Abstract
T cell-mediated allergy to Ni++ is one of the most common forms of allergic contact dermatitis, but how the T-cell receptor (TCR) recognizes Ni++ is unknown. We studied a TCR from an allergic patient that recognizes Ni++ bound to the MHCII molecule DR52c containing an unknown self-peptide. We identified mimotope peptides that can replace both the self-peptide and Ni++ in this ligand. They share a p7 lysine whose εNH2 group is surface-exposed when bound to DR52c. Whereas the TCR uses germ-line complementary-determining region (CDR)1/2 amino acids to dock in the conventional diagonal mode on the mimotope–DR52c complex, the interface is dominated by the TCR Vβ CDR3 interaction with the p7 lysine. Mutations in the TCR CDR loops have similar effects on the T-cell response to either the mimotope or Ni++ ligand. We suggest that the mimotope p7 lysine mimics Ni++ in the natural TCR ligand and that MHCII β-chain flexibility in the area around the peptide p7 position forms a common site for cation binding in metal allergies.
Keywords: antigen presentation, hypersensitivity, peptide display library, crystal structure, metal recognition
T-cell receptors (TCRs) made up of α- and β-chains have a very large number of ligands, including self- and foreign peptides, superantigens, lipids, small organic haptens, and metal ions. These ligands usually react with TCRs when they are bound to major histocompatibility complex (MHC) proteins. TCRs usually bind MHC/peptide combinations in similar orientations such that the TCR is aligned diagonally across the MHC/peptide, with the Vβ region of the TCR placed over the α1 α-helix of the MHCI heavy chain or of the MHCII α-chain and the Vα region placed over the α2 α-helix of the MHCI heavy chain or the β1 α-helix of the MHCII β-chain (1, 2). We (1, 3–5) and others (6–8) have suggested that germ line-encoded residues in the complementary-determining region (CDR)1 and CDR2 loops of the TCR variable regions are evolutionarily conserved for interaction with the MHC. These residues are also involved when natural killer T (NKT) cells recognize glycolipids bound to the nonclassical MHCI protein CD1d, and even when T cells recognize the non-MHC protein CD155 (9).
It is noteworthy, however, that the presence of these evolutionarily conserved amino acids does not assure the conventional diagonal docking mode for TCRs on MHC molecules. For example, the NKT TCRs have a unique docking mode on CD1d (10, 11): Protein superantigens bridge TCRs to MHCII in a manner that prevents conventional TCR–MHCII interaction (12), and TCRs often dock to autoantigen peptides bound to MHCII in unusual orientations (13, 14). Thus, in cases in which the structural details of the interaction between the TCR and its target are not known, we cannot be certain that the interaction will occur with the TCR in its usual orientation on MHC or not. Such is the case for the recognition of metal ions by TCRs (15). Although it is assumed that ions such as Be++, Ni++, Co++, Cu++, Cr+++, and Au++ are recognized by TCRs in combination with MHC bound to a particular self-peptide, very little is known about the structures of the TCR/MHC/metal ion combinations. This applies, for example, to the T cells that mediate allergy to Ni++, a common contact allergen affecting up to 20% of humans (16).
To begin to understand TCR interaction with metal ions, we decided to study the TCR of one particular CD4+, Ni++-reactive human T-cell ANi2.3. ANi2.3 belongs to a group of TCRs that contain human Vβ17 that others have found are overrepresented among Ni++-specific CD4+ TCRs of patients suffering from particularly severe Ni++ contact hypersensitivity (17). We have previously reported that ANi2.3 reacts with the human MHCII protein DR52c (HLA-DRA, HLA-DRB3*0301) and Ni++ when a particular unknown peptide(s) is bound to DR52c (18). Here, using a display library method we have previously described (19, 20), we screened libraries of DR52c bound to partially randomized peptides in attempts to identify the specific peptide recognized by ANi2.3 in the presence of Ni++. Instead, we isolated many peptide mimotopes that, when bound to DR52c, engaged this TCR and activated the ANi2.3 T cell in the absence of Ni++. The C-terminal halves of these mimotopes had highly related amino acid sequences that included an almost invariant lysine at the p7 position, which we considered might mimic Ni++.
We solved the structures of two of these mimotope peptides bound to DR52c and the structure of the ANi2.3 TCR bound to one of the mimotope complexes. In each of the three structures, the peptide p7 lysine assumes a conformation that puts its εNH2 group on the surface of the molecule. Interactions between the TCR CDR1/2 loops and the DR52c helices help dock it in the conventional diagonal mode, but the area of strongest TCR contact surrounds and involves the mimotope p7 lysine εNH2. Mutagenesis of the TCR CDR loops shows that the TCR engages the natural Ni++ and mimotope ligands very similarly. We discuss the likelihood that the positive charge of this lysine side chain mimics Ni++ in the interaction with the ANi2.3 TCR. Our structure also provides insight into how the area between the MHC-bound peptide and the arched helix of the MHCII β1 helix might be a common site for cation binding in metal allergies.
Results
Common Motif Among Peptide Mimotopes for the ANi2.3 T Cell.
