Abstract
Mitochondrial dysfunction has been implicated in the pathogenesis of type 2 diabetes. Identifying novel regulators of mitochondrial bioenergetics will broaden our understanding of regulatory checkpoints that coordinate complex metabolic pathways. We previously showed that Nur77, an orphan nuclear receptor of the NR4A family, regulates the expression of genes linked to glucose utilization. Here we demonstrate that expression of Nur77 in skeletal muscle also enhances mitochondrial function. We generated MCK-Nur77 transgenic mice that express wild-type Nur77 specifically in skeletal muscle. Nur77-overexpressing muscle had increased abundance of oxidative muscle fibers and mitochondrial DNA content. Transgenic muscle also exhibited enhanced oxidative metabolism, suggestive of increased mitochondrial activity. Metabolomic analysis confirmed that Nur77 transgenic muscle favored fatty acid oxidation over glucose oxidation, mimicking the metabolic profile of fasting. Nur77 expression also improved the intrinsic respiratory capacity of isolated mitochondria, likely due to the increased abundance of complex I of the electron transport chain. These changes in mitochondrial metabolism translated to improved muscle contractile function ex vivo and improved cold tolerance in vivo. Our studies outline a novel role for Nur77 in the regulation of oxidative metabolism and mitochondrial activity in skeletal muscle.
Keywords: Nr4a, nuclear receptor, mitochondria
The mitochondrion is an indispensable component for cellular respiration. To meet ongoing energy demands, cells up-regulate oxidative metabolism to boost ATP production. In skeletal muscle, a number of transcriptional factors have been shown to affect mitochondrial function and biogenesis. Peroxisome proliferator activated receptor γ coactivator 1 α (Pgc1α), for instance, coactivates a number of nuclear receptors and transcription factors (estrogen-related receptor α, nuclear respiratory factor [Nrf]1, and Nrf2) to promote mitochondrial biogenesis (1–4). Overexpression of peroxisome proliferator activated receptor delta in skeletal muscle also increases the abundance of slow-twitch muscle fiber and mitochondrial biogenesis (5). Uncovering novel biologic regulators that control mitochondrial function is fundamental to unraveling the complex metabolic pathways that control substrate utilization and energy metabolism.
Nur77 is a member of the NR4A subgroup of nuclear receptors. Structural studies have shown that these receptors lack an accessible ligand-binding pocket. The activity of these receptors in biological context appears to be primarily modulated by factors that increase abundance of the NR4A transcripts (e.g., cAMP signaling) and posttranslational modifications. Previous studies have suggested that three NR4A receptors (NR4A1, NR4A2, and NR4A3) regulate overlapping target genes and may be functionally redundant in certain contexts. Thus, depending on the tissue in question, the phenotypic consequences of deleting a single NR4A isoform may be subtle.
NR4As have been shown to regulate a diverse array of biologic processes, including thymocyte apoptosis, tumor suppression, and regulation of adrenal steroidogenesis (6–8). We and others have identified the NR4A receptors as transcriptional regulators of glucose metabolism and downstream effectors of β-adrenergic receptor signaling (9–14). NR4A transcripts are rapidly and potently induced by β-adrenergic stimulation in C2C12 myotubes and skeletal muscle in vivo (9, 11). Nur77 promotes the expression of genes controlling glucose uptake (Glut4) and glycolysis in skeletal muscle (9). Conversely, loss of Nur77 expression predisposes mice to diet-induced insulin resistance (14). We therefore hypothesized that sustained Nur77 expression in skeletal muscle would alter systemic glucose homeostasis and energy metabolism.
Here we report the generation and characterization of transgenic mouse that selectively overexpresses Nur77 in skeletal muscle from the MCK promoter. Unexpectedly, skeletal muscle from these mice was strikingly redder in color, suggestive of changes in mitochondria. Accordingly, the transgenic muscle contained more mitochondrial DNA and exhibited increased oxidative metabolism. In addition, the intrinsic respiratory control of mitochondria isolated from Nur77 transgenic muscle was improved. Finally, these changes in muscle mitochondria function and metabolism translated to increased fatigue resistance and cold tolerance in Nur77 transgenic mice. These data identify Nur77 as a novel modulator of skeletal muscle oxidative metabolism.
MATERIALS AND METHODS
Generation of MCK-Nur77 mice and animal husbandry
The murine Nur77 coding sequence was PCR amplified and cloned into the EcoRI site of the pCK4800 plasmid (gift of Eric Olson). The transgene was linearized with BssHII digestion and microinjected into pronuclei of C57BL/6J embryos by the Transgenic Mouse Facility at University of California, Irvine. Transgene genotyping was confirmed by PCR amplification of a 723 bp product using the following primers: forward, 5′ ATGAAGGAGGGTGCTGGCTACAAT 3′; reverse, 5′ TCCTCAAACTTGAAGGACGCCGAA 3′. Mice were fed ad libitum and maintained on a 12-h light-dark cycle and were age- and gender matched for all experiments. Animal studies were conducted in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and in accordance with UCLA Animal Research Committee guidelines.
In vivo studies
Male mice (4–5 months old) were fed a 45% fat diet (D12451; Research Diets, New Brunswick, NJ) for 4 weeks before the intraperitoneal glucose tolerance test. The intraperitoneal glucose tolerance test (1 g/kg of glucose) was performed after a 6-h fast during the light cycle. Body composition was determined by EchoMRI 3-in-1 System (Houston, TX). Grip strength and wire tests were performed as previously described (15). For the cold-exposure study, mice were fed ad libitum the night before and then housed individually in the morning at 4°C without bedding or food. Rectal body temperature was measured hourly with a BAT-10 digital thermometer with a Ret-3 rectal probe (Physitemp Instruments, Clifton, NJ).
Plasma and tissue chemistry
Plasma insulin was measured by Mouse Serum Adipokine Panel 7-plex kit (Millipore, Billerica, MA). Determination of fatty acylcarnitine and organic acid concentrations from gastrocnemius muscle was as previously described (16). For muscle glycogen quantitation, 30 to 100 mg of quadriceps muscle was digested in 300 μl of 30% KOH at 100°C for 10 min. Glycogen was precipitated by the addition of 60 μl of 20% Na2SO4 and 720 μl of ethanol. After incubation at −20°C for 1 h, samples were spun down and washed once with 70% ethanol. The dried glycogen pellet was digested in 100 μl 4 N H2SO4 for 10 min at 100°C. After cooling on ice, the sample was neutralized with 100 μl 4 N NaOH. Ten microliters of aliquot was removed for quantitation by glucose oxidase assay (G3660; Sigma).
