Abstract
The HLA region shows diversity concerning the number and content of DRB genes present per haplotype. Similar observations are made for the equivalent regions in other primate species. To elucidate the evolutionary history of the various HLA-DRB genes, a large panel of intron sequences obtained from humans, chimpanzees, rhesus macaques, and common marmosets has been subjected to phylogenetic analyses. Special attention was paid to the presence and absence of particular transposable elements and/or to their segments. The sharing of different parts of the same long interspersed nuclear element-2 (LINE2, L2) and various Alu insertions by the species studied demonstrates that one precursor gene must have been duplicated several times before the Old World monkey (OWM) and hominid (HOM) divergence. At least four ancestral DRB gene families appear to have been present before the radiation of OWM and HOM, and one of these even predates the speciation of Old and New World primates. Two of these families represent the pseudogenes DRB6/DRB2 and DRB7, which have been locked in the genomes of various primate species over long evolutionary time spans. Furthermore, all phylogenies of different intron segments show consistently that, apart from the pseudogenes, only DRB5 genes are shared by OWM and HOM, and they demonstrate the common history of certain DRB genes/lineages of humans and chimpanzees. In contrast, the evolutionary history of some other DRB loci is difficult to decipher, thus illustrating the complex history of the evolution of DRB genes due to a combination of mutations and recombination-like events. The selected approach allowed us to shed light on the ancestral DRB gene pool in primates and on the evolutionary relationship of the various HLA-DRB genes.
Keywords: MHC, HLA, evolution, transposable elements, introns, primates
Introduction
HLA-DR (Human Leukocyte Antigen-D-related) specificities have originally been described as the serological equivalents of HLA-D that had earlier been defined by cellular assays (van Rood et al. 1977). Subsequent sequencing, transfection studies, and reactivity profiles with alloantisera pinpointed which major histocompatibility complex (MHC) class II molecules were responsible for the reactivity with these antisera. Later on, molecular studies demonstrated the presence of one DRA gene, encoding the alpha chain, and multiple DRB genes, encoding the beta chain, on different haplotypes, which display, in addition, copy number variation. Similar analyses have been performed with nonhuman primates allowing the identification of multiple DRB genes as well. Although the DRA gene in humans is highly conserved, nine different HLA-DRB genes have been described. HLA-DRB1, -B3, and -B5 encode functional gene products, whereas -B2, -B6, -B7, -B8, and -B9 represent pseudogenes as manifested by various insertions/deletions (indels) and deleterious mutations. HLA-DRB4 may represent a pseudogene, depending on the haplotype on which it is embedded. HLA-DRB genes show different degrees of allelic variation, of which DRB1 is the most polymorphic. HLA-DRB6 is an example of a highly variable pseudogene, whereas -DRB7 is more or less monomorphic. In humans and other primate species, each haplotype contains a DRA gene in concert with various numbers of DRB genes. Within the human population, five major region configurations have been described, characterized by the presence of a unique combination of DRB genes/pseudogenes per haplotype. Region configuration polymorphism may be even more pronounced in other primate species. For the chimpanzee (Pan troglodytes) and some macaque species (Macaca mulatta and M. fascicularis), nine or even more than 30 different DRB haplotypes have been described, respectively, whereas such polymorphism is absent in common marmosets (Callithrix jacchus), a New World primate species (Antunes et al. 1998; de Groot et al. 2008; Doxiadis, de Groot N, de Groot, et al. 2008). The paralogous or orthologous relationship of the various DRB genes in primates appears to be complex and has long been a matter of debate.
The divergence time of Old World monkey (OWM)-hominid (HOM) on the one hand and OWM-New World monkeys (NWM) on the other hand have been calculated to be ∼25 My, and ∼36 Ma, respectively (Glazko and Nei 2003; Perelman et al. 2011). Most primate DRB alleles appear to be relatively young entities, as sharing is only observed for closely related species (Blancher et al. 2006; Doxiadis et al. 2006; O’Connor et al. 2007). Comparative studies, mainly based on exon 2 analyses, demonstrate that orthologs of different HLA-DRB genes may be present in other species such as the chimpanzee, gorilla, orangutans, and macaques (Klein et al. 1991; Brandle et al. 1992; Schonbach et al. 1993; Slierendregt et al. 1994). In NWM, however, similarity of DRB exon 2 segments appears to be based on convergent evolution (Antunes et al. 1998; Kriener, O'HUigin, Tichy, et al. 2000). Subsequent intron analyses, indeed, showed that the majority of contemporary HLA-DRB1 alleles have been generated during the past 250,000 years, whereas many DRB lineages and loci predate the divergence of the hominoid species (Erlich et al. 1996; Bergstrom et al. 1998). A recent comparison of exon 2 and introns obtained from HOMs and OWMs indicated that DRB5 and DRB6 loci are >25-My-old entities (Doxiadis, de Groot N, de Groot, et al. 2008). However, the number of DRB genes that have been present in the ancestral pool of primates at given speciation events is not yet known.
In addition to intron sequences being studied, the sharing of certain transposable elements (TE) has been employed for the deciphering of the evolutionary relationships of genes. TE, first described in 1956 (McClintock 1956), were regarded for a long time as “junk” DNA but appear to make up more than 40% of the mammalian genome (Bannert and Kurth 2004, 2006; Goodier and Kazazian 2008; Oliver and Greene 2011). In the past decade, TE have been increasingly recognized as powerful facilitators of evolution by promoting recombination and gene transfer (Deininger and Batzer 2002; Bannert and Kurth 2004; Kazazian 2004; Belancio et al. 2008; Goodier and Kazazian 2008; Xing et al. 2009; Britten 2010; Iskow et al. 2010; Levin and Moran 2011; Munoz-Lopez et al. 2011; Oliver and Greene 2011).
There are mainly three groups of class I retro elements, all moving in a “copy and paste” manner and involving the reverse transcription of an RNA intermediate and an insertion of its copy into a new site in the genome. The first group consists of retrotransposons characterized by long terminal repeats (LTR) and includes, for instance, the human endogenous retroviruses (HERV) comprising ∼8% of the human genome. These are relics of past rounds of germline infections caused by viruses that lost their ability to reinfect and became trapped in the host genome (Goodier and Kazazian 2008; Oliver and Greene 2011). Several ERV families are also present in both OWM and NWM, indicating that the primary infection event occurred more than 25 Ma (Bannert and Kurth 2006).
