Abstract
Activating transcription factor 5 (ATF5) is a member of the ATF/cAMP response element-binding family of transcription factors, which compose a large group of basic region leucine zipper proteins whose members mediate diverse transcriptional regulatory functions. ATF5 has a well-established prosurvival activity and has been found to be overexpressed in several human cancers, in particular glioblastoma. However, the role(s) of ATF5 in development and normal physiology are unknown. Here we address this issue by deriving and characterizing homozygous Atf5 knockout mice. We find that Atf5−/− pups die neonatally, which, as explained below, is consistent with an olfactory defect resulting in a competitive suckling deficit. We show that Atf5 is highly expressed in olfactory sensory neurons (OSNs) in the main olfactory epithelium starting from embryonic stage 11.5 through adulthood. Immunostaining experiments with OSN-specific markers reveal that ATF5 is expressed in some immature OSNs and in all mature OSNs. Expression profiling and immunostaining experiments indicate that loss of Atf5 leads to a massive reduction in mature OSNs resulting from a differentiation defect and the induction of apoptosis. Ectopic expression of Atf5 in neural progenitor cells induces expression of multiple OSN-specific genes. Collectively, our results suggest a model in which Atf5 is first expressed in immature OSNs and the resultant ATF5 functions to promote differentiation into mature OSNs. Thus, ATF5 is required for terminal differentiation and survival of OSNs.
Keywords: hyposmia, olfaction, neonatal death, olfactory marker protein, TUJ1
Activating transcription factor 5 (ATF5; also known as ATFx) is a member of the ATF/cAMP response element-binding (CREB) family of transcription factors, which compose a large group of basic region leucine zipper (bZIP) proteins whose members mediate diverse transcriptional regulatory functions (reviewed in ref. 1). A role for ATF5 in promoting cell survival was first suggested from expression profiling experiments, which revealed that Atf5 was the gene most down-regulated in interleukin-3–dependent murine hematopoietic cells following apoptosis induction elicited by cytokine deprivation (2). Functional experiments demonstrated that the prosurvival function of ATF5 results from its ability to inhibit apoptosis (3).
A variety of subsequent studies have shown that ATF5 is overexpressed in, and contributes to the survival of, several human cancer cell lines and solid tumors, in particular glioblastoma (4, 5). The high level of ATF5 in brain tumors, coupled with the absence of ATF5 expression in mature neurons, has raised the possibility that ATF5 may be a therapeutic target for treatment of glioblastoma. Consistent with this idea, inhibiting ATF5 function, using either a dominant-negative mutant or a small interfering RNA, causes massive apoptotic death of rodent and human glioblastoma cell lines. By contrast, comparable loss of ATF5 function does not affect survival of cultured neurons or glial cells (6, 7).
A rigorous evaluation of the role of ATF5 as a therapeutic target requires a clear understanding of normal ATF5 function. However, the role of ATF5 in development and physiology has not been determined. Here we address this issue by deriving and characterizing Atf5−/− mice.
Results
Derivation of Atf5−/− Mice.
To investigate the physiological role of ATF5, we derived Atf5−/− mice. The gene-targeting strategy involved replacing almost the entire coding region of Atf5 with a LacZ-Neo cassette, such that the LacZ gene is under the control of the endogenous Atf5 promoter (Fig. S1A). The genomic structure of the Atf5 locus in Atf5+/+, Atf5+/−, and Atf5−/− mice was confirmed by Southern blot (Fig. S1A) and PCR (Fig. S1B) analyses. As expected, Atf5 expression was lost in mouse embryonic fibroblasts derived from Atf5−/− mice and in the liver (Fig. S1C) and olfactory epithelium (OE) of Atf5−/− mice (Figs. S1D and S2C).
Atf5−/− Mice Undergo Neonatal Death.
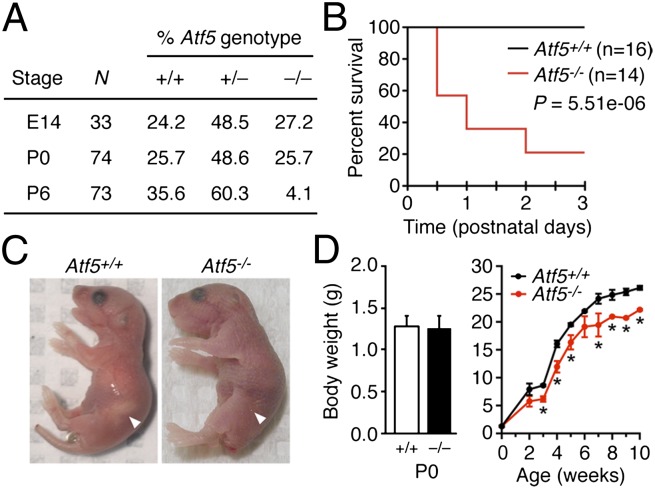
At birth (postnatal day 0, P0), no major phenotypic differences were observed between Atf5+/+, Atf5+/−, and Atf5−/− pups, and the ratio of all three genotypes was consistent with the predicted Mendelian distribution (Fig. 1A). Likewise, as expected, the genotypes of embryonic stage day 14 (E14) embryos were consistent with the predicted Mendelian distribution.
Fig. 1.
Atf5−/− mice undergo neonatal death. (A) Percentage of viable Atf5 genotypes among E14 embryos and newborn (P0) and 6-d-old (P6) pups. n, number of pups examined. (B) Kaplan–Meier survival curve of Atf5+/+ and Atf5−/− mice up to 3 d after birth. (C) Images showing Atf5+/+ and Atf5−/− mice 24 h after birth. In Atf5+/+ mice, the arrow points to the milk spot, which is absent in Atf5−/− mice. (D) Body-weight analysis of Atf5+/+ and surviving Atf5−/− pups at P0 (Left) and over the first 10 wk (Right). *P < 0.05.
