A novel motif in the Insert B domain of the mitochondrial dynamin Dnm1 is essential for interaction with its adaptor Mdv1 during mitochondrial fission.
Abstract
To initiate mitochondrial fission, dynamin-related proteins (DRPs) must bind specific adaptors on the outer mitochondrial membrane. The structural features underlying this interaction are poorly understood. Using yeast as a model, we show that the Insert B domain of the Dnm1 guanosine triphosphatase (a DRP) contains a novel motif required for association with the mitochondrial adaptor Mdv1. Mutation of this conserved motif specifically disrupted Dnm1–Mdv1 interactions, blocking Dnm1 recruitment and mitochondrial fission. Suppressor mutations in Mdv1 that restored Dnm1–Mdv1 interactions and fission identified potential protein-binding interfaces on the Mdv1 β-propeller domain. These results define the first known function for Insert B in DRP–adaptor interactions. Based on the variability of Insert B sequences and adaptor proteins, we propose that Insert B domains and mitochondrial adaptors have coevolved to meet the unique requirements for mitochondrial fission of different organisms.
Introduction
Eukaryotic cells possess a family of dynamin-related proteins (DRPs), each of which is responsible for a specific cellular membrane-remodeling event. For example, the dynamin-related GTPase Drp1 (Dnm1 in yeast) mediates mitochondrial (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999) and peroxisomal fission (Koch et al., 2003; Li and Gould, 2003; Kuravi et al., 2006), Atlastin (Hu et al., 2009; Orso et al., 2009; Moss et al., 2011) and Mitofusins (Hales and Fuller, 1997; Hermann et al., 1998; Rapaport et al., 1998; Chen et al., 2003; Eura et al., 2003) play roles in ER and mitochondrial membrane fusion, respectively, and ARC5 (Gao et al., 2003) facilitates chloroplast membrane division. Like classical dynamin, DRPs self-assemble into highly ordered oligomers that use the energy of GTP hydrolysis to remodel lipid bilayers (Praefcke and McMahon, 2004).
Dynamin and DRPs have conserved GTPase, middle, and GTPase effector domains (see Fig. 1 A; van der Bliek, 1999). These domains mediate self-assembly and modulate GTPase activity. Dynamin and DRPs also contain nonconserved domains that, in some cases, have been shown to determine their cellular distribution and heterotypic interactions. For example, a proline-rich domain at the C terminus of dynamin facilitates its binding to a variety of actin-binding proteins. Dynamin also harbors a pleckstrin homology (PH) domain between its GTPase and GTPase effector domain, which is essential for interactions with the plasma membrane. In place of the PH domain, DRPs contain a region called Insert B (InsB) whose length and sequence varies. Whether or not there is a conserved function for InsB is not clear.
Figure 1.
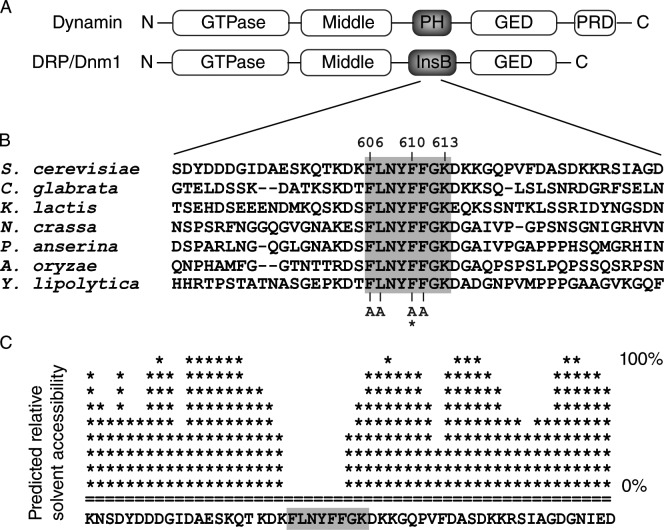
Dnm1 InsB contains a sequence of eight strictly conserved residues (F610–K613) predicted to be solvent inaccessible. (A) Domain structures of Dynamin and the DRP/Dnm1. GED, GTPase effector domain; PRD, proline-rich domain. (B) Alignment of a segment of InsB from the indicated fungal Dnm1 homologues. The asterisk marks the position of the dnm1F610A mutation. (C) Plot showing the predicted relative solvent accessibility of a section of Dnm1 InsB (PredictProtein server). Shaded boxes indicate residues that are conserved in fungi.
Although dynamin interacts directly with the lipid bilayer via its PH domain, DRPs do not initially interact with the lipid bilayers they remodel. Instead, they are recruited to specific cellular sites via interactions with membrane-associated adaptor proteins. In most cases, the DRP domains necessary for adaptor interactions have not been identified. However, structural studies of dynamin and several DRPs (Mears et al., 2007, 2011; Faelber et al., 2011; Ford et al., 2011) suggest that InsB is a likely candidate because it is predicted to reside at the base of the DRP oligomer, closest to the membrane.
We used the yeast mitochondrial fission machinery as a model to directly test the function of InsB in DRP–adaptor interactions and DRP1 membrane recruitment. In vivo, Dnm1 (the yeast DRP) is recruited to the mitochondrial outer membrane by Mdv1 (the adaptor; Tieu and Nunnari, 2000; Cerveny et al., 2001), which in turn associates with membrane-anchored Fis1 (Mozdy et al., 2000). Once on the membrane, Dnm1 coassembles with Mdv1 into spirals that encircle and divide the mitochondrial tubule (Bhar et al., 2006; Naylor et al., 2006). We identified a motif in the Dnm1 InsB domain that is required to recruit Dnm1 to mitochondria. The amino acid sequence of this motif is strictly conserved among fungi and is predicted to form a solvent-inaccessible helix (PSIPRED v2.6). Amino acid substitutions in this InsB helix inhibit the recruitment of Dnm1 to mitochondria and block fission. Importantly, these mutations do not impair Dnm1 self-assembly. Instead, they specifically disrupt the Dnm1–Mdv1 interaction. The disrupted interaction is rescued by suppressor mutations in the Mdv1 β-propeller domain to which Dnm1 binds. These findings identify a new functional motif in the Dnm1 InsB domain required for Mdv1 binding and recruitment of Dnm1 from the cytoplasm to its site of action on mitochondria.
