Abstract
We have used microarray analysis to study the transcriptome of the bacterial pathogen Bordetella bronchiseptica over the course of five time points representing distinct stages of biofilm development. The results suggest that B. bronchiseptica undergoes a coordinately regulated gene expression program similar to a bacterial developmental process. Expression and subsequent production of the genes encoding flagella, a classical Bvg− phase phenotype, occurs and is under tight regulatory control during B. bronchiseptica biofilm development. Using mutational analysis, we demonstrate that flagella production at the appropriate stage of biofilm development, i.e. production early subsequently followed by repression, is required for robust biofilm formation and maturation. We also demonstrate that flagella are necessary and enhance the initial cell-surface interactions, thereby providing mechanistic information on the initial stages of biofilm development for B. bronchiseptica. Biofilm formation by B. bronchiseptica involves the production of both Bvg-activated and Bvg-repressed factors followed by the repression of factors that inhibit formation of mature biofilms.
Introduction
Bordetella bronchiseptica is a gram negative bacterial pathogen with a broad host range that naturally infects a wide variety of farm and companion animals [1], [2], [3]. It is the etiological agent or a co-contributor to a number of veterinary syndromes such as kennel cough in dogs, atrophic rhinitis (AR) and pneumonia in pigs and bronchopneumonia in guinea pigs, rabbits, horses, rats, mice, cats and nonhuman primates. B. bronchiseptica is also being increasingly isolated from humans mainly from immunocompromised patients. In many of these cases, the infections are caused by exposure to pets with B. bronchiseptica [4], [5].
A hallmark of B. bronchiseptica infections is long-term to life-long asymptomatic carriage. Despite vaccination, these bacteria continue to circulate and persist in animals. B. bronchiseptica is frequently isolated from the nasal cavities of vaccinated animals suggesting that vaccines fail to protect animals from infection [1], [2]. Additionally, B. bronchiseptica is capable of establishing a chronic and asymptomatic infection and can be harvested from the nasal cavities of rats, mice, and swine for extended periods [6], [7], [8], [9], [10]. We have isolated B. bronchiseptica from the rat nasopharynx even after 85 days of inoculation (our unpublished results) and the nasal cavity of laboratory mice remains colonized by B. bronchiseptica for the life of the animal [11].
A principal impediment towards the development of improved vaccines and interventions for Bordetella Spp. is a gap in our understanding of the mechanisms that contribute to persistence or the carrier state. A convincing and frequently proposed hypothesis to explain the survival and continued persistence of bacterial pathogens is that these organisms exist as biofilms. Recent studies have supported this hypothesis for members of the Bordetella genus by demonstrating that both B. pertussis and B. bronchiseptica are capable of forming biofilms on abiotic surfaces [12], [13], [14], [15], [16] and in the mouse respiratory tract [12], [17], [18], [19]. Biofilms are defined as a community of microorganisms enveloped in a hydrated extracellular polymeric matrix that adheres to the interface of a liquid and a surface. Numerous studies have documented increased resistance of biofilms to antibiotic treatments and the components of the host immune system [20], [21], [22], [23], [24]. Thus, bacterial biofilms are increasingly recognized as important contributors to chronic or persistent infections [25], [26].
We and others have previously shown that biofilm formation in Bordetella is regulated by BvgAS, a two-component sensory transduction system [13], [15]. This locus comprises a sensor kinase protein, BvgS, and a DNA-binding response-regulator protein, BvgA. In response to environmental cues, BvgAS controls the expression of a spectrum of phenotypic phases transitioning between a virulent (Bvg+) phase and a non-virulent (Bvg− phase), a process referred to as phenotypic modulation. During the virulent Bvg+ phase, the BvgAS system is fully active and many of the known virulence factors are expressed [27]. Conversely, BvgAS is inactive during the Bvg− phase, resulting in the maximal expression of motility loci, virulence-repressed genes (vrg), and genes required for the production of urease [28], [29], [30]. Previous studies have demonstrated that the Bvg+ phase is required for respiratory tract colonization [6], [31], [32], [33], [34], while the Bvg− phase of B. bronchiseptica likely promotes survival outside of the mammalian host [31], [32].
Biofilm formation initiates with planktonic bacteria attaching to a surface leading to the formation of a monolayer, followed sometimes by formation of clusters and microcolonies and subsequent development of differentiated structures in which individual bacteria as well as the entire community are surrounded by an extracellular matrix [35], [36], [37]. For B. bronchiseptica, we have shown that the Bvg-mediated control of biofilm development is exerted subsequent to the initial attachment of the bacterial cells to a surface. We have also shown that the B. bronchiseptica Bps polysaccharide, which is not regulated by BvgAS, promotes biofilm formation at steps post-attachment, specifically in the development of three-dimensional structures [14], [18], [38]. Thus, for B. bronchiseptica, while some information has been obtained about the factors and the regulatory mechanisms that mediate later stages of biofilm development, nothing is known regarding early biofilm stages.
Biofilm cells differ from their planktonic cell counterparts in the genes and proteins they express, resulting in distinct physiological states and phenotypes [24], [39], [40]. Microscopic and genetic analysis has indicated that biofilm formation occurs in a sequential stage-specific and coordinated manner similar to microbial development, such as sporulation by Bacillus species and swarmer-to-stalk cell transition in Caulobacter cresentus [41]. The purpose of this study was to gain insights into the gene-expression profile and the transcriptional control operative in the biofilm associated B. bronchiseptica as a basis for understanding the contribution of individual genes in the various stages of biofilm development. Our analysis led to the surprising discovery that the expression of the genes encoding flagella, a classical Bvg− phase phenotype, occurs early and is under tight regulatory control during B. bronchiseptica biofilm development. We have obtained convincing evidence that flagella are critical during the initial stages of biofilm formation. Our data suggest that the regulatory mechanism coordinating biofilm development in B. bronchiseptica results in the production of a classical Bvg− phase phenotype under Bvg+ phase conditions.
Materials and Methods
Bacterial Strains and Plasmids
The strains and plasmids utilized in this study are listed in Table 1. Bordetella strains were grown in Stainer-Scholte (SS) broth [42] for both biofilm and planktonic cultures. The bacteria were maintained on Bordet Gengou (BG) agar (Difco, Sparks, MD) supplemented with 7.5% defibrinated sheep blood to determine hemolysis and colony morphology. The green fluorescent protein (GFP) expression vector pTac-GFP [14] was mobilized in strain RB50, Rev1ΔflaA, WTΔfrlAB, or Bvg− ΔflaA by triparental mating as described previously [43], and exconjugates were selected on BG agar containing 50 µg/mL chloramphenicol and 50 µg/mL streptomycin. Randomly picked colonies containing pTac-GFP were grown in SS broth containing 50 µg/mL chloramphenicol and were analyzed for GFP expression utilizing a Nikon Eclipse TE300 inverted microscope. One of the GFP-expressing clones corresponding to each of the strains was chosen for experimental analysis. Comparison of the GFP-expressing strains with the respective parental strains not containing the pTac-GFP plasmid revealed no differences in growth in batch cultures or colony morphology on BG agar containing blood. Strains containing the pTac-GFP plasmid were grown in the presence 50 µg/mL chloramphenicol.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source |
| Strains | ||
| RB50 | Wild type B. bronchiseptica isolate | [31] |
| RB54 | Bvg− phase-locked; ΔbvgS derivative of RB50 | [31] |
| Rev1 | RB50 frlr; Replacement of the frlAB with the fhaB promoter resulting inexpression of flagella in the Bvg+ phase; | [6] |
| Rev1ΔflaA | nonflagellated; ΔflaA derivative of Rev1 | [6] |
| Bvg− ΔflaA | nonflagellated; ΔflaA derivative of RB54 | [59] |
| WTΔfrlAB | nonmotile; ΔfrlAB derivative of RB50 | [6] |
| Plasmids | ||
| pTAC-GFP | CMr derivative of pBBR1-GFP | [14] |
| pKK233-2 | hybrid trp/lac (tac) promoter | [60] |
| pBBR1-GFP | promoterless GFP, CAT vector | [61] |
RNA Isolation
RNA was extracted using a previously described method [44]. Briefly, single colony was inoculated in SS broth containing 40 µg/mL streptomycin at 37°C with shaking. Bacteria were then subcultured at a starting optical density at 600 nm (OD600) of 0.05–0.1 into 150×15 mm petri dishes (Fisher Scientific, Pittsburgh, PA) and incubated at 37°C for the indicated amount of time. A 10 mL aliquot of cells was removed for preparation of planktonic RNA, centrifuged, and then lysed with 1 ml RLT (Qiagen, Valencia, CA). The remaining media was removed, attached cells were washed three times with cold PBS and the remaining cells were lysed in situ with 1 mL of RLT buffer (Qiagen, Valencia, CA) and then isolated using the Qiagen RNeasy Kit (Qiagen, Valencia, CA).