Significantly, however, following birth mice lacking Atf5 failed to thrive, as evidenced by the drastic reduction in the percentage of Atf5−/− pups between P0 and P6 (Fig. 1A). The results of Fig. 1B show that within 48 h after birth the majority (11/14) of Atf5−/− pups died, whereas no neonatal death was observed in Atf5+/+ mice. Further analysis of the dead Atf5−/− mice revealed a lack of milk in their stomach (Fig. 1C), consistent with the possibility of a competitive suckling deficit. Despite similar weights at birth, the weight of the few surviving Atf5−/− pups was less than that of their wild-type siblings throughout 10 wk after birth (Fig. 1D).
High Selective Expression of Atf5 in the Olfactory System.
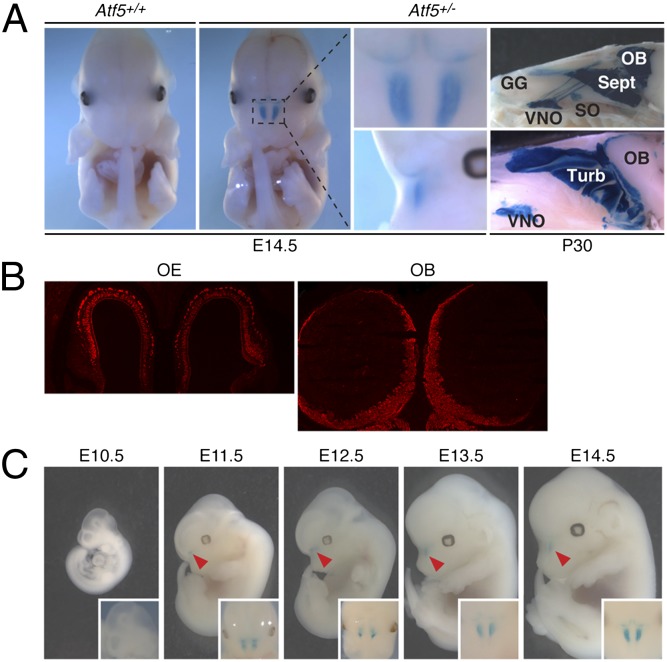
As a first step toward determining why loss of Atf5 results in neonatal lethality, we performed a series of experiments to analyze Atf5 expression. In Atf5+/− and Atf5−/− mice, LacZ transcription is directed by the Atf5 promoter (Figs. S1A and S2A), and thus β-galactosidase (β-gal) activity provides a convenient readout of Atf5 expression. The whole-mount β-gal activity assay of Fig. 2A shows that in an Atf5+/− mouse at E14.5, Atf5 was highly and specifically expressed in the nasal cavity. To confirm that the β-gal expression pattern faithfully recapitulated that of endogenous Atf5, we performed in situ hybridization. The results in Fig. S2B show that, like β-gal, Atf5 was highly and specifically expressed in the nasal cavity.
Fig. 2.
High, selective expression of Atf5 in the olfactory system. (A) Whole-mount β-gal activity assay. (Left) Front views of β-gal activity in Atf5+/+ and Atf5+/− E14.5 embryos. Close-up views of β-gal activity in the Atf5+/− embryo (dashed box) are shown from front and side. (Far Right) Whole-mount β-gal activity assay on two different sagittal head sections of a P30 Atf5+/− mouse. GG, Grueneberg ganglion; OB, olfactory bulb; Sept, septum; SO, septal organ; Turb, turbinates; VNO, vomeronasal organ. Magnification 5×. In the Upper but not Lower image at Far Right, the surface of the OB has been maintained, accounting for the apparent differences in β-gal staining. Enlarged images are shown in Fig. S8A. (B) Immunofluorescence monitoring β-gal staining in the OE and OB of a P0 Atf5+/− pup. Magnification 50×. (C) Whole-mount β-gal activity assay on Atf5+/− E10.5–E14.5 embryos (arrowheads point to β-gal staining) (Insets, front view).
The nasal cavity is composed of two types of epithelia, respiratory and olfactory epithelia (reviewed in ref. 8). To determine which epithelial type is the site of Atf5 expression, we performed a whole-mount β-gal activity assay on sagittal head sections of an Atf5+/− P30 mouse. Fig. 2A (Right) shows that β-gal activity was restricted to the olfactory system, including the both the turbinates and the septum of the main OE (hereafter referred to as the OE), the vomeronasal organ (VNO), the Grueneberg ganglion, the septal organ, and the olfactory bulb (OB). In situ hybridization confirmed that Atf5 was expressed in the OE (Fig. S2C) and VNO (Fig. S2D). Expression of Atf5 in the OE and OB was also detected by immunostaining for β-gal (Fig. 2B and see Fig. S3 below).
We next analyzed Atf5 expression at a series of different developmental stages. We found that Atf5 was selectively expressed in the olfactory system from as early as E11.5 (Fig. 2C). Expression of Atf5 in the olfactory system remained strong through adulthood (up to 10 mo tested so far; Fig. S2E). In addition, we found that Atf5 expression was restricted to the olfactory system through adulthood. Taken together, these results demonstrate that Atf5 is highly expressed in the olfactory system from early development (E11.5) through adulthood, suggesting an important role in olfactory function.
Atf5 Is Selectively Expressed in Olfactory Sensory Neurons.