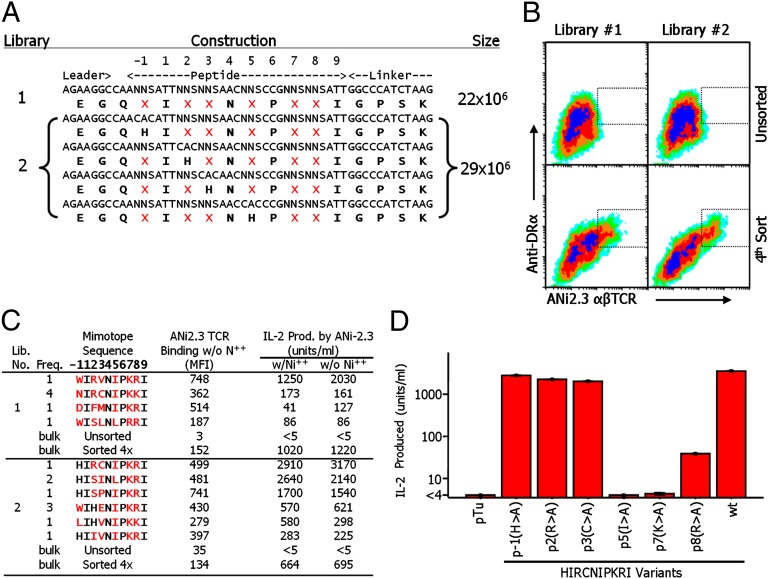
In previous studies, we and others have shown that the human T-cell clone ANi2.3, isolated from a patient with nickel hypersensitivity, reacts with Ni++ presented by the human MHCII protein DR52c bearing an unknown peptide (17, 18). Our efforts to find this peptide among the peptides naturally processed and bound to DR52c failed, suggesting that it may be present in very low abundance. Therefore, we turned to the use of baculovirus-expressed MHC–peptide display libraries as described previously (19, 20). We produced two types of libraries (Fig. 1A). In both, based on the crystal structure of DR52c bound to the Tu peptide (21), we fixed the anchor amino acids at p1, p4, p6, and p9 to those of the Tu peptide (I, N, P, and I). In the first library, six potential TCR contact amino acids (p1, p2, p3, p5, p7, and p8) were randomized via an NNS codon mixture. The second library was a group of four pooled sublibraries, prepared like the first library, except that, in each, either p1, p2, p3, or p5 was fixed to H as well in order to test the idea that a histidine somewhere in the N-terminal half of the peptide might cooperate with DR52c β 81H in binding Ni (18).
Fig. 1.
Identification of ANi2.3-stimulating, Ni++-independent peptide mimotopes in baculovirus DR52c–peptide libraries. (A) The designs and sizes of DR52c–peptide libraries are shown. Positions randomized to all 20 amino acids in the libraries are marked with a red X and were encoded by NNS, where N = A, C, T, or G and S = G or C. The preparation and characterization of library 1 have been previously described (19, 20). The four libraries that were pooled to form library 2 were produced and characterized identically, except that in each an additional position was fixed to histidine. (B) SF9 cells infected with the libraries were analyzed and sorted by flow cytometry at day 3 after infection in the presence of 100 μM NiCl2 using the combination of a phycoerythrin-labeled anti-DR Mab (20LC.11) and an Alexa 647-labeled multivalent fluorescent version of soluble ANi2.3 TCR. The dotted boxes show the gate used to sort DR+/TCR+-infected cells from which expanded viral stocks were prepared for the next cycle of analysis and/or sorting. (C) Single infected SF9 cells were sorted based on high DR expression and ANi2.3 TCR binding and then used to prepare expanded viral stocks. Each stock was used to prepare infected SF9 cells, which were retested for DR expression and TCR binding and whose viral DNA was used to determine the encoded peptide. The stocks were also used to infect ICAM/B7.1+ SF9 cells, which were tested for stimulation of IL-2 production by ANi2.3 T cells in the presence or absence of Ni++. The figure shows the sequences of the best five stimulating mimotopes from each library (red amino acids were selected at the randomized positions) and the number of times that the sequence was found among the total sequences. The mean fluorescence intensity (MFI) of binding with the multivalent ANi2.3 reagent is shown for cells gated to express equally high staining with the anti-DR mAb. Also shown is the response of ANi2.3 T cells to each peptide with or without Ni++. For comparison, TCR binding and IL-2 production are shown for SF9 cells infected with the bulk unsorted or four times-enriched libraries. (D) The figure shows the effect of mutating six positions of the pHIR (HIRCNIPKRI) mimotope to alanine on the response of the ANi2.3 T cell. The responses to the control Tu peptide and the unmutated (wt) peptide are also shown. Results are the average of three separate experiments with SEM.
The two libraries were separately screened. In each case, SF9 insect cells infected with the library were sorted by flow cytometry for simultaneous binding of a multimeric, fluorescent version of the ANi2.3 TCR in the presence of Ni++ and of a fluorescent anti-DR mAb. Within four rounds of sorting, in both cases a population blossomed out that bound both the TCR and anti-DR reagent strongly (Fig. 1B, Lower).