Results
Dnm1 InsB contains a conserved sequence motif predicted to be solvent inaccessible
In a previous genetic screen (Bhar et al., 2006), we identified an InsB mutation (F610L) that interfered with Dnm1 function in mitochondrial fission. Alignment of Saccharomyces cerevisiae Dnm1 InsB with its fungal homologues revealed a motif of eight strictly conserved residues encompassing F610 (Fig. 1, A and B). These residues are predicted to form a short helix (PSIPRED v2.6) that is less accessible to solvent than surrounding residues (Fig. 1 C), suggesting that they are part of an interaction interface.
InsB motif residues are important for Dnm1 function in mitochondrial fission
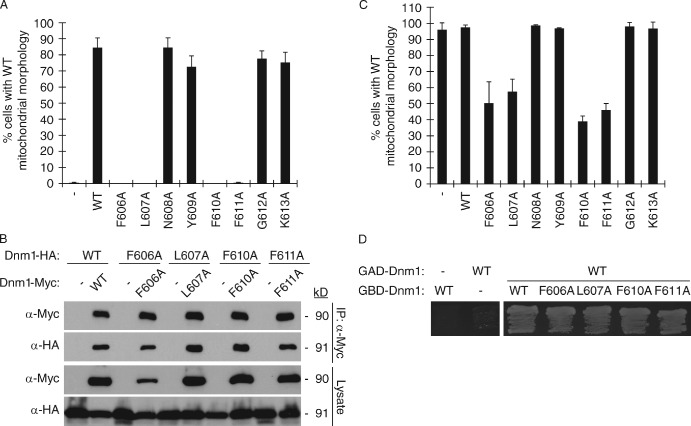
Yeast mitochondria form multiple tubular and branched structures that are continually remodeled by fission and fusion events. When fission is disrupted or blocked, mitochondria fuse to form netlike structures or a single interconnected tubule that often collapses to one side of the cell (Bleazard et al., 1999). To evaluate Dnm1 InsB motif function, we individually replaced the eight conserved residues with alanine and quantified the ability of each mutant protein to rescue fission defects in cells lacking the wild-type (WT) Dnm1 protein (dnm1Δ). As shown in Fig. 2 A, expression of WT Dnm1 rescued fission in up to 90% of dnm1Δ cells. In contrast, four Dnm1 mutant proteins (F606A, L607A, F610A, and F611A) failed to rescue mitochondrial morphology in this strain. Hereafter, these four loss-of-function proteins will be collectively referred to as Dnm1InsBmut. Western blotting of whole cell extracts indicated that the Dnm1InsBmut proteins were expressed at levels similar to WT Dnm1 (Fig. S1 A). Thus, mutations in conserved residues of InsB block Dnm1 function in mitochondrial fission but do not affect protein stability.
Figure 2.
Dnm1 InsB conserved residues are important for Dnm1 function in mitochondrial fission. (A) Quantification of mitochondrial morphology in dnm1Δ cells expressing indicated Dnm1 variants. (B) Lysates from cells expressing the indicated C-terminal HA- or Myc-tagged Dnm1 proteins were used for immunoprecipitation with anti-Myc agarose beads. Immunoprecipitated fractions (top) and lysates (bottom) were analyzed by SDS-PAGE and Western blotting with anti-Myc and anti-HA antibodies. (C) Quantification of mitochondrial morphology in WT cells expressing indicated Dnm1 variants. (A and C) Black columns and error bars represent the mean and standard deviation of at least three independent experiments (n = 100). (D) pGBD and pGAD plasmids expressing the indicated fusion proteins were cotransformed into the Y187 yeast two-hybrid reporter strain and grown on S-Dextrose minus adenine plates at 30°C for 3 d.
In the absence of WT Dnm1, the inability of Dnm1InsBmut proteins to support fission could be caused by their failure to self-assemble. To test this possibility, we coexpressed HA- and Myc-tagged versions of the same InsB mutant proteins in dnm1Δ cells and performed coimmunoprecipitation (coIP) assays. As shown in Fig. 2 B, each form of the HA-tagged Dnm1InsBmut protein was efficiently coprecipitated with the Myc-tagged version of itself. Thus, the F606, L607, F610, and F611 residues in InsB are not essential for Dnm1 self-assembly.
We also observed that coexpression of WT and Dnm1InsBmut proteins caused dominant-negative fission defects (Fig. 2 C). Although expression of a second copy of WT DNM1 in WT cells did not cause significant changes in mitochondrial morphology, expression of Dnm1InsBmut proteins in WT cells blocked fission in up to 60% of the population. One explanation for these dominant-negative fission phenotypes is that Dnm1InsBmut proteins are able to assemble with WT Dnm1. Consistent with this idea, we observed an interaction between WT Dnm1 and Dnm1InsBmut proteins in a two-hybrid assay (Fig. 2 D). The interaction of Dnm1InsBmut with WT Dnm1 could interfere with multiple steps in mitochondrial fission including fission complex formation and Dnm1–adaptor interactions.
The InsB motif is essential for Dnm1 binding to the Mdv1 adaptor
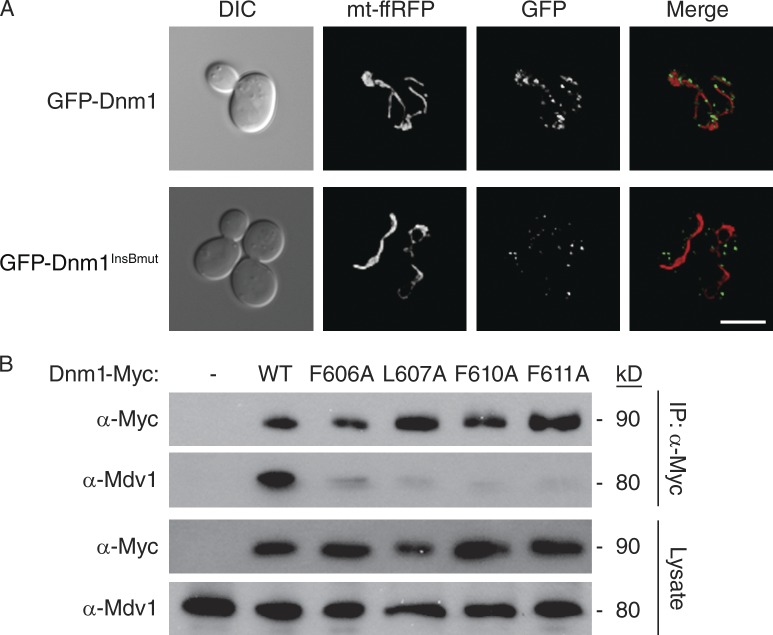
After mitochondrial membrane recruitment and self-assembly, the majority of GFP-tagged Dnm1 can be visualized as puncta on mitochondrial tubules (Fig. 3 A; Otsuga et al., 1998). In contrast, all of the GFP-Dnm1InsBmut proteins failed to associate with mitochondria (Fig. 3 A, a representative cell is shown). Instead, GFP-Dnm1InsBmut proteins assembled into punctuate structures that moved rapidly through the cytoplasm and could not be captured by digital imaging. In a few cells, GFP-Dnm1InsBmut also formed larger, immobile aggregates in the cytoplasm (Fig. 3 A). Importantly, this localization of GFP-Dnm1InsBmut proteins is indistinguishable from that observed for GFP-Dnm1 in cells lacking Fis1 or fission adaptor proteins (Mozdy et al., 2000; Griffin et al., 2005). This observation raised the possibility that mutations in Dnm1InsBmut proteins were disrupting interactions with the Mdv1 adaptor.