Preparation of Labeled cDNA and Microarray Analysis
A 2-color hybridization format was used for the microarray analysis. For each biological replicate, prepared RNA was first treated with DNase1 (Promega, Madison, WI) to remove genomic DNA and then RNA extracted from biofilm cells and used to generate Cy5-labeled cDNA and RNA extracted from planktonic cells was used to generate Cy3-labeled cDNA. Additionally, dye-swap experiments were performed analogously, in which the fluorescent labels were exchanged to ensure that uneven incorporation did not confound our results. Fluorescently-labeled cDNA copies of the total RNA pool were prepared and the two differentially labeled reactions to be compared were then mixed, followed by buffer exchange, purification, and concentration as described [45], [46], [47], [48]. The two differentially labeled reactions to be compared were combined and hybridized to a B. bronchiseptica strain RB50 specific long-oligonucleotide microarray [48]. Slides were then scanned using a GenePix 4000B microarray scanner and analyzed with GenePix Pro software (Axon Instruments, Union City, CA). Spots were assessed visually to identify those of low quality and arrays were normalized so that the median of ratios across each array was equal to 1.0. Spots of low quality were identified and were filtered out prior to analysis. Ratio data from the biological replicates were compiled and normalized based on the total Cy3% intensity and Cy5% intensity to eliminate slide to slide variation. Gene expression data were then normalized to 16 S rRNA. The statistical significance of the gene expression changes observed was assessed by using the significant analysis of microarrays (SAM) program [49]. A one-class unpaired SAM analysis using a false discovery rate of 0.001% was performed. Hierarchical clustering of microarray data using Pearson Correlation metrics and Average Linkage clustering was performed using MeV software from TIGR [50]. All microarray data are available in the supplementary text and have been deposited in ArrayExpress under accession number E-MEXP-3295.
cDNA Preparation and Quantitative RT-PCR
Prepared RNA was first treated with DNase1 (Promega, Madison, WI) to remove genomic DNA and then reverse transcribed using Super Script III (Invitrogen, Carlsbad, CA). A mixture of 10 mM dNTPs (Invitrogen, Carlsbad, CA), 300 ng/µL random primers (Invitrogen, Carlsbad, CA), and 20 µL of RNA was incubated at 65°C for 5 min. After incubation first strand buffer, 10 mM DTT, and RNase OUT were added and incubated at room temperature for 10 minutes. Super Script III was subsequently added and incubated at 42°C for 50 minutes followed by inactivation at 70°C for 15 min. Mock reactions were preformed in parallel, without the addition of reverse transcriptase, to check for genomic contaminants. cDNA was then stored at −20°C until needed. Real time reactions were carried out in a final volume of 25 µL containing 2 µL of cDNA, Taqman mix (Applied Biosystems, Foster City, CA), and the primer pair. Primers were designed using Primer Express (Applied Biosystems, Foster City, CA). The probe was labeled using d-Fam as the quencher and d-Tam as the fluorophore. The PCR parameters were an initial cycle of 50°C for 2 minutes followed by 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Calculations for comparison between samples were performed using genomic standard curve analysis with rpoD being used as the standardization control. Results were analyzed for significance using the Student’s t test and a P value less than 0.005 was considered significant.
Microtiter Plate Assay of Bordetella Biofilm Formation
Biofilm development was examined using a microtiter dish assay as previously described [14]. Bordetella strains were inoculated at an OD600 of 0.05 into 100 µL of SS broth in a 96-well polyvinylchloride (PVC) microtiter plate. The plates were sealed with tape and then incubated at 37°C. At indicated time points the media and planktonic cells were removed and the plates were vigorously washed to remove loosely adherent cells. The remaining cells were stained with 0.1% crystal violet (CV) and incubated at room temperature for 30 minutes and then washed. CV staining the cells was solubilized with 200 µL of 95% ethanol. The solubilized CV was quantitated by transferring 125 µL to a new microtiter dish and determining the OD540. Results were analyzed for significance using the Student’s t test and a P value less than 0.05 was considered significant.
Confocal Scanning Laser Microscopy
Mid log phase (OD600∼0.7–1.0) grown cultures of B. bronchiseptica strains carrying the pTac-GFP construct were inoculated in two chambered coverslips and cultured in SS broth with chloramphenicol. 12 mm glass coverslips were partially submerged in the culture and allowed to incubate for 48 hours at 37°C under static conditions. Biofilms formed at the air liquid interface of the coverslips were gently rinsed with PBS to remove any unattached bacteria and mounted with ProLong gold antifade reagent (Invitrogen, Carlsbad, CA), followed by visualization using a Zeiss LSM 510 confocal scanning laser microscope.
Scanning Electron Microscopy
Bordetella strains were cultured statically on glass coverslips vertically submerged in SS broth, resulting in an air-liquid interface. At the indicated time points, the coverslips were removed, washed with sterile water and fixed using 2.5% glutaraldehyde for 1 hour. Samples were then processed for scanning electron microscopy as previously described [15], [18].
Results
B. bronchiseptica Exhibits a Coordinately Regulated Gene Expression Program During Biofilm Formation Similar to a Developmental Process
Previously, we have demonstrated that biofilm development in B. bronchiseptica is a microscopically observable sequential process that can be separated into distinct stages [14], [15]. Based on these data, we hypothesized that B. bronchiseptica undergoes a series of adaptive changes that allow it to successfully transition into sessile growth. A corollary of this hypothesis is that B. bronchiseptica cells within each biofilm stage exhibit gene expression patterns distinct from bacteria grown planktonically. To identify global stage-specific gene sets unique to biofilm formation for B. bronchiseptica, whole-genome transcriptome analysis was used to measure mRNA abundance under biofilm and planktonic growth conditions after 6, 12, 24, 36, and 48 hours of growth. A previously utilized static model of bacterial biofilm growth in petri dishes was used for these analyses [38], [44]. As shown in Figure S1, B. bronchiseptica cells adhered to the plastic surface were observed by SEM at 6 hours. At 12 hours, more cells adhered to the plastic surface; however these sessile cells lacked any apparent structural organization. By 24 hours, cell clusters separated by individual cells were apparent and by 48 hours cells were present mainly as mature macrocolonies encased in an opaque white matrix-like material (Figure S1). Therefore, this static biofilm system replicates the developmental stages of bacterial biofilms and allows transcriptome analysis during planktonic and biofilm growth under identical growth conditions.
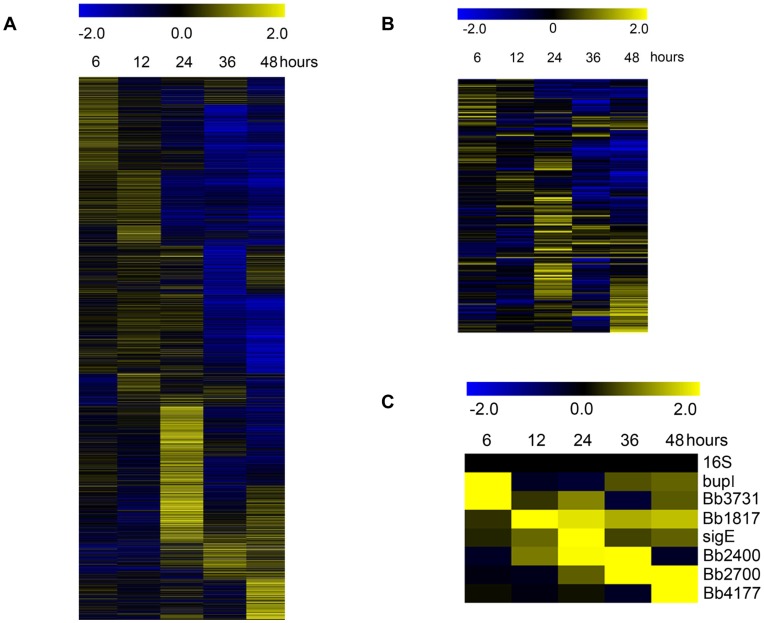
More than 33% of the B. bronchiseptica genes exhibited statistically significant transcriptional activation or repression during biofilm growth. A self-organizing map technique was applied to the biofilm regulated expression profiles to identify gene sets with similar expression patterns. As shown in Figure 1A, this clustering map reveals a periodicity or a cascade of continuous expression lacking clear boundaries or sharp transitions and demonstrates an orderly timing of gene expression extremely similar to global gene expression profiles occurring during bacterial development (Table S1) [41], [51], [52].
