Abstract
Cook's (Cook, 1977) distance is one of the most important diagnostic tools for detecting influential individual or subsets of observations in linear regression for cross-sectional data. However, for many complex data structures (e.g., longitudinal data), no rigorous approach has been developed to address a fundamental issue: deleting subsets with different numbers of observations introduces different degrees of perturbation to the current model fitted to the data and the magnitude of Cook's distance is associated with the degree of the perturbation. The aim of this paper is to address this issue in general parametric models with complex data structures. We propose a new quantity for measuring the degree of the perturbation introduced by deleting a subset. We use stochastic ordering to quantify the stochastic relationship between the degree of the perturbation and the magnitude of Cook's distance. We develop several scaled Cook's distances to resolve the comparison of Cook's distance for different subset deletions. Theoretical and numerical examples are examined to highlight the broad spectrum of applications of these scaled Cook's distances in a formal influence analysis.
Keywords: Cook's distance, Perturbation, Relative influential, Conditionally scaled Cook's distance, Scaled Cook's distance, Size issue
1. Introduction
Influence analysis assesses whether a modification of a statistical analysis, called a perturbation (see Section 2.2), seriously affects specific key inferences, such as parameter estimates. Such perturbation schemes include the deletion of an individual or a subset of observations, case weight perturbation, and covariate perturbation among many others [8, 9, 30]. For example, for linear models, a perturbation measures the effect on the model of deleting a subset of the data matrix. In general, perturbation measures do not depend on the data directly, but rather on its structure via the model. If a small perturbation has a small effect on the analysis, our analysis is relatively stable, while if a large perturbation has a small effect on the analysis, we learn that our analysis is robust [11, 16]. If a small perturbation seriously influences key results of the analysis, we want to know the cause [9, 11]. For instance, in influence analysis, a set of observations is flagged as ‘influential’ if its removal from the dataset produces a significant difference in the parameter estimates or equivalently a large value of Cook's distance for the current statistical model [8, 5].
Since the seminal work of Cook [8] on Cook's distance in linear regression for cross-sectional data, considerable research has been devoted to developing Cook's distance for detecting influential observations (or clusters) in more complex data structures under various statistical models [8, 10, 6, 1, 12, 23, 15, 29, 14]. For example, for longitudinal data, Preisser and Qaqish [19] developed Cook's distance for generalized estimating equations, while Christensen, Pearson and Johnson [7], Banerjee and Frees [4], and Banerjee [3] considered case deletion and subject deletion diagnostics for linear mixed models. Furthermore, in the presence of missing data, Zhu et al. [29] developed deletion diagnostics for a large class of statistical models with missing data. Cook's distance has been widely used in statistical practice and can be calculated in popular statistical software, such as SAS and R.
A major research problem regarding Cook's distance that has been largely neglected in the existing literature is the development of Cook's distance for general statistical models with more complex data structures. The fundamental issue that arises here is that the magnitude of Cook's distance is positively associated with the amount of perturbation to the current model introduced by deleting a subset of observations. Specifically, a large value of Cook's distance can be caused by deleting a subset with a larger number of observations and/or other causes such as the presence of influential observations in the deleted subset. To delineate the cause of a large Cook's distance for a specific subset, it is more useful to compute Cook's distance relative to the degree of the perturbation introduced by deleting the subset [11, 30].
The aim of this paper is to address this fundamental issue of Cook's distance for complex data structures in general parametric models. The main contributions of this paper are summarized as follows.
-
a.1
We propose a quantity to measure the degree of perturbation introduced by deleting a subset in general parametric models. This quantity satisfies several attractive properties including uniqueness, non-negativity, monotonicity, and additivity.
-
a.2
We use stochastic ordering to quantify the relationship between the degree of the perturbation and the magnitude of Cook's distance. Particularly, in linear regression for cross-sectional data, we first show the stochastic relationship between the Cook's distances for any two subsets with possibly different numbers of observations.
-
a.3
We develop several scaled Cook's distances and their first-order approximations in order to compare Cook's distance for deleted subsets with different numbers of observations.
The rest of the paper is organized as follows. In Section 2, we quantify the degree of the perturbation for set deletion and delineate the stochastic relationship between Cook's distance and the degree of perturbation. We develop several scaled Cook's distances and derive their first-order approximations. In Section 3, we analyze simulated data and a real dataset using the scaled Cook's distances. We give some final remarks in Section 4.
2. Scaled Cook's Distance
2.1. Cook's distance
Consider the probability function of a random vector , denoted by p(Y|θ), where θ = (θ1, . . . , θq)T is a q × 1 vector in an open subset Φ of Rq and Yi = (yi,1, . . . , yi,mi), in which the dimension of Yi, denoted by mi, may vary significantly across all i. Cook's distance and many other deletion diagnostics measure the distance between the maximum likelihood estimators of θ with and without Yi [10, 8]. A subscript ‘[I]’ denotes the relevant quantity with all observations in I deleted. Let Y[I] be a subsample of Y with YI = {Y(i,j) : (i, j) ∈ I} deleted and p(Y[I]|θ) be its probability function. We define the maximum likelihood estimators of θ for the full sample Y and a subsample Y[I] as
| (2.1) |
respectively. Cook's distance for I, denoted by CD(I), can be defined as follows:
| (2.2) |
where Gnθ is chosen to be a positive definite matrix. The matrix Gnθ is not changed or re-estimated when a subset of the data is deleted. Throughout the paper, Gnθ is set as or its expectation, where represents the second-order derivative with respect to θ. For clustered data, the observations within the same cluster are correlated. A sensible model p(Y|θ) should explicitly model the correlation structure in the clustered data and thus implicitly incorporates such a correlation structure.
More generally, suppose that one is interested in a subset of θ or q1 linearly independent combinations of θ, say LTθ, where L is a q × q1 matrix with rank(L) = q1 [4, 10]. The partial inuence of the subset I on , denoted by CD(I|L), can be defined as
| (2.3) |
For notational simplicity, even though we may focus on a subset of θ, we do not distinguish between CD(I|L) and CD(I) throughout the paper.
Based on (2.2), we know that Cook's distance CD(I) is explicitly determined by three components including the current model fitted to the data, denoted by , the dataset Y, and the subset I itself. Cook's distance is also implicitly determined by the goodness of fit of to Y for I, denoted by , and the degree of the perturbation to introduced by deleting the subset I, denoted by . Thus, we may represent CD(I) as follows:
| (2.4) |
where F1(·, ·, ·) and F2(·,·) represent nonlinear functions.
We may use the value of CD(I) to assess the influential level of the subset I. We may regard a subset I as influential if either the value of CD(I) is relatively large compared with other Cook's distances or the magnitude of CD(I) is greater than the critical points of the χ2 distribution [10]. However, for complex data structures, we will show that it is useful to compare Cook's distance relative to its associated degree of perturbation.
2.2. Degree of perturbation
Consider the subset I and the current model . We are interested in developing a measure to quantify the degree of the perturbation to introduced by deleting the subset I regardless of the observed data Y. We emphasize here that the degree of perturbation is a property of the model, unlike Cook's distance which is also a property of Y. Abstractly, should be defined as a mapping from a subset I and to a nonnegative number. However, according to the best of our knowledge, no such quantities have ever been developed to define a workable for an arbitrary subset I in general parametric models due to many conceptual difficulties [11]. Specifically, even though [11] placed the Euclidean geometry on the perturbation space for one-sample problems, such a geometrical structure cannot be easily generalizable to general data structures (e.g., correlated data) and related parametric models. For instance, for correlated data, a sensible model should model the correlation structure and a good measure should explicitly incorporate the correlation structure specified in and the subset I. However, the Euclidean geometry proposed by [11] cannot incorporate the correlation structure in the correlated data.
Our choice of is motivated by five principles as follows.
(P.a) (non-negativity) For any subset I, is always non-negative.
(P.b) (uniqueness) if and only if I is an empty set.
(P.c) (monotonicity) If , then .