Figure 3.
InsB conserved residues are critical for Dnm1–Mdv1 interaction. (A) Representative images of GFP-Dnm1 and GFP-Dnm1InsBmut (F606A, L607A, F610A, or F611A) localization in dnm1Δ cells. Differential interference contrast microscopy (DIC), mitochondrial matrix-targeted dsRed (mt-ffRFP), GFP, and merged images are shown. Bar, 5 µm. (B) Lysates from cells expressing the indicated C-terminal Myc-tagged Dnm1 were used for immunoprecipitation with anti-Myc agarose beads. Immunoprecipitated fractions (top) and lysates (bottom) were analyzed by SDS-PAGE and Western blotting with anti-Myc and anti-Mdv1 antibodies.
In vitro pull-down assays using purified GST-Mdv1 β-propeller and His-tagged Dnm1 showed that the Mdv1 β-propeller domain binds directly to Dnm1 (Fig. S1 B). To determine whether InsB mutations affected the Dnm1–Mdv1 interaction, we performed coIP experiments from dnm1Δ cells expressing Myc-tagged Dnm1InsBmut proteins. Although full-length Mdv1 was efficiently coprecipitated by WT Dnm1-Myc, Mdv1 interaction with all of the Dnm1InsBmut-Myc proteins was dramatically reduced (Fig. 3 B). These combined results identify a novel motif in InsB essential for Dnm1–Mdv1 interaction and Dnm1 recruitment to mitochondria.
Suppressors of a dnm1InsBmut mutation cluster in the Mdv1 β-propeller
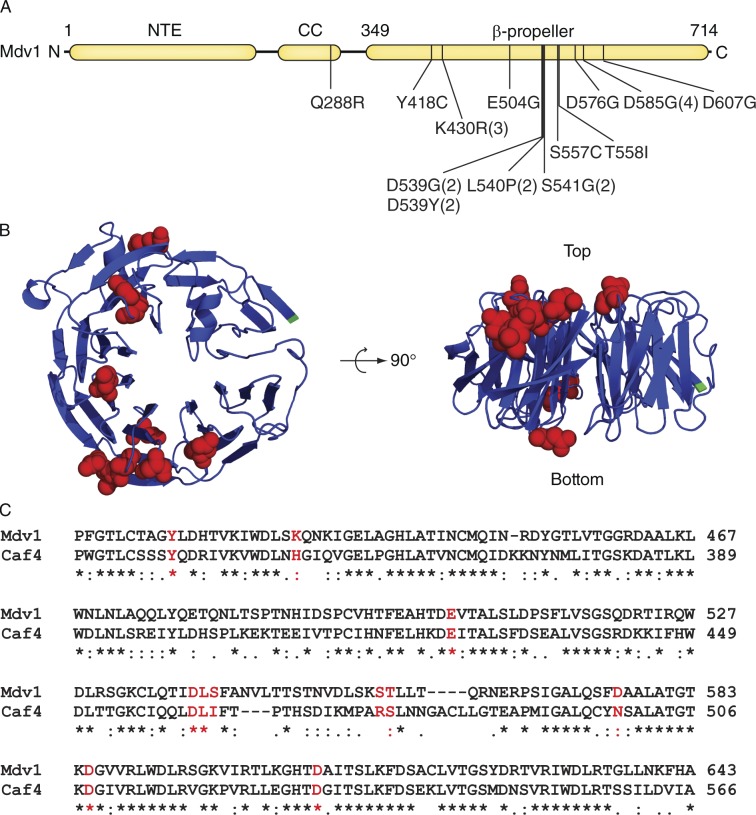
Using an integrated form of the dnm1F610A mutation, we performed a suppressor screen to identify residues in Mdv1 important for Dnm1–Mdv1 interaction (see Materials and methods and Table S1). Although the screen covered 84% of the MDV1 coding sequence, all but one suppressor mutation fell in the Mdv1 β-propeller domain (Fig. 4 A). Most of the affected residues localized to the top of the Mdv1 β-propeller model (Fig. 4 B, right), suggesting that they are part of an interaction interface. Suppressors affecting residues on the bottom of the structure may define an additional binding interface. Two additional mutations, S557C and T558I, lay in a short sequence that was eliminated during homology modeling. These results are consistent with our finding that the Mdv1 β-propeller is sufficient for direct interaction with Dnm1 (Fig. S1 B). The final suppressor mutation altered residue Q288 in the Mdv1 coiled-coil domain. This suppressor was not analyzed further, as the structure and function of the coiled-coil domain has been extensively studied (Koirala et al., 2010; Zhang et al., 2012).
Figure 4.
Suppressors of a dnm1InsB mutation cluster in the Mdv1 β-propeller. (A) Domain structure of Mdv1 with indicated Dnm1F610A suppressor mutations (the number of mutations obtained for each allele is in parentheses). NTE, N-terminal Extension; CC, coiled coil. (B) Top and side views of the Mdv1 β-propeller model (blue). Red indicates residues changed by suppressor mutations. The N terminus is in green. (C) Alignment of the portions of the Mdv1 (aa 408–643) and Caf4 (aa 329–566) β-propeller sequences. Residues affected by suppressor mutations are shown in red. Symbols below the sequence alignment indicate identity (*), strong similarity (:), and weak similarity (.) of amino acids.