Figure 1. Hierarchical clustering of the transcriptional response of B. bronchiseptica strain RB50 throughout biofilm development identified by comparing cDNA from planktonic cells to biofilm cells at 6, 12, 24, 36, and 48 hours of growth.
A) Expression profiles representing global transcriptional changes B) Expression profiles of annotated B. bronchiseptica transcription factors. C) Expression profiles of annotated B. bronchiseptica transcription factors maximally expressed under biofilm growth conditions at 6, 12, 24, 36, and 48 hours. Data are mean centered for each array element and averaged from three biological replicates. All expression profiles of genes are in row and are represented using the color scale at top. Yellow, indicates increased expression in biofilm cells; blue, decreased gene expression in biofilm cells; black, no significant change in gene expression.
Gene expression occurring throughout bacterial development is governed by the production of transcription factors at the appropriate stages [51]. Therefore, the same self-organizing map technique was applied to the annotated B. bronchiseptica transcription factors revealing a parallel temporally regulated expression pattern as the global B. bronchiseptica gene expression profiles (Figure 1B and Table S1). To highlight this tight temporal expression pattern, transcription factors used in Figure 1B were trimmed to contain only those present in the greatest abundances at each point time (Figure 1C). Specifically, bupI Bb4742 and Bb3731, were found to be maximally expressed at 6 hours (Figure 1C). Bb1817 was found to be maximally expressed at 12 hours, while sigE Bb3752 was found to be maximally expressed at 24 hours (Figure 1C). Bb2400 was found to be maximally expressed at 24 hours and 36 hours, and Bb2700 was found to be maximally expressed at 36 hours and 48 hours (Figure 1C). Lastly, Bb4177 was found to be maximally expressed at 48 hours (Figure 1C). This rigid expression pattern suggests that specific transcription factors or regulators are needed at distinct stages or times during biofilm development. A complete list of gene expression measurements during biofilm growth can be found in Table S1. Overall, the transcriptional profiles for both global gene expression and transcription factor-specific gene expression suggest that B. bronchiseptica undergoes a coordinately regulated gene expression program similar to a bacterial developmental process.
Bvg−Phase Phenotype is Favored During Initial Stages of Biofilm Formation
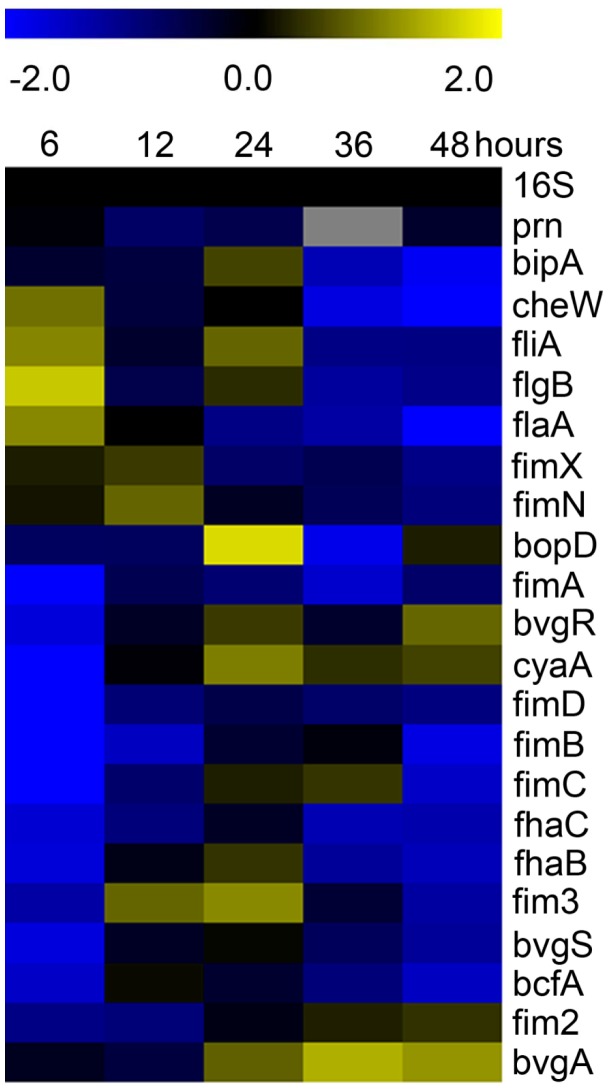
We and others have previously demonstrated that BvgAS is required for B. bronchiseptica biofilm development [13], [15]. However, gene expression occurring at distinct stages of biofilm development has not been investigated for many BvgAS-regulated factors. To address this, we applied a self-organizing map technique to genes encoding classical BvgAS-regulated factors, such as adhesins, toxins, and those contributing to flagella synthesis and motility (Figure 2). This analysis revealed that many of the positively regulated BvgAS genes were repressed at 6 hours, while many negatively regulated BvgAS genes were found to be induced. Specifically, genes involved in motility (cheW, fliA, flgB, flaA) were upregulated, while bvgR and the genes positively regulated by BvgAS (bopD, fimA, cyaA, fimD, fimB, fimC, fhaC, fhaB, fim3, bvgS, bcfA, and fim2) were repressed under biofilm growth conditions at 6 hours (Figure 2). Of the known Bvg-activated genes, only prn, bipA and bvgA were found to be expressed at similar levels under biofilm and planktonic growth conditions at 6 hours (Figure 2). The gene expression profiles of these classical BvgAS-regulated factors suggest that a Bvg− phase phenotype is favored during early or initial stages of biofilm formation.
Figure 2. Hierarchical clustering of the transcriptional response of BvgAS-regulated genes throughout biofilm development identified by comparing cDNA from planktonic cells to biofilm cells at 6, 12, 24, 36, and 48 hours of growth.
Expression profiles representing transcriptional changes of genes regulated by BvgAS, along with 16S. Data are mean centered for each array element and averaged from three biological replicates. All expression profiles of genes are in rows and are represented using the color scale at top with gray indicating missing data. Yellow, indicates increased expression in biofilm cells; blue, decreased gene expression in biofilm cells; black, no significant change in gene expression.
The BvgAS-repressed Gene flaA is Differentially Expressed During B. bronchiseptica Biofilm Development
Perhaps the best characterized phenotype of the Bvg− phase is motility. The expression of genes involved in the synthesis of flagella is tightly regulated through a complex hierarchy requiring the presence of the regulatory proteins, FrlA and FrlB, and the production of the flagellin monomer, FlaA. BvgA negatively regulates the production of flagella and subsequent motility in B. bronchiseptica by repressing the expression of frlA and frlB [6], [28], [29]. frlA and frlB encode activators of flagellar genes and have been hypothesized to regulate flaA expression through a fliA analogue in Bordetella [6].