Quantitative real-time PCR
Total RNA preparation and quantitative real-time PCR were performed as described (9). Expression was normalized to 36B4 expression. Primer sequences are listed in supplementary Table I and as previously described. Mitochondrial DNA content was determined by quantitative PCR amplification of mitochondrial-encoded D-loop DNA and normalized to nuclear-encoded telomerase (Tert). The template was prepared by solubilizing muscle tissue in lysis buffer (100 mM Tris [pH 8.0], 5 mM EDTA, 0.2% SDS, 200 mM NaCl) overnight followed by phenol:chloroform extraction, and ethanol precipitation. Total DNA (10 ng) was used for each reaction. Primer sequences are listed in supplementary Table I.
Immunoblotting
Total muscle lysate was prepared in a dounce homogenizer in 80 mM Tris (pH 6.8), 2% SDS, 10% glycerol, 1 mM PMSF, PhosSTOP, and Complete EDTA-free protease inhibitor cocktail (Roche). Antibody dilutions are as follows: PDH-E1α (pSer293) 1:10,000 (AP1062; Calbiochem), PDH-E1α 1:50,000 (GTX104015; GeneTex), Ppp1r1a 1:50,000 (ab40877, ABCAM), porin 1:2,000 (MSA03; MitoSciences), fission1 (Fis1) 1:5,000 (GTX111010; GeneTex), Lamin A/C 1:1,000 (Santa Cruz Biotechnology), optic atrophy protein 1 (Opa1) 1:2,000 (BD612606, BD), mitofusin (Mfn)1 1:1,000 (Clone N111/24, UC Davis/NIH NeuroMab Facility), Mfn2 1:2,000 (Ab56889, ABCAM), PSer637-dynamin-related protein (Drp)1 1:2,000 (CS6319; Cell Signaling), Drp1 1:1,000 (Wh0010059M1, clone 3B5; Sigma), and OXPHOS Rodent WB antibody cocktail 1:2,500 (MS604; MitoSciences). Antibody binding is detected by enhanced chemiluminescence. Images were captured by ImageQuant LAS4000 (GE). ImageJ V.10.2 software was used for quantification of band intensity.
Histochemical staining
Muscle was snap frozen in liquid nitrogen chilled isopentane. All staining was performed on 10 μM frozen gastrocnemius sections. For NADH-TR staining, a muscle section was incubated for 30 min at room temperature in a NADH solution prepared by combining 8 mg NADH/5 ml in Tris 0.05 M (pH 7.6) with 5 ml of freshly made nitro-blue tetrazolium (10 mg/5 ml Tris 0.05 M [pH 7.6]). Succinate dehydrogenase staining was performed by incubating the sections for 15 min at room temperature in 0.2 M sodium phosphate buffer (pH 7.6) solution with 270 mg sodium succinate and 10 mg nitro-blue tetrazolium per 10 ml. Cytochrome c oxidase staining solution consisted of 5 mg 3,3′-diaminobenzidine tetrahydrochloride, a few crystals of catalase, 10 mg cytochrome c, and 750 mg sucrose in 10 ml of 0.05 M sodium phosphate buffer (pH 7.4). Sections were stained for 1 h at room temperature. Periodic acid-Schiff staining was done by the UCLA Department of Pathology, Division of General Histology.
In vitro fatty acid oxidation
C2C12 myoblasts were plated in V7 plates (Seahorse Biosciences, MA) 1 day before adenovirus infection as described previously (9). Three days after infection, the rate of cellular oxidation of fatty acid was measured using a final concentration of 200 μM palmitate (17). Mix, wait, and measure times were 5, 2, and 1.5 min, respectively. Fatty-acid oxidation was terminated by the addition to 50 μM etomoxir. Palmitate-induced oxygen consumption is calculated by subtracting the post-etomoxir oxygen consumption rate from the peak oxygen consumption rate.
Mitochondrial respiration
Mitochondria were isolated from hamstring muscle of mice euthanized by cervical dislocation. Samples and buffers were kept on ice at all times. Hamstring muscle was rapidly isolated and minced in ice-cold IBm1 buffer (1 M sucrose, 1 M Tris/HCl [pH 7.4], 1 M KCl, 0.5 M EDTA [pH 8], 10% fatty-acid free BSA) at pH 7.4 for 2 min. The slurry was then transferred to a 15 ml conical tube on ice. The supernatant was removed, and the tissue was digested with 0.05% trypsin in IBm1 for 30 min on ice. The sample was then spun down at 200 g for 3 min, refreshed with 15 ml of IBm1 buffer, and homogenized on ice in a Potter-Elvehjam tube for nine passes at 80 rpm. The homogenate was spun down at 700 g for 10 min. The supernatant was next spun at 8,000 g for 10 min. The pelleted mitochondria were first resuspended in 500 μl IBm2 (1 M sucrose, 0.1 M EGTA, 1 M Tris/HCl [pH 7.4]) before adding an additional 4.5 ml of IBm2. The sample was spun again at 8,000 g for 10 min. The supernatant was removed, and the sample was resuspended in 75 μl of IBm2. Mitochondria concentration was determined by Bradford protein assay. Mitochondrial respiration assay was performed using the Seahorse XF24 analyzer as described with the following modifications (18). A total of 2.5 μg of mitochondria was seeded per well. The assay buffer contains 0.5 M sodium pyruvate and 0.5 M malate, each adjusted to pH ∼7.2. State III respiration was initiated with the addition of 4 mM ADP and terminated with 2 μM oligomycin. Uncoupled respiration was initiated with 4 μM carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP) and terminated with 4 μM antimycin A.
Glutathione assay
Muscle glutathione concentration was measured by GSH-Glo Glutathione Assay (Promega). Total GSH was measured by adding 500 μM tris(2-carboxyethyl)phosphine to the reaction.
Ex vivo muscle contraction
Isolated muscle stimulation was performed as previously described (19).
Tissue culture
C2C12 myoblasts were transduced with adenovirus at an MOI of 250 (9). Cells were harvested 3 days later.
Statistical analysis
Student t-test was used to determine statistical significance. Error bars represent SEM unless otherwise noted.
RESULTS
Transgenic expression of Nur77 in skeletal muscle
We previously showed that Nur77 regulates the expression of a battery of glucose utilization genes in fast-twitch muscle fibers (9). To further explore the role of Nur77 in skeletal muscle, we generated transgenic mice that expressed Nur77 from the muscle creatine kinase (MCK) enhancer (20). We generated two lines of MCK-Nur77 transgenic mice that reproduced following Mendelian ratios, and we observed similar phenotypes in both lines. We subsequently focused the majority of our efforts on progeny from the higher expressing line B.