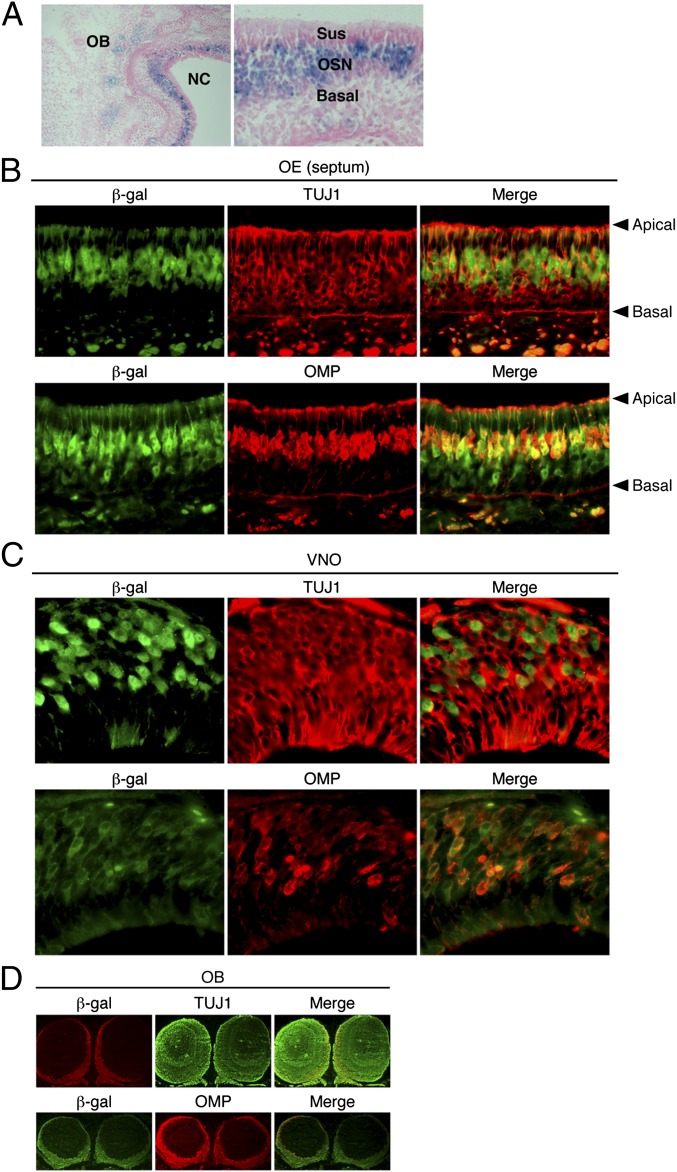
To identify the cell type that expresses Atf5, a β-gal activity assay was performed on horizontal sections of the nasal cavity. Fig. 3A shows that Atf5 expression was localized between the sustentacular and the basal cell layers, an area that corresponds to the region where olfactory sensory neurons (OSNs), both immature and mature, reside.
Fig. 3.
Atf5 is selectively expressed in OSNs. (A) β-Gal activity assay on a horizontal section of the nasal cavity of a P0 Atf5−/− pup, followed by eosin staining. Magnification 100× (Left) and 200× (Right). OB, olfactory bulb; NC, nasal cavity; Sus, sustentacular cells; OSN, olfactory sensory neurons; Basal, globose basal and horizontal basal cells. (B–D) Immunofluorescence monitoring β-gal, TUJ1, and OMP staining in coronal sections of the septum (B), VNO (C), and OB (D) of a P0 Atf5+/− pup. Merged images are shown on the Right. Arrowheads indicate the apical and basal layers of the OE. Magnification 200× (B), 400× (C), and 50× (D). Enlarged images of D are shown in Fig. S8B.
To delineate the specific cell type(s) that expresses Atf5, we performed double immunostaining for β-gal and specific OE markers. TUJ1 (also called TUBB3) is a well-established marker that is strongly expressed in immature OSNs and weakly expressed in mature OSNs (9). Fig. 3B (Upper) shows that, in the OE, many TUJ1+ cells, particularly those located near the basal layer, did not costain with β-gal (Fig. S3 A and B). Significantly, however, some β-gal+ cells also stained strongly for TUJ1. The staining pattern of GAP43, an alternative marker of immature OSNs (10), was very similar to that observed for TUJ1 (Fig. S4).
We also performed coimmunostaining for OMP, a characteristic marker of mature OSNs (9). We found that in the OE, all OMP+ neurons, which reside near the apical surface of the OE, were also β-gal+ (Fig. 3B, Lower, and Fig. S3 A and C). Similar results were obtained in adult mice (Fig. S3D). Taken together, the results described above demonstrate that Atf5 is expressed in some immature (TUJ1+) and in all mature (OMP+) OSNs.
We performed comparable double immunostaining experiments for the VNO and OB. For the VNO, we found that Atf5 expression, as evidenced by β-gal staining, was also found in both TUJ1+ and OMP+ vomeronasal sensory neurons (VSNs) (Fig. 3C). For the OB, strong β-gal+ staining was confined to the outer, glomerular layer, which contains axons from OSNs that reside in the OE (Fig. 3D and Fig. S3E).
ATF5 Is Required for Terminal Differentiation of OSNs.
