Abstract
Cilia are found on most human cells and exist as motile cilia or non-motile primary cilia. Primary cilia play sensory roles in transducing various extracellular signals, and defective ciliary functions are involved in a wide range of human diseases. Centrosomes are the principal microtubule-organizing centers of animal cells and contain two centrioles. We observed that DNA damage causes centriole splitting in non-transformed human cells, with isolated centrioles carrying the mother centriole markers CEP170 and ninein but not kizuna or cenexin. Loss of centriole cohesion through siRNA depletion of C-NAP1 or rootletin increased radiation-induced centriole splitting, with C-NAP1-depleted isolated centrioles losing mother markers. As the mother centriole forms the basal body in primary cilia, we tested whether centriole splitting affected ciliogenesis. While irradiated cells formed apparently normal primary cilia, most cilia arose from centriolar clusters, not from isolated centrioles. Furthermore, C-NAP1 or rootletin knockdown reduced primary cilium formation. Therefore, the centriole cohesion apparatus at the proximal end of centrioles may provide a target that can affect primary cilium formation as part of the DNA damage response.
Keywords: centrosome, DNA damage response, kizuna, C-NAP1, NEK2
Introduction
The structures that form the mitotic spindle poles in animal somatic cells, the centrosomes, consist of two cylindrical microtubule assemblies, the centrioles, arranged at right angles to one another and embedded in the pericentriolar material (PCM) (reviewed in ref. 1). Centrioles lose their orthogonal arrangement when they disengage late in mitosis prior to their duplication,2 which occurs predominantly in S phase. Once they disengage, each acts as a “mother,” nucleating a procentriole to serve as the foundation of a daughter centriole. The daughter centrioles lengthen and mature during G2 phase.3 Importantly, the mother centrioles remain tethered to one another until late G2 phase (reviewed in refs. 4 and 5).
Centrosome abnormalities and amplification are frequently observed in tumor cells, with aneuploidy and chromosomal instability being closely correlated with excess centrosomes.6 One cause of centrosome amplification is DNA damage.7 Centrioles normally disengage prior to duplication in a process that is controlled by separase and Plk1,8 but premature centriole splitting/ disengagement has been observed after ionizing radiation (IR) in various human cell types and after incomplete DNA replication in rodent cells.9,10 The mechanism that allows centriole disengagement and overduplication after DNA damage is not yet clear, although one target of the normal centriole disengagement process is the cohesin component, Scc1.11
During the normal cell cycle, centrosome separation in late G2 involves the regulated removal of the physical linkage that tethers the duplicated centrosomes together.3,12,13 Maintaining this centrosome cohesion is rootletin, which interacts with the large coiled-coil protein, C-NAP1, to connect parental centrioles.14,15 C-NAP1 phosphorylation by NEK2 kinase leads to its dissociation from centrioles, the loss of rootletin and centrosome separation.15-18 Furthermore, RNAi depletion of either C-NAP1 or rootletin causes centrosomes to separate.14 Another target of NEK2 is β-catenin, a mitotically stabilized form of which mediates centrosome separation.19 NEK2 overexpression or protein phosphatase 1α (PP1α) inhibition drives centrosome splitting,18,20 while ATM-mediated activation of PP1α to oppose NEK2 activity inhibits centrosome separation after by IR-induced DNA damage.21
Serving another role at the plasma membrane, the mother centriole can serve as the basal body from which a primary cilium forms (reviewed in refs. 22 and 23). Cilia are found on most human cells and exist as motile cilia or non-motile primary cilia. Primary cilia play a sensory role and regulate cell signaling through the Hedgehog and Wnt pathways.24,25 Defects in ciliary functions are involved in a range of human diseases, from polycystic kidney disease to cancer (reviewed in ref. 26). The distal ends of mature centrioles carry subdistal and distal appendages, which anchor cytoplasmic microtubules and contribute to primary cilium formation.12,13,27,28 Furthermore, the PCM associated with the mother centriole differs from that associated with the daughter centriole and is more effective at the anchoring of radial microtubule arrays, so that after centriole disengagement, the daughter is highly mobile in the cytoplasm until S phase.2 The PLK1 substrate, kizuna (kiz), stabilizes the PCM for its mitotic functions and associates mainly with the mother centriole.29
Here, we explore whether proteins involved in centrosome tethering are also involved in DNA damage responses of the centrosome. We found that IR-induced centriole splitting is restrained by the centrosome cohesion apparatus. Given the involvement of centrioles in primary ciliogenesis, we also tested how DNA damage might affect this process. We found that the centrosome cohesion proteins C-NAP1 and rootletin positively regulate ciliogenesis, so that DNA damage-induced centriole splitting may contribute to the DNA damage response through the primary cilium.