SF9 cells infected with a single virus were sorted for sequencing. Despite the presence of Ni++ during the screening, both libraries produced only related peptides that stimulated ANi2.3 in the absence of Ni++. A list of the properties of the 10 best ANi2.3-stimulating mimotopes is shown in Fig. 1C. The most striking feature of these mimotopes was a very strong selection of I, K, or R at p5, p7, and p8, respectively, in both libraries, with substitutions of L, R, and K occasionally allowed at these positions. There appeared to be no selection of a particular amino acid at p1, p2, or p3. We performed the screen again after predepleting the libraries of Ni++-independent mimotopes for three cycles, as in Fig. 1B, but in the absence of Ni++, before rescreening again in the presence of Ni++. This experiment also did not yield Ni++-dependent mimotopes, but did, however, yield a few Ni++-independent mimotopes similar in sequence to those listed in Fig. 1C (Table S1).
Therefore, we decided to examine examples of the related Ni++-independent mimotopes in Fig. 1 to see whether they might offer some clues as to how the natural self-peptide in normal DR52c+ antigen-presenting cells presents Ni++ to ANi2.3 T cells. First, in baculovirus, we made a series of mutations in the best-stimulating mimotope, HIRCNIPKRI (pHIR), such that individual amino acids at p1, p2, p3, p5, p7, and p8 were changed to alanine. ICAM/B7.1+ SF9 cells infected with mutant viruses were tested for their ability to stimulate ANi2.3 (Fig. 1D). As predicted from the variability of the N-terminal sequences of the enriched peptides (Fig. 1C), substitution of A at p1, p2, and p3 had no effect on the response of the ANi2.3 T cell. However, substitution of A for the amino acids at p5, p7, or p8 dramatically reduced or eliminated the response. Thus, this alanine scan and the heavy selection of p5I, p7K, and p8R in the libraries indicated that the ANi2.3 T cell focuses on the C-terminal end of the peptide mimotopes.
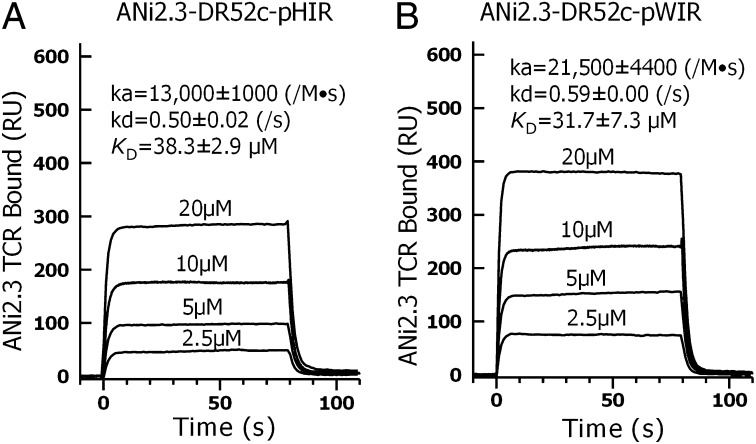
We performed surface plasmon resonance binding studies with the ANi2.3 TCR and soluble versions of two of the DR52c–mimotope complexes produced with baculovirus. The TCR was immobilized in the flow cell of a Biacore sensor chip. Various concentrations of the pHIR or WIRVNIPKRI (pWIR)-containing DR52c complexes (Fig. 2 A and B, respectively) were injected and the binding kinetics were recorded. Both complexes bound the ANi2.3 TCR with modest on rates and very fast off rates, resulting in kds (dissociation rates) in the micromolar range typical of TCR interactions with MHCII ligands.
Fig. 2.
ANi2.3 TCR binds to mimotope–DR52c complexes with typical TCR kinetics. The results of surface plasmon resonance studies are shown for the binding of soluble (A) DR52c–pHIR and (B) DR52c–pWIR complexes to immobilized ANi2.3 TCR. The binding kinetics were recorded in resonance unit (RU) when four different concentrations of DR52c–mimotope complexes were injected for 80 s through a flow cell of a Biacore CM5 biosensor chip containing immobilized ANi2.3 TCR. Identical injections in a second flow cell with an immobilized control TCR (Yae62) were used to correct for the fluid phase signal. Association rates (kas), dissociation rates (kds), and overall (affinity) KD (dissociation constant) with SEM were calculated from the kinetic data using instrument software.
Surface Exposure of the p7 Lysine εNH2 Group of the Mimotopes and Potential Ni++ Binding Site.
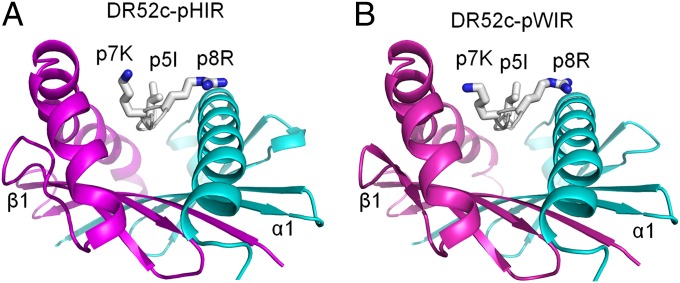
The heavy selection of lysine, and to a lesser extent arginine, at p7 in the mimotopes and the loss of activity when this position was mutated to alanine suggested to us that the positive charge of these amino acids might mimic Ni++ in the interaction with the ANi2.3 TCR, thus accounting for the lack of their dependence on Ni++ for ANi2.3. In TCR–MHCII structures reported thus far, p7 amino acids with short side chains most often point into the wall of the MHCII β-chain helix; however, p7 amino acids with large/long side chains most often assume rotamers that expose at least part of the side chain on the surface (Table S2). In order to see whether the p7K side chain can be surface-exposed well, we solved the structures of the pHIR and pWIR complexes to resolutions of 2.2 and 2.5 Å, respectively (Table S3). The structures of the two mimotopes bound to DR52c were very similar (Fig. 3). Importantly, the side chain of p7K, as well as those of p5I and p8R, was exposed on the surface of the DR52c protein and was, therefore, well-positioned to interact with the ANi2.3 TCR, lending support to the idea that the εNH2 group of p7K could mimic the Ni++ cation.