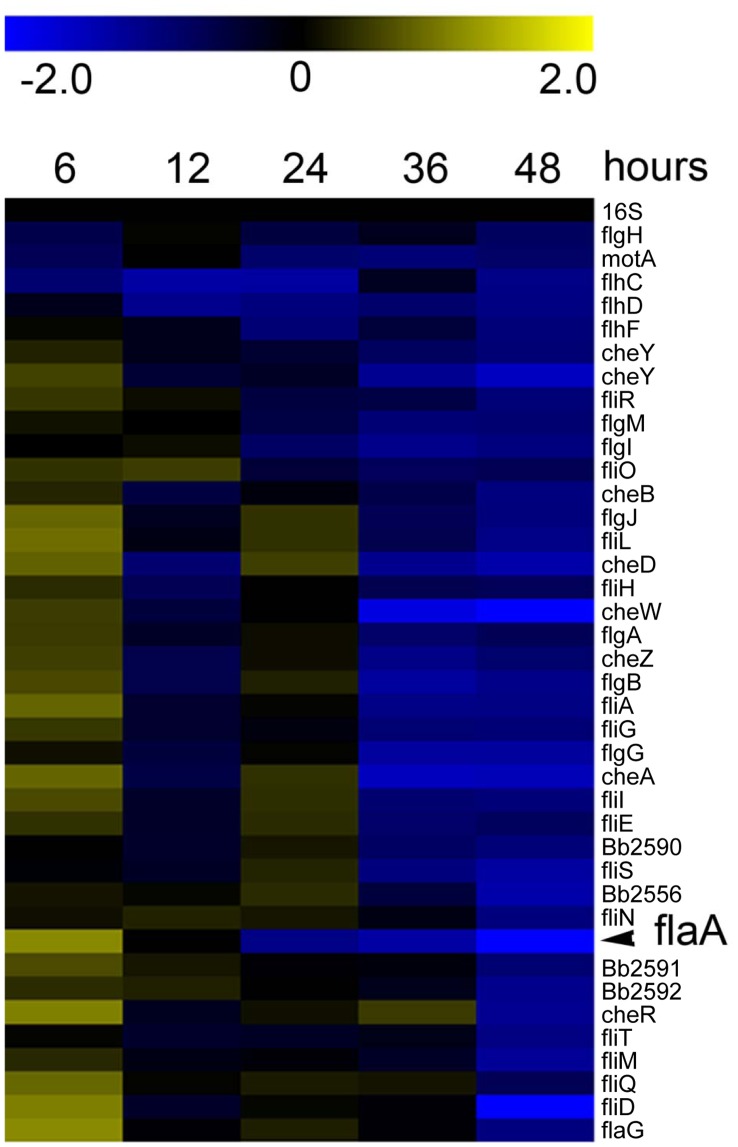
As shown in Figure 3A, we found that the majority of genes located within the motility locus exhibited transcriptional changes throughout B. bronchiseptica biofilm development. In general, these transcriptional changes were inversely related to later time points such that the expression of many of these genes were upregulated or increased early at 6 hours and then decreased late at 48 hours (Figure 3). The flagella structural gene flaA is marked as an example of this inverse or differential gene expression pattern (Figure 3). However, frlA and frlB were observed to be similarly expressed in both planktonic and biofilm conditions at 6 hours followed by a subsequent repression at 48 hours (Table S1).
Figure 3. Hierarchical clustering of the transcriptional response of genes located within the motility locus throughout biofilm development identified by comparing cDNA from planktonic cells to biofilm cells at 6, 12, 24, 36, and 48 hours of growth.
Expression profiles representing transcriptional changes of genes located within the motility locus, along with 16S. Data are mean centered for each array element and averaged from three biological replicates. All expression profiles of genes are in row and are represented using the color scale at top. Yellow, indicates increased expression in biofilm cells; blue, decreased gene expression in biofilm cells; black, no significant change in gene expression. The flagella structural gene flaA is indicated.
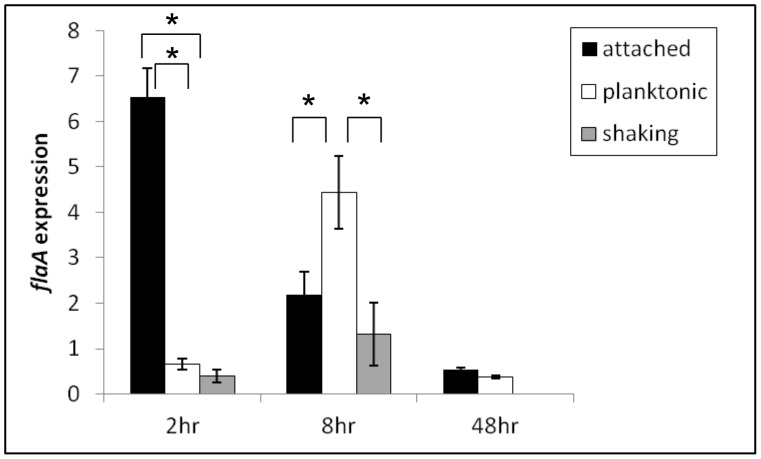
To independently verify the gene expression pattern of flaA, encoding the flagellin monomer, quantitative RT-PCR was performed using RNA isolated from cells adhered to polystyrene plates during early (2 and 8 hours) and late (48 hours) stages of biofilm development. We additionally included RNA isolated from cells grown in static planktonic and from cells grown in shaking culture conditions. flaA expression was highest in bacteria attached to the plate at 2 hours (Figure 4). At this time point, the expression of flaA in either the planktonic or the shaking cultures was comparatively lower (Figure 4). The expression of flaA is repressed in attached cells at 8 hours followed by very low expression at 48 hours (Figure 4). In contrast, flaA expression increases to reach maximum levels at 8 hours in cells grown in static planktonic and shaking culture conditions (Figure 4). Similar to that observed for the sessile cells, cells grown in static planktonic and shaking culture conditions for 48 hours, expressed flaA at comparatively lower levels (Figure 4). By 48 hours, there are viable bacteria in all the three populations as determined by plating on BG agar plates and by the observation that rpoD transcript levels are still detectable. These data confirm the results from the microarray analysis by revealing the differential expression pattern of flaA during B. bronchiseptica biofilm development. Overall, the pattern of flaA expression indicates that surface-adherent bacteria preferentially express flagella at early times and repress the expression at later stages of biofilm formation.
Figure 4. Differential expression of flaA during biofilm development.
Gene expression of the flagella structural gene flaA as determined by quantitative RT-PCR in attached, planktonic, or cells grown in shaking culture conditions at 2, 8, and 48 hours of growth. Gene expression standardized to rpoD is plotted along the y-axis and attached, planktonic, and shaking culture conditions are shown along the x-axis. Data shown are averages obtained from triplicate cultures. The error bars represent +/− the standard deviation. Asterisks represent a p value less than 0.005.
Flagella are Necessary for Initial Surface Contact but are not Necessary at the Later Stages of Biofilm Development
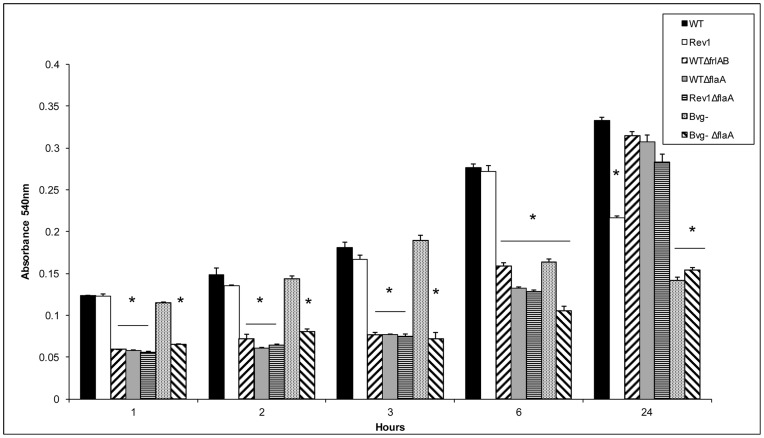
The upregulation of genes involved in motility and the flagellar synthesis, including the flagellin monomer flaA, at early time points of biofilm development suggested that flagella serve a critical role in Bordetella biofilm formation. We therefore hypothesized that flagella are necessary for establishing the initial interactions with the surface. To examine the role of flagella in initial surface contact, we utilized mutants containing deletions in either the frlAB or the flaA genes (Table 1). Biofilm formation was quantified by staining the cells attached to a polystyrene plate with crystal violet over a time course of 1, 2, 3 and 6 hours. Compared to the WT strain, which is capable of producing flagella, the strains containing deletions in either frlAB or flaA (WTΔfrlAB, WTΔflaA) and therefore lacking the ability to produce flagella were defective in forming biofilms for all the early time points examined (Figure 5). These data thus demonstrate that flagella are necessary for establishing initial surface interactions by the WT strain.