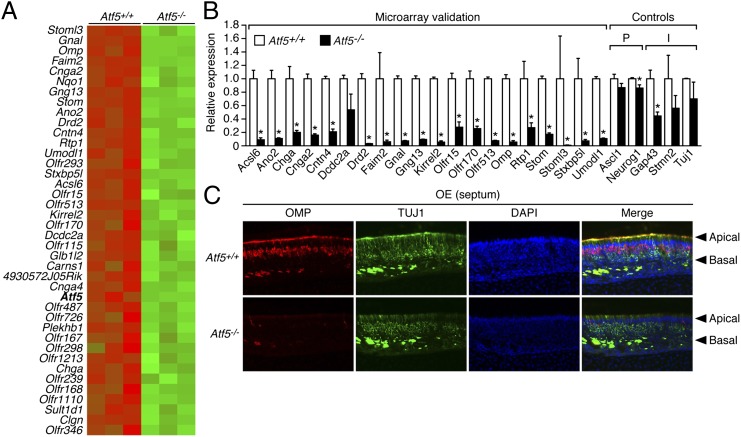
The highly restricted expression pattern of Atf5 in the olfactory system strongly suggested that Atf5 has an important role in OSNs. To test this possibility, we first compared gene expression profiles of OE from Atf5+/+ and Atf5−/− mice. Briefly, olfactory turbinates and septum, where the main OE is located, were dissected and collected from three different litters of Atf5+/+ and Atf5−/− P0 pups, and total RNA was isolated from each sample and subjected to microarray analysis. Using ≥1.5-fold as a threshold and P < 0.001 as a cutoff, the microarray results revealed that the expression levels of 274 genes were significantly decreased in Atf5−/− OE (Fig. S5 and Dataset S1). Fig. 4A presents a heat map of the 40 genes whose expression levels were decreased most substantially in Atf5−/− OE. Notably, among the 274 genes with decreased expression levels, ∼85% were associated with neuronal function including essential components of the canonical olfactory signal transduction pathway, such as multiple olfactory receptor (Olfr) genes, the olfactory-specific G protein Gnal (also known as Golf), the guanine nucleotide-binding protein Gng13, the adenylate cyclase Adcy3, the cyclic nucleotide gated channel Cnga2, and the calcium-activated chloride channel Ano2 (11).
Fig. 4.
ATF5 is required for terminal differentiation of OSNs. (A) Heat map displaying the 40 genes whose expression levels were most decreased in Atf5−/− OE (green) relative to that in Atf5+/+ OE (red). (B) qRT-PCR monitoring gene expression in OE of P0 Atf5+/+ and Atf5−/− mice. The results were normalized to the expression level in Atf5+/+ mice, which was set to 1. P, progenitor markers; I, immature OSN markers. *P < 0.05. (C) Immunofluorescence monitoring OMP (red) and TUJ1 (green) staining in OE (from the septum) of P0 Atf5+/+ and Atf5−/− mice. Sections were counterstained with DAPI (blue). Merged images are shown on the Right. Arrowheads indicate the apical and basal layers of the OE. Magnification 200×.
To validate the microarray results, we selected 20 genes that were among the most affected by loss of Atf5 and analyzed their expression by quantitative RT-PCR (qRT-PCR). The results of Fig. 4B show that the expression levels of 19 of the 20 genes were significantly decreased in Atf5−/− OE. Several of these validated genes, including Cnga2, Gnal, Gng13, and Rtp1, are known to be essential for olfactory function (11). Notably, previous studies have shown that mice lacking Gnal (12), Cnga2 (13), or Adcy3 (14) are profoundly hyposmic and have a neonatal death phenotype comparable to that of Atf5−/− mice. qRT-PCR analysis of a representative set of OSN-specific genes indicated that their expression levels in Atf5+/− and Atf5+/+ OE were comparable (Fig. S6A).
For comparison, we also analyzed expression of the neuronal progenitor markers Ascl1 (also called Mash1) and Neurog1 (also called Ngn1) and the immature OSN markers Gap43, Stmn2 (also called Scg10), and Tuj1 (Fig. 4B). Expression levels of Ascl1 and Neurog1 were not substantially reduced in Atf5−/− OE, indicating that OSN progenitors are not affected by loss of Atf5. Expression levels of Gap43, Stmn2, and Tuj1 were modestly reduced, suggesting that some immature OSNs are affected by loss of ATF5, consistent with the immunostaining results demonstrating expression of Atf5 in some but not all immature OSNs (Fig. 3B).
To confirm the expression profiling and qRT-PCR results, we performed immunostaining experiments. As expected, OMP staining was readily detectable in OE from Atf5+/+ (Fig. 4C) as well as from Atf5+/− (Fig. S6B) mice. Fig. 4C shows that, in Atf5−/− OE, OMP staining was almost completely abolished, indicating a dramatic loss of mature OSNs (Fig. S6B). By contrast, the TUJ1 staining pattern of Atf5+/+ and Atf5−/− OE was roughly comparable (Fig. 4C). Likewise, the GAP43 staining pattern of Atf5+/− and Atf5−/− OE was similar (Fig. S4).
ATF5 Is Critical for Survival of Mature OSNs.
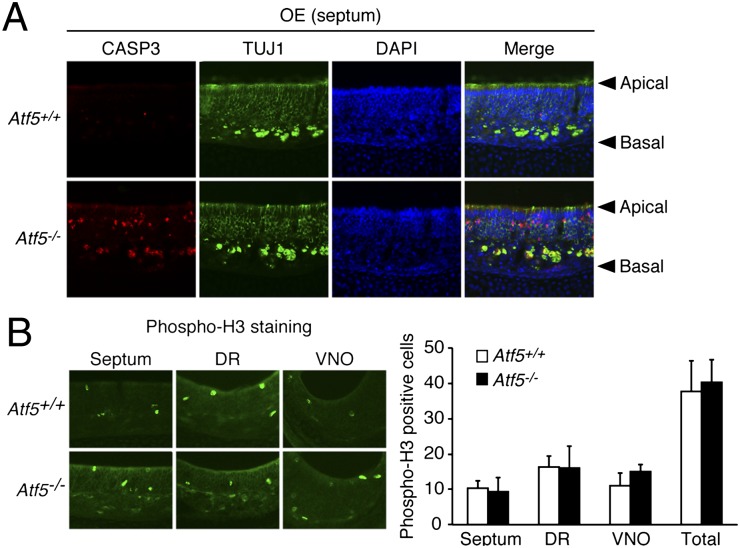
As described above, ATF5 is an anti-apoptotic factor, raising the possibility that ATF5 functions, at least in part, by preventing apoptosis and thereby promoting survival of OSNs. To investigate this possibility, we analyzed apoptosis in OE from Atf5+/+ and Atf5−/− mice by immunostaining with an antibody to cleaved caspase 3. The results show that, in Atf5+/+ (Fig. 5A) and Atf5+/− (Fig. S6C), OE apoptosis was virtually undetectable, consistent with previous reports (15, 16), whereas Atf5−/− OE contained a large number of apoptotic cells (Fig. 5A and Fig. S6C). Double immunostaining with TUJ1 revealed that apoptosis occurred in some immature OSNs, but most apoptotic cells did not stain strongly for TUJ1 and were in the apical region of OE that is normally occupied by mature OSNs (Fig. 5A). In contrast to the apoptosis results, there was no difference in cellular proliferation between Atf5+/+ and Atf5−/− OE as evidenced by equivalent phospho-histone H3 staining (Fig. 5B).