In addition to Mdv1, yeast encodes a second fission adaptor protein called Caf4 (Griffin et al., 2005). Although these two adaptors are paralogues and have similar domain structures, Caf4 is not essential for mitochondrial fission. Sequence alignment revealed that most of the suppressor mutations affected conserved or similar amino acids in the Mdv1 and Caf4 β-propeller domains (Fig. 4 C). This conservation is consistent with an important functional role for these amino acids in vivo.
All but one of the residues (S541) identified in our suppressor screen differ from those reported by others (Cerveny and Jensen, 2003; Naylor et al., 2006). The mutations in these previous studies were selected based on homology modeling with known β-propeller structures, whereas the mutations we identified here were selected by the organism in a suppressor screen and are specifically relevant to defects caused by InsB mutations. The exception, S541, was reported not to have a phenotype when replaced by glutamine (Q) in the full-length Mdv1 protein (Naylor et al., 2006). In contrast, we found that an Mdv1 S541G mutant protein was unable to rescue mitochondrial fission defects in an mdv1Δ strain (Table 1). This difference may be because of the different yeast strain backgrounds used in these two studies. It is also possible that mutation to glycine has a more significant effect on local protein structure and flexibility than mutation to glutamine.
Table 1.
Characterization of Mdv1 suppressors of the dnm1F610A allele
| Mutations | WT mitochondrial morphologya | GFP mitochondrial punctab | Interaction with Dnm1c | |||
| DNM1 mdv1Δ | dnm1F610A mdv1Δ | Dnm1 | Dnm1F610A | Dnm1 | Dnm1F610A | |
| % | % | |||||
| Mdv1 | 67 ± 7 | 0 ± 0 | + | + | + | − |
| Y418C | 6 ± 3 | 41 ± 16 | + | + | ++ | + |
| K430R | 9 ± 8 | 28 ± 4 | + | + | + | − |
| E504G | 19 ± 8 | 32 ± 16 | + | + | ++ | − |
| D539G | 9 ± 2 | 68 ± 11 | + | + | ++ | + |
| D539Y | 11 ± 1 | 61 ± 14 | + | + | ++ | + |
| L540P | 12 ± 5 | 58 ± 14 | + | + | ++ | + |
| S541G | 8 ± 4 | 38 ± 12 | + | + | ++ | + |
| S557C | 9 ± 8 | 45 ± 8 | + | + | ++ | + |
| T558I | 10 ± 8 | 34 ± 6 | + | + | ++ | + |
| D576G | 68 ± 17 | 53 ± 9 | + | + | − | − |
| D585G | 45 ± 3 | 30 ± 8 | + | + | − | − |
| D607G | 15 ± 6 | 31 ± 12 | + | + | ++ | − |
Numbers are the mean and standard deviation of at least three independent experiments, n = 300.
+, GFP puncta localized to mitochondrial tubules.
Yeast two-hybrid assays. −, No growth on His or Ade minus medium; +, growth on His but not Ade minus medium; ++, growth on both His and Ade minus media. Growth on Ade minus medium is the more stringent indicator of protein–protein interaction.
Suppressor mutations in the Mdv1 β-propeller rescue mitochondrial fission defects caused by Dnm1F610A
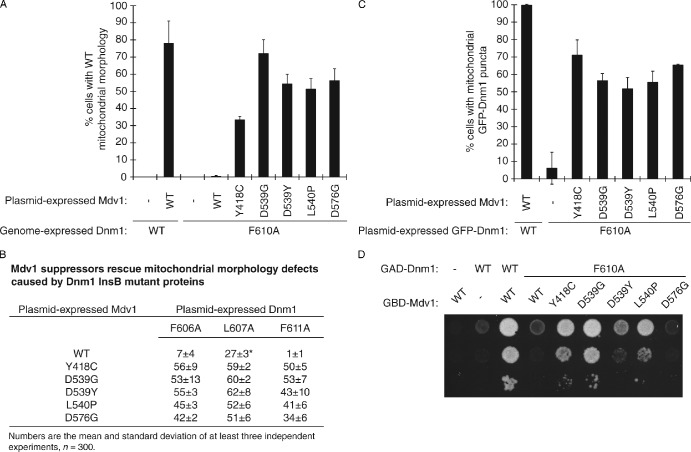
In control studies, the Mdv1 suppressor proteins were all stably expressed in vivo (Fig. S1 C). To verify that the Mdv1 suppressor proteins were capable of rescuing fission, we expressed them from a plasmid in cells lacking WT Mdv1 and expressing Dnm1F610A protein from the genome. All of the Mdv1 suppressors rescued mitochondrial fission defects in this strain (up to 70% rescue). Representative results are shown in Fig. 5 A for the five best rescuing Mdv1 suppressors (Y418C, D539G, D539Y, L540P, and D576G). The same Mdv1 suppressors also rescued mitochondrial morphology defects caused by other Dnm1InsBmut proteins (F606A, L607A, and F611A; Fig. 5 B).
Figure 5.
Suppressor mutations in the Mdv1 β-propeller rescue mitochondrial fission defects caused by Dnm1F610A. (A) Quantification of mitochondrial morphology in mdv1Δ DNM1 and mdv1Δ dnm1::dnm1F610A strains expressing the indicated Mdv1 suppressor proteins. (B) Quantification of mitochondrial morphology in mdv1Δdnm1Δ strains expressing indicated Mdv1 and Dnm1 proteins from plasmids. The mitochondria were visualized with MitoFluor Red 589 (Molecular Probes). (*) Plasmid-expressed WT Mdv1 rescued Dnm1L607A better than genome-expressed WT Mdv1 (Fig. 2 A). This is likely because of the higher steady-state abundance of the plasmid-expressed protein. (C) Quantification of GFP-Dnm1 localization in mdv1Δ dnm1Δ cells expressing the indicated Mdv1 and GFP-Dnm1 variants. (A and C) Black columns and error bars represent the mean and standard deviation of at least three independent experiments (A, n = 300; C, n = 150). (D) pGBD and pGAD plasmids expressing the indicated fusion proteins were cotransformed into the Y187 yeast two-hybrid reporter strain and grown on S-Dextrose minus histidine plates at 30°C for 3 d.