Figure 5. Flagella are necessary for initial surface contact but deleterious for biofilm maturation.
Kinetics of biofilm development for the different strains was analyzed by the microtiter assay. Optical densities (OD540) of solubilized crystal violet from surface associated cells are plotted along the y-axis and time in hours, is shown along the x-axis. Data shown are averages obtained from at least 6 wells each time from 2 independent experiments. The error bars represent +/− the standard error. Asterisks represent a p value less than 0.05.
In addition to examining biofilm formation at early time points, we also analyzed the biofilms formed by these strains at 24 hours. Strains containing deletions in either frlAB or flaA genes (WTΔfrlAB, WTΔflaA) were able to form biofilms as robust as the WT by 24 hours, suggesting that the presence of flagella is not absolutely required for continued biofilm formation at later time (Figure 5).
Previously, we have shown that while the BvgAS signal transduction system is required for efficient biofilm formation in Bordetella, it is not essential for initial surface interactions [15]. Specifically, we showed that there were no significant differences in the adherence of the WT and the Bvg− phase-locked strains to the microtiter plates at the early time points (Figure 5 and [15]). We also found that the Bvg+ and the Bvgi phase locked strains were also defective in biofilm formation at early time points compared to the WT strain [15]. The Bvg+ phase-locked strain encodes a constitutively active BvgS protein that is insensitive to modulators, whereas the Bvgi phase-locked strain is locked in the intermediate phase [32]. Both the Bvg+ and the Bvgi strains are non-motile and do not produce flagella, whereas the Bvg− phase locked strain is motile and produces flagella. We hypothesized that flagella are responsible for the observed early biofilm forming ability of the Bvg− phase-locked strain. Compared to the Bvg− phase-locked strain, the Bvg− ΔflaA strain formed less biofilms at 1, 2, 3 and 6 hours (Figure 5). These results suggest that flagella can contribute to the initial biofilm formation by B. bronchiseptica in the absence of Bvg-activated factors. At 6 and 24 hours, compared to the WT strain, the Bvg− phase locked strain was defective in biofilm formation (Figure 5). This suggests that expression of flagella in the absence of Bvg-activated factors is not sufficient for continued formation of efficient biofilms.
Repression of Flagella is Essential for Biofilm Formation
Based on the inverse regulation of flagellar and motility gene expression during biofilm development, we hypothesized that while initial expression of flagella is necessary for enhancing initial surface contact, continued expression of flagella in the WT strain would be deleterious for biofilm development. To begin addressing the requirement for repression of flagella during later stages of biofilm development, we utilized a strain (Rev1) that ectopically expresses flagella in the wild-type background. Rev1 was previously generated by replacing the Bvg-repressed frlAB promoter with the Bvg-activated fhaB promoter [6].
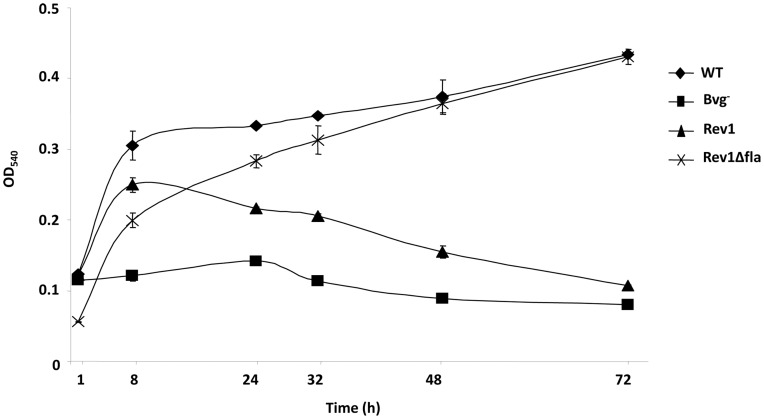
To quantify biofilm formation, cells attached to the polystyrene plate were stained with crystal violet over a time course of 1, 2, 3, 6, 8, 24, 32, 48, and 72 hours. Consistent with the requirement of flagella at early stages of biofilm formation, the biofilms formed by the Rev1 strain was similar to that of the WT strain for up to 6 hours (Figures 5 and 6). At later time points, e.g. at 8 hours or later, the Rev1 strain formed decreased biofilms relative to the WT strain until reaching that of the Bvg− phase-locked strain by 72 hours (Figure 6).
Figure 6. Flagella are deleterious for biofilm maturation.
Kinetics of biofilm development for different strains was analyzed by the microtiter assay. Optical densities (OD540) are plotted along the y-axis and time in hours, is shown along the x-axis. Data shown are averages obtained from at least 6 wells each time from 2 independent experiments. The error bars represent +/− the standard error. All differences are statistically significant (p<0.05, student’s t test), except among the strains at 1 h time point and between the strains Rev1Δfla and WT at 48 and 72 hours.
In the strain Rev1, the frlAB locus is ectopically expressed from the fhaB promoter. Since frlAB encodes transcriptional regulatory proteins, activation of this locus may lead to the expression of yet unidentified frlAB-regulated motility-independent genes, which may influence biofilm development. In order to address this caveat and to demonstrate that the ectopic expression of flagella is sufficient for the biofilm defect associated with the Rev1 strain, the Rev1 strain containing a deletion of the flaA gene (Rev1ΔflaA) (Table 1) was utilized. At early stages of biofilm formation, up to 8 hours, the adherence capacity of the Rev1Δfla strain, lacking the ability to produce flagella, was lower than that of the WT strain and the Rev1 strain (Figures 5 and 6). However, after 8 hours, the Rev1Δfla strain started to display increased biofilm formation compared to that formed by the Rev1 strain. At 48 and 72 hours, both the WT and the Rev1ΔflaA strain formed biofilms to similar levels (Figure 6). Taken together, these data demonstrate that ectopic expression of flagella is detrimental for continued biofilm formation at later time points.
Microscopic Analysis of Biofilms Formed by the WT and Different Isogenic Mutants
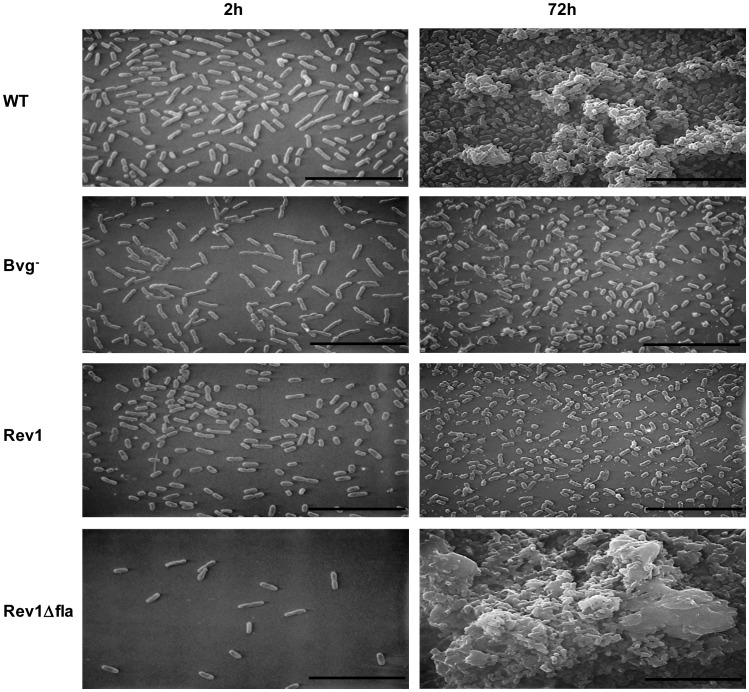
To further define the role that flagella play in B. bronchiseptica biofilm development, we visualized architectural features of biofilms by scanning electron microscopy (SEM). Glass coverslips were vertically submerged in a bacterial suspension creating an air-liquid interface that was visualized using SEM. After 2 hours of incubation, only strains capable of producing flagella, WT, Bvg−, and Rev1 were found to adhere in significant numbers to the surface (Figure 7). Comparatively, the Rev1ΔflaA strain lacking the ability to produce flagella adhered sporadically to the surface after 2 hours with a substantial decrease in the number of cells adhering relative to the WT strain and the isogenic parent, Rev1 (Figure 7). At later stages of biofilm development i.e. at 72 hours, strains incapable of flagella repression, Rev1 and Bvg−, were unable to cluster together and form three dimensional structures. In contrast, the WT and the Rev1Δfla strains formed three dimensional structures encased in a matrix like material (Figure 7). These data demonstrate that flagella are needed to initiate contact with a surface. However, this defect can be compensated at later time points in strains that can repress or lack flagella production. The similar biofilm forming ability of the Rev1 and the WT strains further demonstrates that the potential frlAB-regulated motility-independent genes are not responsible for the biofilm defect exhibited by the Rev1 strain. Thus, the inability of the Rev1 strain to form mature biofilms is due to the failure to repress flagella production during later stages of biofilm formation.