Fig. 5.
ATF5 is critical for survival of mature OSNs. (A) Immunofluorescence monitoring cleaved caspase 3 (CASP3) (red) and TUJ1 (green) staining in OE (from the septum) of P0 Atf5+/+ and Atf5−/− mice. Sections were counterstained with DAPI (blue). Merged images are shown on the Right. Arrowheads indicate the apical and basal layers of the OE. Magnification 200×. (B) (Upper) Immunofluorescence monitoring phosphorylated histone H3 (Phospho-H3) staining in the septum, dorsal region (DR), and VNO of newborn Atf5+/+ and Atf5−/− mice. (Right) The average number of phospho-H3–positive cells in three different microscopic fields is indicated for each region.
ATF5 Can Direct OSN-Specific Gene Expression.
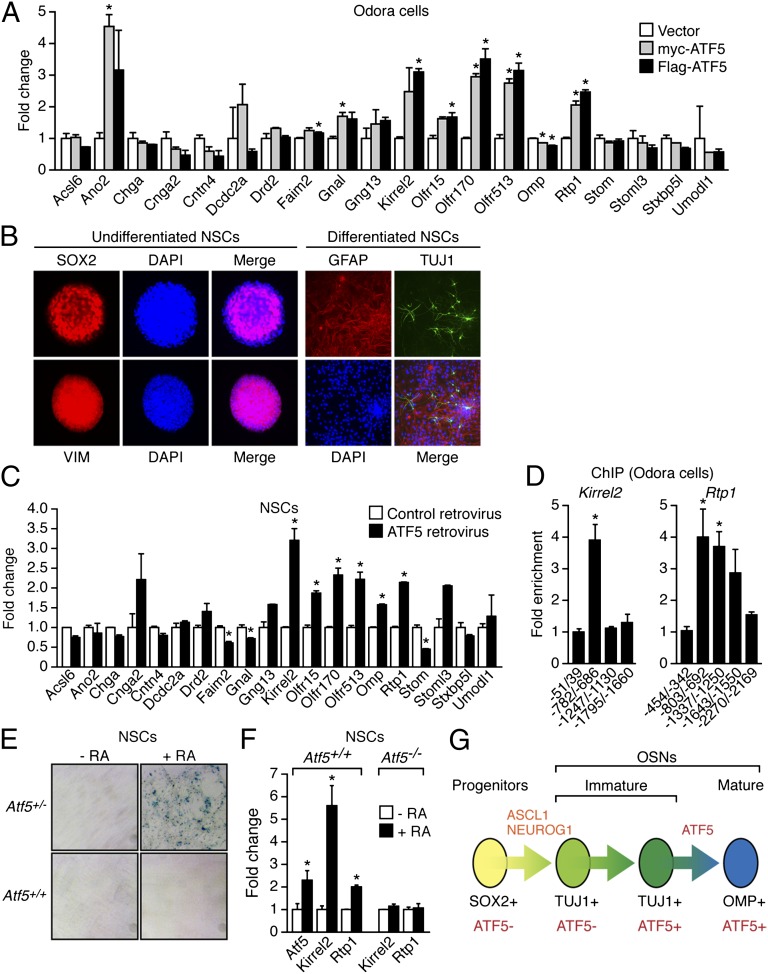
Because ATF5 is a transcription factor, it seemed likely that ATF5 functions by stimulating expression of genes required for OSN maturation and survival. To test this idea, we asked whether ectopic expression of Atf5 in rat Odora cells, which are derived from OSN progenitor cells and are an experimental model of OSN progenitors (17), would result in the transcriptional activation of OSN-specific genes. We tested a panel of 20 genes previously found to have decreased expression levels in Atf5−/− OE (Fig. 4 A and B). Myc- or Flag-tagged ATF5 were transiently expressed in Odora cells (Fig. S7A) and after 6 d qRT-PCR analysis revealed the up-regulation of a number of OSN-specific genes including Ano2, Kirrel2, Olfr15, Olfr170, Olfr513, and Rtp1 (Fig. 6A).
Fig. 6.
ATF5 can direct OSN-specific gene expression. (A) qRT-PCR expression analysis of 20 genes in rat Odora cells transfected with myc-ATF5, Flag-ATF5, or empty vector. Fold change is relative to the vector control, which was set at a value of 1. (B) Immunofluorescence monitoring SOX2 and VIM staining in undifferentiated neural stem cells (NSCs) (Left) and GFAP and TUJ1 staining in differentiated NSCs (Right). Cells were counterstained with DAPI (blue). Merged images are shown. Magnification 200×. (C) qRT-PCR analysis monitoring expression of 20 genes in murine NSCs transduced with a retrovirus that expresses ATF5 or empty retrovirus. Fold change is relative to the empty retrovirus control, which was set at a value of 1. (D) ChIP analysis monitoring ATF5 binding to the promoters of Kirrel2 and Rtp1 in Odora cells. Fold enrichment is specified relative to nonspecific recruitment to a control gene desert locus, which was set to 1. Primer coordinates are given relative to the AUG start codon. (E) β-Gal staining of Atf5+/+ or Atf5+/− NSCs induced to differentiate in the presence of 25 µM retinoic acid (RA). Magnification 20×. (F) qRT-PCR analysis monitoring expression of Atf5 and Kirrel2 in Atf5+/+ and Atf5−/− NSCs in the presence and absence of RA. Fold change in expression is relative to the expression level in the absence of RA, which was set at 1. (G) Model for ATF5-directed differentiation of OSNs in the OE. *P < 0.05.