The second and most numerous group comprises the long interspersed nuclear repeats (LINE), which make up approximately 20% of the human genome. These elements lack LTR structures. The oldest element, LINE2 (L2), represents an autonomous retrotransposon that was spread before the mammalian radiation and is now present as a truncated molecular fossil at 3% in our genome (Smith A, personal communication). L2 has a maximum length of 5 kb; however, the only available consensus sequence is 3-kb long and has only one open reading frame for pol (Kohany et al. 2006; Oliver and Greene 2011). Far more abundant are LINE1 (L1) elements, which comprise 17% of the human DNA and have been active in the mammalian genomes since the marsupial–eutherian split and have expanded over the past 100–150 My (Smit 1996). However, the expansion has slowed since 25 Ma, and most human L1 insertions are molecular fossils that cannot retrotranspose to new genomic locations.
The third group of TE comprises the short interspersed nuclear elements (SINE). SINEs are small, nonautonomous elements, because they have no protein coding capacity. They are therefore dependent on LINEs for their amplification and retrotransposition. In humans, Alu elements are the most abundant SINEs, with ∼1 million copies, making up more than 10% of the genome. Full-length Alu elements are ∼300 bp long and commonly found in introns and intergenic segments. Most of the Alu elements duplicated more than 40 Ma (AluSx and AluJ), whereas others are far younger, such as AluSg (∼25 My) and AluY, which expanded during the period of the OWM and HOM divergence (Jurka and Smith 1988; Batzer and Deininger 2002).
In the past, TE have also been used to decipher the evolutionary relationships of Mhc class I genes (Sawai et al. 2004) and class II DRB genes (Andersson et al. 1987; Mayer et al. 1993; Mnukova-Fajdelova et al. 1994; Klein and O’HUigin 1995; Satta et al. 1996; Gongora, Figueroa, Klein, et al. 1997; Andersson et al. 1998; Fernandez-Soria et al. 1998; Bontrop et al. 1999; Kriener, O'HUigin, Klein, et al. 2000; Klein et al. 2007; Doxiadis, de Groot, Bontrop, et al. 2008). However, because of the limited number of TE and DRB sequences that were available for analysis, there is as yet no clear picture of the evolution of these genes. Thus, the aim of this study was to analyze the integration of the various TE into the introns of DRB genes of common marmosets, rhesus macaques, chimpanzees, and humans. To gain a complete as possible insight into the evolution of the DRB genes in primates, we compared the resulting data with the phylogenies of the diverse DRB intron sequences of the species studied.
Materials and Methods
Nomenclature
Most nonhuman primate DRB genes and alleles have been named on the basis of the similarity of exon 2 to HLA counterparts. In the instance in which these similarities are lacking, DRB sequences received a “W” (Workshop) designation (Robinson et al. 2003). Allele names are given according to the nomenclature for factors of the HLA system (Marsh et al. 2010; de Groot et al. 2012).
Long Polymerase Chain Reaction of Patr-DRB Alleles
Genomic DNA has been extracted from immortalized B-cell lines of two chimpanzees that were known to encode for Patr-DRB1*10:01 (Lady, Pan troglodytes verus) and Patr-DRB4*02:01 (Brigitte, Pan troglodytes troglodytes; de Groot et al. 2009) using a standard salting-out procedure. For the amplification of intron 1 to exon 5 of Patr-DRB1*10:01, two different amplifications were performed with the primer sets Patr-DRB1*1001F AAG CTC CCT GGA GGC TCC TGC ATG GCA GCG and Patr-DRB1*1001-R′ GCG CAC GTC CTC CTC CTG GTT ATG GAT GCG for the amplification of intron 1 and DRB1*10_ex2-F2 AGC GGG TGC GGT TGC TGG AAA GAC GCG TCC and 3′Mamu-DRB1*0306/1007-ex5_R CCT GTT GGC TGA AGT CCA GAG TGT CCT GGG for the amplification of exon 2 to exon 5, respectively. Polymerase chain reactions (PCRs) were performed in a 50 µl reaction volume containing 2.5 units of Long PCR Enzyme mix with 0.6 µM of each primer, 5 mM MgCl2, 1× PCR buffer, 0.2 µM dNTPs (Fermentas, Germany), and 10 µl of 50 ng/µl genomic DNA.
The cycling parameters for both reactions consisted of a denaturation step of 2 min at 94°C initial, followed by 10 cycles with a denaturation step of 10 s at 94°C, an annealing step of 30 s at 60°C, and an extension step of 7 min at 68°C, then followed by 20 cycles with a denaturation step of 10 s at 94°C, an annealing step of 30 s at 58 °C, and an extension step of 8 min at 68°C. A final extension step was performed at 68°C for 10 min. Accordingly, two different PCR reactions were performed for the amplification of Patr-DRB4*02:01. Primer pair 5′Mamu-DRB1*0306-ex1_F TGG AGG CTC CTG CAT GGC AGC GCT GAC AGT and Patr-DRB7*0101-ex2_R ATA GTT GTG TCT GCA GTA GGT GTC CAC CGC were used for the amplification of intron 1, and primer pair Patr-DRB4*0201_ex2_F1 ACA GCA CGT TTC TTG GAG CAG CTT AAG TTT and 5′DRB1*0306/1007-ex5_R CCT GTT GGC TGA AGT CCA GAG TGT CCT GGG for the amplification of exon 2 to intron 4 (partial). The PCR reactions were performed in a 50 µl reaction volume containing 2.5 units of Long PCR Enzyme mix with 0.6 µM of each primer, 5 mM MgCl2, 1× PCR buffer, 0.2 µM dNTPs (Fermentas), and 10 µl of 50 ng/µl genomic DNA. The cycling parameters for the amplification of intron 1 were an initial denaturation step of 2 min at 94°C, followed by 10 cycles with a denaturation step of 10 s at 94°C, an annealing step of 30 s at 60°C, and an extension step of 14 min at 68°C, followed by 20 cycles with a denaturation step of 10 s at 94°C, an annealing step of 30 s at 58°C, and an extension step of 15 min at 68°C. A final extension step was performed at 68°C for 10 min. The cycling parameters for the amplification of exon 2 to intron 4 (partial) were an initial denaturation step for 2 min at 94°C, followed by 10 cycles with a denaturation step of 10 s at 94°C, an extension step of 30 s at 64°C, and an annealing step of 7 min at 68°C, followed by 25 cycles with a denaturation step of 10 s at 94°C, an annealing step of 30 s at 63°C, and an extension step of 8 min at 68°C. A final extension step was performed at 68°C for 10 min.
