the functional significance of findings in this study with a view to using the protein networks as indicators of pluripotent state and as a foundation for identification of factors that can be used to manipulate the different pluripotent states.
Supplementary Material
Footnotes
* This work was supported in part by a pilot grant from the Center for Proteomics and Bioinformatics at Case Western Reserve University to P. J. T and R. M. E. R. M. E acknowledges funds from the Cleveland Foundation used in part to fund this study.
 This article contains supplemental Tables S1 to S7.
This article contains supplemental Tables S1 to S7.
1 The abbreviations used are:
- mESC
- mouse embryonic stem cell
- mEpiSC
- mouse epiblast stem cell.
REFERENCES
- 1. Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., Vallier L. (2007) Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- 2. Brook F. A., Gardner R. L. (1997) The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. U.S.A. 94, 5709–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Evans M. J., Kaufman M. H. (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 4. Martin G. R. (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tesar P. J. (2005) Derivation of germ-line-competent embryonic stem cell lines from preblastocyst mouse embryos. Proc. Natl. Acad. Sci. U.S.A. 102, 8239–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. G. (2007) New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- 7. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 8. Kim Y. Y., Ku S. Y., Rosenwaks Z., Liu H. C., Oh S. K., Moon S. Y., Choi Y. M. (2011) Red ginseng extract facilitates the early differentiation of human embryonic stem cells into mesendoderm lineage. Evidence-Based Complementary Alternative Med. 167376, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gundry R. L., Tchemyshyov I. S. S., Tarasova Y., Raginski K., Boheler K. R., Van E. Y. K., Jennifer E. (2010) Expanding the mouse embryonic stem cell proteome: Combining three proteomic approaches. Wiley-VCH: Weinheim, ALLEMAGNE, Vol. 10, p 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Hoof D., Passier R., Ward-Van Oostwaard D., Pinkse M. W., Heck A. J., Mummery C. L., Krijgsveld J. (2006) A Quest for Human and Mouse Embryonic Stem Cell-specific Proteins. Mol. Cell. Proteomics 5, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 11. Ho L., Jothi R., Ronan J. L., Cui K., Zhao K., Crabtree G. R. (2009) An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U.S.A. 106, 5187–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., Orkin S. H. (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- 13. Edwards Y. J., Bryson K., Jones D. T. (2008) A meta-analysis of microarray gene expression in mouse stem cells: redefining stemness. PLoS ONE 3, e2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Assou S., Le Carrour T., Tondeur S., Ström S., Gabelle A., Marty S., Nadal L., Pantesco V., Réme T., Hugnot J. P., Gasca S., Hovatta O., Hamamah S., Klein B., De Vos J. (2007) A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells 25, 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Müller F. J., Laurent L. C., Kostka D., Ulitsky I., Williams R., Lu C., Park I. H., Rao M. S., Shamir R., Schwartz P. H., Schmidt N. O., Loring J. F. (2008) Regulatory networks define phenotypic classes of human stem cell lines. Nature 455, 401–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ho L., Crabtree G. R. (2010) Chromatin remodelling during development. Nature 463, 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ho L., Ronan J. L., Wu J., Staahl B. T., Chen L., Kuo A., Lessard J., Nesvizhskii A. I., Ranish J., Crabtree G. R. (2009) An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 106, 5181–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kidder B. L., Palmer S., Knott J. G. (2009) SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27, 317–328 [DOI] [PubMed] [Google Scholar]
- 19. Schaniel C., Ang Y. S., Ratnakumar K., Cormier C., James T., Bernstein E., Lemischka I. R., Paddison P. J. (2009) Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells 27, 2979–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chenoweth J. G., McKay R. D., Tesar P. J. (2010) Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev. Growth Differ. 52, 293–301 [DOI] [PubMed] [Google Scholar]
- 21. Najm F. J., Chenoweth J. G., Anderson P. D., Nadeau J. H., Redline R. W., McKay R. D., Tesar P. J. (2011) Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell 8, 318–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chenoweth J. G., Tesar P. J. (2010) Isolation and maintenance of mouse epiblast stem cells. Methods Mol. Biol. 636, 25–44 [DOI] [PubMed] [Google Scholar]
- 23. Jimenez C. R., Huang L., Qiu Y., Burlingame A. L. (1998) Current Protocols in Protein Science. John Wiley and Sons Inc, New York [Google Scholar]
- 24. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Meth. 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 25. Searle B. C. (2010) Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 26. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 27. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 28. Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010) Proteome scale characterization of human s-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Storey J. (2002) A direct approach to false discovery rates. J. Roy. Stati. Soc. B 64, 479–498 [Google Scholar]
- 30. Som A., Harder C., Greber B., Siatkowski M., Paudel Y., Warsow G., Cap C., Schöler H., Fuellen G. (2010) The PluriNetWork: An Electronic Representation of the network underlying pluripotency in mouse, and its applications. PLoS ONE 5, e15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blake J. A., Bult C. J., Kadin J. A., Richardson J. E., Eppig J. T., the Mouse Genome Database G. (2011) The mouse genome database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 39, D842–D848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gassmann M., Grenacher B., Rohde B., Vogel J. (2009) Quantifying Western blots: Pitfalls of densitometry. Electrophoresis 30, 1845–1855 [DOI] [PubMed] [Google Scholar]