Fig. 3.
The side chains of critical mimotope amino acids p5I, p7K, and p8R are surface-exposed on the mimotope–DR52c complexes. The structures of two of the mimotopes in Fig. 1C, (A) pHIR and (B) pWIR, bound to DR52c were solved to resolutions of 2.2 and 2.5 Å, respectively. The peptide binding grooves are viewed from the C terminus of the peptides. Ribbon representations of the α1 (cyan) and β1 (magenta) DR52c helices and the peptide backbone (white) are shown with wire-frame representations of surface-exposed side chains of p5I, p7K, and p8R [CPK (Corey, Pauling, Koltun) coloring].
The analysis of the two structures presented here and a previous structure of DR52c bound to a Tu-derived peptide show that the width of the space between the DR52c α1 helix and the DR52c β1 helix (measured as the distance between the α65 and β67 α-carbons flanking the p7 position of the peptide) varies from 15.34 to 15.81 Å (average 15.56 Å), putting it among the widest seen in various mouse and human MHCIIs (Table S4). This wide space between the peptide and the β1 helix may allow Ni++ to bind in this region when DR52c contains the natural peptide that presents Ni++.
Extensive Interaction of the ANi2.3 TCR with p7K and Surrounding Area of the DR52c–pHIR Complex.
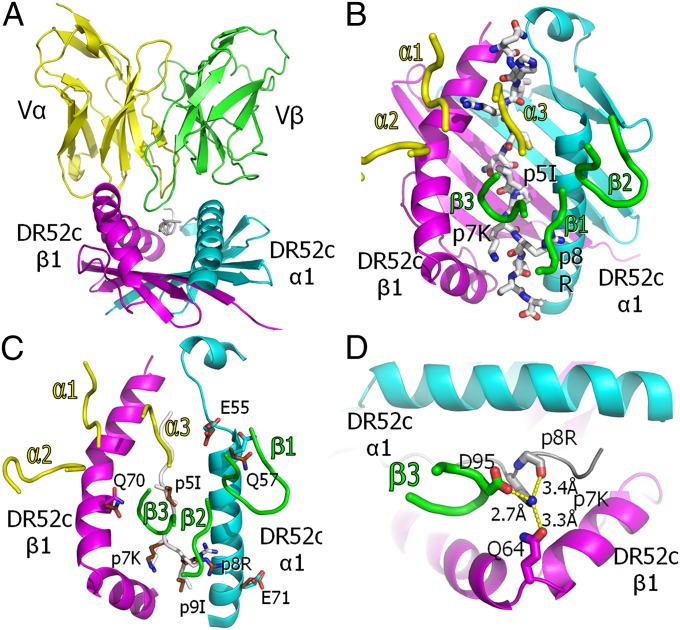
For crystallography, we produced a single-chain version of the ANi2.3 TCR (Fig. S1) containing only the Vα and Vβ domains connected by a glycine-rich linker (scFv) (22). A complex of this TCR with DR52c–pHIR was crystallized and the structure was solved to a resolution of 3.3 Å (Table S3). The TCR engages the MHCII–peptide complex in the usual diagonal mode, with the CDR3 regions of Vα and Vβ centered over the peptide and the Vα CDR1 and CDR2 loops poised over the DR52c β1 helix and the Vβ CDR1 and CDR2 loops over the α1 helix (Fig. 4 A and B). An overlay of the free versus ANi2.3 TCR-bound DR52c–pHIR shows relatively little adjustment of the MHCII or peptide amino acid side chains during the interaction (Fig. 4C).
Fig. 4.
Conventional orientation of the ANi2.3 TCR on pHIR–DR52c and Vβ CDR3 coordination of the highly selected p7K. (A) Ribbon representations of the Vα (yellow) and Vβ (green) domains of the ANi2.3 TCR bound to the DR52c–pHIR complex: DR52c α1, cyan; β1, magenta; pHIR, white. The view is from the peptide C terminus down the peptide binding groove. (B) Ribbon representations of DR52c α1 (cyan) and β1 (magenta) domains, and a wire-frame representation of the pHIR peptide (O, red; N, blue; C, white). The ANi2.3 TCR CDR loops are presented as tubes (Vα, yellow; Vβ, green). (C) DR52c and the ANi2.3 CDR3 loops are shown as in B, and the pHIR backbone is shown as a white tube. Side chains of the eight amino acids of the ligand that change their conformation upon ANi2.3 binding are shown in wire-frame representation (O, red; N, blue; α1: C, cyan; β1: C, magenta; peptide: C, white). Superimposed are the same side chains in the unbound DR52c–pHIR structure (O, red; N, blue; C, brown). (D) Top view of salt bridge/H bond coordination of the εNH2 of pHIR p7K with the carboxylate of ANi2.3 VβCR3 95D, the carbonyl oxygen of DR52c β1 Q64, and the backbone oxygen of p8R.