(P.d) (additivity) If , and p(YI1·2|Y[I1], θ) = p(YI1·2|Y[I1·2], θ) for all θ, then we have .
(P.e) should naturally arise from the current model , the data Y, and the subset I.
Principles (P.a) and (P.b) indicate that deleting any nonempty subset always introduces a positive degree of perturbation. Principle (P.c) indicates that deleting a larger subset always introduces a larger degree of perturbation. Principle (P.d) presents the condition for ensuring the additivity property of the perturbation. Since Y[I1·2] is the union of Y[I1] and YI2, p(YI1·2|Y[I1], θ) = p(YI1·2], θ) is equivalent to that of YI1·2 being independent of YI2 given Y[I1]. The additivity property has important implications in cross-sectional, longitudinal, and family data. For instance, in longitudinal data, the degree of perturbation introduced by simultaneously deleting two independent clusters equals the sum of their degrees of individual cluster perturbations.
Principle (P.e) requests that should depend on the triple . We propose based on the Kullback-Leibler divergence between the fitted probability function p(Y|θ) and the probability function of a model for characterizing the deletion of YI, denoted by p(Y|θ, I). Note that p(Y|θ) = p(Y[I]|θ)p(YI|Y[I], θ), where p(YI|Y[I], θ) is the conditional density of YI given Y[I]. Let θ* be the true value of θ under [24, 25]. We define p(Y|θ, I) as follows:
| (2.5) |
in which p(YI|Y[I], θ*) is independent of θ. In (2.5), by fixing θ = θ* in p(YI|Y[I], θ), we essentially drop the information contained in YI as we estimate θ. Specifically, is the maximum likelihood estimate of θ under p(Y|θ, I). If is correctly specified, then p(YI|Y[I], θ*) is the true data generator for YI given Y[I]. The Kullback-Leibler distance between p(Y|θ) and p(Y|θ, I), denoted by KL(Y, θ|θ*, I), is given by
| (2.6) |
We use KL(Y, θ|θ*, I) to measure the effect of deleting YI on estimating θ without knowing that the true value of θ is θ*. If YI is independent of Y[I], then we have
which is independent of Y[I]. In this case, the effect of deleting YI on estimating θ only depends on {p(YI|θ) : θ ∊ Θ}.
A conceptual difficulty associated with KL(Y,θ|θ*, I) is that both θ and θ* are unknown. Although θ* is unknown, it can be assumed to be a fixed value from a frequentist viewpoint. For the unknown θ, we can always use the data Y and the current model to calculate an estimator in a neighborhood of θ*. Under some mild conditions [24, 25], one can show that is asymptotically normal and thus should be centered around θ*. Moreover, since Cook's distance is to quantify the change of parameter estimates after deleting a subset, we need to consider all possible θ around θ* instead of focusing on a single θ. Specifically, we consider θ in a neighborhood of θ* by assuming a Gaussian prior for θ with mean θ* and positive definite covariance matrix Σ* (e.g., the Fisher information matrix), denoted by p(θ|θ*, Σ*). Finally, we define as the weighted Kullback-Leibler distance between p(Y|θ) and p(Y|θ, I) as follows:
| (2.7) |
This quantity can also be interpreted as the average effect of deleting YI on estimating θ with the prior information that the estimate of θ is centered around θ*. Since is directly calculated from the model and the subset I, it can naturally account for any structure specified in . Furthermore, if we are interested in a particular set of components of θ and treat others as nuisance parameters, we may fix these nuisance parameters in their true value.
To compute in a real data analysis, we only need to specify and (θ*, Σ*). Then, we may use some numerical integration methods to compute . Although (θ*, Σ*) are unknown, we suggest substituting θ* by an estimator of θ, denoted by , and Σ* by the covariance matrix of . Throughout the paper, since is a consistent estimator of θ* [24, 25], we set and Σ* as the covariance matrix of .
We obtain the following theorems, whose detailed assumptions and proofs can be found in the Appendix.
Theorem 1. Suppose that L({Y : p(YI|Y[I], θ) = p(YI|Y[I], θ*)}) > 0 for any θ = θ*, where L(A) is the Lebesgue measure of a set A. Then, defined in (2.7) satisfies the four principles (P.a)-(P.d).
As an illustration, we show how to calculate under the standard linear regression model for cross-sectional data as follows.
Example 1. Consider the linear regression model , where xi is a p × 1 vector and the εi are independently and identically distributed (i.i.d) as . Let Y = (y1, . . . , yn)T and X be an n × p matrix of rank p with i-th row . In this case, θ = (βT, σ2)T . Recall that , , , and , where In is an n × n identity matrix and Hx = (hij) = X(XTX)–1XT. We first compute the degree of the perturbation for deleting each (yi; xi). We consider two scenarios: fixed and random covariates. For the case of fixed covariates, assumes . After some algebraic calculations, it can be shown that equals
| (2.8) |
where Eθ is taken with respect to . Moreover, the right hand side of (2.8) contains only terms involving n and X, since perturbation is defined only in terms of the underlying model . This is also at core why only stochastic ordering is possible for Cooks distance, which is a function of both perturbation and data. See Section 2.3 for detailed discussions. Furthermore, if β is the parameter of interest in θ and σ2 is a nuisance parameter, then and 1/(2n) can be dropped from in (2.8).
Furthermore, for the case of random covariates, we assume that the xi's are independently and identically distributed with mean μx and covariance matrix Σx. It can be shown that equals
| (2.9) |
If β is the parameter of interest in θ and σ2 is a nuisance parameter, then reduces to p/(2n). Furthermore, consider deleting a subset of observations {(yik, xik) : k = 1, · · · , n(I)} and I = {i1, . . . , in(I)}. It follows from Theorem 1 that . Furthermore, for the case of random covariates, we have for any subset I with n(I) observations. Thus, in this case, deleting any two subsets I1 and I2 with the same number of observations, that is n(I1) = n(I2), has the same degree of perturbation. An important implication of these calculations in real data analysis is that we can directly compare CD(I1) and CD(I2) when n(I1) = n(I2).
2.3. Cook's distance and degree of perturbation
To understand the relationship between and CD(I) in (2.4), we temporarily assume that the fitted model is the true data generator of Y. To have a better understanding of Cook's distance, we consider the standard linear regression model for cross-sectional data as follows.
Example 1 (continued). We are interested in β and treat σ2 as a nuisance parameter. We first consider deleting individual observations in linear regression. Cook's distance [8] for case i, (yi, xi), is given by
| (2.10) |
where is the least squares estimate of β, is a consistent estimator of σ2, and , in which . It should be noted that except for a constant p, CD({i}) is almost the same as the original Cook's distance (Cook, 1977). As shown in (2.8) and (2.9), regardless of the exact value of (yi, xi), deleting any (yi, xi) has the approximately same degree of perturbation to . Moreover, the CD({i}) are comparable regardless of i. Specifically, if εi N(0, σ2), then follows the χ2(1) distribution for all i. For the case of random covariates, if xi are identically distributed, then all CD({i}) are truly comparable, since they follow the same distribution.
We consider deleting multiple observations in the linear model. Cook's distance for deleting the subset I with n(I) is given by
| (2.11) |
where êI is an n(I) × 1 vector containing all êi for i ∊ I and , in which XI is an n(I) × p matrix whose rows are for all i ∊ I. Similar to the deletion of a single case, deleting any subset with the same number of observations introduces approximately the same degree of perturbation to , and the CD(I) are comparable among all subsets with the same n(I). We will make this statement precise in Theorem 2 given below.
Generally, we want to compare CD(I1) and CD(I2) for any two subsets with n(I1) ≠ n(I2). As shown in Example 1, when n(I1) > n(I2), deleting I1 introduces a larger degree of perturbation to model compared to deleting I2. To compare Cook's distances among arbitrary subsets, we need to understand the relationship between and CD(I) for any subset I. Surprisingly, in linear regression for cross-sectional data, we can show the stochastic relationship between and CD(I) as follows.