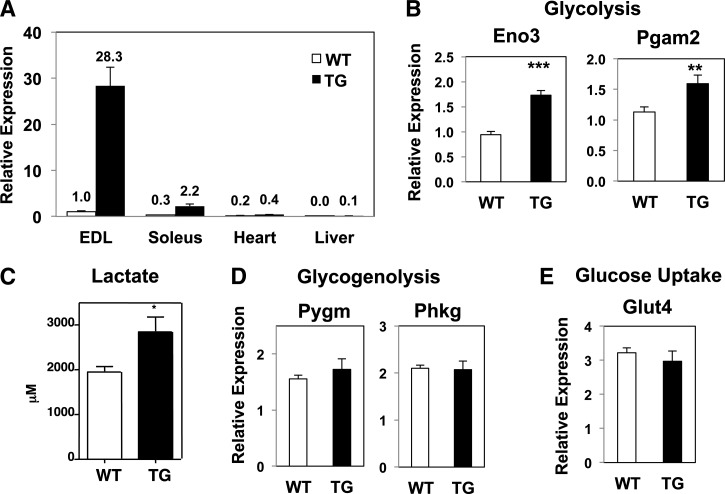
Relative to littermate controls, the level of Nur77 expression was 28-fold higher in the extensor digitorum longus (EDL) of transgenic mice (Fig. 1A). Consistent with previous characterization of the MCK promoter, Nur77 overexpression in the transgenic mouse was observed predominantly in skeletal muscle. Basal Nur77 expression in the heart was 5-fold lower than in the EDL and was increased by just 2-fold in the transgenic mice (20). We observed no change in hepatic Nur77 expression. Enhanced Nur77 activity was confirmed by induction of its target gene fructose bisphosphatase 2 in the gastrocnemius muscle (supplementary Fig. IA) (9). As expected, Nur77 was expressed at much higher levels in fast-twitch (EDL) than in slow-twitch muscle fiber (soleus) (Fig. 1A). Nur77 overexpression had no effect on body weight (supplementary Fig. IIA) or body composition (supplementary Fig. IIB). In the transgenic muscle, we confirmed up-regulation of the glycolytic genes enolase 3 and phosphoglycerate mutase 2, two previously identified targets of Nur77, although the fold-induction was modest (Fig. 1B) (9). We observed higher lactate concentrations in muscle lysates from MCK-Nur77 transgenic mice (Fig. 1C), suggesting that the modest induction in glycolytic genes was sufficient to increase glycolytic flux. Nevertheless, Nur77 overexpression in skeletal muscle did not protect the mice from diet-induced glucose intolerance (supplementary Fig. IIC). The expression of glycogenolysis genes, muscle glycogen phosphorylase and phosphorylase kinase γ 1, and the gene encoding the insulin-sensitive glucose transporter (Glut4) was not different between groups (Fig. 1D, E). We postulate that Nur77-mediated induction of gene expression in these lines may be limited by the fact that Nur77, as well as its target genes involved in glucose utilization, are already abundantly expressed in skeletal muscle, such that additional transcriptional input may have limited impact on augmenting expression.
Fig. 1.
Nur77 expression in skeletal muscle. A, B, D, E: Expression of genes from EDL muscle (except as specified in panel A) measured by quantitative PCR. Numbers in inset of panel A denote fold-change relative to wild-type EDL. C: Lactate concentration of gastrocnemius lysate. n = 4–8 for panel A; n = 7–9 for all other panels. All mice were male. All expression was normalized to 36B4. WT, wild-type; TG, transgenic. *P < 0.05; **P < 0.01; ***P < 0.001.
Nur77 increases the expression of slow-twitch muscle markers
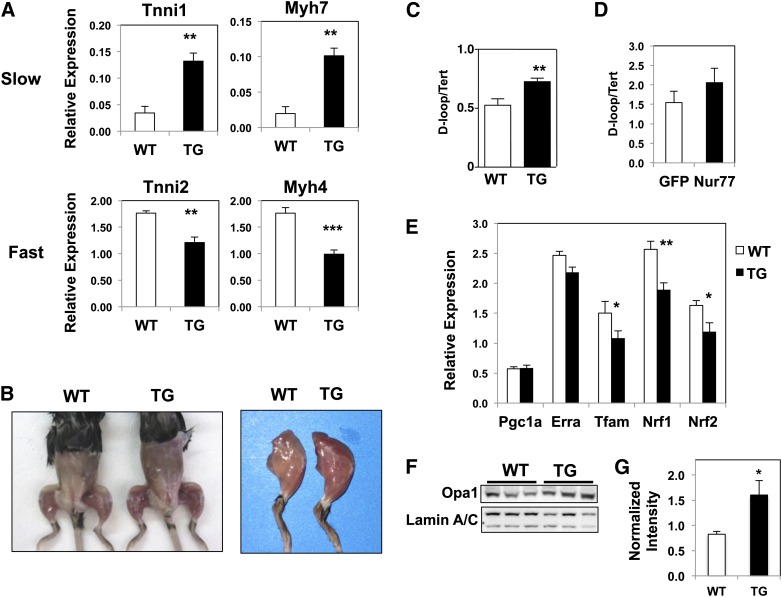
Based on the preferential expression of Nur77 in white glycolytic muscle fibers, we hypothesized that Nur77 overexpression would shift the muscle composition toward fast-twitch glycolytic fibers. Quantitative PCR analysis of fiber-type markers revealed the contrary, however. MCK-Nur77 transgenic muscle showed higher expression of troponin I type 1 and myosin heavy chain (Myh) 7, two markers of slow-twitch skeletal muscle fibers. Conversely, the expression of fast-twitch fiber markers, troponin I type 2 and Myh4, was lower in transgenic muscle (Fig. 2A). The change in fiber type composition was more pronounced in muscles that contain mixed fiber types, such as the gastrocnemius and plantaris, and less apparent in muscles that are predominantly fast-twitch, glycolytic in nature (data not shown). There was no difference in the expression of Myh1 and Myh2, which encode the myosin heavy chain for type 2d and type 2a fibers, respectively (supplementary Fig. Ib). This constellation of troponin and myosin heavy chain markers is suggestive of a shift toward slow-twitch oxidative muscle fibers in MCK-Nur77 transgenic mice.
Fig. 2.
Muscle fiber composition and mitochondrial content of MCK-Nur77 transgenic muscle. A: Expression of marker genes for fast- versus slow-twitch fibers in plantaris muscle measured by quantitative PCR (n = 7–9, male). B: Representative image of age- and gender-matched wild-type versus transgenic mice. Mice shown here are female and 12 months old. C: Mitochondrial (D-loop) DNA content normalized to nuclear DNA (Tert) content from white gastrocnemius muscle (n = 6–7; male). D: Mitochondrial DNA content from C2C12 myotubes transduced with control (GFP) or Nur77 adenovirus. Error bar represents SD. E: Expression of mitochondrial biogenesis genes in plantaris. F: Immunoblot of plantaris lysate for Opa1 and loading control Lamin A/C, with densitometry quantification of seven mice per group in panel G (n = 7–9, male). WT, wild-type; TG, transgenic. *P < 0.05; **P < 0.01; ***P < 0.001.