A complete analysis of the contacts between ANi2.3 and the DR52–pHIR complex is shown in Table S5, with a summary shown in Table 1. The TCR Vβ CDR3 contributes half of the total contacts with the ligand and almost two-thirds of the contacts with the peptide, nearly all of which are with p5I, p7K, and p8R. These results are consistent with those of the mutational analysis of pHIR (Fig. 1D) and the finding that the ANi2.3 TCR heavily selected these three amino acids in the mimotopes whereas tolerated many different amino acids at p1, p2, and p3 (Fig. 1C). The TCR Vβ CDR3 loop reaches deeply into the peptide binding groove (Fig. 4A) to contact the side chains of all three of these amino acids (Fig. 4B). Most importantly, the carboxylate of Vβ 95D at the tip of the CDR3 loop makes a salt bridge to the εNH2 group of p7K, which also forms hydrogen bonds to the peptide backbone at p8 and to Q64 on the DR52c β1 helix (Fig. 4D). Despite the 3.3-Å resolution of the structure, electron density for these amino acid side chains is clear in the map (Fig. S2).
Table 1.
Summary of contacts between ANi23 TCR and DR52c–pHIR
| CDR3 loop | Amino acid | Atom-to-atom contacts to DR52c–pHIR | |||
| Total | To DR52c α1 | To pHIR | To DR52c β1 | ||
| Vα CDR1 | A28 | 3 | — | 2 | 1 |
| T29 | 32 | — | 7 | 25 | |
| P30 | 1 | — | 1 | — | |
| Y31 | 18 | — | 3 | 15 | |
| Vα CDR2 | F50 | 21 | — | — | 21 |
| S51 | 4 | — | — | 4 | |
| Vα CDR3 | S95 | 5 | — | 5 | — |
| G96 | 16 | — | 16 | — | |
| N97 | 3 | — | 3 | — | |
| T98 | 1 | 1 | — | — | |
| Vβ CDR1 | D28 | 5 | — | 5 | — |
| Vβ CDR2 | Y46 | 3 | 3 | — | — |
| Q48 | 7 | 7 | — | — | |
| I49 | 9 | 9 | — | — | |
| N51 | 1 | 1 | — | — | |
| D52 | 14 | 14 | — | — | |
| Q54 | 20 | 20 | — | — | |
| Vβ CDR3 | R94 | 31 | 18 | 13 | — |
| D95 | 45 | — | 44 | 1 | |
| G96 | 24 | — | 13 | 11 | |
| Y97 | 53 | — | 5 | 48 | |
| T98 | 2 | — | 2 | — | |
| Total | 318 | 73 | 119 | 126 | |
Similarities Between ANi2.3 TCR Interaction with the Mimotope and Ni++.
If the binding of ANi2.3 to DR52c–pHIR mimics the way in which ANi2.3 reacts with Ni++ presented by DR52c plus the natural unknown peptide, then the TCR amino acids important in the interaction with the two ligands should be similar, especially in the vicinity of the p7 position. To test this idea, we used retroviral transduction (23, 24) to create a series of T-cell transfectomas bearing human CD4 and either the wild-type ANi2.3 TCR or a version with an alanine substitution at one of 29 positions in the Vα and Vβ CDR loops of the TCR. Each transduced T-cell population was sorted in bulk for equally high TCR and CD4 expression, and each was compared with the wild-type T cell for its ability to respond to either the DR52c+ human lymphoblastoid B-cell line HO301 in the presence of optimal Ni++ or to the chicken B-cell line DT40 transduced with retrovirus encoding DR52c with the HIR mimotope covalently attached. The results are shown in Fig. 5A and schematically mapped onto the pHIR–DR52 surface along with the ANi2.3 TCR footprint in Fig. 5B.
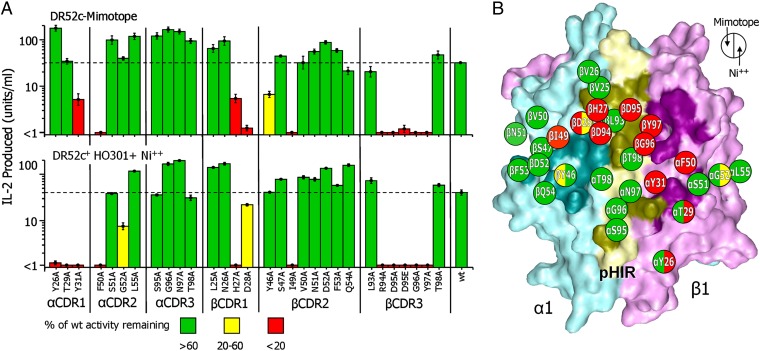
Fig. 5.