To test whether Mdv1 suppressors could rescue the mitochondrial recruitment of Dnm1F610A, Mdv1 suppressors and GFP-Dnm1F610A were coexpressed in an mdv1Δdnm1Δ strain. GFP mitochondrial puncta were visible in up to 70% of these cells (Fig. 5 C). The mitochondrial recruitment and puncta formation by GFP-Dnm1F610A suggested that Dnm1 interaction with the Mdv1 suppressor proteins had been restored. However, this interaction could not be detected in coIP assays, even after chemical cross-linking (unpublished data). Thus, the restored interaction between Dnm1F610A and Mdv1 suppressors is less robust than WT.
As an alternative, we evaluated Dnm1F610A–Mdv1 suppressor interactions using the yeast two-hybrid growth assay. Previous studies established that WT Dnm1 and WT Mdv1 interact in this assay (Fig. 5 D; Tieu and Nunnari, 2000; Cerveny and Jensen, 2003; Karren et al., 2005). Although the interaction was severely disrupted when Dnm1F610A was paired with WT Mdv1, the interaction was partially (D539Y and L540P) or completely (Y418C and D539G) restored by substituting an Mdv1 suppressor protein for WT Mdv1 (Fig. 5 D). Surprisingly, the Mdv1D576G suppressor protein reproducibly rescued both mitochondrial morphology and GFP-Dnm1F610A localization (Fig. 5, A–C), but was unable to restore growth in the two-hybrid assay (Fig. 5 D). In the morphology rescue and localization studies, binding of Dnm1F610A to this Mdv1 suppressor may be sufficient to recruit Dnm1F610A to the membrane, after which cooligomerization of both proteins into mitochondrial fission complexes could further stabilize the interaction. Such stabilizing forces may be compromised by the spheroplasting/lysis required for coIP studies or by the fusion of both proteins to nuclear targeting sequences used in the two-hybrid assay. This interpretation is supported by our finding that GFP-Mdv1D576G assembled into punctate fission complexes in the presence of Dnm1F610A (Fig. S1 D).
Discussion
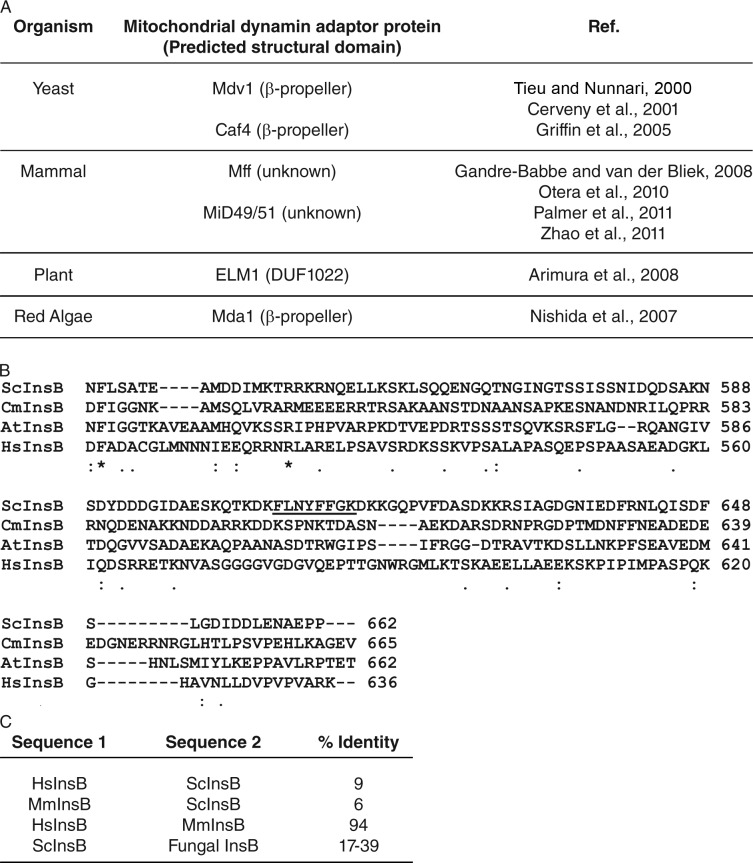
Although Dnm1 binding to Mdv1 and recruitment to the mitochondrial membrane is essential for fission, the Dnm1 domains required for this interaction were not known. Here we identify a novel motif in Dnm1 InsB that is specifically required for interaction with the Mdv1 β-propeller. Second-site suppression studies and cell-based assays confirm that the InsB–β-propeller interaction is critical for Dnm1–Mdv1 binding and Dnm1 membrane recruitment. The Dnm1 InsB motif and Mdv1 adaptor sequences required for this interaction are conserved in fungi but not in mammals or plants (Fig. 6). Thus, different InsB domains and adaptors may have coevolved in different organisms to mediate membrane targeting of mitochondrial dynamin-related GTPases.
Figure 6.
Comparison of InsB domains and mitochondrial fission adaptors in different organisms. (A) Mitochondrial fission adaptors and their predicted structural domains are listed for the indicated organisms. (B) Alignment of Dnm1 InsB sequences from Saccharomyces cerevisiae (Sc), Cyanidioschyzon merolae (Cm), Arabidopsis thaliana (At), and Homo sapiens (Hs). The conserved motif in S. cerevisae Dnm1 InsB is underlined. Symbols below the sequence alignment indicate identity (*), strong similarity (:), and weak similarity (.) of amino acids. (C) The percentage of amino acid identity between the pairs of InsB sequences in each row is indicated. mM, Mus musculus. The range of identities for the fungal homologues includes pairwise comparisons of ScInsB with InsB from all the fungal species listed in Fig. 1 B.
Our mutational analysis demonstrates that four hydrophobic residues in the Dnm1 InsB motif are necessary for interaction with Mdv1. However, the purified Dnm1 InsB domain (aa 535–639) is not sufficient to bind the Mdv1 β-propeller in vitro (unpublished data). Because the conserved motif in InsB is hydrophobic, it may not adopt a functional conformation when InsB is expressed on its own. In full-length Dnm1, the InsB motif may normally be buried in the interior of the protein. In this case, a conformational change in Dnm1 would be required to expose critical residues in InsB for adaptor binding. A conformational change of this type could be stimulated by GTP binding to Dnm1. This scenario would be consistent with the previous finding that Mdv1 preferentially binds to the GTP-bound form of Dnm1 (Cerveny and Jensen, 2003; Naylor et al., 2006; Lackner et al., 2009).