Sanger Sequencing
PCR fragments were purified using a gel extraction kit (Fermentas) according to the manufacturer’s guidelines. Purified PCR products were sequenced on an ABI 3130xl Genetic analyser (Applied Biosystems, Foster City, CA, USA) with the help of several internal primers synthesized by Invitrogen (Paisley, Scotland). The sequencing reaction was performed by using 0.2 µM of the respective forward or reverse primer, 1 µl BigDye terminator (Applied Biosystems), and 2 µl of 5× dilution buffer (400 mM Tris-HCl and 10 mM MgCl2) in a total volume of 10 µl. The resulting sequences were analyzed using the SeqManPro 9.04 (DNASTAR, Madison, WI, USA) software.
Alignment of DRB Sequences and Detection of TE
The recently published C. jacchus (Caja)-, M. mulatta (Mamu)-, P. troglodytes (Patr)-, and HLA-DRB genomic sequences (Doxiadis, de Groot, Bontrop, et al. 2008; Doxiadis, de Groot N, de Groot, et al. 2008) were compared with the two newly described Patr-DRB1*10:01 (Accession number HE800526, Kitty) and Patr-DRB4*02:01 (accession number: HE800525, Brigitte) sequences and to diverse full-length DRB sequences obtained from the National Center for Biotechnology Information database (Caja-DRB: AC242730; Mamu-DRB; AM910414, AM910415, AM 910417, AC148706, AM910419, AM91042, AM910410, AC148697, AM910411, AC148700, AM910413, AM910412, AC148663, AM910421, AM910422, AM910423; Patr-DRB: AM910425, NW_01236523, AP006503, AM910426, AM910428, AM910424, AM910429; HLA-DRB: AY663400, AL662842, NG_002433, AL137064, CR75330, NW_923051, AM910430, AY66341, AY663415, AY663405, AL713966, AL929581, AL662842, BX927235, AL713966, NW_923051) intron–exon boundaries of all DRB sequences were annotated (data not shown). For phylogenetic analyses of noncoding sequences, various TE and repeats were removed by using the RepeatMasker version: open-3.3.0 (Smit AFA, Hubley R, and Green P, unpublished data; http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker, accessed August 6, 2012). The remaining intron sequences were aligned per intron using the MacVec 12.0.3 software with manual interference, and major indels were removed. For the detection of TE, all full-length DRB sequences were analyzed using repeat masking from the Repbase database (http://www.girinst.org/censor/index.php, accessed August 6, 2012; Kohany et al. 2006).
Phylogenetic Analyses
Phylogenetic trees were reconstructed using the maximum likelihood method, Phyml (Guindon and Gascuel 2003), whereby for each of the DRB intron sequence alignments, the employed evolutionary model was chosen based on jModelTest (Posada 2008). In each case, the underlying model is specified in the respective figure legend as either the generalized time-reversible (GTR) model (Rodriguez et al. 1990) or the Hasegawa-Kishino-Yano model (Hasegawa et al. 1985). For each phylogeny, 1,000 bootstrap runs were performed. For the presence/absence clustering, each of the DRB alleles was binary encoded based on the presence (1) or absence (0) of each of the identified TE given in table 1 and supplementary table S1, Supplementary Material online. The distance between two alleles was defined as the percentage of matching bits, also known as the Hamming distance. The resulting distance matrix was then used to perform a neighbor joining clustering using PHYLIP version 3.68. All trees were visualized using the software Dendroscope (Huson et al. 2007).
Table 1.
Integration of L2 Parts 1–3 in Intron 1 of Various DRB Alleles.
| L2 Part 3 | L2 Part 2 | L2 Part 1 | |
|---|---|---|---|
| Caja-DRB1*03:03 | |||
| Caja-DRB*W12:01 | 2,275–2,560 | 2,032–2,207 | |
| Caja-DRB*W16:05 | |||
| Mamu-DRB1*03:03:01 | 2,020–2,194 | ||
| Mamu-DRB1*03:06 | 2,253–2,560 | 2,020–2,194 | |
| Mamu-DRB1*03:09 | 2,061–2,194 | ||
| Mamu-DRB1*03:10 | 2,875–3,080 | 2,253–2,790 | 2,020–2,194 |
| Mamu-DRB1*04:04 | 2,253–2,797 | 2,020–2,194 | |
| Mamu-DRB1*10:07 | 2,037–2,194 | ||
| Mamu-DRB*W2:01 | 2,253–2,810 | 2,060–2,195 | |
| Mamu-DRB*W3:03:01 | 2,255–2,560 | 2,060–2,203 | |
| Mamu-DRB*W3:05 | 2,253–2,560 | 2,078–2,194 | |
| Mamu-DRB*W4:01 | 2,253–2,811 | 2,060–2,194 | |
| Mamu-DRB*W25:07 | 2,253–2,810 | 2,060–2,194 | |
| Mamu-DRB*W6:03 | |||
| Mamu-DRB*W6:02 | |||
| Mamu-DRB*W6:09:01 | 2,255–2,560 | 2,060–2,194 | |
| Mamu-DRB3*04:05 | 2,253–2,790 | 2,020–2,194 | |
| Mamu-DRB3*04:08 | 2,253–2,790 | 2,020–2,194 | |
| Mamu-DRB5*03:01 | 2,873–3,071 | 2,038–2,194 | |
| Mamu-DRB6*01:14 | 2,489–3,008 | 2,090–2,194 | |
| Mamu-DRB6*01:18 | 2,489–3,008 | 2,090–2,194 | |
| Patr-DRB1*02:01:01 | 2,253–2,560 | 2,020–2,193 | |
| Patr-DRB1*02:04 | 2,253–2,560 | 2,020–2,193 | |
| Patr-DRB1*07:01 | 2,253–2,797 | 2,060–2,194 | |
| Patr-DRB1*10:01 | 2,262–2,560 | 2,078–2,203 | |
| Patr-DRB2 | 2,489–3,008 | 2,090–2,194 | |
| Patr-DRB3*01:02:01/02 | 2,262–2,557 | 2,078–2,194 | |
| Patr-DRB3*01:03 | 2,253–2,557 | 2,078–2,194 | |
| Patr-DRB3*02:08 | 2,871–3,009 | 2,262–2,557 | 2,078–2,194 |
| Patr-DRB3*07:01 | 2,253–2,557 | 2,078–2,194 | |
| Patr-DRB4*02:01 | 2,871–3,009 | ||
| Patr-DRB5*01:02 | 2,871–3,008 | 2,258–2,369 | 2,142–2,203 |
| Patr-DRB6*01:09N | 2,489–3,002 | 2,090–2,194 | |
| Patr-DRB7*01:01N | Not tested | ||
| HLA-DRB1*01:02:01 | 2,279–2,532 | 2,037–2,194 | |
| HLA-DRB1*03:01:01:01 | 2,253–2,810 | 2,064–2,194 | |
| HLA-DRB1*04:01:01 | 2,887–3,009 | 2,253–2,579 | 2,043–2,194 |