Increased mitochondrial content in Nur77-transgenic muscle
One defining characteristic of slow-twitch oxidative muscle is its abundance in mitochondria and myoglobin, which accounts for its reddish appearance. Accordingly, Nur77-transgenic muscle appeared strikingly redder in color and contained more mitochondria (Fig. 2B, C). We determined mitochondrial content by measuring the relative abundance of mitochondrial-encoded gene D-loop to the nuclear-encoded telomerase reverse transcriptase (Tert). Indeed, white quadriceps muscle from Nur77-transgenic mice had increased mitochondrial DNA content compared with littermate controls (Fig. 2C). To determine if the change in mitochondria abundance was directly attributable to Nur77-driven mitochondrial biogenesis, we measured mitochondrial content in C2C12 myotubes acutely transduced with adenovirus-encoding Nur77. However, unlike the transgenic muscle, acute overexpression of Nur77 in cultured myocytes did not alter mitochondrial DNA content, suggesting that the changes observed in vivo were mediated by an indirect pathway (Fig. 2D). Consistent with this hypothesis, Nur77 did not increase the expression of a number of transcriptional factors known to drive mitochondrial biogenesis (Fig. 2E). Specifically, the expression of Pgc1α and estrogen-related receptor α did not change, whereas the expression of mitochondrial transcription factor A, Nrf1, and Nrf2 (also known as Gabpa) was down-regulated in Nur77-transgenic muscle. These findings support the hypothesis that Nur77-mediated changes in mitochondrial density occur through an indirect pathway and are not the result of direct transcriptional up-regulation of mitochondrial biogenesis genes.
Changes in mitochondrial DNA content likely relate to alterations in mitochondrial dynamics. In Nur77-transgenic muscle, the expression of mitochondrial fission genes Drp1 and Fis1 homolog was reduced, whereas the expression of mitochondrial fusion genes Opa1 and Mfn2 was unchanged (supplementary Fig. IIIA). At the protein level, there was a reduced level of Fis1 and an increased abundance of Opa1 and Mfn2 (Fig. 2F, G; supplementary Fig. IIIB, C), which supports a program that promotes mitochondrial fusion at the expense of reducing mitochondrial fission. Recent studies suggest that impaired mitochondrial fusion is associated with reduced mitochondrial DNA content (21, 22). We propose that enhanced fusion and diminished mitochondrial fission has the converse effect of increasing mitochondrial DNA content. In an environment of high energy demand, such as in skeletal muscle, enhanced mitochondrial fusion can minimize local accumulation of defective or abnormal mitochondria and to increase ATP production and respiratory capacity (23).
Enhanced oxidative metabolism in Nur77-overexpressing muscle
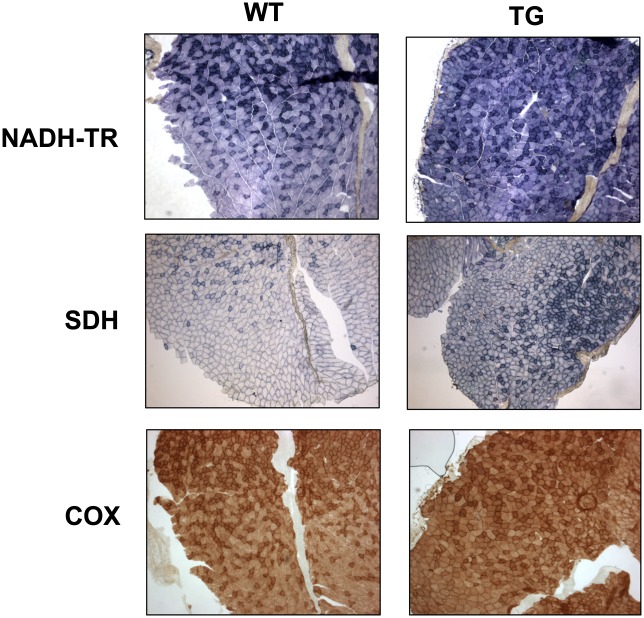
Based on the changes in mitochondrial abundance and fiber composition, we performed histochemical staining on gastrocnemius sections to determine if there was a concordant change in oxidative metabolism. The enzymatic activities of NADH-TR, succinate dehydrogenase, and cytochrome c oxidase are commonly used as markers of oxidative metabolism. Fig. 3 shows that Nur77-transgenic muscle exhibited increased pigment retention in all three assays, reflecting increased oxidative metabolism. This result is concordant with the phenotypic changes predicted on the basis of the increased mitochondrial abundance in transgenic muscle fibers.
Fig. 3.
Increased activities of oxidative metabolism markers in MCK-Nur77 muscle. Histochemical staining of 10 μM gastrocnemius cryosections for NADH-TR diaphorase, succinate dehydrogenase (SDH), and cytochrome oxidase (COX) activities. Images represent 50× magnification. WT, wild-type; TG, transgenic.
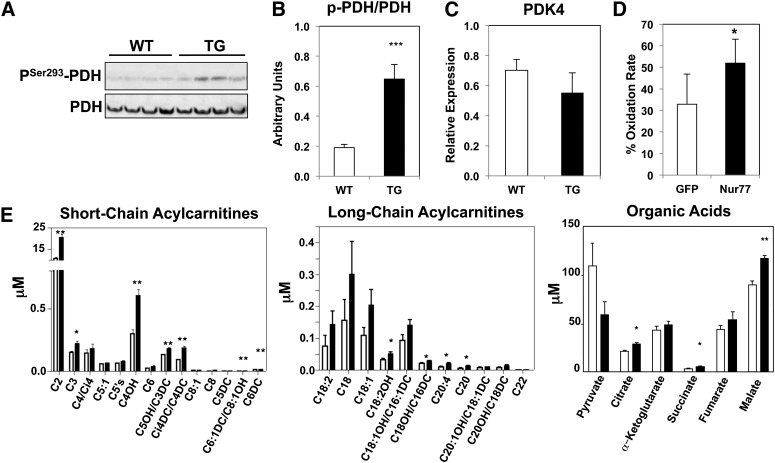
We next sought to determine whether the shift in oxidative metabolism was selective for glucose or fatty acid oxidation. Phosphorylation of pyruvate dehydrogenase by pyruvate dehydrogenase kinase (PDK)4 inhibits its enzymatic activity and limits pyruvate entry into the Krebs cycle. This mode of regulation effectively shifts the balance of substrate utilization toward fatty acid oxidation, which is indeed what we observed in the Nur77-transgenic muscle. We detected enhanced phosphorylation of pyruvate dehydrogenase at serine 293 (Fig. 4A, B). The fact that the mRNA level of PDK4 was unchanged suggests that alteration of pyruvate dehydrogenase phosphorylation occurred at the level of PDK4 activity (Fig. 4C). Our hypothesis that Nur77 facilitates fatty acid oxidation was confirmed by measuring the rate of palmitate-induced oxygen consumption in C2C12 myoblasts acutely overexpressing Nur77 (Fig. 4D).
Fig. 4.