Similar pattern of ANi2.3 TCR interaction with the pHIR mimotope and Ni++. (A) The bar graph shows the sensitivity of the response of ANi2.3 to mutation of 29 amino acids of its six CDR3 loops to alanine or mutation of Vβ D95 to E. T cells bearing the wild-type or each mutant TCR were tested for response to DT40 chicken cells bearing DR52c covalently attached to the pHIR mimotope (Upper) or to HO301 DR52c+ lymphoblastoid cells plus 100 µM Ni++ (Lower) as measured by IL-2 secretion. The average ± SEM of three experiments is shown. The bars for responses that were less than 20% of the wild-type response are colored red, yellow (between 20% and 60%), and green (greater than 60%). (B) The solvent-accessible surface of the DR52 α1 (cyan) and β1 (magenta) and pHIR (yellow) is shown with the footprint of the ANi2.3 TCR in darker versions of the same colors. The 29 mutated residues of the ANi2.3 TCR are schematically mapped onto the pHIR–DR52 surface with their side-chain positions labeled within a circle. The left half of each circle is colored the same as the bars in A, Upper and the right half is colored the same as the bars in A, Lower.
The effects of the mutations on the responses to the two ligands were very similar, especially in the area interacting with p7K of pHIR. Many of the mutations that dramatically reduced both responses clustered in this region. Most dramatically, mutation of any of the four amino acids (R94, D95, G96, and Y97) at the tip of Vβ CDR3 eliminated both responses. In the structure of the complex, these amino acids are not only in extensive contact with the critical pHIR amino acids p5I, p7K, and p8R, as mentioned above, but they also interact with the adjacent DR52c α1 and β1 helices (Table 1). In a previous study of another Ni++-reactive TCR whose Vα and Vβ CDRs were nearly identical to those of ANi2.3 and that also had RDGY at the tip of Vβ CDR3, mutation of the D to A was also shown to eliminate the Ni++ response, but so did the very conservative mutation of D to E (25, 26). Therefore, we also assessed the effect of this Vβ D95-to-E mutation on the ANi2.3 T cell (Fig. 5A). The ANi2.3 response both to the mimotope and to Ni++ was eliminated by this mutation, suggesting that a very precise geometry is required for both responses, supplied by the D but not by the E, rather than the simple presence of a negatively charged amino acid at this position. In the case of Ni++, this would be consistent with the participation of this D in the precise coordination of the Ni++ cation in this location.
H27 and D28 of the Vβ CDR1 loop are two other important amino acids in this cluster, because the mutation of either one affects both responses (Fig. 5 A and B). In the ANi2.3–pHIR structure, D28 interacts with the heavily selected p8R of pHIR (Table 1 and Table S5), but H27 makes no contact with the DR52c or pHIR. Rather, its side chain points up toward the center of the Vβ CDR1 loop and interacts closely with the Vβ β-strand leading up to CDR3 and with the Jβ β-strand leading away from CDR3. Its mutation to A could potentially indirectly affect binding to DR52c–pHIR by altering the Vβ CD1 shape or the shape and position of the critical Vβ CDR3 loop.
No mutation in Vα CDR3 had any effect on either response (Fig. 5 A and B), despite considerable contact with p1H and p2R of pHIR in the DR52c–mimotope structure (Table 1 and Table S5). Likewise, mutation of Vα CDR1 amino acids Y26 and T29 had no effect on the response of ANi2.3 to the mimotope, despite considerable contact of T29 with both p2R of pHIR and the DR52c β2 helix (Table 1 and Table S5). In contrast, the mutation of either Y26 or T29 eliminated the response to Ni++, the most striking difference in the effects of the mutations on the two responses.
Taken together, these results support the conclusion that the ANi2.3 TCR recognizes the mimotopes and Ni++ bound to the unknown self-peptide similarly in the conventional diagonal binding mode but that the p7K of the mimotopes is likely a mimic of Ni++, both of which dominate the TCR interaction via Vβ CDR3.
Discussion
The more than 20 y of published MHC and TCR–MHC structures have taught us a great deal about how peptides bind to classical MHCI and MHCII and how TCRs recognize this complex in the response to foreign peptide antigens presented by self-MHC or to self-peptides bound to foreign MHC alleles. However, we understand little about the nature of the T-cell ligands that form when small chemical moieties, such as plant urushiols or penicillin (27, 28) or metal cations (15), attach to self-peptide–MHC complexes to form a hapten-like target for T cells. For metal cations, these range from Ni++, mediating the widespread hypersensitivity to nickel-containing jewelry (16), to Be++, which causes the rarer, but much more serious, fulminating lung inflammation seen in chronic beryllium disease (CBD) (29). At present, we do not know the structure of the MHC–peptide–cation complex ligand involved in any of these diseases. The structures we present here offer insight into how Ni++ becomes part of such a ligand.
We have found a series of peptide mimotopes that, when bound to DR52c, activate the Ni++-reactive ANi2.3 T cell in the absence of Ni++. A common feature of these mimotopes was a lysine at the p7 position whose side chain assumed a rotamer that brought its εNH2 group to the surface of the DR52c–peptide complex to form a salt bridge to an aspartic acid in the ANi2.3 TCR Vβ CDR3 loop. This TCR aspartic acid was essential for recognition of both the mimotope and the natural Ni++ epitope. This and other similarities between the interaction of the mimotope and the natural Ni++ ligand with the ANi2.3 TCR have led us to propose that the positively charged lysine occupies the same position in the mimotope complex as the Ni++ cation does in the natural complex.