- 33. Tan H. Y., Ng T. W. (2008) Accurate step wedge calibration for densitometry of electrophoresis gels. Optics Commun. 281, 3013–3017 [Google Scholar]
- 34. van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010) An Oct4-Centered Protein Interaction Network in Embryonic Stem Cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chinnadurai G. (2002) CtBP, an Unconventional Transcriptional Corepressor in Development and Oncogenesis. Mol. Cell 9, 213–224 [DOI] [PubMed] [Google Scholar]
- 36. Sánchez-Tilló E., Lázaro A., Torrent R., Cuatrecasas M., Vaquero E. C., Castells A., Engel P., Postigo A. (2010) ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 29, 3490–3500 [DOI] [PubMed] [Google Scholar]
- 37. Ueda J., Tachibana M., Ikura T., Shinkai Y. (2006) Zinc Finger Protein Wiz Links G9a/GLP Histone Methyltransferases to the Co-repressor Molecule CtBP. J. Biol. Chem. 281, 20120–20128 [DOI] [PubMed] [Google Scholar]
- 38. Shanmugam K., Green N. C., Rambaldi I., Saragovi H. U., Featherstone M. S. (1999) PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol. Cell. Biol. 19, 7577–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moens C. B., Selleri L. (2006) Hox cofactors in vertebrate development. Developmental Biology 291, 193–206 [DOI] [PubMed] [Google Scholar]
- 40. Saladi S. V., Keenen B., Marathe H. G., Qi H., Chin K. V., de la Serna I. L. (2010) Modulation of extracellular matrix/adhesion molecule expression by BRG1 is associated with increased melanoma invasiveness. Mol. Cancer 9, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oh J., Sohn D. H., Ko M., Chung H., Jeon S. H., Seong R. H. (2008) BAF60a Interacts with p53 to Recruit the SWI/SNF Complex. J. Bio. Chem. 283, 11924–11934 [DOI] [PubMed] [Google Scholar]
- 42. Hsiao P. W., Fryer C. J., Trotter K. W., Wang W., Archer T. K. (2003) BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol. Cell. Biol. 23, 6210–6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han D. W., Greber B., Wu G., Tapia N., Araúzo-Bravo M. J., Ko K., Bernemann C., Stehling M., Scholer H. R. (2011) Direct reprogramming of fibroblasts into epiblast stem cells. Nat Cell Biol. 13, 66–71 [DOI] [PubMed] [Google Scholar]
- 44. Aapola U., Mäenpää K., Kaipia A., Peterson P. (2004) Epigenetic modifications affect Dnmt3L expression. Biochem. J. 380 (Pt 3), 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu Y. G., Hirasawa R., Hu J. L., Hata K., Li C. L., Jin Y., Chen T., Li E., Rigolet M., Viegas-Pequignot E., Sasaki H., Xu G. L. (2008) Regulation of DNA methylation activity through Dnmt3L promoter methylation by Dnmt3 enzymes in embryonic development. Hum. Mol. Genet. 17(17), 2654–2664 [DOI] [PubMed] [Google Scholar]
- 46. Chen Z. X., Mann J. R., Hsieh C. L., Riggs A. D., Chédin F. (2005) Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 95, 902–917 [DOI] [PubMed] [Google Scholar]
- 47. Li J. Y., Pu M. T., Hirasawa R., Li B. Z., Huang Y. N., Zeng R., Jing N. H., Chen T., Li E., Sasaki H., Xu G. L. (2007) Synergistic Function of DNA Methyltransferases Dnmt3a and Dnmt3b in the Methylation of Oct4 and Nanog. Mol. Cell. Biol. 27, 8748–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsumura A., Hayakawa T., Kumaki Y., Takebayashi S. I., Sakaue M., Matsuoka C., Shimotohno K., Ishikawa F., Li E., Ueda H. R., Nakayama J. I., Okano M. (2006) Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes to Cells 11, 805–814 [DOI] [PubMed] [Google Scholar]
- 49. Datta J., Majumder S., Bai S., Ghoshal K., Kutay H., Smith D. S., Crabb J. W., Jacob S. T. (2005) Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells. Cancer Res. 65, 10891–10900 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Euskirchen G. M., Auerbach R. K., Davidov E., Gianoulis T. A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M. B., Snyder M. (2011) Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7, e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang J., Tam W. L., Tong G. Q., Wu Q., Chan H. Y., Soh B. S., Lou Y., Yang J., Ma Y., Chai L., Ng H. H., Lufkin T., Robson P., Lim B. (2006) Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 8, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 53. Maldonado-Saldivia J., van den Bergen J., Krouskos M., Gilchrist M., Lee C., Li R., Sinclair A. H., Surani M. A., Western P. S. (2007) Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells 25, 19–28 [DOI] [PubMed] [Google Scholar]
- 54. Okuda A., Fukushima A., Nishimoto M., Orimo A., Yamagishi T., Nabeshima Y., Kuro-o M., Nabeshima Y. I., Boon K., Keaveney M., Stunnenberg H. G., Muramatsu M. (1998) UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 17, 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. In Stem Cells: Scientific Progress And Future Research Directions, National Institutes of Health, University Press of the Pacific: 2001; Vol. ISBN-10: 1410218961, pp 1–224 [Google Scholar]
- 56. Wolf D., Goff S. P. (2007) TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57 [DOI] [PubMed] [Google Scholar]
- 57. Hata K., Okano M., Lei H., Li E. (2002) Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993 [DOI] [PubMed] [Google Scholar]
- 58. Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M. M., Choudhary J. (2010) An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell. Stem Cell. Proteomic 6, 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liao H. F., Tai K. Y., Chen W. S., Cheng L. C., Ho H. N., Lin S. P. (2012) Functions of DNA methyltransferase 3-like in germ cells and beyond. Biol. Cell. June 4 doi: 10.1111/boc.201100109 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.