Figure 7. Scanning electron microscopy (SEM) of biofilms formed at the air-liquid interface.
Different strains were grown on glass cover slips for 2 hours (left panels) and 72 hours (right panels) followed by processing for SEM as described in the Materials and Methods. The scale bar represents 10 µm.
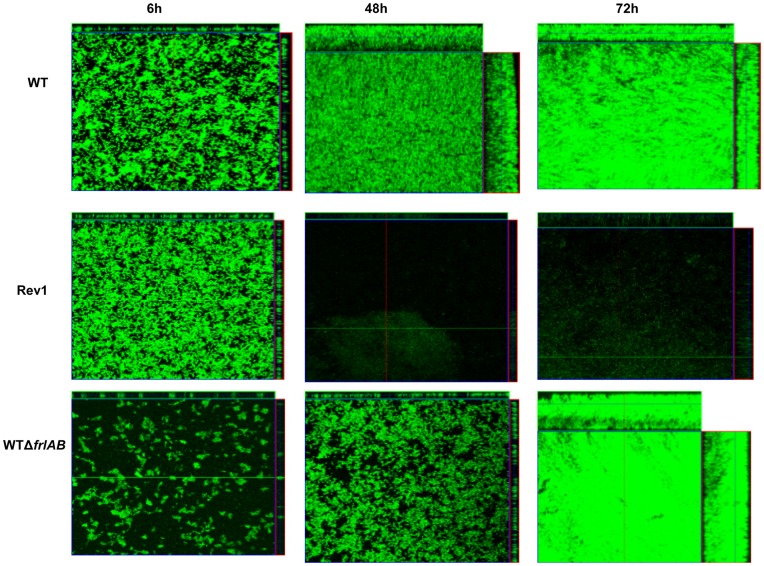
To further examine the contribution of flagella on impacting biofilm structure, we continued this experiment with GFP-expressing derivatives of the WT and the mutant strains. After 6 hours of incubation, the WT strain attached and covered a large area of the coverslip, which appeared to be completely occupied by 48 hours (Figure 8). By 72 hours, the entire coverslip had been extensively covered by the WT strain, resulting in the visualization of a thick layer of cells at the air-liquid interface (Figure 8). In contrast, the WTΔfrlAB strain was defective in surface-adherence at 6 hours as demonstrated with large areas of the coverslips remaining devoid of the bacterial cells. At 72 hours, the WTΔfrlAB strain had recovered from the initial defect in surface attachment and colonized the glass cover slip similar to the WT strain (Figure 8). Consistent with above results, while the Rev1 strain did not appear to be defective in attaching to the coverslip at 6 hours, the majority of the coverslip was essentially devoid of bacterial cells at 48 and 72 hours (Figure 8). These data confirm that the inability to repress flagella is detrimental for continued biofilm formation at later time points.
Figure 8. Confocal scanning laser micrographs (CSLM) of B. bronchiseptica biofilm development.
CSLM of biofilms formed at the air-liquid interface of different strains grown on glass coverslips after 6 hours (left panels), 48 hours (middle panels), and 72 hours (right panels). The cells were tagged with GFP and thus are green. For each micrograph, the middle panel represents the x–y plane, and the adjacent top and side panels represent the x–z and y–z planes, respectively.
Discussion
Biofilm formation has been proposed to be a process of microbial development similar to that observed in cell cycle-controlled swarmer-to-stalk cell transition in Caulobacter crescentus, sporulation in Bacillus subtilis, and fruiting-body formation by Myxococcus xanthus [41], [51]. This view is gaining acceptance as an increasing number of studies reveal alterations in bacterial cell physiology, along with the transcriptional program underlying these physiologies, which occur throughout the progression from planktonic to the biofilm state [36], [37], [44], [52], [53]. Despite the almost widespread acceptance of this model, it has been heavily criticized due to the lack of experimental evidence directly linking a stage-specific biofilm developmental phenotype with a temporal genetic program or mechanism. Recently, the existence of a regulatory program for stage-specific biofilm development in Pseudomonas aeruginosa has been demonstrated by Petrova and Sauer [52]. We have used microarray analysis to study the transcriptome of B. bronchiseptica over the course of five time points representing distinct stages of biofilm development (Figure S1 and [15]). Our data revealed prominent shifts in the global transcriptional program that occur at distinct stages of biofilm development. Annotated B. bronchiseptica transcription factors were additionally found to have a similar temporally regulated expression pattern as the global B. bronchiseptica gene expression profiles. This temporally regulated expression pattern suggests that specific transcription factors are needed at distinct stages or times during biofilm development providing experimental evidence linking stage-specific biofilm phenotypes to a sequential genetic mechanism. Overall, both the transcription factor and global gene expression profiles suggest that B. bronchiseptica undergoes a coordinately regulated gene expression program during biofilm development similar to a bacterial developmental process.
Global transcriptome analysis led to the surprising discovery that many of the Bordetella genes previously known to be positively regulated by the BvgAS signal transduction system were repressed during early stages of biofilm development, while many negatively regulated BvgAS genes were found to be induced. This finding was unexpected since biofilm formation and maturation requires BvgAS activation and well documented Bvg+ phase conditions, such as growth at 37°C and the absence of chemical modulators [15], [17], [18]. The observed gene expression pattern further suggested that a Bvg− phase phenotype is favored during initial stages of biofilm formation. Microarray analysis additionally led to the discovery that expression of flagella occurs and is under tight regulatory control during B. bronchiseptica biofilm development such that the expression of many of the genes located within the motility locus were either upregulated early at 6 hours, including the flagellin monomer, flaA, or similarly expressed in both planktonic and biofilm conditions at 6 hours, such as frlA and frlB, followed by a subsequent repression late at 48 hours.
Using mutational analysis in a combination of several different assays, we confirmed the microarray data and demonstrated that B. bronchiseptica expresses flagella early during biofilm development and then subsequently represses the expression of flagella. B. bronchiseptica strains unable to produce flagella failed to bind to a surface at early stages of biofilm formation. However, this defect is ameliorated with time since strains that lack flagella production were able to form biofilms as robust as the WT strain by late biofilm stages. Together, the presented data demonstrate that flagella are needed to contact a surface; however, they are not required for biofilm maturation.
The expression of B. bronchiseptica flagellar synthesis genes are thought to be regulated through a complex hierarchy similar to the cascade of transcriptional events that occur in E. coli [54]; however there are many details of this regulatory hierarchy that remain undetermined. To summarize our current understanding, BvgAS lies at the top of the hierarch and negatively regulates the expression of frlA and frlB [6], [29]. FrlAB is required for the production of flagella and has been hypothesized to regulate fliA, which in turn upregulates flaA, encoding the flagellin monomer, required for the production of flagella [6], [28]. In other microorganisms, genes required for the synthesis of flagella have been found to be repressed in the biofilm cells leading to suggestions that flagella repression is a key step in biofilm development [35], [55], [56]. However, to our knowledge, the requirement to repress flagella production during biofilm development has not been directly demonstrated. Using a series of mutants that allowed us to alter the known mechanisms within the regulatory hierarchy for B. bronchiseptica flagella production, we demonstrated that flagella repression is absolutely required for formation of mature and structured biofilms.