Fatty-acid oxidation as the preferred fuel source in transgenic muscle. A: Immunoblot of plantaris lysate for PSer293-PDH and PDH. Quantitation is shown in B. C: Expression of PDK4 in plantaris. D: Fatty acid oxidation rate of C2C12 myoblasts overexpressing adeno-GFP or adeno-Nur77, corrected for non-fatty acid mediated oxygen consumption. Error bar represents SD. E: Acylcarnitine and organic acid concentrations from gastrocnemius lysate. Open bar: wild-type; Closed bar: transgenic (n = 7–9, male). WT, wild-type; TG, transgenic. *P < 0.05; **P < 0.01; ***P < 0.001.
Our hypothesis of preferential utilization of fatty acids in Nur77-transgenic muscle was further supported by acylcarnitine profiling by mass spectrometry. We observed increased abundance of short-chain acylcarnitine species such as C2-, C4-cognates, downstream byproducts of fatty acid oxidation, in Nur77-transgenic lysates (Fig. 4E). There was also a small increase in the concentration of select long-chain fatty acylcarnitines, which may occur with large increases in the flux of fatty-acid oxidation. Importantly, the increase in fatty-acid oxidation and obligate metabolism of acetyl-CoA did not deplete the Krebs cycle intermediates. In fact, transgenic muscle contained higher concentrations of anaplerotic intermediates such as succinate, fumarate, and malate, consistent with increased amino acid metabolism (Fig. 4E). Additional evidence for increased protein metabolism was the elevated abundance of C3 and C5 acylcarnitines, which are byproducts of branched-chain amino acid metabolism. Overall, the constellation of metabolomic analysis supports a model of substrate utilization in the Nur77-transgenic mice that mimics the fasting state, wherein the muscle preferentially metabolizes fatty acids and protein to fuel energy demands.
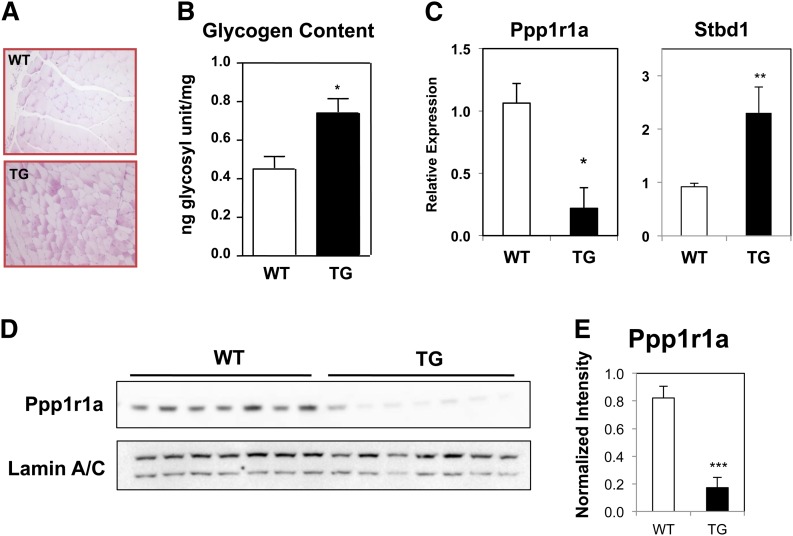
Increased glycogen content of Nur77-overexpressing muscle
Slow-twitch oxidative fibers typically have lower glycogen content because they primarily rely on triglyceride as the fuel source. Based on the shift in expression of myosin heavy chains and substrate utilization, we hypothesized that glycogen content would be reduced in Nur77-transgenic muscle. On the contrary, however, we observed increased periodic acid-Schiff staining in transgenic quadriceps sections (Fig. 5A). Increased glycogen content was confirmed by quantitation of glycosyl units released by potassium hydroxide digestion, which revealed a 60% increase in muscle glycogen content (Fig. 5B). The increase in muscle glycogen content was attributable to reduced levels of Ppp1r1a, a well-characterized inhibitor of protein phosphatase 1 (PP1) (Fig. 5C–E). PP1 inactivates glycogen phosphorylase, thereby limiting glycogenolysis. Reduced expression of Ppp1r1a would thus be predicted to increase the activity of PP1 and diminish glycogen breakdown. The increased glycogen content was accompanied by higher expression of starch-binding domain-containing protein 1, which colocalizes with glycogen macromolecule (24). The expression of starch-binding domain-containing protein 1 was previously shown to correlate positively with glycogen abundance (24), consistent with our observation in Nur77-transgenic muscle. In the context of increased abundance of mitochondria and C2 and C4 acylcarnitines, glycogenolysis in the Nur77-overexpressing muscle was likely blunted because the energy needs were met with increased fatty acid oxidation. The incongruence between glycogen content and fiber composition in our mouse model also suggests that Nur77 directs fiber-type-independent pathways to alter metabolism in skeletal muscle.
Fig. 5.
Increased glycogen content of Nur77-overexpressing muscle. A: Periodic acid-Schiff stain of formalin-fixed quadriceps section. B: Glycogen content quantitation of white gastrocnemius muscle (n = 4–8, male). C: Gene expression of plantaris muscle (n = 7–9, male). D: Immunoblot of plantaris lysate for Ppp1r1a expression. E: Quantitation of Ppp1r1a protein level relative to Lamin A/C loading control. WT, wild-type; TG, transgenic. *P < 0.05; **P < 0.01; ***P < 0.001.
Improved mitochondrial function in isolated mitochondria from Nur77-transgenic muscle
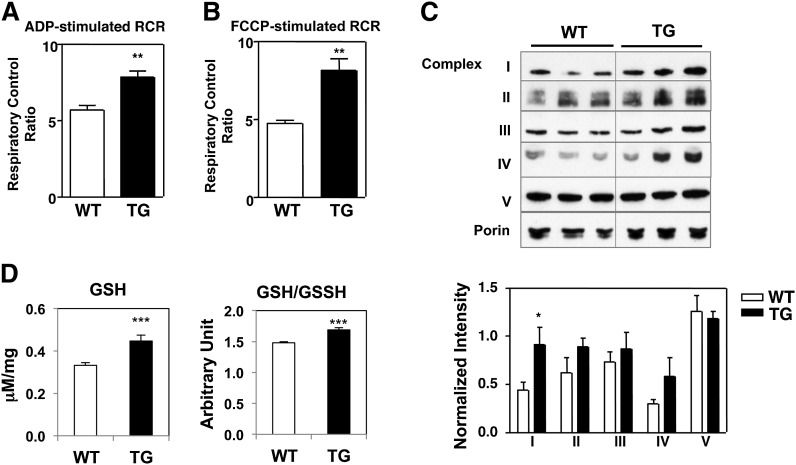
The data presented above demonstrated that Nur77 overexpression in skeletal muscle increased mitochondrial abundance and oxidative metabolism. We next sought to determine if Nur77 overexpression altered intrinsic mitochondrial function by measuring mitochondrial respiration. We isolated intact mitochondria from hamstring muscle of Nur77-transgenic mice and their littermate controls and measured the rate of oxygen consumption upon ADP-stimulation (state III respiration) and after oligomycin-mediated termination of ATP synthesis (state IVo, a measure of basal respiration). The ratio of state III to state IV respiration, also known as respiratory control ratio, is a measure of coupled respiration or of mitochondrial capacity for substrate oxidation and ATP turnover (18). Fig. 6A shows that Nur77-transgenic mitochondria exhibited a higher respiratory control ratio, consistent with enhanced coupled respiration and mitochondrial function. We observed a similar result with FCCP-stimulated respiration, where respiration is controlled exclusively by substrate oxidation (Fig. 6B). These findings demonstrate that, in addition to increasing mitochondrial content, Nur77 overexpression in skeletal muscle increases intrinsic mitochondrial oxidative capacity.