| HLA-DRB1*04:03:01 | 2,887–3,009 | 2,253–2,579 | 2,043–2,194 |
| HLA-DRB1*07:01:01:01 | 2,871–3,009 | 2,253–2,597 | 2,060–2,194 |
| HLA-DRB1*09:01:02 | 2,871–3,009 | 2,253–2,597 | 2,060–2,194 |
| HLA-DRB1*10:01:01 | 2,253–2,810 | 2,078–2,194 | |
| HLA-DRB1*11:01:01 | 2,260–2,810 | 2,064–2,194 | |
| HLA-DRB1*13:01:01 | 2,253–2,810 | 2,078–2,194 | |
| HLA-DRB1*14:01:01 | 2,253–2,810 | 2,078–2,194 | |
| HLA-DRB1*15:01:01:01 | 2,257–2,560 | 2,020–2,194 | |
| HLA-DRB2*01:01 | 2,489–3,008 | 2,090–2,194 | |
| HLA-DRB3*01:01:02:01 | 2,871–3,009 | 2,253–2,557 | 2,078–2,194 |
| HLA-DRB3*02:02:01:01 | 2,253–2,797 | 2,078–2,194 | |
| HLA-DRB4*01:01:01:01 | 2,882–3,079 | 2,253–2,810 | 2,043–2,194 |
| HLA-DRB5*01:01:01 | 2,871–3,008 | 2,254–2,369 | 1,918–2,194 |
| HLA-DRB6*02:01 | 2,489–3,008 | 2,090–2,194 | |
| HLA-DRB7*01:01:01 | 2,875–3,080 | 1,918–2,866 | |
| HLA-DRB8*01:01 | Deletion | ||
Results
TE within DRB Intron Sequences
The intron sequences of 28 human, 14 chimpanzee, 22 rhesus macaque, and 3 common marmoset DRB genes have been analyzed for the presence/absence and location of TE (listed in fig. 1 and supplementary fig. S1, Supplementary Material online). As could be expected from the high variable lengths of intron 1 DRB sequences (Doxiadis, de Groot N, de Groot, et al. 2008), most of the TEs map within this intron (fig. 1). Many different sorts of TE have been observed, and examples will be given for the use of these TE as evolutionary markers. Special attention will be paid to the L2 element that was incorporated into the ancestral mammalian genome more than 80 Ma.
Fig. 1.
Presence of TE in intron 1 of various DRB alleles. The two first two rows describe the TE, and the third row gives the orientation of the respective element (s, sense and as, antisense). A gray or colored filling indicates the presence of the respective element. A colored filling is chosen if the respective element is discussed further in the text. The contiguous L2 parts 2 + 3 (table 1) of DRB2 and DRB6 alleles are colored in dark blue in the L2 part 2 column.
Defining the Ancestral DRB Gene Pool
The data illustrate that somewhere in the evolutionary past, an L2 element must have been integrated into intron 1 of an ancestral primate DRB gene. Footprints of this intron are present in nearly all DRB genes, but the element itself has not been stable. As can be seen, based on size and composition, three major antisense integrated segments of an L2 element can be defined (table 1; fig. 1). Thus, by comparing the presence and absence of these segments in different species belonging to various infra orders, one can start to determine the age of the different genes. The first segment is present in nearly all DRB alleles including Caja-DRB*W12:01 (table 1 and fig. 1). This testifies to the fact that the original L2 integration took place more than 25 Ma. Most of the DRB alleles of all four species share a second L2 part, which is, though, separated from part 1 on the chromosome. Nevertheless, this is not the case for HLA-DRB7, which harbors a contiguous L1 + L2 part. The third part of the L2 element can be detected only in some of the Mamu-, Patr-, and HLA-DRB alleles but not in the Caja-DRB alleles. According to the presence or absence of the three L2 segments, a scheme for the evolution of the DRB genes has been developed (fig. 2). An ancient DRB precursor has been duplicated several times, leading to at least four DRB precursor genes that should have been present in primates before the OWM–HOM divergence. One of these is the DRB7 pseudogene, which contains the longest segments of the L2 element: namely, the contiguous parts 1 and 2, along with part 3, which may have become separated by the incorporation of an AluS element after one duplication round (fig. 1). The second branch of DRB precursors may have undergone a separation of the L2 parts 1 and 2 + 3 by the incorporation of an unknown TE. The result of a second round of duplication seems to be, on the one hand, the DRB2/DRB6 pseudogene, which has remained up to the present in OWM and HOM, and, on the other hand, a second precursor gene with separated L2 parts 1, 2, and 3, probably caused by an AluSq integration (fig. 1). However, another duplication seems to have given rise to the precursor of Mamu/Patr/HLA-DRB5, Patr/HLA-DRB4, and Patr/HLA-DRB1*04/07/09 and most probably to Patr/HLA-DRB3 genes. Although some Patr/HLA-DRB3 genes have apparently lost their L2 part 3, the clustering of all DRB3 genes/alleles of these species on one branch in the NJ tree—illustrating the presence/absence of all TEs—supports this interpretation (table 1 and supplementary fig. S2, Supplementary Material online). The other precursor gene has an integration of an AluY, at least in HLA intron 1 sequences, which may have caused the loss of the third L2 part. In the other species, the loss of L2 part 3 may have been caused by another yet unknown agent. This latter DRB precursor includes all other Mamu, Patr, and HLA-DRB genes, as well as the Caja-DRB*W12 lineage. Thus, it is highly probable that a common ancestor of OWM and HOM already possessed at least four DRB genes.
Fig. 2.
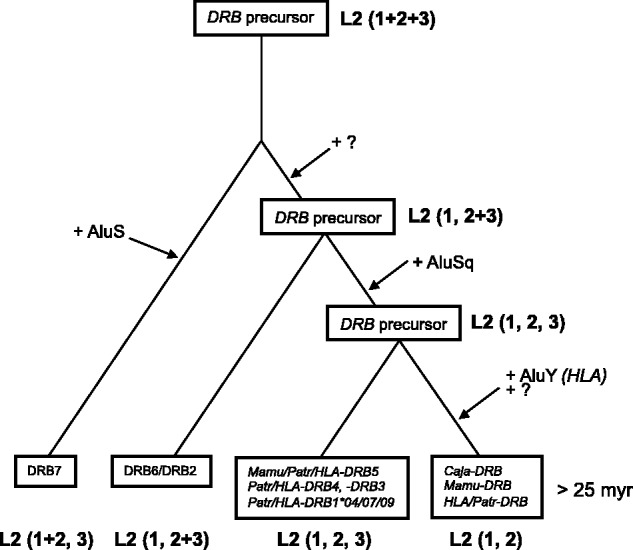
Evolution of Mhc-DRB genes based on the presence of L2 segments. “?” indicates the integration of an Alu/TE element of which the nature remained unknown.