Our GFP localization and two-hybrid studies established that suppressor mutations in the Mdv1 β-propeller partially restore the interaction of the adaptor with Dnm1F610A. However, this suppression is not allele specific because the suppressor mutations also rescue defects caused by dnm1F606A, dnm1L607A, and dnm1F611A (Fig. 5 B). These results suggest that suppression is not occurring by a classical “lock and key” model. It is possible that the suppressor mutations in the Mdv1 β-propeller increase protein flexibility, allowing them to bind the mutant Dnm1 proteins more efficiently. An alternative explanation is that the suppressor mutations establish new contacts that enhance the interaction between the Mdv1 β-propeller and Dnm1F610A. Although all twelve of the Mdv1 suppressor mutations in the β-propeller were able to recruit and assemble GFP-tagged WT Dnm1 into mitochondrial puncta, ten failed to rescue mitochondrial fission (Table 1, <20% rescue in a DNM1 mdv1Δ strain). Interestingly, these ten Mdv1 suppressor proteins exhibit more robust interaction with WT Dnm1 in a two-hybrid assay (Table 1), likely because of the new contacts. When WT Dnm1 is substituted for Dnm1F610A, these new interactions may interfere with post-recruitment and/or assembly steps of mitochondrial fission. The substituted amino acids in the Mdv1 suppressors must be physically close enough to Dnm1 to establish contacts. Thus, the residues affected by the suppressor mutations likely delineate regions of the Mdv1 β-propeller in close proximity to Dnm1.
As new mitochondrial fission components were identified, it became clear that different organisms express unrelated adaptor proteins (Fig. 6 A; Tieu and Nunnari, 2000; Cerveny et al., 2001; Griffin et al., 2005; Nishida et al., 2007; Arimura et al., 2008; Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010; Palmer et al., 2011; Zhao et al., 2011). We propose that the InsB domain provides some of the variability needed to mediate these diverse DRP–adaptor interactions. As shown in Fig. 6 (B and C), sequence alignments reveal little identity between InsB domains of fungi, algae, plants, and mammals. Conversely, high identity is observed among InsB sequences of representative mammals (Fig. 6 C, 94%). The functional interaction we observe between Dnm1 InsB and the Mdv1 β-propeller in yeast may be recapitulated for DRP–adaptor interactions in other organisms. However, there are almost certainly additional DRP–adaptor interfaces that remain to be identified, especially in mammals, where a single DRP is able to interact with several structurally distinct adaptors (Fig. 6 A; Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010; Palmer et al., 2011; Zhao et al., 2011). A recent study showed that mutation of a conserved residue in the Drp1 middle domain disrupts interaction with a mitochondrial adaptor called Mff (Strack and Cribbs, 2012). The model that InsB domains mediate DRP interactions in other organisms can be directly tested in genetic and cellular studies, as well as structural studies of DRPs bound to their cognate adaptor proteins.
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study include JSY5740 (MATa leu2Δ1 his3Δ200 trp1Δ63 ura3-52 lys2Δ202), JSY1361 (MATα leu2Δ1 his3Δ200 trp1Δ63 ura3-52 lys2Δ202 dnm1::HIS3), JSY9134 (MATα leu2Δ1 his3Δ200 trp1Δ63 ura3-52 lys2Δ202 mdv1::HIS3 dnm1::HIS3), JSY9744 (MATα leu2Δ1 his3Δ200 trp1Δ63 ura3-52 lys2Δ202 mdv1::HIS3 dnm1::dnm1F610A fzo1-1), JSY9983 (MATα leu2Δ1 his3Δ200 trp1Δ63 ura3-52 lys2Δ202 mdv1::HIS3 dnm1::dnm1F610A), JSY3903 (MATα leu2Δ1 his3Δ200 trp1Δ63 ura3-52 lys2Δ202 mdv1::HIS3), and JSY5148 (MATa trp1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3, GAL2-ADE2 met2::GAL7-lacZ).
A list of plasmids used in this study is provided in Table S2, except for the His-Dnm1 expression plasmid, which was received from J. Nunnari (University of California, Davis, Davis, CA). For pRS415-dnm1InsBmut, site-directed mutagenesis of a pRS415-DNM1 template was used to introduce sequences encoding single substitutions F606A, L607A, N608A, Y609A, F610A, F611A, G612A, and K613A. Similar methods were used to generate pRS415MET25-GFP-dnm1InsBmut, pRS425-dnm1InsBmut-MYC, and pRS426- dnm1InsBmut-3xHA. To create pGAD-C1-DNM1 and pGAD-C1-dnm1F610A, BamHI-DNM1-SalI and BamHI-dnm1F610A-SalI fragments were cloned between the BamHI and SalI sites of the pGAD-C1 vector. Similarly, BamHI-MDV1-SalI and BamHI-mdv1suppressor-SalI fragments were cloned between the BamHI and SalI sites of the pGBD-C1 vector to generate pGBD-C1-MDV1 and pGBD-C1-mdv1suppressor.
Fluorescence microscopy
Mitochondrial morphologies were quantified in WT, dnm1Δ, and dnm1Δmdv1Δ strains expressing the indicated proteins. The WT morphology category includes unbudded or budded cells with more than two free tubule ends in the mother cell. The formation of GFP-Dnm1 mitochondrial puncta was quantified by analysis of deconvolved epifluorescence images of random fields of cells. Phenotypic quantification is reported as the mean and standard deviation of three independent experiments (total n ≥ 300 cells unless noted). Unless specified in the figure legend, the mitochondria were visualized by expressing mitochondrial-targeted fast-folding RFP (mt-ffRFP). Dnm1 InsB variants were expressed from the DNM1 promoter in the pRS415 vector. GFP-tagged Dnm1 variants were expressed from the pRS415-MET25 vector. Mdv1 variants were expressed from the pRS416-MET25 plasmid. Yeast cells were grown at 30°C in selective synthetic dextrose medium containing 0.1 mg/ml methionine. Overnight cultures were diluted to 0.2 OD600 and grown for 3–5 h (OD600, 0.5–1.0). The methionine-repressible MET25 promoter is leaky under these conditions and expresses approximately fourfold more protein at steady state than that expressed from the endogenous DNM1 or MDV1 promoters (Karren et al., 2005).
A microscope (Axioplan 2; Carl Zeiss) equipped with a 100× oil immersion objective was used to observe and image cells. For mitochondria and GFP-Dnm1 puncta, 0.275 µm optical sections encompassing the entire yeast cells were deconvolved and analyzed using Axiovision version 4.6 (Carl Zeiss). All slices were projected on the transparency setting and the three-dimensional projections were converted to a single image. Final images were assembled using Photoshop and Illustrator (Adobe). Linear brightness and contrast adjustments were applied to the entire image.