Theorem 2. For the standard linear model, where y = Xβ + ε and , we have the following results:
for any , CD(1) is stochastically larger than CD(I2) for any X, that is, holds for any t > 0.
Suppose that the components of XI and XI’ are identically distributed for any two subsets I and I’ with n(I) = n(I’). Thus, CD(I) and CD(I’) follow the same distribution when n(I) = n(I’) and CD(I1) is stochastically larger than CD(I2) for any two subsets I2 and I1 with n(I1) > n(I2).
Theorem 2 (a) shows that the Cook's distances for two nested subsets satisfy the stochastic ordering property. Theorem 2 (b) indicates that for random covariates, the Cook's distances for any two subsets also satisfy the stochastic ordering property under some mild conditions.
According to Theorem 2, for more complex data structures and models, it may be natural to use the stochastic order to stochastically quantify the positive association between the degree of the perturbation and the magnitude of Cook's distance. Specifically, we consider two possibly overlapping subsets I1 and I2 with . Although I1) may not be greater than CD(I2) for a fixed dataset Y, CD(I1), as a random variable, should be stochastically larger than CD(I2) if is the true model for Y. We make the following assumption:
Assumption A1. For any two subsets I1 and I2 with ,
| (2.12) |
holds for any t > 0, where the probability is taken with respect to .
Assumption A1 is essentially saying that if is the true data generator, then CD(I1) stochastically dominates CD(I2) whenever . According to the definition of stochastic ordering [20], we can now obtain the following proposition.
Proposition 1. Under Assumption A1, for any two subsets I1 and I2 with , Cook's distance satisfies
| (2.13) |
holds for all increasing functions h(·). In particular, we have and is greater than the α-quantile of for any α ∊ [0, 1], where denotes the α–quantile of the distribution of CD(I) for any subset I.
Proposition 1 formally characterizes the fundamental issue of Cook's distance. Specifically, for any two subsets I1 and I2 with , CD(I1) has a high probability of being greater than CD(I2) when is the true data generator. Thus, Cook's distance for subsets with different degrees of perturbation are not directly comparable. More importantly, it indicates that CD(I) cannot be simply expressed as a linear function of . Thus, the standard solution, which standardizes CD(I) by calculating the ratio of CD(I) over , is not desirable for controlling for the effect of .
2.4. Scaled Cook's distances
We focus on developing several scaled Cook's distances for I, denoted by SCD(I), to detect relatively influential subsets, while accounting for the degree of perturbation . Since we have characterized the stochastic relationship between and CD(I) when is the true data generator, we are interested in reducing the effect of the difference among for different subsets I on the magnitude of CD(I). A simple solution is to calculate several features (e.g., mean, median, or quantiles) of CD(I) and match them across different subsets under the assumption that is the true data generator. Throughout the paper, we consider two pairs of features including (mean, Std) and (median, Mstd), where Std and Mstd, respectively, denote the standard deviation and the median standard deviation. By matching any of the two pairs, we can at least ensure that the centers and scales of the scaled Cook's distances for different subsets are the same when is the true data generator.
We introduce two scaled Cook's distance measures, called scaled Cook's distances, as follows.
Definition 1. The scaled Cook's distances for matching (mean, Std) and (median, Mstd) are, respectively, defined as
where both the expectation and the quantile are taken with respect to .
We can use SCD1(I) and SCD2(I) to evaluate the relatively influential level for different subsets I. A large value of SCD1(I) (or SCD2(I)) indicates that the subset I is relatively influential. Therefore, for any two subsets I1 and I2, the probability of observing the event SCD(I1) > SCD(I2) and that of the event SCD(I1) < SCD(I2) should be reasonably close to each other. Thus, the SCD(I) are roughly comparable. Note that the scaled Cook's distances do not provide a “per unit” effect of removing one observation within the set I, whereas they measure the standardized influential level of the set I when is true. Moreover, the standardization in Definition 1 still implies that higher than average values of CD(I) still correspond with high positive values of SCD(I), even though for some deletions, it is possible for SCD(I) to be negative unlike CD(I).
The next task is how to compute , , , and for each subset I under the assumption that is the true data generator. Computationally, we suggest using the parametric bootstrap to approximate the four quantities of CD(I) as follows.
Step 1. We use to approximate the model , generate a random sample Ys from and then calculate for each s and each subset I.
Step 2. By repeating Step 1 S times, we can obtain a sample {CD(I)(s) : s = 1, · · ·, S} and then we use its empirical mean to approximate E[CD(I)|].
Step 3. We approximate Std[CD(I)|], QCD(I)(0.5|), and Mstd[CD(I)|] by using their corresponding empirical quantities of {CD(I)(s) : s = 1, · · ·, S}.
In this process, we have used to approximate [24] and simulated data Ys from in the standard parametric bootstrap method. If Y truly comes from , then the simulated data Ys should resemble Y. Since is a consistent estimate of θ*, and thus is a consistent estimate of . Similar arguments hold for the other three quantities of CD(I). In Steps 2 and 3, we can use a moderate S, say S = 100, in order to accurately approximate all four quantities of CD(I). According to our experience, such an approximation is very accurate even for small n. See the simulation studies in Section 3.1 for details. However, for most statistical models with complex data structures, it can be computationally intensive to compute for each Ys. We will address this issue in Section 2.6.
As an illustration, we consider how to calculate SCD1(I) for any subset I in the linear regression model.
Example 1 (continued). In (2.11), since all CD(I) share , we replace by . Thus, we approximate CD(I) by , where and
To compute SCD1(I), we just need to calculate the two quantities and Std[CD*(I)|]. Since CD*(I) is a quadratic form, it can be shown that
where denotes the expectation taken with respect to X.
2.5. Conditionally scaled Cook's distances
In certain research settings (e.g., regression), it may be better to perform influence analysis while fixing some covariates of interest, such as measurement time. For instance, in longitudinal data, if different subjects can have different numbers of measurements and measurement times, which are not covariates of interest in an influence analysis, it may be better to eliminate their effect in calculating Cook's distance. We are interested in comparing Cook's distance relative to while fixing some covariates.
To eliminate the effect of some fixed covariates, we introduce two conditionally scaled Cook's distances as follows.
Definition 2. The conditionally scaled Cook's distances (CSCD) for matching (mean, Std) and (median, Mstd) while controlling for Z are, respectively, defined as
where Z is the set of some fixed covariates in Y and the expectation and quantiles are taken with respect to given Z.
According to Definition 2, these conditionally scaled Cook's distances can be used to evaluate the relative influential level of different subsets I given Z. Similar to SCD1(I) and SCD2(I), a large value of CSCD1(I, Z) (or CSCD2(I, Z)) indicates a large influence of the subset I after controlling for Z. It should be noted that because Z is fixed, the CSCDk(I, Z) do not reflect the influential level of Z and the CSCDk(I, Z) may vary across different Z. The conditionally scaled Cook's distances measure the difference of the observed influence level of the set I given Z to the expected influence level of a set with the same data structure when is true and Z is fixed.
The next problem is how to compute , , , and for each subset I when is the true data generator and Z is fixed. Similar to the computation of the scaled Cook's distances, we can essentially use almost the same approach to approximate the four quantities for CSCD1(I, Z) and CSCD2(I, Z). However, a slight difference occurs in the way that we simulate the data. Specifically, let YZ be the data Y with Z fixed. We need to simulate random samples from and then calculate for each subset I.
As an illustration, we consider how to calculate CSCD1(I, Z) for any subset I in the linear regression model.
Example 1 (continued). We set Z = X to calculate CSCD1(I, Z). We need to compute and . Since CD*(I) is a quadratic form, it is easy to show and .
2.6. First-order approximations
We have focused on developing the scaled Cook's distances and their approximations for the linear regression model. More generally, we are interested in approximating the scaled Cook's distances for a large class of parametric models for both independent and dependent data.
We obtain the following theorem.
Theorem 3. If Assumptions A2-A5 in the Appendix hold and n(I)/n → γ ∊ [0, 1), where n(I) denotes the number of observations of I, then we have the following results:
- Let and , CD(I) can be approximated by
(2.14) ;
.