Evolution of Primate DRB Genes Currently Present: Intron Analyses and TE
Phylogenetic analyses of all introns have been performed separately (figs. 3 and 4; supplementary figs. S3 and S4, Supplementary Material online) in addition to the phylogenetic analysis of an AluJb element present in intron 1 of most DRB alleles (supplementary fig. S5, Supplementary Material online). Furthermore, a hierarchical clustering based on the presence/absence pattern of TEs within introns 1–5 has been performed (supplementary fig. S2, Supplementary Material online). Several clades with deep branch lengths supported by high bootstrap values can be observed consistently for all phylogenies. Concerning the different intron analyses, intron 2 comprises the most informative sequences, and therefore its phylogeny is illustrated in figure 3, whereas figure 4 shows the phylogenetic analysis of intron 3 as an example for the shorter introns 3–5. The other intron phylogenies are available as supplementary material (supplementary figs. S3 [intron 1] and S4 [intron 4], Supplementary Material online, and intron 5, data not shown).
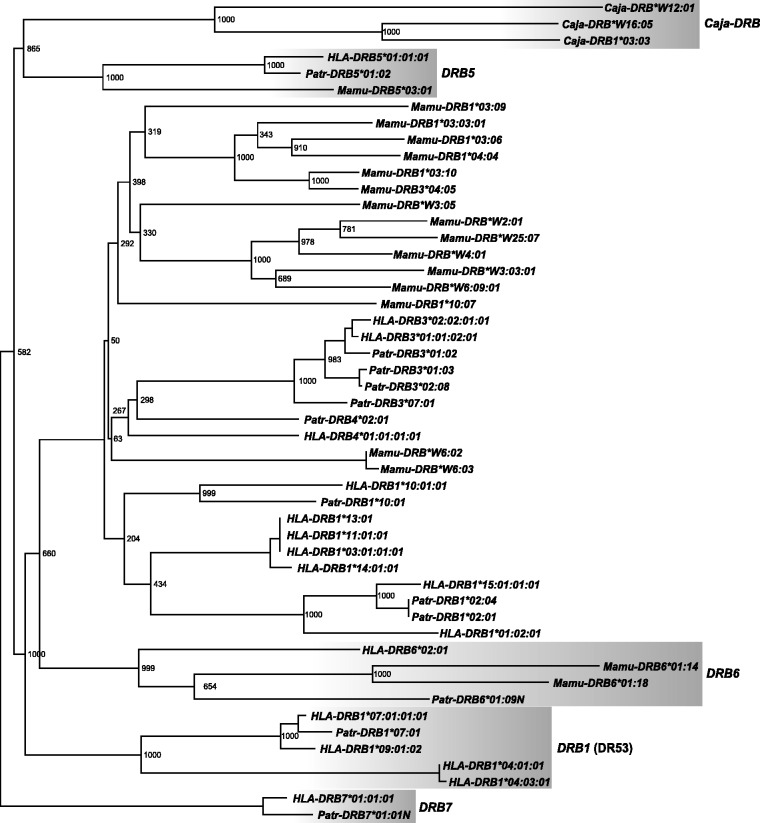
Fig. 3.
Phylogeny of DRB intron 2 sequences. The phylogeny was reconstructed using a maximum-likelihood approach, employing the GTR model of nucleotide substitution with gamma-distributed rate heterogeneity. The depicted phylogram is a consensus of 1,000 bootstrap runs. Bootstrap values are given as absolute numbers. DRB7 sequences were used as outgroup. The five ancestral DRB gene families are highlighted.
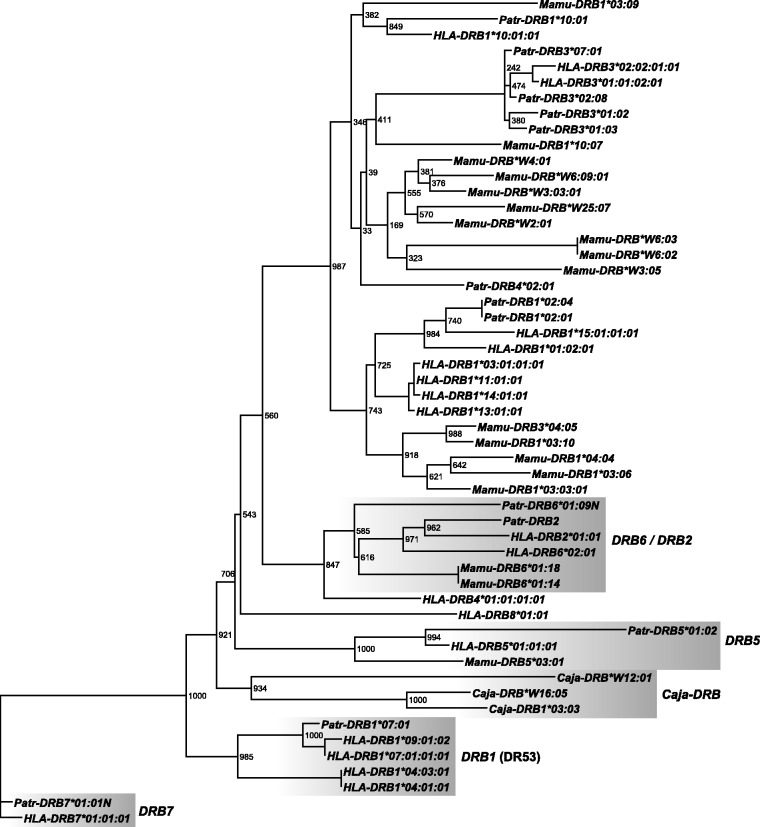
Fig. 4.
Phylogeny of DRB intron 3 sequences. The phylogeny was reconstructed using a maximum-likelihood approach, employing the GTR model of nucleotide substitution with gamma-distributed rate heterogeneity. The depicted phylogram is a consensus of 1,000 bootstrap runs. Bootstrap values are given as absolute numbers. DRB7 sequences were used as outgroup. The five ancestral DRB gene families are highlighted.