CoIP assays
For Dnm1–Dnm1 interaction experiments, functional HA- and Myc-tagged Dnm1 variants were expressed in dnm1Δ cells. CoIPs with anti–c-Myc agarose-conjugated beads (Sigma-Aldrich) were performed as described previously (Koirala et al., 2010). 30 OD600 cell equivalents were harvested and resuspended in 500 µl IP buffer (0.5% Triton X-100, 150 mM NaCl, 1 mM EDTA, 50 mM Tris, pH 7.4, and 1:500 protease inhibitor cocktail set III [EMD]). Cells were lysed with glass bead and cleared by centrifugation at 18,000 g for 10 min. 400 µl of supernatant was incubated with 40 µl of anti–c-Myc–conjugated agarose beads (Sigma-Aldrich) for 1 h at 4°C. Agarose beads were collected and washed in immunoprecipitation buffer. The bound proteins were released by incubating the beads in 60 µl SDS-PAGE sample buffer lacking β-mercaptoethanol at 60°C for 8 min. 3.2 µl β-mercaptoethanol was added to the samples before boiling. Immunoprecipitated proteins were analyzed by SDS-PAGE and Western blotting with anti-Myc (Santa Cruz Biotechnology, Inc.) and anti-HA (University of Utah Core Facility) antibodies.
Mdv1–Dnm1 interaction was analyzed by coIP after dithiobis(succinimidyl propionate) (DSP) cross-linking in dnm1Δ cells expressing endogenous Mdv1 and plasmid-borne Myc-tagged Dnm1 variants (Koirala et al., 2010). 50 OD600 cell equivalents were spheroplasted by treating with 0.2 mg/ml zymolase for 60 min at 30°C followed by treatment with 2.5 mM DSP (Thermo Fisher Scientific) at 30°C for 30 min. 50 mM glycine was added to the cell suspensions and all subsequent buffers to quench DSP. Spheroplasts disrupted using a dounce homogenizer (Wheaton) were spun at 18,000 g for 10 min. Pellets were solubilized for 10 min at 4°C in 500 µl of immunoprecipitation buffer (1% Triton X-100, 150 mM NaCl, 30 mM Hepes-KOH, pH 7.4, and 1:500 protease inhibitor cocktail set III) and centrifuged at 18,000 g for an additional 10 min. 400 µl of supernatant was incubated for 1 h at 4°C with anti-HA–conjugated agarose beads (Sigma-Aldrich). Proteins released from agarose beads were separated by SDS-PAGE and analyzed by ECL Western blotting with anti-Myc and anti-Mdv1 antibodies.
Screen for mdv1 suppressors of the dnm1F610A allele
The growth phenotypes of strains used for this screen are summarized in Table S1. PCR amplification with Taq DNA polymerase was used to introduce random mutations into the MDV1 coding region. The PCR products were introduced into linearized pRS416-MET25 using gap repair (Orr-Weaver et al., 1983) in mdv1Δ dnm1::dnm1F610A fzo1-1 cells. In temperature-sensitive fzo1-1 cells, ongoing mitochondrial fission causes fragmentation, mitochondrial genome loss, and inability to grow on glycerol medium at 37°C (Hermann et al., 1998). Disrupting fission in this strain by introducing an mdv1Δ mutation and expressing dnm1F610A from the endogenous DNM1 locus (dnm1::dnm1F610A) prevents mitochondrial fragmentation and genome loss, allowing mdv1Δ dnm1::dnm1F610A fzo1-1 strains to grow on glycerol at the elevated temperature. Expression of WT Mdv1 from a plasmid does not restore the temperature-sensitive glycerol growth defect in this strain. In contrast, mdv1Δ dnm1::dnm1F610A fzo1-1 cells expressing Mdv1suppressor from a plasmid fail to grow on glycerol at 37°C, indicating that Mdv1suppressor restores mitochondrial fission. Cells containing Mdv1suppressor-expressing plasmids were identified by their ability to grow on glycerol at 25°C, but not at 37°C. Candidate clones with verified phenotypes were sequenced to identify MDV1 mutations. In alleles with multiple amino acid changes, mutations were separated by site-directed mutagenesis. Mutations contributing to growth phenotypes were analyzed for mitochondrial morphology and GFP-Dnm1 localization.
Mdv1 β-propeller modeling
The β-propeller model shown in Fig. 4 was generated from the crystal structure of the Cdc4 WD40 repeat (PDB accession no. 1NEX) using the PHYRE Protein Fold Recognition server (Kelley and Sternberg, 2009). Residues 349–713 of Mdv1 are variably modeled as a seven- or an eight-bladed β-propeller, depending on the structures most recently deposited in the PDB. The eight-bladed β-propeller model shown in Fig. 4 includes the majority of residues identified in the second-site suppressor analysis described here.
Yeast two-hybrid analysis
Yeast two-hybrid studies to analyze Dnm1–Mdv1 and Dnm1 self-interactions were performed in the Y187 S. cerevisiae strain background (Takara Bio Inc.) via a growth assay as described previously (Guthrie and Fink, 2002). pGAD and pGBD plasmid expressing the indicated fusion proteins were cotransformed into the Y187 reporter strain. Interaction between two fusion proteins leads to expression of one of several reporter genes in this strain, allowing the yeast cells to grow on S-dextrose minus histidine or minus adenine. WT Dnm1–Dnm1InsBmut interactions were performed in cells coexpressing GAD-Dnm1 WT and GBD-Dnm1InsBmut. The Dnm1–Mdv1 interaction was tested in both directions. However, the interaction was only detected when Dnm1 and Mdv1 were fused with the GAD and GBD domains, respectively.
Online supplemental material
Table S1 shows a screen for mdv1 suppressors of dnm1F610A. Table S2 shows the plasmids used in this study. Fig. S1 shows expression, interaction, and assembly properties of Dnm1 and Mdv1 variants. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207079/DC1.
Supplementary Material
Acknowledgments
We thank Jane Macfarlane for expertise in mutagenesis and plasmid construction, members of the Shaw laboratory for critical discussions, and J. Nunnari for the His-Dnm1 expression plasmid.
Research support was provided by National Institutes of Health grants GM53466 and GM84970 to J.M. Shaw. Sequencing and oligonucleotide synthesis services were provided by University of Utah Core Facilities.