Theorem 3 (a) establishes the first order approximation of Cook's distance for a large class of parametric models for both dependent and independent data. This leads to a substantial savings in computational time, since it is computationally easier to calculate , , and compared to CD(I). Theorem 3 (b) and (c) give an approximation of and , respectively. Generally, it is difficult to give a simple approximation to and , since it involves the fourth moment of which does not have a simple form.
Based on Theorem 3, we can approximate the scaled Cook's distance measures as follows.
Step (i). We generate a random sample Ys from and calculate based on the simulated sample Ys and fixed Z, denoted by . Explicitly, to calculate , we replace Y in , , and by Ys. The computational burden involved in computing is very minor.
Compared to the exact computation of the scaled Cook's distances, we have avoided computing the maximum likelihood estimate of θ based on Ys, which leads to great computational savings in computing for large S, say S > 100. Theoretically, since is a consistent estimate of θ*, is a consistent estimate of . Compared with reestimating for each Ys, a drawback of using in calculating is that does not account for the variability in . Similar arguments hold for the other three quantities of CD(I).
Step (ii). By repeating Step (i) S times, we can use the empirical quantities of to approximate , , , and . Subsequently, we can approximate CSCD1(I, Z) and CSCD2(I, Z) and determine their magnitude based on .
For instance, let and be, respectively, the sample mean and standard deviation of . We calculate
We use to approximate and then compare across different I in order to determine whether a specific subset I is relatively influential or not. Moreover, since can be regarded as the ‘true’ scaled Cook's distance when is true, we can either compare with for all subsets Ĩ and s or compare with for all s. Specifically, we calculate two probabilities as follows:
| (2.15) |
| (2.16) |
where #(Î) is the total number of all possible sets and 1(·) is an indicator function of a set. We regard a subset I as influential if the value of PA(I, Z) (or PB(I, Z)) is relatively large. Similarly, we can use the same strategy to quantify the size of CSCD2(I, Z), SCD1(I), and SCD2(I).
Another issue is the accuracy of the first order approximation to the exact CD(I). For relatively influential subsets, even though the accuracy of the first-order approximation may be relatively low, can easily pick out these influential points. Thus, for diagnostic purposes, the first-order approximation may be more effective at identifying influential subsets compared to the true Cook's distance. We conduct simulation studies to investigate the performance of the first-order approximation relative to the exact CD(I). Numerical comparisons are given in Section 3.
We consider cluster deletion in generalized linear mixed models (GLMM). Example 2. Consider a dataset that is composed of a response yij, covariate vectors xij(p × 1) and cij(p1 × 1), for observations j = 1, . . . , mi within clusters i = 1, . . . , n. The GLMM assumes that conditional on a p1 × 1 random variable bi, yij follows an exponential family distribution of the form [18]
| (2.17) |
where in which β = (β1, . . . , βp)T and k(·) is a known continuously differentiable function. The distribution of bi is assumed to be N(0, Σ), where Σ = Σ(γ) depends on a p2 × 1 vector γ of unknown variance components. In this case, we fix all covariates xij and cij and all mi and include them in Z. For simplicity, we fix (γ, τ) at an appropriate estimate () throughout the example.
We focus here on cluster deletion in GLMMs. After some calculations, the first order approximation of CD(Ii) for deleting the i-th cluster is given by
| (2.18) |
where Ii = {(i, 1), . . . ,(i, mi)}, is the log-likelihood function for the i–th cluster, and . Note that
Then, conditional on all the covariates and {m1, . . . , mn} in Z, we can show that can be approximated by when is true. Moreover, we may approximate by using the fourth moment of . It is not straightforward to approximate and . Computationally, we employ the parametric bootstrap method described above to approximate the conditionally scaled Cook's distances CSCD1(Ii, Z) and CSCD2(Ii, Z).
3. Simulation Studies and A Real Data Example
In this section, we illustrate our methodology with simulated data and a real data example. We also include some additional results in the supplement article [27]. The code along with its documentation for implementing our methodology is available on the first author's website at http://www.bios.unc.edu/research/bias/software.html.
3.1. Simulation Studies
The goals of our simulations were to examine the finite sample performance of Cook's distance and the scaled Cook's distances and their first-order approximations for detecting influential clusters in longitudinal data. We generated 100 datasets from a linear mixed model. Specifically, each dataset contains n clusters. For each cluster, the random effect bi was first independently generated from a distribution and then, given bi, the observations yij (j = 1, . . . , mi; i = 1, . . . , n) were independently generated as and the mi were randomly drawn from {1, . . . 5}. The covariates xij were set as (1, ui, tij)T, among which tij represents time and ui denotes a baseline covariate. Moreover, tij = log(j) and the ui's were independently generated from a N(0, 1) distribution. For all 100 datasets, the responses were repeatedly simulated, whereas we generated the covariates and cluster sizes only once in order to fix the effect of the covariates and cluster sizes on Cook's distance for each cluster. The true value of θ = (βT, σb, σy)T was fixed at (1, 1, 1, 1, 1)T. The sample size n was set at 12 to represent a small number of clusters.
For each simulated dataset, we considered the detection of influential clusters [4]. We fitted the same linear mixed model and used the expectation-maximization (EM) algorithm to calculate and for each cluster I. We treated (σb, σy) as nuisance parameters and β as the parameter vector of interest. We calculated the degree of the perturbation for deleting each subject {i} while fixing the covariates, and then we calculated the conditionally scaled Cook's distances and associated quantities. Let xi be an mi × 3 matrix with the j-th row being . It can be shown that for the case of fixed covariates, we have
| (3.1) |
where Eβ is taken with respect to and , in which and 1mi is an mi × 1 vector with all elements one. We set and substituted β* by .
We carried out three experiments as follows. The first experiment was to evaluate the accuracy of the first-order approximation to CD(I). The explicit expression of is given in Example S2 of the supplementary document. We considered two scenarios. In the first scenario, we directly simulated 100 datasets from the above linear mixed model. In the second scenario, for each simulated dataset, we deleted all the observations in clusters n – 1 and n and then reset (m1, b1) = (1, 4) and (mn, bn) = (5, 3) to generate yi,j for i = 1, n and all j according to the above linear mixed model. Thus, the new first and n-th clusters can be regarded as influential clusters due to the extreme values of b1 and bn. Moreover, the number of observations in these two clusters is unbalanced. We calculated CD(I) and , the average CD(I), and the biases and standard errors of the differences for each cluster {i} (Table 1).
Table 1.
Selected results from simulation studies for n = 12 and the two scenarios: mi, , M, SD, Mdif (×10–2), and SDdif (×10–1) of the three quantities CD(I), , and . mi; denotes the cluster size of subset {i}; denotes the degree of perturbation; M denotes the mean; SD denotes the standard deviation; Mdif and SDdif, respectively, denote the mean and standard deviation of the differences between each quantity and its first-order approximation. In the first scenario, all observations were generated from the linear mixed model, while in the second scenario, two clusters were influential clusters and highlighted in bold. For each case, 100 simulated datasets were used. Results were sorted according to the degree of perturbation for each cluster.