A first clade consists of the HLA- and Patr-DRB7 pseudogenes, which, in addition, share a 6-kb long HERVK14I element in intron 2 (fig. 3; supplementary fig. S1, Supplementary Material online). As has been shown by the existence of the long L2 segment in intron 1 of HLA-DRB7, this pseudogene(s) should be regarded as an evolutionarily old and stable entity that probably got lost in other primate species. Therefore, the tree has been rooted with this cluster.
As expected, representatives of the NWM species—namely, the three Caja-DRB genes—cluster apart in all different phylogenies (figs. 3 and 4, supplementary figs. S3 and S4, Supplementary Material online). However, Caja-DRB*W12:01, which is the most frequent allele of the nearly monomorphic Caja-W12 lineage, is divergent from Caja-DRB1*03:03 and Caja-DRB*W16:05.
A third clade with deep branch lengths is formed by the alleles of the Mamu-, Patr-, and HLA-DRB6/DRB2 pseudogenes, thus confirming that they are orthologs and originated before the divergence of OWM and HOM, as illustrated based on the presence of the same L2 segments (fig. 2). In addition to various LTR sequences, intron 1 of all DRB2/DRB6 pseudogenes is characterized by the integration of a more than 5 kb long sequence of HERVK3I (fig. 1). The existence of identical HERV sequences in all these introns confirms their common evolutionary history of more than 25 My. HLA- and Patr-DRB2 sequences are located closer to each other than to their respective DRB6 paralog in introns 3 and 4 (fig. 4; supplementary fig. S4, Supplementary Material online), verifying that Patr- and HLA-DRB2/DRB6 are indeed orthologs of each other and that the duplication of both (pseudo)genes took place before the divergence of humans and chimpanzees. This conclusion is supported by the observation that the AluJb sequence, which is present in intron 1 of most of the primate DRB genes/alleles, is deleted in Patr- and HLA-DRB2 but is present in Patr-DRB6 (fig. 1). Furthermore, both pseudogenes, DRB2 and DRB6, can be present on the same haplotype in chimpanzees (Brandle et al. 1992).
The DRB5 genes of rhesus, chimpanzee, and humans form a fourth clade with deep branch lengths. Thus, in addition to the DRB6/DRB2 pseudogenes, DRB5 is indeed the only DRB locus shared by OWM and HOM.
A fifth extensive cluster is formed by DRB1 gene alleles of the DRB1 genes typifying DR53, the only region configuration shared by humans and chimpanzees (figs. 3 and 4; supplementary figs. S2–S5, Supplementary Material online). Although located on the same branch, the human DRB1*04 lineage is located at a considerable distance from the HLA/Patr-DRB1*07/09 lineages. Because no ortholog of HLA-DRB1*04 is known in chimpanzees, this lineage must have separated after the divergence of both species or became lost in chimpanzees. Furthermore, the common ancestry of the HLA/Patr-DRB1*07 alleles could be illustrated by the presence of the same part of two LINE 1 elements (fig. 1; supplementary fig. S1, Supplementary Material online).
All other DRB genes/alleles analyzed form a large, widely distributed cluster with far fewer deep branches (figs. 3 and 4). Except for Mamu-DRB5 and -DRB6, Mamu-DRB intron sequences cluster apart from the human and chimpanzee alleles/lineages (fig 3). Some Mamu-DRB alleles are separated from each other by deep branch lengths, indicating that these sequences may represent separate lineages or even loci that appear to be evolutionarily old entities.
Several Patr- and HLA genes/lineages cluster together in the various phylogenies: for instance, DRB3 alleles and the DRB1*15 alleles of humans and the DRB1*02 alleles of chimpanzees. The latter result confirms the earlier observation that Patr-DRB1*02 represents an ortholog of the human DRB1*15/16 (DR2) lineage (Kenter et al. 1992; Mayer et al. 1992). Furthermore, all intron sequences of the DRB1 alleles of the DR52 region configuration—namely, HLA-DRB1*03, *11, *13, and *14 alleles—always form one clade.
In contrast, certain human and chimpanzee DRB loci/alleles cluster dissimilarly in trees based on different introns/intron segments. An example is given for Patr- and HLA-DRB1*10, which cluster together for introns 2–4 (figs. 3 and 4; supplementary fig. S4, Supplementary Material online) but separately in intron 1 (supplementary fig. S3, Supplementary Material online) and the AluJb-intron 1 phylogeny (supplementary fig. S5, Supplementary Material online). The same holds true for HLA-DRB4*01, which is located apart from other DRB lineages in intron 1 (supplementary fig. S3, Supplementary Material online) but next to and/or within the DRB6/DRB2 cluster for its intron 3–5 sequences (fig. 4, supplementary fig. S4, Supplementary Material online; intron 5 not shown). Another example concerns the HLA-DRB8 pseudogene, a member of the DR53 haplotype that is shared in humans and chimpanzees. This pseudogene has lost its exon 1, intron 2, exon 2, and part of its intron 2 sequences. Therefore, only introns 3–5 were available for phylogenetic analyses. Although HLA-DRB8 is located as a separate branch next to HLA-DRB4 in intron 3, it clusters next to Mamu-DRB6 in intron 4, a result that is supported by high bootstrap values (fig. 4, supplementary fig. S4, Supplementary Material online).
Therefore, these human and chimpanzee genes/lineages as well as many of the DRB loci/lineages of rhesus macaques are most probably a result of recombination-like processes.
Discussion
The evolutionary history of the DRB region of nonhuman primates belonging to both the Platyrrhini and the Catarrhini infraorders has been compared by analyzing various intron segments. Because L2 non-LTR retrotransposons in the primates’ genome were active from 200 to ∼80 Ma, the presence and absence of different segments of an L2 element and its nucleotide composition were highly informative concerning the time period before the divergence of both infraorders. In addition, the presence and absence of all TE analyzed in various introns gives a more complete picture than the analysis of parts of the intron sequences alone (Satta et al. 1996; Bergstrom et al. 1998) or of only certain TE sequences (Bontrop et al. 1999; Kriener, O'HUigin, Klein, et al. 2000). Therefore, we have been able to demonstrate that at least four ancestral DRB genes must have been present before the divergence of OWM and HOMs. Moreover, our data indicate that one of the four DRB precursor genes must have existed before the NWM–OWM split. However, it is highly probable that there was more than one precursor gene present at that time point, because DRB6 sequences have also been observed in Strepsirrhines, which diverged from Haplorhines ∼63 Ma (Figueroa et al. 1994). In contrast to earlier publications (Satta et al. 1996; Klein et al. 2007), our data indicate that pseudogenes such as DRB7 and DRB6/DRB2 or their precursors are the ancestors of the present HLA-DRB genes. All OWM and HOM DRB haplotypes analyzed so far contain a remnant gene, DRB9, of which the constancy of its position may be related to the evolutionary stability of the DRA locus; this remnant gene seems to be a remainder of an ancient DRB subregion that may have functioned before the OWM–NWM deviation but later became truncated during contractions of the region (Gongora, Figueroa, O'Huigin, et al. 1997; Klein et al. 2007). Therefore, it is likely that the DRB9 pseudogene was also present before the Catarrhini radiation.