Results
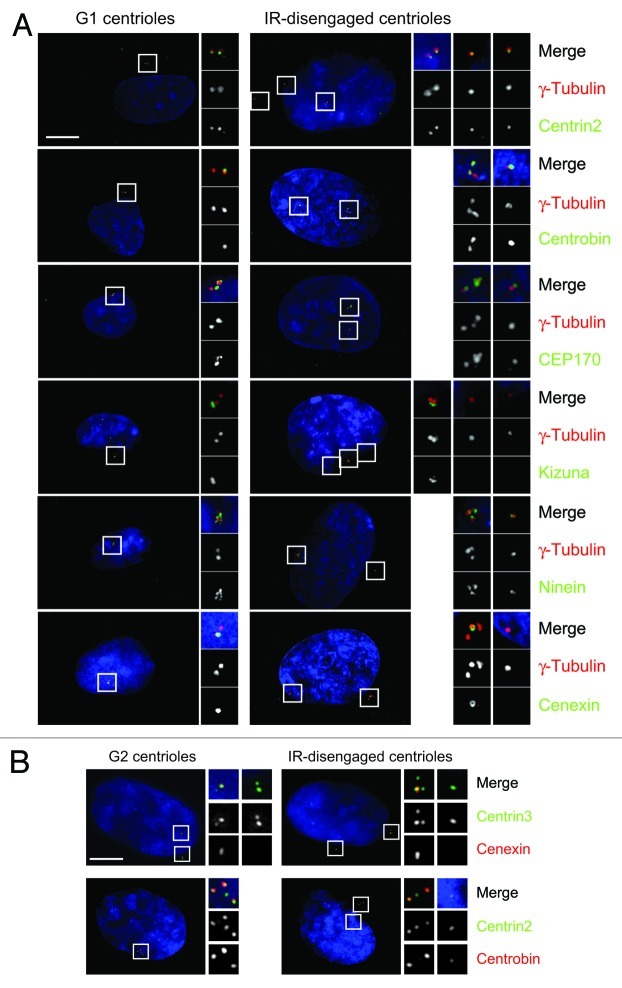
In this study, we have focused on the behavior of individual centrioles, as distinct from the entire centrosome. We refer to “centriole splitting” as single centrioles becoming separated from the main centrosome cluster by > 2.0 μm and thus becoming “isolated.” Centriole splitting precedes G2 phase centrosome reduplication after IR.10 To explore the possible consequences of this splitting in terms of centrosome activities, we examined the composition of the centrioles after irradiation. We used the PCM component, γ-tubulin, as our reference marker in these experiments, because it consistently colocalized with the centriole marker, centrin (Table 1), indicating that no PCM fragmentation occurred after IR, and because it gave a robust signal with no DNA damage-induced granules or background,30 allowing unambiguous identification of centrioles. Isolated centrioles were positive for a number of markers that indicated a normal centriole composition and an associated PCM: centrin,31 centrobin,32 C-NAP1,15 γ-tubulin, glutamylated tubulin,33 NEDD1,34,35 pericentrin36 and rootletin,14 which all localize to centrosomes in untreated cells (Fig. 1A and Table 1; Fig. S1). NEK2 was entirely absent from centrosomes in irradiated cells (Fig. S1), as previously noted.37
Table 1. Composition of engaged and disengaged centrioles following depletion of centrosome cohesion components.
|
Disengaged centrioles |
Marker |
siRNA (+IR) |
siRNA (-IR) |
|||||||
|
None |
Scrambled |
NEK2 |
Rootletin |
C-NAP1 |
Kizuna |
Rootletin |
C-NAP1 |
None, scrambled, NEK2, Kizuna |
||
|
Cenexin |
10 |
0 |
4 |
6 |
4 |
nd |
2 |
8 |
nd |
|
|
CEP170 |
72 |
80 |
86 |
52 |
0 |
50 |
0 |
|||
|
Kizuna |
4 |
6 |
7 |
0 |
4 |
0 |
8 |
|||
|
Ninein |
84 |
76 |
36 |
91 |
8 |
96 |
6 |
|||
|
Rootletin |
100 |
100 |
100 |
// |
36 |
// |
36 |
|||
|
Centrin, Centrobin, C-NAP1, Glutamylated tubulin, NEDD1, Pericentrin, |
88–100 |
94–100 |
||||||||
| Engaged centrioles | Cenexin, Centrin, Centrobin, CEP170, C-NAP1, Glutamylated tubulin, Kizuna, NEDD1, Ninein, Pericentrin, Rootletin | 78–100 | 82–100 | |||||||
hTERT-RPE1 cells were treated with indicated siRNAs for 24 h then, where indicated, treated with 5 Gy and fixed after 24 h, so that all cells were fixed 48 h after siRNA treatment. Cells were stained for γ-tubulin and the indicated markers. Engaged and disengaged centrioles were scored for the presence of the indicated marker. The table shows the % of centrioles that carry the indicated marker. At least 50 cells were counted in each case.