Fig. 6.
Respiratory function of isolated mitochondria from MCK-Nur77 muscle. ADP-stimulated (A) or FCCP-stimulated (B) respiration in isolated mitochondria from hamstring muscle. Result is representative of three independent experiments. C: Immunoblot analysis of electron transport chain complexes from isolated mitochondria. Porin was used as loading control. Quantitation represents average of six mice per genotype. D: Level of reduced glutathione in EDL lysate. WT, wild-type; TG, transgenic. *P < 0.05; **P < 0.01; ***P < 0.001.
We next sought to identify the mechanism by which Nur77 alters mitochondrial respiration. We quantified the abundance of individual complexes of the electron transport chain from isolated mitochondria by immunoblot analysis. As shown in Fig. 6C, Nur77-transgenic muscle contained a higher concentration of complex I (subunit NDUFB8) and a tendency for higher concentration of complex IV (subunit MTCO1). This increased abundance of electron transport chain complexes would be predicted to facilitate substrate oxidation and the respiratory control ratio that we observed in Nur77-overexpressing muscle.
To ensure that increased oxidative metabolism in the transgenic skeletal muscle did not induce oxidative stress, we measured the level of glutathione in muscle lysate. Nur77-transgenic muscle had higher levels of reduced glutathione (GSH) and a higher ratio of reduced to oxidized glutathione (GSH:GSSH), implying that the level of reactive oxygen species was lower in Nur77-overexpressing muscle (Fig. 6D).
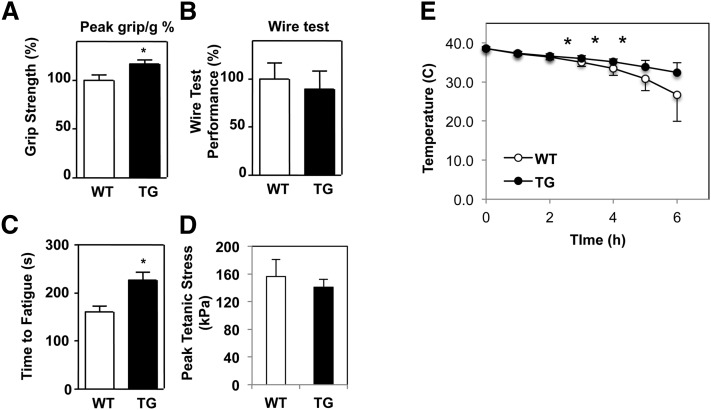
We reasoned that increased abundance of highly functional mitochondria should improve contractile performance of skeletal muscle. Indeed, Nur77-transgenic mice exhibited increased average grip strength compared with wild-type littermates (Fig. 7A). However, there was no difference in hanging time with the wire test (Fig. 7B). Ex vivo, transgenic EDL muscle was more resistant to fatigue in response to tetanic stimulation, consistent with an endurance phenotype predicted by increased muscle mitochondrial content in the transgenic mice (Fig. 7C) (19). There was no difference in contractile force generated between control and Nur77-transgenic mice (Fig. 7D). These findings underscore the functional importance of increased Nur77 expression in skeletal muscle and its ability to augment mitochondrial content and contractile performance.
Fig. 7.
MCK-Nur77 mice have improved muscle performance. A: Peak grip strength out of five trials corrected for body weight and normalized to wild-type control. B: Duration of mice hanging from wire over five trials corrected for body weight and normalized to wild-type control (n = 6, 12 months old, female). C and D: EDL muscle was isolated ex vivo and subjected to isometric tetanic contractions. Time to fatigue (C) was defined as the time it took for the generated force to drop to 60% of the maximal initial force. D: Peak tetanic stress is calculated by normalizing peak tetanic force to the cross-sectional area of the muscle (n = 4–6, 7 months old, female). E: Improved cold tolerance in MCK-Nur77 transgenic mice (n = 7, 6–8 months old, male). WT, wild-type; TG, transgenic. *P < 0.05.
As an additional measure of muscle function, we tested the ability of MCK-Nur77 mice to withstand cold stress. The first line of defense against acute cold exposure is shivering thermogenesis by skeletal muscle (25). We hypothesized that Nur77-transgenic mice, with their increased mitochondrial abundance and function, would have improved cold tolerance. Indeed, over the course of 6 h at 4°C, 6- to 8-month-old transgenic mice were able to better preserve their core body temperature in the fasting state, relative to wild-type littermate controls, demonstrating that Nur77-mediated changes in muscle mitochondria function and metabolism contribute to cold tolerance (Fig. 7E). We posit that increased mitochondrial function and muscle glycogen content enhance energy production in the transgenic mice to facilitate shivering thermogenesis. We verified that the effect observed was not due to expression of the Nur77-transgene in nonmuscle tissue (supplementary Fig. IVA), and there was no difference in cold-induced brown adipose tissue markers between control and transgenic mice (supplementary Fig. IVB).
DISCUSSION
Oxidative metabolism is an efficient mode of cellular energy production that is of particular importance during the fasting state and in endurance exercise. In this study, we identified Nur77 as a novel regulator of oxidative metabolism. Muscle-specific Nur77-overexpression shifted the expression of fiber-specific markers toward type 1 oxidative fibers. We showed that Nur77-transgenic muscle had higher succinate dehydrogenase, NADH-TR, and cytochrome oxidase activity and increased fatty acid oxidation based on abundance of fatty acid oxidation byproducts. The change in oxidative metabolism was attributable to an increase in mitochondrial content and to improved intrinsic mitochondrial function. Functionally, the improvement in oxidative metabolism translated into fatigue resistance and cold tolerance. Our studies thus highlight Nur77 as a novel transcriptional facilitator of oxidative metabolism in skeletal muscle.
We previously implicated Nur77 in the transcriptional regulation of genes involved in glucose utilization downstream of cAMP signaling. Nur77-responsive genes include GLUT4 and numerous glycolytic and glycogenolytic genes. The predicted results of Nur77 activation in this pathway include rapid ATP production through anaerobic metabolism. Other evidence supports the notion that Nur77 is induced by activation of the sympathetic nervous system (10, 26). Under acute physiologic stress, β-adrenergic stimulation may up-regulate Nur77 transcription, which then directs the glycolytic and glycogenolytic programs to increase ATP production in skeletal muscle. Our current finding that stable Nur77 overexpression in skeletal muscle promotes oxidative metabolism implies that Nur77 may also have a role in driving physiologic adaptation during chronic stress.