Data presented in this study were obtained from a variety of in vitro model systems. As with all in vitro model systems, possible artifacts may include high starting bacterial density in nutrient rich broth, oxygen, and nutrient availability. These culture conditions can initially begin with a fully aerated culture subsequently developing an oxygen gradient from the surface to the bottom of the culture. Additionally, gradients within biofilms can often form and result in localized microenvironments. All of these possible environmental parameters are likely to affect gene expression. Also, bacterial products that are either required for or enhance adherence to abiotic surfaces may have a different contributing role in the development and maturation of biofilms formed in the environment or within a host.
In conclusion, it is clear from these studies that biofilm development in Bordetella is under the control of a complex regulatory circuitry requiring the coordinated stage-specific expression of multiple transcription factors. One outcome of this regulatory hierarchy is the transient expression of a Bvg− phase phenotype under Bvg+ phase conditions. B. bronchiseptica is able to survive and grow in soil for extended periods, in a nutrient-limiting environment and at temperatures as low as 10°C [57]. The Bvg− phase of B. bronchiseptica has been demonstrated to promote bacterial survival under conditions of nutrient deprivation [31], [57]. Thus, the expression of Bvg-associated traits such as flagella could lead to efficient biofilm formation in the environment and thus contribute to survival in environmental niches. There may also be a possible infectious benefit of the expression of flagella as well. It has previously been demonstrated that the Rev1 mutant colonized the nasal cavities of rats similarly to wild-type B. bronchiseptica strain, demonstrating that expression of flagella is not deleterious to nasal colonization [6]. The lower temperature (lower than 37°C) of the anterior portions of the upper respiratory tract may also allow expression of flagella by the WT strain during initial stages of infection. Additionally, purified flagella from B. bronchiseptica have been shown to adhere to epithelial cells [58]. Therefore, by initiating biofilm development during early infection, possibly through adherence, flagella may also promote survival within nasal cavities of hosts, thereby increasing transmission dynamics within host populations or between animals.
Supporting Information
Scanning Electron Microscopy (SEM) of biofilms formed on polystyrene Petri plates. Logarithmic phase cultures inoculated into the Petri plate and grown at 37°C for the indicated time followed by processing for SEM as described in the Materials and Methods. The scale bar represents 10 µm.
(TIF)
Sheet 1. Whole Genome Microarray data: Fold-Change expression values during biofilm growth in B. bronchiseptica RB50. DNA microarray analysis was used to measure mRNA levels present in B. bronchiseptica RB50 planktonic cells compared to biofilm cells at 6, 12, 24, 36, and 48 hours of growth. Differences in mRNA levels are listed as mean fold-changes ± standard deviation. Fold-changes were calculated by averaging the data from three biological replicates for 6, 12, and 24 hour time points and two biological replicates for 36 and 48 hour time points. The fluorescent labels were exchanged in dye-swap experiments performed on all biological replicates. Genes identified as significantly regulated by growth phase (“Negative” or “Positive”) were assessed by using the significant analysis of microarrays (SAM) program [49]. A one-class unpaired SAM analysis using a false discovery rate of 0.001% was preformed and the Score(d), q-value, and localdr(%) columns are given. Sheet 2. Trx regulator microarray data: Fold-Change expression values of transcriptional regulators in B. bronchiseptica RB50 during biofilm growth. DNA microarray analysis was used to measure mRNA levels present in B. bronchiseptica RB50 planktonic cells compared to biofilm cells at 6, 12, 24, 36, and 48 hours of growth. Differences in mRNA levels are listed as mean fold-changes ± standard deviation. Fold-changes were calculated by averaging the data from three biological replicates for 6, 12, and 24 hour time points and two biological replicates for 36 and 48 hour time points. The fluorescent labels were exchanged in dye-swap experiments performed on all biological replicates. Genes identified as significantly regulated by growth phase (“Negative” or “Positive”) were assessed by using the significant analysis of microarrays (SAM) program [49]. A one-class unpaired SAM analysis using a false discovery rate of 0.001% was preformed and the Score(d), q-value, and localdr(%) columns are given.
(XLSX)
Acknowledgments
We thank Dr. Jeff F. Miller for a generous gift of the strains. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Funding Statement
Research in the laboratory of R.D. is supported National Institutes of Health (NIH) grant 1R01AI075081. M.C. was supported by an NIH predoctoral training grant (T32 AI07401). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bemis DA (1992) Bordetella and Mycoplasma respiratory infections in dogs and cats. Vet Clin North Am Small Anim Pract 22: 1173–1186. [DOI] [PubMed] [Google Scholar]
- 2.Guerrero RJ (1990) Respiratory disease: An important global problem in the swine industry. Lausanne, Switzerland.
- 3.United States DoA (2008) Swine 2006, Part III: reference of swine health and health management in the United States. Fort Collins.
- 4. Rath BA, Register KB, Wall J, Sokol DM, Van Dyke RB (2008) Persistent Bordetella bronchiseptica pneumonia in an immunocompetent infant and genetic comparison of clinical isolates with kennel cough vaccine strains. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 46: 905–908. [DOI] [PubMed] [Google Scholar]
- 5.Register KB, Sukumar N, Palavecino EL, Rubin BK, Deora R (2012) Bordetella bronchiseptica in a Paediatric Cystic Fibrosis Patient: Possible Transmission from a Household Cat. Zoonoses and public health. [DOI] [PMC free article] [PubMed]
- 6. Akerley BJ, Cotter PA, Miller JF (1995) Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80: 611–620. [DOI] [PubMed] [Google Scholar]
- 7. Kirimanjeswara GS, Mann PB, Harvill ET (2003) Role of antibodies in immunity to Bordetella infections. Infect Immun 71: 1719–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giles CJ, Smith IM, Baskerville AJ, Brothwell E (1980) Clinical bacteriological and epidemiological observations on infectious atrophic rhinitis of pigs in southern England. The Veterinary record 106: 25–28. [DOI] [PubMed] [Google Scholar]
- 9. Rutter JM (1981) Quantitative observations on Bordetella bronchiseptica infection in atrophic rhinitis of pigs. The Veterinary record 108: 451–454. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson TL, Brockmeier SL, Loving CL (2009) Contribution of Bordetella bronchiseptica Filamentous Hemagglutinin and Pertactin to Respiratory Disease in Swine. Infect Immun. [DOI] [PMC free article] [PubMed]
- 11. Harvill ET, Cotter PA, Miller JF (1999) Pregenomic comparative analysis between Bordetella bronchiseptica RB50 and Bordetella pertussis Tohama I in murine models of respiratory tract infection. Infection and immunity 67: 6109–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conover MS, Mishra M, Deora R (2011) Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PloS one 6: e16861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Irie Y, Mattoo S, Yuk MH (2004) The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica . J Bacteriol 186: 5692–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parise G, Mishra M, Itoh Y, Romeo T, Deora R (2007) Role of a putative polysaccharide locus in Bordetella biofilm development. J Bacteriol 189: 750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mishra M, Parise G, Jackson KD, Wozniak DJ, Deora R (2005) The BvgAS signal transduction system regulates biofilm development in Bordetella . J Bacteriol 187: 1474–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Serra D, Bosch A, Russo DM, Rodriguez ME, Zorreguieta A, et al. (2007) Continuous nondestructive monitoring of Bordetella pertussis biofilms by Fourier transform infrared spectroscopy and other corroborative techniques. Anal Bioanal Chem 387: 1759–1767. [DOI] [PubMed] [Google Scholar]
- 17. Sloan GP, Love CF, Sukumar N, Mishra M, Deora R (2007) The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J Bacteriol 189: 8270–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Conover MS, Sloan GP, Love CF, Sukumar N, Deora R (2010) The Bps polysaccharide of Bordetella pertussis promotes colonization and biofilm formation in the nose by functioning as an adhesin. Mol Microbiol 77: 1439–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Serra DO, Conover MS, Arnal L, Sloan GP, Rodriguez ME, et al. (2011) FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One 6: e28811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, et al. (2005) Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun 73: 3210–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, et al. (2003) Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol 171: 4329–4339. [DOI] [PubMed] [Google Scholar]