We next performed similar experiments in murine neural stem cells, which were isolated from the lateral and medial ganglionic eminence of E14 mouse embryos. These cells are not the normal progenitor of OSNs and therefore provide a rigorous test for the ability of ATF5 to direct OSN-specific gene expression. After confirming stemness, as evidenced by expression of SOX2 and VIM (Fig. 6B, Left), and multipotency, as evidenced by the capacity to differentiate into GFAP+ astrocytes and TUJ1+ neurons (Fig. 6B, Right), we transduced neural stem cells with a retrovirus that expresses Atf5 (Fig. S7B). After growth in differentiation medium for 6 d, qRT-PCR was performed to measure expression of the same panel of 20 genes that were analyzed in Fig. 6A. The results show that ectopic expression of Atf5 resulted in up-regulation of a number of OSN-specific genes including Kirrel2, Olfr15, Olfr170, Olfr513, Omp, and Rtp1 (Fig. 6C).
To ask whether some or all of the up-regulated genes were direct targets of ATF5, we performed chromatin-immunoprecipitation (ChIP) experiments. We analyzed four genes—Kirrel2, Olfr170, Olfr513, and Rtp1—that were induced by ectopic expression of ATF5 in both rat Odora and murine neural stem cells. ATF5 occupancy was analyzed by ChIP assays in Odora cells using a series of promoter-specific primers. The ChIP results of Fig. 6D show that ATF5 could be detected on both the Kirrel2 and Rtp1 promoters. By contrast, we were unable to detect ATF5 binding on comparable regions of the Olfr170 and Olfr513 promoters. These collective results support the possibility that ATF5 directs transcription of some OSN-specific genes by direct binding to the target gene promoter.
Retinoic acid promotes olfactory identity and is required for olfactory function (18). Fig. 6E shows that, following treatment with retinoic acid, Atf5+/− murine neural stem cells stained positively for β-gal, indicative of Atf5 expression. Consistent with this result, the qRT-PCR experiment of Fig. 6F shows that treatment of murine neural stem cells with retinoic acid resulted in the up-regulation of Atf5 as well as the OSN-specific genes Kirrel2 and Rtp1. Notably, Kirrel2 and Rtp1 expression was unaffected by retinoic acid treatment of Atf5−/− neural stem cells. Thus, Atf5 expression is activated by retinoic acid, an upstream inducer of OSN differentiation, and is required for retinoic acid-induced transcriptional activation of OSN-specific genes.
Discussion
In this report, we study the physiological role of ATF5 and find that Atf5−/− mice die neonatally, which can be explained by an olfactory defect and a resultant competitive suckling deficit. The neonatal death phenotype that we observe is consistent with that described in previous studies in which genes required for olfactory function have been ablated in mice (12, 13, 19). On the basis of the Atf5 expression pattern and effects of Atf5 loss discussed below, we conclude that ATF5 is required for terminal differentiation and survival of the majority of OSNs in the OE. Whether ATF5 is also required for terminal differentation and survival of all OSN subtypes (reviewed in ref. 11) remains to be determined.
On the basis of β-gal activity in Atf5+/− mice and in situ hybridization we find that Atf5 is highly and selectively expressed in the olfactory system. This conclusion is strongly supported by our observation that the only detectable phenotypic abnormality of Atf5−/− mice is a large reduction of mature OSNs in the OE. Atf5 expression starts at E11.5, when the nose pit emerges, which is about 1 d after the olfactory pit forms and OSNs are first detected (8). Thus, the start of Atf5 expression conincides with the emergence of OSNs from early progenitors. We have also shown that in murine neural stem cells Atf5 expression is induced by retinoic acid, which promotes and is required for OSN differentiation (18). Atf5 continues to be expressed in mature OSNs throughout adulthood, suggesting a continual role for ATF5 in olfactory function. We also detected ATF5 expression in VSNs; however, the role of ATF5 in the VNO will require further investigation.
Our microarray results revealed that the expression levels of many OSN-specific genes were substantially decreased in Atf5−/− OE. These genes include 172 Olfr genes and other genes, such as Gnal (12), Rtp1 (20), and Kirrel2 (21), that are critical for olfactory function. By contrast, genes expressed in OSN progenitors (Ascl1 and Neurog1) were unaffected, and immature OSN-specific genes (Gap43, Stmn2, and Tuj1) were only modestly affected by loss of Atf5. Immunostaining enabled us to attribute the loss of OSN-specific gene expression in Atf5−/− OE to a massive reduction of mature OSNs. By contrast, the number of immature OSNs in OE was relatively unaffected by loss of Atf5. Thus, the reduced number of mature OSNs in Atf5−/− OE can be explained, at least in part, by a failure of immature OSNs to fully differentiate into mature OSNs. In support of this idea, we show that ATF5 can direct neural progenitor cells to express some mature OSN-specific genes. Perhaps not surprisingly, ectopic expression of ATF5 did not result in the transcriptional induction of all mature OSN-specific genes (Fig. 6C), indicating that factors in addition to ATF5 are required to convert neural progenitors into mature OSNs.
Our immunostaining results also revealed that Atf5 is expressed in all mature OSNs and in some, but not all, immature OSNs. Collectively, our results suggest that Atf5 is first expressed in immature OSNs and that the resultant ATF5 functions to promote differentation into mature OSNs (Fig. 6G). Previous studies have identified factors, such as ASCL1 and NEUROG1, that are required for the commitment of neural precursors to OSNs. To our knowledge, ATF5 is the only factor known to specifically act by promoting the conversion of immature to mature OSNs.