The footprint of the ANi2.3 TCR on the mimotope–DR52c complex was dominated by the Vβ CDR3 loop, which adopts an extended conformation to reach into the peptide binding groove while contacting the p7 lysine. Mutation of any of the amino acids at the end of this loop resulted in complete loss of the ANi2.3 response to both Ni++ and the mimotope peptide. For the mimotope, this interaction was so dominant that the ANi2.3 response survived mutation of a number of other TCR amino acids contacting DR52c and the peptide at p1 and p2. This could explain the variety of amino acids that were tolerated at the p1 and p2 positions among the various mimotopes we found for this TCR. On the other hand, the ANi2.3 response to Ni++ was much more sensitive to some mutations in the Vα CDR1 loop than was the response to the mimotope. This could indicate an important role for ANi2.3 contact with C-terminal end of the DR52c β1 helix and/or particular amino acids at the p1 or p2 position of the natural peptide(s) that presents Ni++. This may have contributed to our failure to find Ni++-dependent mimotopes in our libraries, because the dominant, more frequent lysine-containing mimotopes might have won out over the rarer Ni++-dependent mimotopes in the competition for the soluble TCR reagent used to screen our libraries.
Placing the Ni++ cation in a location between the peptide and the arch of the MHCII β1 helix in the vicinity of p7 bears a striking resemblance to the situation with Be++ presentation to T cells in CBD. Genetic susceptibility to CBD is closely linked to MHCII alleles that bear a glutamic acid at position 69 of the β-chain, especially the HLA-DP2 allele (29, 30). We recently solved the structure of DP2, which revealed an acidic pocket on the surface of the molecule that contained the carboxylates of β69Glu as well as two other DP2 acidic amino acids (31). Mutation of any of these amino acids eliminated Be++ presentation to Be++-reactive human T cells. Similar to the position of p7K of pHIR bound to DR52c, this pocket lay in a wide space between the arch of the β1 helix and the side chains of p4 and p7 of the peptide. We have argued that this is the functional site of Be++ binding to DP2. Also, an analysis of over three dozen published MHCII–peptide structures (31) showed that despite the enormous conservation of structure among the many MHCII isotypes and alleles from human and mouse, the space between the peptide and the β1 helix in this region can vary by more than 3 Å. It is tempting to suggest that this flexibility, combined with particular MHCII polymorphisms in the region and particular amino acids at the peptide p4 and p7 positions, can create the ideal site for cation binding in a variety of metal-mediated immune diseases.
In summary, our results show that the ANi2.3 TCR uses a conventional docking mode on the DR52c–pHIR complex and docks very similarly on DR52c containing the natural Ni++–self-peptide ligand. The data also strongly suggest that the εNH2 group of the mimotope p7K mimics the Ni++ in the natural ligand. The focused interaction of its Vβ CDR3 loop with pHIR–p7K or Ni++ in the natural ligand modulates the importance of these MHC contacts in the overall affinity of the TCR for the two ligands.
Materials and Methods
Baculovirus DR52c–Peptide Libraries.
The baculovirus libraries described in Fig. 1 were produced by direct cloning of PCR fragments into baculovirus DNA (19, 20) as described in SI Materials and Methods.
Protein Expression and Purification.
DR52c (extracellular domains) with pHIR or pWIR covalently attached were cloned into a single baculovirus as previously described (21). V regions of the ANi2.3 TCR were fused to mouse C regions and expressed in baculoviruses as previously described (15, 32, 33). For crystallography, the Vα and Vβ portions of the ANi2.3 TCR were fused by GS linker (Vα-linker-Vβ) and were expressed in the periplasmic space of the Rosetta strain of Escherichia coli (22). Details are described in SI Materials and Methods.
Other Materials and Methods.
Other materials and methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff of the Advanced Light Source synchrotron facility (beamline 8.2.2). We also thank Shirley Sobus of the Flow Cytometry Facility and Randy Anselment of the Biomolecular Resource Center at National Jewish Health. The Mopac54 vector was a gift from Dr. Jennifer Maynard, and we received the DT40 cell line from Dr. Anne-Laure Perraud. We thank the Zuckerman Family/Canyon Ranch and Allen Laporte for support of the National Jewish Health Structural Facility. This work was supported in part by US Public Health Service Grants AI-18785 and AI-22295. S.D. is supported by National Institutes of Health Grant KL2 RR025779 and The Boettcher Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4H26, 4H25, and 4H1L).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215928109/-/DCSupplemental.