- 22. Mah TF, O'Toole GA (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9: 34–39. [DOI] [PubMed] [Google Scholar]
- 23. Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, et al. (2004) A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem 279: 54881–54886. [DOI] [PubMed] [Google Scholar]
- 24. Anderson GG, O'Toole GA (2008) Innate and induced resistance mechanisms of bacterial biofilms. Current topics in microbiology and immunology 322: 85–105. [DOI] [PubMed] [Google Scholar]
- 25. Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15: 167–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284: 1318–1322. [DOI] [PubMed] [Google Scholar]
- 27. Cotter PA, Jones AM (2003) Phosphorelay control of virulence gene expression in Bordetella . Trends Microbiol 11: 367–373. [DOI] [PubMed] [Google Scholar]
- 28. Akerley BJ, Miller JF (1993) Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J Bacteriol 175: 3468–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akerley BJ, Monack DM, Falkow S, Miller JF (1992) The bvgAS locus negatively controls motility and synthesis of flagella in Bordetella bronchiseptica . J Bacteriol 174: 980–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McMillan DJ, Shojaei M, Chhatwal GS, Guzman CA, Walker MJ (1996) Molecular analysis of the bvg-repressed urease of Bordetella bronchiseptica . Microb Pathog 21: 379–394. [DOI] [PubMed] [Google Scholar]
- 31. Cotter PA, Miller JF (1994) BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun 62: 3381–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cotter PA, Miller JF (1997) A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol Microbiol 24: 671–685. [DOI] [PubMed] [Google Scholar]
- 33. Martinez de Tejada G, Cotter PA, Heininger U, Camilli A, Akerley BJ, et al. (1998) Neither the Bvg- phase nor the vrg6 locus of Bordetella pertussis is required for respiratory infection in mice. Infect Immun 66: 2762–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Merkel TJ, Stibitz S, Keith JM, Leef M, Shahin R (1998) Contribution of regulation by the bvg locus to respiratory infection of mice by Bordetella pertussis . Infect Immun 66: 4367–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moorthy S, Watnick PI (2004) Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol Microbiol 52: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, et al. (2000) Development and dynamics of Pseudomonas sp. biofilms. J Bacteriol 182: 6482–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG (2002) Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184: 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Conover MS, Redfern CJ, Ganguly T, Sukumar N, Sloan G, et al. (2012) BpsR modulates Bordetella biofilm formation by negatively regulating the expression of the Bps polysaccharide. Journal of bacteriology 194: 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56: 187–209. [DOI] [PubMed] [Google Scholar]
- 40.Petrova OE, Sauer K (2012) Sticky situations - Key components that control bacterial surface attachment. Journal of bacteriology. [DOI] [PMC free article] [PubMed]
- 41. Monds RD, O'Toole GA (2009) The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends in microbiology 17: 73–87. [DOI] [PubMed] [Google Scholar]
- 42. Stainer DW, Scholte MJ (1970) A simple chemically defined medium for the production of phase I Bordetella pertussis . J Gen Microbiol 63: 211–220. [DOI] [PubMed] [Google Scholar]
- 43. Deora R (2002) Differential regulation of the Bordetella bipA gene: distinct roles for different BvgA binding sites. Journal of bacteriology 184: 6942–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moorthy S, Watnick PI (2005) Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol Microbiol 57: 1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buboltz AM, Nicholson TL, Parette MR, Hester SE, Parkhill J, et al. (2008) Replacement of adenylate cyclase toxin in a lineage of Bordetella bronchiseptica . J Bacteriol 190: 5502–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buboltz AM, Nicholson TL, Weyrich LS, Harvill ET (2009) Role of the type III secretion system in a hypervirulent lineage of Bordetella bronchiseptica . Infect Immun 77: 3969–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nicholson TL, Buboltz AM, Harvill ET, Brockmeier SL (2009) Microarray and functional analysis of growth phase-dependent gene regulation in Bordetella bronchiseptica . Infection and immunity 77: 4221–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nicholson TL (2007) Construction and validation of a first-generation Bordetella bronchiseptica long-oligonulceotide microarray by transcriptional profiling the Bvg regulon. BMC Genomics 8: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saeed AI, Sharov V, White J, Li J, Liang W, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378. [DOI] [PubMed] [Google Scholar]
- 51. Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148. [DOI] [PubMed] [Google Scholar]
- 52. Petrova OE, Sauer K (2009) A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS pathogens 5: e1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hickman JW, Harwood CS (2008) Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Molecular microbiology 69: 376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macnab RM (1996) Flagella and Motility. In: Neidhardt FC, III RC, Ingraham JL, Lin ECC, Low KB et al. , editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press. 123–145.
- 55. O'Toole GA, Kolter R (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30: 295–304. [DOI] [PubMed] [Google Scholar]
- 56. Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, et al. (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48: 1511–1524. [DOI] [PubMed] [Google Scholar]
- 57. Porter JF, Wardlaw AC (1993) Long-term survival of Bordetella bronchiseptica in lakewater and in buffered saline without added nutrients. FEMS microbiology letters 110: 33–36. [DOI] [PubMed] [Google Scholar]
- 58. Savelkoul PH, de Kerf DP, Willems RJ, Mooi FR, van der Zeijst BA, et al. (1996) Characterization of the fim2 and fim3 fimbrial subunit genes of Bordetella bronchiseptica: roles of Fim2 and Fim3 fimbriae and flagella in adhesion. Infection and immunity 64: 5098–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cotter PA, Yuk MH, Mattoo S, Akerley BJ, Boschwitz J, et al. (1998) Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infection and immunity 66: 5921–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Amann E, Brosius J (1985) “ATG vectors’ for regulated high-level expression of cloned genes in Escherichia coli. Gene. 40: 183–190. [DOI] [PubMed] [Google Scholar]
- 61. Ouahrani-Bettache S, Porte F, Teyssier J, Liautard JP, Kohler S (1999) pBBR1-GFP: a broad-host-range vector for prokaryotic promoter studies. BioTechniques 26: 620–622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scanning Electron Microscopy (SEM) of biofilms formed on polystyrene Petri plates. Logarithmic phase cultures inoculated into the Petri plate and grown at 37°C for the indicated time followed by processing for SEM as described in the Materials and Methods. The scale bar represents 10 µm.
(TIF)
Sheet 1. Whole Genome Microarray data: Fold-Change expression values during biofilm growth in B. bronchiseptica RB50. DNA microarray analysis was used to measure mRNA levels present in B. bronchiseptica RB50 planktonic cells compared to biofilm cells at 6, 12, 24, 36, and 48 hours of growth. Differences in mRNA levels are listed as mean fold-changes ± standard deviation. Fold-changes were calculated by averaging the data from three biological replicates for 6, 12, and 24 hour time points and two biological replicates for 36 and 48 hour time points. The fluorescent labels were exchanged in dye-swap experiments performed on all biological replicates. Genes identified as significantly regulated by growth phase (“Negative” or “Positive”) were assessed by using the significant analysis of microarrays (SAM) program [49]. A one-class unpaired SAM analysis using a false discovery rate of 0.001% was preformed and the Score(d), q-value, and localdr(%) columns are given. Sheet 2. Trx regulator microarray data: Fold-Change expression values of transcriptional regulators in B. bronchiseptica RB50 during biofilm growth. DNA microarray analysis was used to measure mRNA levels present in B. bronchiseptica RB50 planktonic cells compared to biofilm cells at 6, 12, 24, 36, and 48 hours of growth. Differences in mRNA levels are listed as mean fold-changes ± standard deviation. Fold-changes were calculated by averaging the data from three biological replicates for 6, 12, and 24 hour time points and two biological replicates for 36 and 48 hour time points. The fluorescent labels were exchanged in dye-swap experiments performed on all biological replicates. Genes identified as significantly regulated by growth phase (“Negative” or “Positive”) were assessed by using the significant analysis of microarrays (SAM) program [49]. A one-class unpaired SAM analysis using a false discovery rate of 0.001% was preformed and the Score(d), q-value, and localdr(%) columns are given.
(XLSX)