All phylogenetic analyses of intron sequences and from TE located within the introns confirm that the DRB5 locus is common to the Catarrhini’s DRB region, and it therefore originates before the OWM–HOM deviation. In addition, the DRB1 lineage of DR53 appears to be an old entity although not present in the DRB region of OWM, from which it has apparently been lost, like the DRB7 pseudogene during contraction–expansion events. Furthermore, the data suggest that these loci/lineages have not been subjected to recombination-like procedures as is supported by the observation that these old entities could also be defined by the phylogeny of exon 2 (Doxiadis, de Groot N, de Groot, et al. 2008). In contrast, other DRB genes apparently underwent such recombination-like events during their evolution, a phenomenon also observed by the comparison of intron and exon 2 sequences, which could be explained by the reshuffling of peptide motifs among DRB family members (Doxiadis, de Groot N, de Groot, et al. 2008). An example is given for the HLA-DRB1*10 gene, of which the intron 1 sequence and its TE cluster within or next to the DRB1*03 branch, whereas sequences of the other introns form a separate branch together with Patr-DRB1*10. Therefore, a rearrangement of the DRB1*10 gene should have taken place before the human–chimpanzee divergence, and our data are in accordance with the earlier suggestion that the DRB1*10 gene is composed of different segments (Gongora, Figueroa, Klein, et al. 1997).
TE are useful means to trace these sorts of rearrangements or recombination-like processes and are also discussed as being the originators. In particular, L1 elements and HERV sequences are known to induce gene conversions and/or recombinations and therefore give rise to rearrangements of DNA segments. Both elements are predominantly present not only in DRB pseudogenes such as DRB6, DRB7, and DRB8 but also in the DRB4 gene. Because there are known DRB4 variants of the same HLA-DR53 region configuration, which have been rendered pseudogenes, a role involving these specific TE in the inactivation process of genes may be postulated. Future experiments such as whole genome association studies and the haplotyping of large DNA regions may help to elucidate these postulations.
Supplementary Material
Supplementary figures S1–S5 and table S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Donna Devine for editing the manuscript and Henk van Westbroek for preparing the figures. This study was supported, in part, by NIH/NIAID projects HHSN266200400088C/HHSN272201100013C and 2R24RR01603032-09.
References
- Andersson G, Larhammar D, Widmark E, Servenius B, Peterson PA, Rask L. Class II genes of the human major histocompatibility complex. Organization and evolutionary relationship of the DR beta genes. J Biol Chem. 1987;262:8748–8758. [PubMed] [Google Scholar]
- Andersson G, Svensson AC, Setterblad N, Rask L. Retroelements in the human MHC class II region. Trends Genet. 1998;14:109–114. doi: 10.1016/s0168-9525(97)01359-0. [DOI] [PubMed] [Google Scholar]
- Antunes SG, de Groot NG, Brok H, Doxiadis G, Menezes AA, Otting N, Bontrop RE. The common marmoset: a new world primate species with limited Mhc class II variability. Proc Natl Acad Sci U S A. 1998;95:11745–11750. doi: 10.1073/pnas.95.20.11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A. 2004;101(2 Suppl):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Bergstrom TF, Josefsson A, Erlich HA, Gyllensten U. Recent origin of HLA-DRB1 alleles and implications for human evolution. Nat Genet. 1998;18:237–242. doi: 10.1038/ng0398-237. [DOI] [PubMed] [Google Scholar]
- Blancher A, Tisseyre P, Dutaur M, Apoil PA, Maurer C, Quesniaux V, Raulf F, Bigaud M, Abbal M. Study of Cynomolgus monkey (Macaca fascicularis) MhcDRB (Mafa-DRB) polymorphism in two populations. Immunogenetics. 2006;58:269–282. doi: 10.1007/s00251-006-0102-9. [DOI] [PubMed] [Google Scholar]
- Bontrop RE, Otting N, de Groot NG, Doxiadis GG. Major histocompatibility complex class II polymorphisms in primates. Immunol Rev. 1999;167:339–350. doi: 10.1111/j.1600-065x.1999.tb01403.x. [DOI] [PubMed] [Google Scholar]
- Brandle U, Ono H, Vincek V, Klein D, Golubic M, Grahovac B, Klein J. Trans-species evolution of Mhc-DRB haplotype polymorphism in primates: organization of DRB genes in the chimpanzee. Immunogenetics. 1992;36:39–48. doi: 10.1007/BF00209291. [DOI] [PubMed] [Google Scholar]
- Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci U S A. 2010;107:19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot N, Doxiadis GG, de Vos-Rouweler AJ, de Groot NG, Verschoor EJ, Bontrop RE. Comparative genetics of a highly divergent DRB microsatellite in different macaque species. Immunogenetics. 2008;60:737–748. doi: 10.1007/s00251-008-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Heijmans CM, de Groot N, Doxiadis GG, Otting N, Bontrop RE. The chimpanzee Mhc-DRB region revisited: gene content, polymorphism, pseudogenes, and transcripts. Mol Immunol. 2009;47:381–389. doi: 10.1016/j.molimm.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NG, Otting N, Robinson J, et al. (11 co-authors) Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics. 2012;64:615–631. doi: 10.1007/s00251-012-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, de Groot N, Bontrop RE. Impact of endogenous intronic retroviruses on major histocompatibility complex class II diversity and stability. J Virol. 2008;82:6667–6677. doi: 10.1128/JVI.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxiadis GG, de Groot N, de Groot NG, Doxiadis II, Bontrop RE. Reshuffling of ancient peptide binding motifs between HLA-DRB multigene family members: old wine served in new skins. Mol Immunol. 2008;45:2743–2751. doi: 10.1016/j.molimm.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, Bontrop RE. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics. 2006;58:259–268. doi: 10.1007/s00251-006-0083-8. [DOI] [PubMed] [Google Scholar]