References
- 1.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 3.Dai S, et al. Crossreactive T cells spotlight the germline rules for αβ T cell-receptor interactions with MHC molecules. Immunity. 2008;28(3):324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the αβ T-cell receptor control thymic selection. Nature. 2009;458(7241):1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin L, et al. A single T cell receptor bound to major histocompatibility complex class I and class II glycoproteins reveals switchable TCR conformers. Immunity. 2011;35(1):23–33. doi: 10.1016/j.immuni.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams JJ, et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity. 2011;35(5):681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colf LA, et al. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129(1):135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 8.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon.’. Nat Immunol. 2007;8(9):975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 9.Tikhonova AN, et al. αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity. 2012;36(1):79–91. doi: 10.1016/j.immuni.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448(7149):44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 11.Gapin L. iNKT cell autoreactivity: What is ‘self’ and how is it recognized? Nat Rev Immunol. 2010;10(4):272–277. doi: 10.1038/nri2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, et al. Crystal structure of a complete ternary complex of TCR, superantigen and peptide-MHC. Nat Struct Mol Biol. 2007;14(2):169–171. doi: 10.1038/nsmb1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maynard J, et al. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: Insights into MHC bias and antigen specificity. Immunity. 2005;22(1):81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Wucherpfennig KW, Call MJ, Deng L, Mariuzza R. Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Curr Opin Immunol. 2009;21(6):590–595. doi: 10.1016/j.coi.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veien NK. Systemic contact dermatitis. Int J Dermatol. 2011;50(12):1445–1456. doi: 10.1111/j.1365-4632.2011.05104.x. [DOI] [PubMed] [Google Scholar]
- 16.Schram SE, Warshaw EM, Laumann A. Nickel hypersensitivity: A clinical review and call to action. Int J Dermatol. 2010;49(2):115–125. doi: 10.1111/j.1365-4632.2009.04307.x. [DOI] [PubMed] [Google Scholar]
- 17.Vollmer J, Fritz M, Dormoy A, Weltzien HU, Moulon C. Dominance of the BV17 element in nickel-specific human T cell receptors relates to severity of contact sensitivity. Eur J Immunol. 1997;27(8):1865–1874. doi: 10.1002/eji.1830270808. [DOI] [PubMed] [Google Scholar]
- 18.Lu L, et al. Components of the ligand for a Ni++ reactive human T cell clone. J Exp Med. 2003;197(5):567–574. doi: 10.1084/jem.20021762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford F, Huseby E, White J, Marrack P, Kappler JW. Mimotopes for alloreactive and conventional T cells in a peptide-MHC display library. PLoS Biol. 2004;2(4):E90. doi: 10.1371/journal.pbio.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crawford F, et al. Use of baculovirus MHC/peptide display libraries to characterize T-cell receptor ligands. Immunol Rev. 2006;210:156–170. doi: 10.1111/j.0105-2896.2006.00365.x. [DOI] [PubMed] [Google Scholar]
- 21.Dai S, Crawford F, Marrack P, Kappler JW. The structure of HLA-DR52c: Comparison to other HLA-DRB3 alleles. Proc Natl Acad Sci USA. 2008;105(33):11893–11897. doi: 10.1073/pnas.0805810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maynard J, et al. High-level bacterial secretion of single-chain αβ T-cell receptors. J Immunol Methods. 2005;306(1-2):51–67. doi: 10.1016/j.jim.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 23.Rubtsova K, et al. Many different Vβ CDR3s can reveal the inherent MHC reactivity of germline-encoded TCR V regions. Proc Natl Acad Sci USA. 2009;106(19):7951–7956. doi: 10.1073/pnas.0902728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott-Browne JP, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8(10):1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 25.Vollmer J, et al. Antigen contacts by Ni-reactive TCR: Typical αβ chain cooperation versus α chain-dominated specificity. Int Immunol. 2000;12(12):1723–1731. doi: 10.1093/intimm/12.12.1723. [DOI] [PubMed] [Google Scholar]
- 26.Vollmer J, Weltzien HU, Moulon C. TCR reactivity in human nickel allergy indicates contacts with complementarity-determining region 3 but excludes superantigen-like recognition. J Immunol. 1999;163(5):2723–2731. [PubMed] [Google Scholar]
- 27.Kalish RS, Johnson KL. Enrichment and function of urushiol (poison ivy)-specific T lymphocytes in lesions of allergic contact dermatitis to urushiol. J Immunol. 1990;145(11):3706–3713. [PubMed] [Google Scholar]
- 28.Padovan E. T-cell response in penicillin allergy. Clin Exp Allergy. 1998;28(Suppl 4):33–36. [PubMed] [Google Scholar]
- 29.Fontenot AP, Maier LA. Genetic susceptibility and immune-mediated destruction in beryllium-induced disease. Trends Immunol. 2005;26(10):543–549. doi: 10.1016/j.it.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Falta MT, Bowerman NA, Dai S, Kappler JW, Fontenot AP. Linking genetic susceptibility and T cell activation in beryllium-induced disease. Proc Am Thorac Soc. 2010;7(2):126–129. doi: 10.1513/pats.201002-022RM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai S, et al. Crystal structure of HLA-DP2 and implications for chronic beryllium disease. Proc Natl Acad Sci USA. 2010;107(16):7425–7430. doi: 10.1073/pnas.1001772107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huseby ES, et al. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122(2):247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Kappler J, White J, Kozono H, Clements J, Marrack P. Binding of a soluble αβ T-cell receptor to superantigen/major histocompatibility complex ligands. Proc Natl Acad Sci USA. 1994;91(18):8462–8466. doi: 10.1073/pnas.91.18.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.