| CD (I) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Scenario I | Scenario II | ||||||||||
| mi | M | Mdif | SD | SDdif | mi | M | Mdif | SD | SDdif | ||
| 1 | 0.10 | 0.11 | 0.01 | 0.09 | 0.03 | 1 | 0.08 | 0.37 | 1.01 | 0.18 | 0.18 |
| 2 | 0.11 | 0.12 | 0.32 | 0.12 | 0.15 | 2 | 0.11 | 0.10 | 0.08 | 0.09 | 0.12 |
| 2 | 0.11 | 0.15 | 1.24 | 0.18 | 0.64 | 1 | 0.11 | 0.08 | 0.02 | 0.11 | 0.02 |
| 2 | 0.13 | 0.18 | 0.87 | 0.19 | 0.36 | 2 | 0.13 | 0.13 | 0.08 | 0.12 | 0.12 |
| 2 | 0.15 | 0.17 | 0.25 | 0.19 | 0.20 | 2 | 0.16 | 0.13 | -0.13 | 0.12 | 0.08 |
| 3 | 0.16 | 0.23 | 0.55 | 0.19 | 0.50 | 2 | 0.20 | 0.20 | 0.08 | 0.19 | 0.12 |
| 2 | 0.19 | 0.26 | -0.02 | 0.32 | 0.25 | 3 | 0.23 | 0.21 | -0.06 | 0.18 | 0.22 |
| 3 | 0.22 | 0.34 | 2.97 | 0.35 | 0.99 | 4 | 0.25 | 0.23 | 0.37 | 0.23 | 0.26 |
| 4 | 0.27 | 0.41 | 3.35 | 0.38 | 1.77 | 5 | 0.28 | 0.78 | 18.59 | 0.61 | 4.71 |
| 5 | 0.40 | 0.70 | 5.43 | 0.60 | 1.90 | 5 | 0.37 | 0.38 | 0.90 | 0.32 | 0.46 |
| 4 | 0.57 | 1.15 | 1.57 | 1.29 | 1.73 | 5 | 0.54 | 0.70 | 1.32 | 0.68 | 0.82 |
| 5 | 0.60 | 1.21 | 3.62 | 1.49 | 1.62 | 4 | 0.56 | 0.65 | 1.06 | 0.69 | 0.54 |
| Scenario I | Scenario II | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mi | M | Mdif | SD | SDdif | mi | M | Mdif | SD | SDdif | ||
| 1 | 0.10 | 0.12 | 0.22 | 0.02 | 0.05 | 1 | 0.08 | 0.09 | 0.43 | 0.01 | 0.04 |
| 2 | 0.11 | 0.12 | 0.41 | 0.01 | 0.03 | 2 | 0.11 | 0.12 | 0.45 | 0.02 | 0.04 |
| 2 | 0.11 | 0.13 | 0.46 | 0.02 | 0.04 | 1 | 0.11 | 0.13 | 0.09 | 0.02 | 0.03 |
| 2 | 0.12 | 0.15 | 0.40 | 0.02 | 0.07 | 2 | 0.13 | 0.15 | 0.38 | 0.02 | 0.04 |
| 2 | 0.15 | 0.17 | 0.34 | 0.03 | 0.08 | 2 | 0.16 | 0.18 | 0.26 | 0.02 | 0.04 |
| 3 | 0.16 | 0.18 | 0.77 | 0.02 | 0.08 | 2 | 0.20 | 0.23 | 0.12 | 0.03 | 0.05 |
| 2 | 0.19 | 0.22 | 0.21 | 0.04 | 0.09 | 3 | 0.23 | 0.27 | 0.46 | 0.03 | 0.07 |
| 3 | 0.22 | 0.26 | 0.62 | 0.04 | 0.09 | 4 | 0.25 | 0.29 | 1.13 | 0.03 | 0.13 |
| 4 | 0.26 | 0.32 | 1.63 | 0.03 | 0.15 | 5 | 0.28 | 0.36 | 1.94 | 0.04 | 0.18 |
| 5 | 0.40 | 0.55 | 2.58 | 0.07 | 0.29 | 5 | 0.37 | 0.48 | 1.86 | 0.05 | 0.18 |
| 4 | 0.57 | 0.97 | 2.21 | 0.12 | 0.21 | 5 | 0.53 | 0.82 | 4.26 | 0.10 | 0.34 |
| 5 | 0.60 | 1.03 | 5.87 | 0.16 | 0.99 | 4 | 0.56 | 0.93 | 1.64 | 0.11 | 0.17 |
| Scenario I | Scenario II | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mi | M | Mdif | SD | SDdif | mi | M | Mdif | SD | SDdif | ||
| 1 | 0.10 | 0.18 | 1.48 | 0.04 | 0.20 | 1 | 0.08 | 0.13 | 1.05 | 0.04 | 0.22 |
| 2 | 0.11 | 0.14 | 1.16 | 0.03 | 0.10 | 2 | 0.11 | 0.14 | 1.18 | 0.03 | 0.12 |
| 2 | 0.11 | 0.15 | 1.37 | 0.03 | 0.16 | 1 | 0.11 | 0.18 | 0.78 | 0.04 | 0.10 |
| 2 | 0.13 | 0.18 | 1.72 | 0.05 | 0.35 | 2 | 0.13 | 0.18 | 1.15 | 0.03 | 0.13 |
| 2 | 0.15 | 0.21 | 2.02 | 0.05 | 0.25 | 2 | 0.16 | 0.23 | 1.28 | 0.04 | 0.14 |
| 3 | 0.16 | 0.19 | 2.05 | 0.03 | 0.25 | 2 | 0.20 | 0.30 | 1.07 | 0.06 | 0.16 |
| 2 | 0.19 | 0.29 | 2.36 | 0.07 | 0.24 | 3 | 0.23 | 0.31 | 1.72 | 0.06 | 0.22 |
| 3 | 0.22 | 0.30 | 2.55 | 0.07 | 0.32 | 4 | 0.25 | 0.30 | 1.96 | 0.05 | 0.42 |
| 4 | 0.26 | 0.35 | 2.84 | 0.06 | 0.39 | 5 | 0.28 | 0.39 | 4.06 | 0.09 | 0.66 |
| 5 | 0.40 | 0.58 | 2.13 | 0.11 | 0.71 | 5 | 0.37 | 0.50 | 2.67 | 0.09 | 0.52 |
| 4 | 0.57 | 1.16 | 1.17 | 0.18 | 0.55 | 5 | 0.53 | 0.89 | 0.60 | 0.14 | 0.68 |
| 5 | 0.60 | 1.14 | -4.18 | 0.25 | 2.29 | 4 | 0.56 | 1.13 | 0.94 | 0.21 | 0.41 |
Inspecting Table 1 reveals three findings as follows. First, when no influential cluster is present in the first scenario, the average CD(I) is an increasing function of , whereas it is only positively proportional to the cluster size n(I) with a correlation coefficient of 0.83. This result agrees with the results of Proposition 1. Secondly, in the second scenario, the average CD(I) for the true ’good’ clusters is positively proportional to with a correlation coefficient of 0.76, while that for the influential clusters is associated with both and the amount of influence that we introduced. Thirdly, for the true ‘good’ clusters, the first-order approximation is very accurate and leads to small average biases and standard errors. Even for the influential clusters, is relatively close to CD(I). For instance, for cluster {n}, the bias of 0.19 is relatively small compared with 0.78, the mean of CD({n}).
In the second experiment, we considered the same two scenarios as the first experiment. Specifically, for each dataset, we approximated and by setting S = 200 and using their empirical ones, and calculated their first approximations and . Across all 100 data sets, for each cluster I, we computed the averages of and , and the biases and standard errors of the differences and .
Table 1 shows the results for each scenario. First, in both scenarios, the average is an increasing function of , whereas it is only positively proportional to the cluster size n(I) with a correlation coefficient (CC) of 0.80. This is in agreement with the results of Proposition 1. The average of are positively proportional to mi (CC=0.76) and (CC=0.99). Secondly, for all clusters, the first-order approximations of and are very accurate and lead to small average biases and standard errors.
The third experiment was to examine the finite sample performance of Cook's distance and the scaled Cook's distances for detecting influential clusters in longitudinal data. We considered two scenarios. In the first scenario, for each of the 100 simulated datasets, we deleted all the observations in cluster n and then reset mn = 1 and varied bn from 0.6 to 6.0 to generate yn,j according to the above linear mixed model. The second scenario is almost the same as the first scenario except that we reset mn = 10. Note that when the value of bn is relatively large, e.g., bn = 2:5, the n-th cluster is an influential cluster, whereas the n-th cluster is not influential for small bn. A good case-deletion measure should detect the n-th cluster as truly influential for large bn, whereas it does not for small bn. For each data set, we approximated CSCD1(I, Z), CSCD2(I, Z), , and by setting S = 100. Subsequently, we calculated PA(I, Z) and PB(I, Z) in (2.15) and . Finally, across all 100 datasets, we calculated the averages and standard errors of all diagnostic measures for the n-th cluster for each scenario.