- Erlich HA, Bergstrom TF, Stoneking M, Gyllensten U. HLA sequence polymorphism and the origin of humans. Science. 1996;274:1552b–1554b. doi: 10.1126/science.274.5292.1552b. [DOI] [PubMed] [Google Scholar]
- Fernandez-Soria VM, Morales P, Castro MJ, Suarez B, Recio MJ, Moreno MA, Paz-Artal E, Arnaiz-Villena A. Transcription and weak expression of HLA-DRB6: a gene with anomalies in exon 1 and other regions. Immunogenetics. 1998;48:16–21. doi: 10.1007/s002510050395. [DOI] [PubMed] [Google Scholar]
- Figueroa F, O'HUigin C, Tichy H, Klein J. The origin of the primate Mhc-DRB genes and allelic lineages as deduced from the study of prosimians. J Immunol. 1994;152:4455–4465. [PubMed] [Google Scholar]
- Glazko GV, Nei M. Estimation of divergence times for major lineages of primate species. Mol Biol Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- Gongora R, Figueroa F, Klein J. Complex origin of the HLA-DR10 haplotype. J Immunol. 1997;159:6044–6051. [PubMed] [Google Scholar]
- Gongora R, Figueroa F, O'Huigin C, Klein J. HLA-DRB9—possible remnant of an ancient functional DRB subregion. Scand J Immunol. 1997;45:504–510. doi: 10.1046/j.1365-3083.1997.d01-428.x. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 1988;85:4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Kenter M, Otting N, Anholts J, Jonker M, Schipper R, Bontrop RE. Mhc-DRB diversity of the chimpanzee (Pan troglodytes) Immunogenetics. 1992;37:1–11. doi: 10.1007/BF00223539. [DOI] [PubMed] [Google Scholar]
- Klein D, Vincek V, Kasahara M, Schonbach C, O'HUigin C, Klein J. Gorilla major histocompatibility complex-DRB pseudogene orthologous to HLA-DRBVIII. Hum Immunol. 1991;32:211–220. doi: 10.1016/0198-8859(91)90058-h. [DOI] [PubMed] [Google Scholar]
- Klein J, O'HUigin C. Class II B Mhc motifs in an evolutionary perspective. Immunol Rev. 1995;143:89–111. doi: 10.1111/j.1600-065x.1995.tb00671.x. [DOI] [PubMed] [Google Scholar]
- Klein J, Sato A, Nikolaidis N. MHC, TSP, and the origin of species: from immunogenetics to evolutionary genetics. Annu Rev Genet. 2007;41:281–304. doi: 10.1146/annurev.genet.41.110306.130137. [DOI] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriener K, O'HUigin C, Klein J. Alu elements support independent origin of prosimian, platyrrhine, and catarrhine Mhc-DRB genes. Genome Res. 2000;10:634–643. doi: 10.1101/gr.10.5.634. [DOI] [PubMed] [Google Scholar]
- Kriener K, O'HUigin C, Tichy H, Klein J. Convergent evolution of major histocompatibility complex molecules in humans and New World monkeys. Immunogenetics. 2000;51:169–178. doi: 10.1007/s002510050028. [DOI] [PubMed] [Google Scholar]
- Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SG, Albert ED, Bodmer WF, et al. (24 co-authors) Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer WE, O'HUigin C, Klein J. Resolution of the HLA-DRB6 puzzle: a case of grafting a de novo-generated exon on an existing gene. Proc Natl Acad Sci U S A. 1993;90:10720–10724. doi: 10.1073/pnas.90.22.10720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer WE, O'HUigin C, Zaleska-Rutczynska Z, Klein J. Trans-species origin of Mhc-DRB polymorphism in the chimpanzee. Immunogenetics. 1992;37:12–23. doi: 10.1007/BF00223540. [DOI] [PubMed] [Google Scholar]
- McClintock B. Intranuclear systems controlling gene action and mutation. Brookhaven Symp Biol. 1956:58–74. [PubMed] [Google Scholar]
- Mnukova-Fajdelova M, Satta Y, O'HUigin C, Mayer WE, Figueroa F, Klein J. Alu elements of the primate major histocompatibility complex. Mamm Genome. 1994;5:405–415. doi: 10.1007/BF00357000. [DOI] [PubMed] [Google Scholar]
- Munoz-Lopez M, Macia A, Garcia-Canadas M, Badge RM, Garcia-Perez JL. An epi [c] genetic battle: LINE-1 retrotransposons and intragenomic conflict in humans. Mob Genet Elements. 2011;1:122–127. doi: 10.4161/mge.1.2.16730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor SL, Blasky AJ, Pendley CJ, Becker EA, Wiseman RW, Karl JA, Hughes AL, O'Connor DH. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59:449–462. doi: 10.1007/s00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. Mobile DNA and the TE-Thrust hypothesis: supporting evidence from the primates. Mob DNA. 2011;2:8. doi: 10.1186/1759-8753-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, et al. (11 co-authors) A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez F, Oliver JL, Marin A, Medina JR. The general stochastic model of nucleotide substitution. J Theor Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- Satta Y, Mayer WE, Klein J. Evolutionary relationship of HLA-DRB genes inferred from intron sequences. J Mol Evol. 1996;42:648–657. doi: 10.1007/BF02338798. [DOI] [PubMed] [Google Scholar]
- Sawai H, Kawamoto Y, Takahata N, Satta Y. Evolutionary relationships of major histocompatibility complex class I genes in simian primates. Genetics. 2004;166:1897–1907. doi: 10.1534/genetics.166.4.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonbach C, Vincek V, Mayer WE, Golubic M, O'HUigin C, Klein J. Multiplication of Mhc-DRB5 loci in the orangutan: implications for the evolution of DRB haplotypes. Mamm Genome. 1993;4:159–170. doi: 10.1007/BF00352232. [DOI] [PubMed] [Google Scholar]
- Slierendregt BL, Otting N, van Besouw N, Jonker M, Bontrop RE. Expansion and contraction of rhesus macaque DRB regions by duplication and deletion. J Immunol. 1994;152:2298–2307. [PubMed] [Google Scholar]
- Smit AF. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 1996;6:743–748. doi: 10.1016/s0959-437x(96)80030-x. [DOI] [PubMed] [Google Scholar]
- van Rood JJ, van Leeuwen A, Keuning JJ, Termijtelen A. Evidence for two series of B-cell antigens in man and their comparison with HLA-D. Scand J Immunol. 1977;6:373–384. doi: 10.1111/j.1365-3083.1977.tb02092.x. [DOI] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, et al. (11 co-authors) Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.