In the absence of Atf5, some OSNs were found to undergo apoptosis, which likely also contributes to the loss of mature OSNs in Atf5−/− OE. The ability of ATF5 to promote survival of mature OSNs may result from ATF5-directed transcriptional stimulation of anti-apoptotic gene (see, for examples, refs. 5 and 22). In support of this possibility, we found that the expression level of Faim2, which encodes a neuron-specific inhibitor of apoptosis (23), was dramatically decreased in Atf5−/− OE (Fig. 4B). However, previous studies have found that disruption of normal OSN differentiation can result in apoptosis (24–26). Thus, apoptosis induction in the absence of Atf5 may also be explained by the inability of immature OSNs to fully differentiate into mature OSNs.
Materials and Methods
Mice.
An Atf5+/LacZ embryonic stem cell clone was obtained from the National Institutes of Health Knockout Mouse Project repository, and its genomic structure was verified by long-fragment PCR and Southern blotting. The clone was microinjected into C57BL6-albino blastocysts and implanted into a pseudopregnant C57BL6-albino recipient mother to generate chimeric F0 founders, which were backcrossed to C57BL6 to generate Atf5+/LacZ F1 heterozygotes, which were intercrossed to generate Atf5LacZ/LacZ homozygotes (referred to as Atf5−/− here). Animal protocols were approved by the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School.
Whole-Mount β-Gal Activity Assay.
Tissues were dissected in 100 mM phosphate buffer (pH 7.4) and 2 mM MgCl2 and fixed and stained (27). For Fig. 3A, horizontal tissue sections (16 µm) from a P0 mouse were cryopreserved without fixation, and a β-gal activity assay was performed followed by counterstaining with eosin.
Immunofluorescence.
Dissected mouse heads were cryosectioned followed by immunofluorescence (28). Cells grown on glass coverslips (coated with poly-D-lysine/laminin for neural stem cells) were infected with virus or transfected with plasmids and grown for an additional 60 h. Cells were washed with PBS, fixed in 4% (wt/vol) paraformaldehyde at 4 °C for 10 min, washed three times in PBS at room temperature, and incubated in blocking buffer for 1 h. Coverslips were incubated overnight as described above with the following primary antibodies: OMP (Wako Chemicals); TUJ1 (Covance); GAP43 (Millipore); β-galactosidase (MP Cappel); cleaved caspase-3 (Cell Signaling); phospho-histone H3 (Upstate); SOX2 (R&D System); GFAP (DAKO); and myc, Flag, and Vimentin-Cy3 (Sigma). Secondary antibodies were conjugated with Alexa Fluor 488, Alexa Fluor 594 (Invitrogen), and Cy3 or FITC (Jackson Laboratories) fluorophores. Nuclei were counterstained with DAPI (Molecular Probes). Images were acquired using a Zeiss AxioCam camera mounted to a Zeiss Axioplan immunofluorescence microscope.
Gene Expression Profiling.
Olfactory septa and turbinates were dissected from Atf5+/+ and Atf5−/− P0 pups (n = 3) from different litters and pooled into a single sample. Three independent samples were prepared for each genotype. Total RNA was isolated, and samples were hybridized to GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix) and analyzed using Affymetrix software. RMA method (29, 30) in the Affy package from Bioconductor was used in R to summarize the probe-level data and normalize the dataset to remove across-array variation. A moderated t test in Limma package (31) was used to determine whether a gene’s expression level differed between Atf5+/+ and Atf5−/−. Genes with a fold change ≥1.5 and a P value <0.001 were considered significant. The data discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (32) and are accessible through GEO accession no. GSE37609 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37609).
qRT-PCR.
Total RNA was extracted using TRIzol (Invitrogen). First-strand cDNA was synthesized using the ImProm-II Reverse Transcription System (Promega). qRT-PCR was performed with gene-specific primers using the ABI Prism 7700 Sequence Detector System (Applied Biosystems) (28) using Gapdh or Rpl41 as an internal control.
Embryonic Neural Stem Cell Culture.
Lateral and medial ganglionic eminences were dissected from the forebrains of E14 mouse embryos and collected in PBS with 2% (wt/vol) glucose on ice. Tissues were dissociated by triturating with a micropipette. Undifferentiated neural stem cells (NSCs) were maintained as neurospheres in NSC growth medium (DMEM/F-12, 100 µg/mL transferrin, 5 µg/mL insulin, 20 nM progesterone, 10 µM putrescine, 30 nM selentine) in the presence of 20 ng/mL EGF (StemCell Technologies). NSCs were differentiated in NSC Basal Medium plus Differentiation Supplement (StemCell Technologies). For retinoic acid treatment, NSCs were grown on plates coated with poly(D) lysine and aminin (Sigma) and incubated with 20 µM retinoic acid in differentiation medium for 48 h. Cells were fixed and stained for β-gal activity or harvested for total RNA extraction and qRT-PCR.
Ectopic Atf5 Expression.
Odora cells were cultured in DMEM with 10% (vol/vol) FBS and 7% (vol/vol) CO2 at 33 °C. Cells were transfected with pFLAG-CMV-ATF5 or pCS2-Myc-ATF5, constructed by amplifying Atf5 by RT-PCR and cloning it into p3XFlag-myc-CMV-26 (Sigma) or pCS2-MT (33), respectively, or empty vector using Nucleofector Kit V and program T-028 (Lonza). After 72 h, cells were processed for gene expression, ChIP, or immunofluorescence.
ChIP Assays.
pCS2-Myc-ATF5–transfected Odora cells were fixed with 1% (wt/vol) formaldehyde for 10 min at 25 °C and then stopped with 0.125 M glycine, followed by a standard ChIP assay (34) using nonimmune mouse IgG (Santa Cruz) or anti-myc mouse monoclonal antibody (Sigma). ChIP DNA and input DNA were used as templates for qPCR.