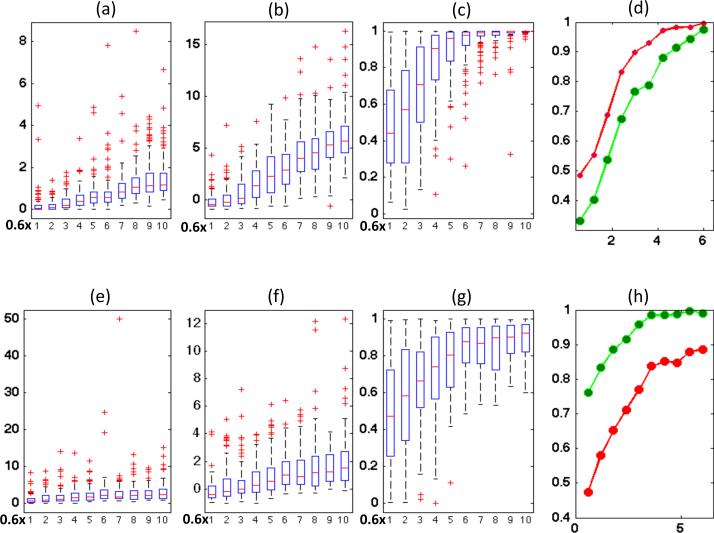
Inspecting Figure 1 reveals some findings as follows. First, deleting the n-th cluster with 10 observations causes a larger effect than that with 1 observation (Fig 1 (a) and (e), (d) and (h)). As expected, the distributions of CD({n}) and shift up as bn increases (Fig 1 (a), (b), (e), and (f)). Secondly, in the first scenario, CD({n}) is stochastically smaller than most other CD(I)s, when the value of bn is relatively small (Fig. 1 (d)). However, in the second scenario, CD({n}) is stochastically larger than most other CD(I)s (Fig. 1 (h)) even for small values of bn. Specifically, when mn = 1, the average PC({n}, Z) is smaller than 0.4 as bn = 0.6 and bn = 1.2, whereas when mn = 10, the average PC({n}, Z) is higher than 0.75 even as bn = 0.6. In contrast, in the two scenarios, the value of PB({n}, Z) is close to 0.5 as bn = 0.6 (Fig. 1 (d) and (h)). It indicates that the cluster size does not have a big effect on the distribution of (Fig. 1 (c) and (g)).
Fig 1.
Simulation results from 100 datasets simulated from a linear mixed model in the two scenarios. The first row corresponds to the first scenario, in which m12 = 1 and b12 varies from 0.6 to 6.0. The second row corresponds to the second scenario, in which m12 = 10 and b12 varies from 0.6 to 6.0. Panels (a) and (e) show the box plots of Cook's distances as a function of b12; panels (b) and (f) show the box plots of CSCD1(I, Z) as a function of b12; panels (c) and (g) show the box plots of PB(I, Z) as a function of b12; panels (d) and (h) show the mean curve of PB(I, Z) based on CSCD1(I, Z) (red line) and the mean curve of PC(I, Z) based on CD(I) (green line) as functions of b12.
3.2. Yale Infant Growth Data
The Yale infant growth data were collected to study whether cocaine exposure during pregnancy may lead to the maltreatment of infants after birth, such as physical and sexual abuse. A total of 298 children were recruited from two subject groups (cocaine exposed group and unexposed group). One feature of this dataset is that the number of observations per children mi varies significantly from 2 to 30 [22, 21]. The total number of data points is . Following Zhang [26], we considered two linear mixed models given by , where yi,j is the weight (in kilograms) of the j-th visit from the i-th subject, xi,j = (1, di,j, (di,j – 120)+, (di,j – 200)+, (gi – 28)+, di,j(gi – 28)+, di,j(gi – 28)+, (di,j – 60)+ (gi – 28)+, (di,j – 490)+(gi – 28)+, sidi,j, si(di,j – 120)+)T, in which di,j and gi (days) are the age of visit and gestational age, respectively, and si is the indicator for gender. In addition, we assumed εi = (εi,1, . . . , εi,mi)T ~ Nmi (0, Ri(α)), where α is a vector of unknown parameters in Ri(α). We first considered . We refer to this model as model M1. Then, it is assumed that variance and autocorrelation parameters are, respectively, given by V(d) = exp(α0 + α1d + α2d2 + α3d3) and ρ(l) = α4 + α5l, where l is the lag between two visits. We refer to this model as model M2.
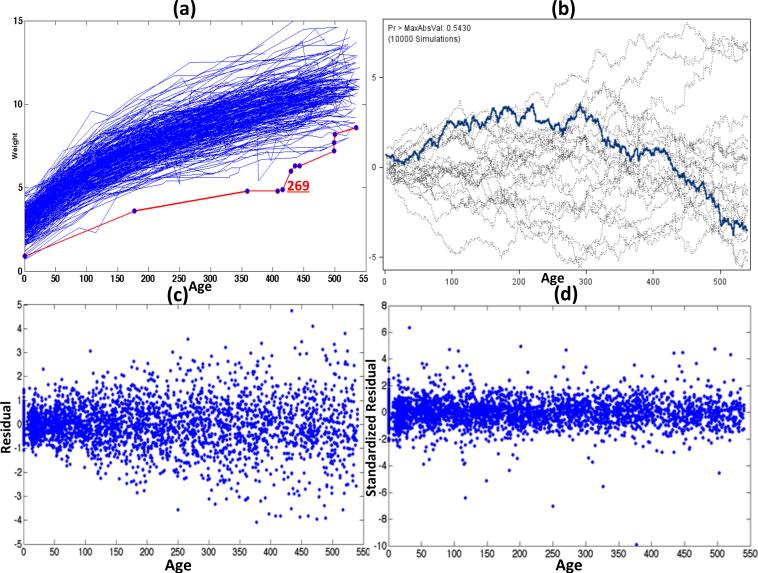
We systematically examined the key assumptions of models M1 and M2 as follows. (i) We presented a cumulative residual plot and calculated the cumulative sums of residuals over the age of the visit to test [17], whose p-value is greater than 0.543. It may suggest that the mean structure is reasonable. The cumulative residual plot is given in Figure 2 (b).
Fig 2.
Yale infant growth data. Panel (a) presents the line plot of infant weight against age, in which the observations of subject 269 are highlighted; panel (b) shows the cumulative residual curve versus age, in which the observed cumulative residual curve is highlighted in blue; and panels (c) and (d), respectively, present age versus raw residual and age versus standardized residual for cluster deletion.
(ii) For model M1, inspecting the plot of raw residuals against age in Figure 2 (c) reveals that the variance of the raw residuals appears to increase with the age of visit. As pointed by Zhang [26], it may be more sensible to use model M2. Let be the vector of standardized residuals of M2, where ri = (ri,1, . . . , ri,mi)T. The standardized residuals under M2 do not have any apparent structure as age increases (Figure 2 (d)).
(iii) Under each model, we calculated CD(I) for each child [4]. We treated β as parameters of interest and all elements of α as nuisance parameters. For model M1, we obtained a strong Pearson correlation of 0.363 between Cook's distance and the cluster size. This indicates that the bigger the cluster size, the larger the Cook's distance measure. Figure 4 (b) highlights the top ten influential subjects. Compared with model M1, we observed similar findings by using CD(I) under model M2, which were omitted for space limitations.
There are several difficulties in using Cook's distance under both models M1 and M2 [19, 7, 4, 3]. First, cluster size varies significantly across children and deleting a larger cluster may have a higher probability of having a larger influence as discussed in Section 2.3. For instance, we observe (m285, CD({285})) = (8, 0.738) and (m274, CD({274})) = (22, 1.163). A larger CD({274}) can be caused by a larger m274 = 22 and/or influential subject 274, among others. Since m274 is much larger than m285, it is difficult to claim that subject 274 is more influential than subject 285. Secondly, there is no rule for determining whether a specific subject is influential relative to the fitted model. Specifically, it is unclear whether the subjects with larger CD({i}) are truly influential or not. Thirdly, inspecting Cook's distance solely does not seem to delineate the potential misspecification of the covariance structure under model M1. We will address these three difficulties by using the new case-deletion measures.