Footnotes
Abbreviations used in this paper:
- coIP
- coimmunoprecipitation
- DRP
- dynamin-related protein
- DSP
- dithiobis(succinimidyl propionate)
- InsB
- Insert B
- mt-ffRFP
- mitochondrial-targeted fast-folding RFP
- PH
- pleckstrin homology
- WT
- wild-type
References
- Arimura S., Fujimoto M., Doniwa Y., Kadoya N., Nakazono M., Sakamoto W., Tsutsumi N. 2008. Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites. Plant Cell. 20:1555–1566 10.1105/tpc.108.058578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhar D., Karren M.A., Babst M., Shaw J.M. 2006. Dimeric Dnm1-G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J. Biol. Chem. 281:17312–17320 10.1074/jbc.M513530200 [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J.M. 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1:298–304 10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., Jensen R.E. 2003. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol. Biol. Cell. 14:4126–4139 10.1091/mbc.E03-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., McCaffery J.M., Jensen R.E. 2001. Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol. Biol. Cell. 12:309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160:189–200 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura Y., Ishihara N., Yokota S., Mihara K. 2003. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 134:333–344 10.1093/jb/mvg150 [DOI] [PubMed] [Google Scholar]
- Faelber K., Posor Y., Gao S., Held M., Roske Y., Schulze D., Haucke V., Noé F., Daumke O. 2011. Crystal structure of nucleotide-free dynamin. Nature. 477:556–560 10.1038/nature10369 [DOI] [PubMed] [Google Scholar]
- Ford M.G., Jenni S., Nunnari J. 2011. The crystal structure of dynamin. Nature. 477:561–566 10.1038/nature10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S., van der Bliek A.M. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19:2402–2412 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Kadirjan-Kalbach D., Froehlich J.E., Osteryoung K.W. 2003. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA. 100:4328–4333 10.1073/pnas.0530206100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E.E., Graumann J., Chan D.C. 2005. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 170:237–248 10.1083/jcb.200503148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. 2002. Guide to yeast genetics and molecular biology. Methods in enzymology series. Vol. 350 San Diego: Academic Press, Inc [Google Scholar]
- Hales K.G., Fuller M.T. 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 90:121–129 10.1016/S0092-8674(00)80319-0 [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143:359–373 10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., Blackstone C. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karren M.A., Coonrod E.M., Anderson T.K., Shaw J.M. 2005. The role of Fis1p–Mdv1p interactions in mitochondrial fission complex assembly. J. Cell Biol. 171:291–301 10.1083/jcb.200506158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M.A., Schrader M. 2003. Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278:8597–8605 10.1074/jbc.M211761200 [DOI] [PubMed] [Google Scholar]
- Koirala S., Bui H.T., Schubert H.L., Eckert D.M., Hill C.P., Kay M.S., Shaw J.M. 2010. Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes. J. Cell Biol. 191:1127–1139 10.1083/jcb.201005046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi K., Nagotu S., Krikken A.M., Sjollema K., Deckers M., Erdmann R., Veenhuis M., van der Klei I.J. 2006. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119:3994–4001 10.1242/jcs.03166 [DOI] [PubMed] [Google Scholar]
- Labrousse A.M., Zappaterra M.D., Rube D.A., van der Bliek A.M. 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 4:815–826 10.1016/S1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- Lackner L.L., Horner J.S., Nunnari J. 2009. Mechanistic analysis of a dynamin effector. Science. 325:874–877 10.1126/science.1176921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gould S.J. 2003. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278:17012–17020 10.1074/jbc.M212031200 [DOI] [PubMed] [Google Scholar]
- Mears J.A., Ray P., Hinshaw J.E. 2007. A corkscrew model for dynamin constriction. Structure. 15:1190–1202 10.1016/j.str.2007.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears J.A., Lackner L.L., Fang S., Ingerman E., Nunnari J., Hinshaw J.E. 2011. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18:20–26 10.1038/nsmb.1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T.J., Andreazza C., Verma A., Daga A., McNew J.A. 2011. Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc. Natl. Acad. Sci. USA. 108:11133–11138 10.1073/pnas.1105056108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151:367–380 10.1083/jcb.151.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor K., Ingerman E., Okreglak V., Marino M., Hinshaw J.E., Nunnari J. 2006. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 281:2177–2183 10.1074/jbc.M507943200 [DOI] [PubMed] [Google Scholar]
- Nishida K., Yagisawa F., Kuroiwa H., Yoshida Y., Kuroiwa T. 2007. WD40 protein Mda1 is purified with Dnm1 and forms a dividing ring for mitochondria before Dnm1 in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. USA. 104:4736–4741 10.1073/pnas.0609364104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Szostak J.W., Rothstein R.J. 1983. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 101:228–245 10.1016/0076-6879(83)01017-4 [DOI] [PubMed] [Google Scholar]
- Orso G., Pendin D., Liu S., Tosetto J., Moss T.J., Faust J.E., Micaroni M., Egorova A., Martinuzzi A., McNew J.A., Daga A. 2009. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 460:978–983 10.1038/nature08280 [DOI] [PubMed] [Google Scholar]
- Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., Mihara K. 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191:1141–1158 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., Keegan B.R., Brisch E., Thatcher J.W., Hermann G.J., Bleazard W., Shaw J.M. 1998. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143:333–349 10.1083/jcb.143.2.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Osellame L.D., Laine D., Koutsopoulos O.S., Frazier A.E., Ryan M.T. 2011. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12:565–573 10.1038/embor.2011.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G.J., McMahon H.T. 2004. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 5:133–147 10.1038/nrm1313 [DOI] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273:20150–20155 10.1074/jbc.273.32.20150 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147:699–706 10.1083/jcb.147.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack S., Cribbs J.T. 2012. Allosteric modulation of Drp1 mechanoenzyme assembly and mitochondrial fission by the variable domain. J. Biol. Chem. 287:10990–11001 10.1074/jbc.M112.342105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. 2000. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151:353–366 10.1083/jcb.151.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek A.M. 1999. Functional diversity in the dynamin family. Trends Cell Biol. 9:96–102 10.1016/S0962-8924(98)01490-1 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chan N.C., Ngo H.B., Gristick H., Chan D.C. 2012. Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (Mdv1) coiled coil. J. Biol. Chem. 287:9855–9861 10.1074/jbc.M111.329359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlén P., Tomilin N., Shupliakov O., Lendahl U., Nistér M. 2011. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30:2762–2778 10.1038/emboj.2011.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.