Statistical Analysis.
All experiments were performed at least three times. Mean values for individual experiments are expressed as mean ± SD. P values were calculated using the log-rank test in the survival package in R (35) (Fig. 1B) or the Welch two-sample t test (Figs. 1D, 4B, 5B; and 6 A, C, D, and F; and Figs. S1D, S6A, and S7B). A P value <0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank James Schwob for providing Odora cells. This work is supported by National Institutes of Health Grant R01CA115817 (to M.R.G.). M.R.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE37609).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210479109/-/DCSupplemental.
References
- 1.Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis. 2003;8(3):225–228. doi: 10.1023/a:1023633704132. [DOI] [PubMed] [Google Scholar]
- 2.Devireddy LR, Teodoro JG, Richard FA, Green MR. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science. 2001;293(5531):829–834. doi: 10.1126/science.1061075. [DOI] [PubMed] [Google Scholar]
- 3.Persengiev SP, Devireddy LR, Green MR. Inhibition of apoptosis by ATFx: A novel role for a member of the ATF/CREB family of mammalian bZIP transcription factors. Genes Dev. 2002;16(14):1806–1814. doi: 10.1101/gad.992202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monaco SE, Angelastro JM, Szabolcs M, Greene LA. The transcription factor ATF5 is widely expressed in carcinomas, and interference with its function selectively kills neoplastic, but not nontransformed, breast cell lines. Int J Cancer. 2007;120(9):1883–1890. doi: 10.1002/ijc.22469. [DOI] [PubMed] [Google Scholar]
- 5.Sheng Z, et al. A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat Med. 2010;16(6):671–677. doi: 10.1038/nm.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelastro JM, et al. Downregulation of activating transcription factor 5 is required for differentiation of neural progenitor cells into astrocytes. J Neurosci. 2005;25(15):3889–3899. doi: 10.1523/JNEUROSCI.3447-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason JL, et al. ATF5 regulates the proliferation and differentiation of oligodendrocytes. Mol Cell Neurosci. 2005;29(3):372–380. doi: 10.1016/j.mcn.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Treloar HB, Miller AM, Ray A, Greer CA. In: Development of the Olfactory System: The Neurobiology of Olfaction. Menini A, editor. Boca Raton, FL: CRC Press; 2010. [PubMed] [Google Scholar]
- 9.Carter LA, MacDonald JL, Roskams AJ. Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. J Neurosci. 2004;24(25):5670–5683. doi: 10.1523/JNEUROSCI.0330-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng LE, Reed RR. Zfp423/OAZ participates in a developmental switch during olfactory neurogenesis. Neuron. 2007;54(4):547–557. doi: 10.1016/j.neuron.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munger SD, Leinders-Zufall T, Zufall F. Subsystem organization of the mammalian sense of smell. Annu Rev Physiol. 2009;71:115–140. doi: 10.1146/annurev.physiol.70.113006.100608. [DOI] [PubMed] [Google Scholar]
- 12.Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20(1):69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 13.Brunet LJ, Gold GH, Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17(4):681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 14.Wong ST, et al. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27(3):487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 15.Voyron S, et al. Apoptosis in the development of the mouse olfactory epithelium. Brain Res Dev Brain Res. 1999;115(1):49–55. doi: 10.1016/s0165-3806(99)00055-3. [DOI] [PubMed] [Google Scholar]
- 16.Cowan CM, Roskams AJ. Apoptosis in the mature and developing olfactory neuroepithelium. Microsc Res Tech. 2002;58(3):204–215. doi: 10.1002/jemt.10150. [DOI] [PubMed] [Google Scholar]
- 17.Murrell JR, Hunter DD. An olfactory sensory neuron line, odora, properly targets olfactory proteins and responds to odorants. J Neurosci. 1999;19(19):8260–8270. doi: 10.1523/JNEUROSCI.19-19-08260.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawson NE, LaMantia AS. Once and again: retinoic acid signaling in the developing and regenerating olfactory pathway. J Neurobiol. 2006;66(7):653–676. doi: 10.1002/neu.20236. [DOI] [PubMed] [Google Scholar]
- 19.Guillemot F, et al. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75(3):463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 20.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119(5):679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 21.Serizawa S, et al. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127(5):1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 22.Dluzen D, Li G, Tacelosky D, Moreau M, Liu DX. BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. J Biol Chem. 2011;286(9):7705–7713. doi: 10.1074/jbc.M110.207639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reich A, et al. Fas/CD95 regulatory protein Faim2 is neuroprotective after transient brain ischemia. J Neurosci. 2011;31(1):225–233. doi: 10.1523/JNEUROSCI.2188-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duggan CD, DeMaria S, Baudhuin A, Stafford D, Ngai J. Foxg1 is required for development of the vertebrate olfactory system. J Neurosci. 2008;28(20):5229–5239. doi: 10.1523/JNEUROSCI.1134-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray RC, Navi D, Fesenko J, Lander AD, Calof AL. Widespread defects in the primary olfactory pathway caused by loss of Mash1 function. J Neurosci. 2003;23(5):1769–1780. doi: 10.1523/JNEUROSCI.23-05-01769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci USA. 2004;101(23):8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gogos JA, Osborne J, Nemes A, Mendelsohn M, Axel R. Genetic ablation and restoration of the olfactory topographic map. Cell. 2000;103(4):609–620. doi: 10.1016/s0092-8674(00)00164-1. [DOI] [PubMed] [Google Scholar]
- 28.Wang SZ, et al. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133(17):3389–3398. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- 29.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 30.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smyth GK. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3.
- 32.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115(3):587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng Z, Wang SZ, Green MR. Transcription and signalling pathways involved in BCR-ABL-mediated misregulation of 24p3 and 24p3R. EMBO J. 2009;28(7):866–876. doi: 10.1038/emboj.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Statist. 1996;5(3):299–314. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.