Several lines of evidence support the postulate that Nur77 may orchestrate effector pathways in chronic stress. First, NGFI-B (rat NR4A1) plays a role in the development of the rat adrenal cortex (27). Second, in mouse Y1 adrenocortical cells, Wilson et al. (8) showed that NGFI-B mediates adrenocorticotropic hormone (ACTH)-induced expression of 21-hydroxylase, the enzyme that catalyzes the final step of cortisol biosynthesis. In AtT-20 pituitary corticotroph cells, corticotropin-releasing hormone induces the expression of Nur77 (28–31). Finally, Nur77 blocks the negative feedback of glucocorticoid on ACTH expression, thereby permitting continuous ACTH synthesis and secretion to direct cortisol secretion during sustained physiologic stress (28, 31). We propose that Nur77-mediated increase in oxidative metabolism in skeletal muscle is an additional pathway by which Nur77 supports the metabolic needs during chronic stress.
Consistent with our findings, Pearen et al. (32) recently reported that skeletal muscle overexpression of supraphysiologically active Nor1-VP16 fusion protein promoted a switch toward oxidative fiber type, albeit the Nor1-VP16 transgenic mice exhibited increased abundance of type 2a, not type 1, fibers. The precise mechanism by which Nr4a alters fiber composition remains unclear. One plausible scenario is Nr4a-mediated activation of the calcineurin-nuclear factor of activated T cell signaling cascade, which then promotes formation of slow-twitch muscle fibers (33). The calcineurin-nuclear factor of activated T cell pathway, by way of MEF2, also plays a role in activation of Pgc1α, which then promotes mitochondrial biogenesis (34). It is also intriguing to consider the notion that Nur77-mediated changes in mitochondrial dynamics may secondarily influence changes in fiber composition.
The Nor1-VP16 mouse also displayed evidence of increased mitochondrial fusion as well as mitochondrial DNA content. These observations raise the interesting question of the function of Nur77 in the regulation of mitochondrial dynamics. De novo synthesis of mitochondria requires significant energy expenditure and time delay. Emerging evidence supports the model that, under conditions of high energy demand, mitochondrial fusion is an efficient and rapid way to maximize the utility of suborganelles (35). Mitochondrial fusion has the additional benefit of preventing local accumulation of defective mitochondria (23). In conjunction with mitochondrial turnover, altering the balance of mitochondrial dynamics provides a defense mechanism by limiting the oxidative stress of defective mitochondria (36). By enhancing mitochondrial fusion, Nur77 has thus provided another means to improve respiratory efficiency and fuel production to meet the energy demand of skeletal muscle and the organism at large. Additional studies are ongoing to address the mechanism by which Nur77 regulates mitochondrial dynamics and its relationship to mitophagy.
The observation that Nur77 and Nor1-VP16 overexpression in skeletal muscle resulted in similar phenotypes provides further evidence for the functional redundancy of these receptors in many biological contexts. This redundancy likely explains the absence of prominent deficits in oxidative metabolism or changes in mitochondrial content in muscles of Nur77 knockout mice (data not shown). Because Nur77/Nor1 double knockout mice succumb to acute myeloid leukemia before weaning (7), complex breeding strategies to generate tissue-specific deletion of Nur77 and Nor1 are needed to determine if mitochondrial biogenesis is impaired in compound mutant mice. We did not observe improvement in systemic glucose metabolism in Nur77-transgenic mice, in contrast to what was reported for the Nor1-VP16 transgenic mouse (32). The differences in our findings may be explained by the use of a viral transcription-activating domain to augment physiologic Nor1 activity in the study of Pearen et al (32).
In summary, we have identified Nur77 as a novel regulator of oxidative metabolism in skeletal muscle. Our demonstration that acute overexpression of Nur77 induces glycolytic programming, whereas sustained overexpression promotes oxidative metabolism, positions Nur77 as an integral mediator of physiologic stress.
Supplementary Material
Acknowledgments
The authors thank Eric Olson for the gift of the MCK enhancer plasmid and Dr. Claudio Villanueva for helpful discussions.
Footnotes
Abbreviations:
- ACTH
- adrenocorticotropic hormone
- Drp
- dynamin-related protein
- EDL
- extensor digitorum longus
- FCCP
- carbonylcyanide p-trifluoromethoxyphenylhydrazone
- Fis1
- fission1
- GSH
- glutathione
- MCK
- muscle creatine kinase
- Mfn
- mitofusin
- Myh
- myosin heavy chain
- Nrf
- nuclear respiratory factor
- Opa1
- optic atrophy protein 1
- PDK
- pyruvate dehydrogenase kinase
- Pgc1α
- peroxisome proliferator activated receptor γ coactivator 1 α
- PP1
- protein phosphatase 1
This work was funded by National Institutes of Health grants DK081683, HL030568, HL028481, and 5P30DK063491. The project was supported partially through funding from the UCLA Muscular Dystrophy Core Center grant 5P30AR057230 and the National Center for Research Resources grant S10RR026744. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures and one table.