(iv) Under each model, we calculated for deleting each subject {i} for fixed covariates, and then we calculated the conditionally scaled Cook's distances and associated quantities. We then used 1000 bootstrap samples to approximate CSCD1(I, Z) and CSCD2(I, Z). Subsequently, we calculated PA(I, Z) and PB(I, Z) in (2.15).
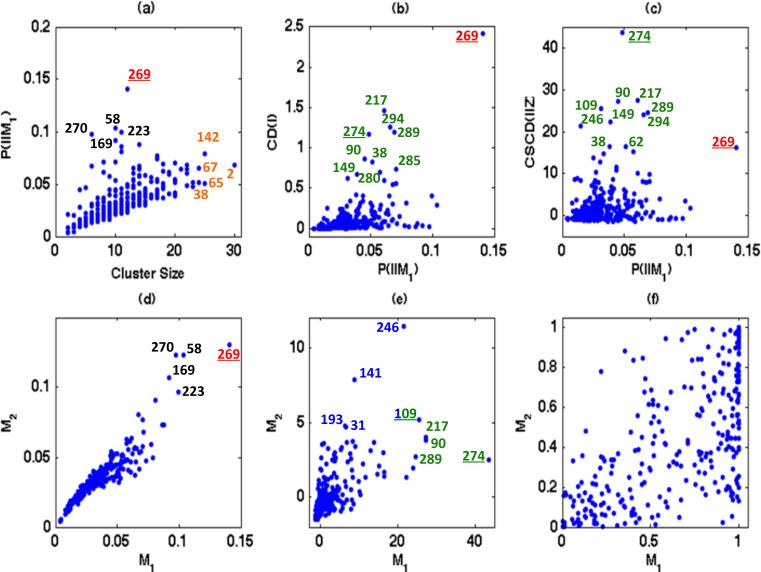
We observed several findings. First, under model M1, we observed a strong positive correlation between and mi (Fig. 3 (a)). Secondly, even though m269 = 12 is moderate, subject 269 has the largest degree of perturbation. Inspecting the raw data in Figure 2 (a) reveals that subject 269 is of older age during visits compared with other subjects. Thirdly, we also observed a strong positive correlation between and the Cook's distance (Fig. 3 (b)), which may indicate their stochastic relationship as discussed in Section 2.3. Fourthly, we observed a positive correlation between Cook's distance and the conditionally scaled Cook's distance (Fig. 3(b) and (c)), but their levels of influence for the same subject are quite different. For instance, the magnitude of CSCD1({269}, Z) is only moderate, whereas CD1({269}, Z) is the highest one. We observed similar findings under model and presented some findings in Figure 3 (d) and (e).
Fig 3.
Yale infant growth data. Panel (a) shows mi versus , in which the ten subjects with the largest values of degree of perturbation or cluster size are highlighted; panel (b) shows versus CD(I), in which the top ten influential subjects are highlighted; panel (c) shows versus CSCD1(I, Z), in which the top eleven influential subjects are highlighted; and panels (d), (e), and (f), respectively, show , CSCD1(I, Z), and PB(I, Z) for models and .
We used PB(I, Z) to quantify whether a specific subject is influential relative to the fitted model (Fig. 3 (f)). For instance, since CD({246}) = 0.253, it is unclear whether subject 246 is influential or not according to CD, whereas we have CSCD1({246}, Z) = 21.443 and PB({246}, Z) = 1.0. Thus, subject 246 is really influential after eliminating the effect of the cluster size. Moreover, it is difficult to compare the influential levels of subjects 274 and 285 using CD. All of the conditionally scaled Cook's distances and associated quantities suggest that subject 274 is more influential than subject 285 after eliminating the degree of perturbation difference. We observed similar findings under model and omitted them due to space limitations. See Figure 3 (d) and (e) for details.
We compared the goodness of fit of models and to the data by using the proposed case-deletion measures. First, inspecting Figure 3 (d) reveals a strong similarity between the degrees of perturbation under models and for all subjects. Secondly, by using the conditionally scaled Cook's distance, we observed the different levels of influence for the same subject under and . For instance, CSCD1(I, Z) identifies subjects 246, 141, 109, 193 and 31 as the top five influential subjects under , whereas it identifies subjects 274, 217, 90, 109, and 289 as the top ones under . Finally, examining PB(I, Z) reveals a large percentage of influential points for model , but a small percentage of influential points for model . See Figure 3 (f) for details. This may indicate that model outperforms model . Furthermore, although we may develop goodness-of-fit statistics based on the scaled Cook's distances and show that model outperforms model , this will be a topic of our future research.
In summary, the use of the new case-deletion measures provides new insights in real data analysis. First, explicitly quantifies the degree of perturbation introduced by deleting each subject. Secondly, CSCDk(I, Z) for k = 1, 2 explicitly account for the degree of perturbation for each subject. Thirdly, PB(I, Z) allows us to quantify whether a specific subject is influential relative to the fitted model. Fourthly, inspecting PB(I, Z) and CSCDk(I, Z) may delineate the potential misspecification of the covariance structure under model M1.
4. Discussion
We have introduced a new quantity to quantify the degree of perturbation and examined its properties. We have used stochastic ordering to quantify the relationship between the degree of the perturbation and the magnitude of Cook's distance. We have developed several scaled Cook's distances to address the fundamental issue of deletion diagnostics in general parametric models. We have shown that the scaled Cook's distances provide important information about the relative influential level of each subset. Future work includes developing goodness-of-fit statistics based on the scaled Cook's distances, developing Bayesian analogs to the scaled Cook's distances, and developing user-friendly R code for implementing our proposed measures in various models, such as survival models and models with missing covariate data.
Supplementary Material
Acknowledgments
We thank the Editor Peter Bühlmann, the Associate Editor and two anonymous referees for valuable suggestions, which have greatly helped to improve our presentation.
Appendix
The following assumptions are needed to facilitate the technical details, although they are not the weakest possible conditions. Because we develop all results for general parametric models, we only assume several high-level assumptions as follows.
Assumption A2. for any I is a consistent estimate of θ* ∈ Θ.
Assumption A3. All p(Y[I]|θ) are three times continuously differentiable on Θ and satisfy
in which |R[I](θ)| = op(1) uniformly for all , where and .
Assumption A4. For any I and Z, ,
and .
Assumption A5. For any set I and Z,
Remarks: Assumptions A2-A5 are very general conditions and are generalizations of some higher level conditions for the extremum estimator, such as the maximum likelihood estimate, given in Andrews [2]. Assumption A2 assumes that the parameter estimators with and without deleting the observations in the subset I are consistent. Assumption A3 assumes that the log-likelihood functions for any I and Y[I] admit a second-order Taylor's series expansion in a small neighborhood of θ*. Assumptions A4 and A5 are standard assumptions to ensure that the first- and second-order derivatives of p(Y[I]|θ) and p(YI|Y[I], θ) have appropriate rates of n and nI [2, 28]. Sufficient conditions of Assumptions A2-A5 have been extensively discussed in the literature [2, 28].
Proof of Theorem 1. (P.a) directly follows from the Jensen inequality, (2.6) and (2.7). For (P.b), if I is an empty set, then KL(Y, θ|I) ≡ 0 and thus . On the other hand, if , then KL(Y, θ|I) ≡ 0 for almost every θ. Thus, by using the Jensen inequality, we have p(YI|Y[I], θ) ≡ p(YI|Y[I], θ*) for all θ ∈ Θ. Based on the identifiability condition, we know that I must be an empty set. Let I1·2 = I1 – I2. It is easy to show that
Thus, by substituting the above equation into (2.6), we have
| (4.1) |
in which the second term on the right hand side can be written as
which yield (P.c). Based on the assumption of (P.d), we know that
for all θ. Thus, the second term on the right hand side of (4.1) reduces to , which finishes the proof of (P.d).