REFERENCES
- 1.Arany Z., Lebrasseur N., Morris C., Smith E., Yang W., Ma Y., Chin S., Spiegelman B. M. 2007. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 5: 35–46 [DOI] [PubMed] [Google Scholar]
- 2.Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., et al. 2002. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 418: 797–801 [DOI] [PubMed] [Google Scholar]
- 3.Hock M. B., Kralli A. 2009. Transcriptional control of mitochondrial biogenesis and function. Annu. Rev. Physiol. 71: 177–203 [DOI] [PubMed] [Google Scholar]
- 4.Schreiber S. N., Emter R., Hock M. B., Knutti D., Cardenas J., Podvinec M., Oakeley E. J., Kralli A. 2004. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA. 101: 6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y. X., Zhang C. L., Yu R. T., Cho H. K., Nelson M. C., Bayuga-Ocampo C. R., Ham J., Kang H., Evans R. M. 2004. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2: e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woronicz J. D., Calnan B., Ngo V., Winoto A. 1994. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 367: 277–281 [DOI] [PubMed] [Google Scholar]
- 7.Mullican S. E., Zhang S., Konopleva M., Ruvolo V., Andreeff M., Milbrandt J., Conneely O. M. 2007. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat. Med. 13: 730–735 [DOI] [PubMed] [Google Scholar]
- 8.Wilson T. E., Mouw A. R., Weaver C. A., Milbrandt J., Parker K. L. 1993. The orphan nuclear receptor NGFI-B regulates expression of the gene encoding steroid 21-hydroxylase. Mol. Cell. Biol. 13: 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao L. C., Zhang Z., Pei L., Saito T., Tontonoz P., Pilch P. F. 2007. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Mol. Endocrinol. 21: 2152–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearen M. A., Myers S. A., Raichur S., Ryall J. G., Lynch G. S., Muscat G. E. 2008. The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 149: 2853–2865 [DOI] [PubMed] [Google Scholar]
- 11.Pearen M. A., Ryall J. G., Maxwell M. A., Ohkura N., Lynch G. S., Muscat G. E. 2006. The orphan nuclear receptor, NOR-1, is a target of beta-adrenergic signaling in skeletal muscle. Endocrinology. 147: 5217–5227 [DOI] [PubMed] [Google Scholar]
- 12.Pei L., Castrillo A., Tontonoz P. 2006. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol. Endocrinol. 20: 786–794 [DOI] [PubMed] [Google Scholar]
- 13.Maxwell M. A., Cleasby M. E., Harding A., Stark A., Cooney G. J., Muscat G. E. 2005. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. J. Biol. Chem. 280: 12573–12584 [DOI] [PubMed] [Google Scholar]
- 14.Chao L. C., Wroblewski K., Zhang Z., Pei L., Vergnes L., Ilkayeva O. R., Ding S. Y., Reue K., Watt M. J., Newgard C. B., et al. 2009. Insulin resistance and altered systemic glucose metabolism in mice lacking Nur77. Diabetes. 58: 2788–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudryashova E., Struyk A., Mokhonova E., Cannon S. C., Spencer M. J. 2011. The common missense mutation D489N in TRIM32 causing limb girdle muscular dystrophy 2H leads to loss of the mutated protein in knock-in mice resulting in a Trim32-null phenotype. Hum. Mol. Genet. 20: 3925–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koves T. R., Ussher J. R., Noland R. C., Slentz D., Mosedale M., Ilkayeva O., Bain J., Stevens R., Dyck J. R., Newgard C. B., et al. 2008. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 7: 45–56 [DOI] [PubMed] [Google Scholar]
- 17.Wu M., Neilson A., Swift A. L., Moran R., Tamagnine J., Parslow D., Armistead S., Lemire K., Orrell J., Teich J., et al. 2007. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell Physiol. 292: C125–C136 [DOI] [PubMed] [Google Scholar]
- 18.Rogers G. W., Brand M. D., Petrosyan S., Ashok D., Elorza A. A., Ferrick D. A., Murphy A. N. 2011. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS ONE. 6: e21746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philp A., Chen A., Lan D., Meyer G. A., Murphy A. N., Knapp A. E., Olfert I. M., McCurdy C. E., Marcotte G. R., Hogan M. C., et al. 2011. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) deacetylation following endurance exercise. J. Biol. Chem. 286: 30561–30570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naya F. J., Mercer B., Shelton J., Richardson J. A., Williams R. S., Olson E. N. 2000. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem. 275: 4545–4548 [DOI] [PubMed] [Google Scholar]
- 21.Chen H., Vermulst M., Wang Y. E., Chomyn A., Prolla T. A., McCaffery J. M., Chan D. C. 2010. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 141: 280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Civitarese A. E., MacLean P. S., Carling S., Kerr-Bayles L., McMillan R. P., Pierce A., Becker T. C., Moro C., Finlayson J., Lefort N., et al. 2010. Regulation of skeletal muscle oxidative capacity and insulin signaling by the mitochondrial rhomboid protease PARL. Cell Metab. 11: 412–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romanello V., Sandri M. 2012. Mitochondrial biogenesis and fragmentation as regulators of protein degradation in striated muscles. J. Mol. Cell. Cardiol. In press. [DOI] [PubMed] [Google Scholar]
- 24.Jiang S., Heller B., Tagliabracci V. S., Zhai L., Irimia J. M., DePaoli-Roach A. A., Wells C. D., Skurat A. V., Roach P. J. 2010. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J. Biol. Chem. 285: 34960–34971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannon B., Nedergaard J. 2011. Nonshivering thermogenesis and its adequate measurement in metabolic studies. J. Exp. Biol. 214: 242–253 [DOI] [PubMed] [Google Scholar]
- 26.Myers S. A., Eriksson N., Burow R., Wang S. C., Muscat G. E. 2009. Beta-adrenergic signaling regulates NR4A nuclear receptor and metabolic gene expression in multiple tissues. Mol. Cell. Endocrinol. 309: 101–108 [DOI] [PubMed] [Google Scholar]
- 27.Honkaniemi J., Zhang J. S., Longo F. M., Sharp F. R. 2000. Stress induces zinc finger immediate early genes in the rat adrenal gland. Brain Res. 877: 203–208 [DOI] [PubMed] [Google Scholar]
- 28.Murphy E. P., Conneely O. M. 1997. Neuroendocrine regulation of the hypothalamic pituitary adrenal axis by the nurr1/nur77 subfamily of nuclear receptors. Mol. Endocrinol. 11: 39–47 [DOI] [PubMed] [Google Scholar]
- 29.Parkes D., Rivest S., Lee S., Rivier C., Vale W. 1993. Corticotropin-releasing factor activates c-fos, NGFI-B, and corticotropin-releasing factor gene expression within the paraventricular nucleus of the rat hypothalamus. Mol. Endocrinol. 7: 1357–1367 [DOI] [PubMed] [Google Scholar]
- 30.Stroth N., Liu Y., Aguilera G., Eiden L. E. 2011. Pituitary adenylate cyclase-activating polypeptide controls stimulus-transcription coupling in the hypothalamic-pituitary-adrenal axis to mediate sustained hormone secretion during stress. J. Neuroendocrinol. 23: 944–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okabe T., Takayanagi R., Adachi M., Imasaki K., Nawata H. M. S. 1998. Nur77, a member of the steroid receptor superfamily, antagonizes negative feedback of ACTH synthesis and secretion by glucocorticoid in pituitary corticotrope cells. J. Endocrinol. 156: 169–175 [DOI] [PubMed] [Google Scholar]
- 32.Pearen M. A., Eriksson N. A., Fitzsimmons R. L., Goode J. M., Martel N., Andrikopoulos S., Muscat G. E. 2012. The nuclear receptor, Nor-1, markedly increases type II oxidative muscle fibers and resistance to fatigue. Mol. Endocrinol. 26: 372–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin E. R., Olson E. N., Richardson J. A., Yang Q., Humphries C., Shelton J. M., Wu H., Zhu W., Bassel-Duby R., Williams R. S. 1998. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 12: 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Lluch G., Irusta P. M., Navas P., de Cabo R. 2008. Mitochondrial biogenesis and healthy aging. Exp. Gerontol. 43: 813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soriano F. X., Liesa M., Bach D., Chan D. C., Palacin M., Zorzano A. 2006. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes. 55: 1783–1791 [DOI] [PubMed] [Google Scholar]
- 36.Westermann B. 2010. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11: 872–884 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.