Proof of Theorem 2. (a) Let I3 = I1 \ I2, I1 is a union of two disjoint sets I3 and I2. Without loss of generality, HI1 can be decomposed as
Let λ1,1 ≥ . . . ≥ λ1,n(I1) ≥ 0 and λ2,1 ≥ . . . ≥ λ2,n(I2) ≥ 0 be ordered eigenvalues of HI1 and HI2, respectively, where n(Ik) denotes the number of observations in Ik for k = 1, 2. It follows from Wielandt's eigenvalue inequality [13] that λ1,l ≥ λ2,l for all l = 1, . . . , n(I2). For k = 1, 2, we define as the spectral decomposition of HIk and , where Γk is an orthnormal matrix and Λk = diag(λk,1, . . . , λk,n(Ik)). It can be shown that for k = 1, 2,
Since f(x) = x/(1 – x) is an increasing function of x ∈ (0, 1), this completes the proof of Theorem 2 (a).
Note that , where λj are the eigenvalues of HI and h = (h1, . . . , hn(I))T ~ N(0, σ2In(I)). Moreover, the distribution of λ is uniquely determined by HI. Combining h ~ N(0, σ2In(I)) with the assumptions of Theorem 2 (b) yields that CD(I) and CD(I′) follow the same distribution when n(I) = n(I′). Furthermore, we can always choose an such that and . Following arguments in Theorem 2 (a), we can then complete the proof of Theorem 2 (b).
Proof of Theorem 3. (a) It follows from a Taylor's series expansion and assumption A3 that
where for t ∈ [0, 1]. Combining this with Assumption A4 and the fact that , we get
| (4.2) |
Substituting (4.2) into completes the proof of Theorem 3 (a).
(b) It follows from Assumptions A2-A4 that
Let . Using a Taylor's series expansion along with Assumptions A4 and A5, we get
| (4.3) |
Since ,
It follows from Assumption A4 that for θ in a neighborhood of θ*, Fn(θ) and Fn(θ*) – fI(θ) can be replaced by and , respectively, which completes the proof of Theorem 3 (b).
(c) Similar to Theorem 3 (b), we can prove Theorem 3 (c).
Footnotes
SUPPLEMENTARY MATERIAL
Supplement to ”Perturbation and Scaled Cook's Distance”: (http://www.bios.unc.edu/research/bias/documents/SS-diag_sizeblind.pdf). We include two theoretical examples and additional results obtained from the Monte Carlo simulation studies and real data analysis.
References
- 1.Andersen EB. Diagnostics in Categorical Data Analysis. Journal of the Royal Statistical Society, Series B: Methodological. 1992;54:781–791. [Google Scholar]
- 2.Andrews DWK. Estimation When a Parameter Is on a Boundary. Econometrica. 1999;67:1341–1383. [Google Scholar]
- 3.Banerjee M. Cook's Distance in Linear Longitudinal Models. Communications in Statistics: Theory and Methods. 1998;27:2973–2983. [Google Scholar]
- 4.Banerjee M, Frees EW. Influence Diagnostics for Linear Longitudinal Models. Journal of the American Statistical Association. 1997;92:999–1005. [Google Scholar]
- 5.Beckman RJ, Cook RD. Outlier..........s. Technometrics. 1983;25:119–149. [Google Scholar]
- 6.Chatterjee S, Hadi AS. Sensitivity Analysis in Linear Regression. John Wiley & Sons; 1988. [Google Scholar]
- 7.Christensen R, Pearson LM, Johnson W. Case-deletion Diagnostics for Mixed Models. Technometrics. 1992;34:38–45. [Google Scholar]
- 8.Cook RD. Detection of Influential Observation in Linear Regression. Technometrics. 1977;19:15–18. [Google Scholar]
- 9.Cook RD. Assessment of Local Influence (with Discussion). Journal of the Royal Statistical Society, Series B: Methodological. 1986;48:133–169. [Google Scholar]
- 10.Cook RD, Weisberg S. Residuals and Influence in Regression. Chapman & Hall Ltd.; 1982. [Google Scholar]
- 11.Critchley F, Atkinson RA, Lu G, Biazi E. Influence Analysis Based on the Case Sensitivity Function. Journal of the Royal Statistical Society, Series B: Statistical Methodology. 2001;63:307–323. [Google Scholar]
- 12.Davison AC, Tsai CL. Regression Model Diagnostics. International Statistical Review. 1992;60:337–353. [Google Scholar]
- 13.Eaton ML, Tyler DE. On Wielandt's Inequality and Its Application to the Asymptotic Distribution of the Eigenvalues of a Random Symmetric Matrix. The Annals of Statistics. 1991;19:260–271. [Google Scholar]
- 14.Fung W-K, Zhu Z-Y, Wei B-C, He X. Influence Diagnostics and Outlier Tests for Semiparametric Mixed Models. Journal of the Royal Statistical Society, Series B: Statistical Methodology. 2002;64:565–579. [Google Scholar]
- 15.Haslett J. A Simple Derivation of Deletion Diagnostic Results for the General Linear Model with Correlated Errors. Journal of the Royal Statistical Society, Series B: Statistical Methodology. 1999;61:603–609. [Google Scholar]
- 16.Huber PJ. Robust Statistics. Wiley Series in Probability and Statistics; 1981. [Google Scholar]
- 17.Lin DY, Wei LJ, Ying Z. Model-checking techniques based on cumulative residuals. Biometrics. 2002;58:1–12. doi: 10.1111/j.0006-341x.2002.00001.x. [DOI] [PubMed] [Google Scholar]
- 18.McCullagh P, Nelder JA. Generalized Linear Models. Chapman & Hall Ltd.; 1989. [Google Scholar]
- 19.Preisser JS, Qaqish BF. Deletion Diagnostics for Generalised Estimating Equations. Biometrika. 1996;83:551–562. [Google Scholar]
- 20.Shaked M, Shanthikumar GJ. Stochastic Orders. Springer; 2006. [Google Scholar]
- 21.Stier DM, Leventhal JM, Berg AT, Johnson L, Mezger J. Are Children Born to Young Mothers at Increased Risk of Maltreatment. Pediatrics. 1993;91:642–648. [PubMed] [Google Scholar]
- 22.Wasserman DR, Leventhal JM. Maltreatment of Children Born to Cocaine-Dependent Mothers. American Journal of Diseases of Children. 1993;147:1324–1328. doi: 10.1001/archpedi.1993.02160360066021. [DOI] [PubMed] [Google Scholar]
- 23.Wei B-C. Exponential Family Nonlinear Models. Springer; Singapore: 1998. [Google Scholar]
- 24.White H. Maximum Likelihood Estimation of Misspecified Models. Econometrica. 1982;50:1–26. [Google Scholar]
- 25.White H. Estimation, Inference, and Specification Analysis. Cambridge University Press; 1994. [Google Scholar]
- 26.Zhang H. Analysis of Infant Growth Curves Using Multivariate Adaptive Splines. Biometrics. 1999;55:452–459. doi: 10.1111/j.0006-341x.1999.00452.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhu H, Ibrahim JG. Supplement to “Perturbation and scaled Cook's distance”. 2011. [DOI] [PMC free article] [PubMed]
- 28.Zhu H, Zhang H. Asymptotics for Estimation and Testing Procedures under Loss of Identifiability. Journal of Multivariate Analysis. 2006;97:19–45. [Google Scholar]
- 29.Zhu H, Lee SY, Wei BC, Zhou J. Case Deletion Measures for Models with Incomplete Data. Biometrika. 2001;88:727–737. [Google Scholar]
- 30.Zhu H, Ibrahim JG, Lee S-Y, Zhang H. Perturbation Selection and Influence Measures in Local Influence Analysis. The Annals of Statistics. 2007;35:2565–2588. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.