Figure 1. Composition of split centrioles suggests they are daughters. (A) hTERT-RPE1 cells were fixed 48 h after 5 Gy IR, where indicated, and stained with the indicated antibodies (green) and γ-tubulin (red). DNA was stained with DAPI. Scale bar, 10 μm. (B) Cells were treated as in (A) then stained with the indicated antibodies.
We then examined markers of centriole maturity, cenexin/ ODF2, CEP170 and ninein, which are associated with the appendages on the mother centrioles and, in the case of CEP170 and ninein, with the proximal end of the centrioles.28,38-40 We also tested for kizuna, which is normally associated with mature PCM.29 ≤ 10% of split centrioles carried kizuna or cenexin, while > 70% of them contained CEP170 and ninein (Table 1). Together, these findings indicate that the centrioles that become isolated after IR treatment are structurally intact. Given that CEP170 and ninein can localize to proximal centriole ends, the absence of kizuna and cenexin from the split centrioles suggests that the daughter centriole tends to become isolated after irradiation. To confirm these analyses with centriole-specific markers, we quantitated cenexin and the daughter-associated protein, centrobin,32 at the split centrioles along with centrin2 and centrin3. In 50 cells examined per experiment, the split centriole showed cenexin staining in 12% of cells and centrobin in 84% (Fig. 1B). These data provide robust support for the majority of the split centrioles being daughters.
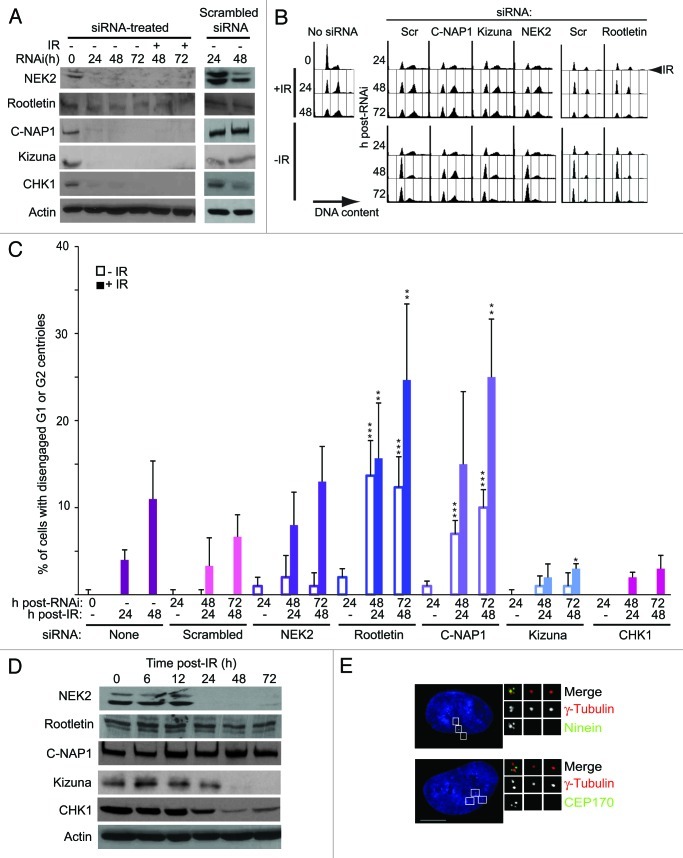
While centriole disengagement, presumably through the activation of separase, is a prerequisite for IR-induced centrosome reduplication, the marked spatial separation of individual centrioles that we see after irradiation is not usually seen during interphase. Therefore, we hypothesized that centrosome cohesion proteins, the tethers that remain even after separase causes disengagement, might control centriole splitting after IR. To test this, we used RNAi to knockdown expression of NEK2, C-NAP1, rootletin or kizuna in RPE1 cells (Fig. 2A). We observed no alteration in cell cycle profile as determined by flow cytometry after NEK2, CROCC (rootletin) or PLK1S1 (kizuna) depletion, although we saw an increase in the 2C fraction in the unirradiated CEP250 (C-NAP1)-knockdown population that likely reflects cell cycle progression difficulties in the absence of C-NAP117 (Fig. 2B). All the knockdown populations showed an arrest profile after IR similar to that seen in control cells, indicating that DNA damage responses were intact after RNAi. Knockdown of NEK2 had no significant effect on centriole splitting (Fig. 2C). However, knockdown of rootletin or C-NAP1 caused marked increases in centriole splitting that were further elevated by IR treatment (Fig. 2C). Checking the cell cycle distribution of splitting, we found that approximately 10% and 15% of the rootletin-depleted cells at 48 h post-IR showed split centrioles in G1 phase and G2 phase, respectively. Kizuna depletion suppressed IR-induced centriole splitting, although the extent of this suppression was at the limit of statistical significance (p = 0.055). These results suggest kizuna as a positive regulator of such splitting. In a control experiment, we found that knockdown of CHK1, a key transducer of the DNA damage response, had only moderate impact on centriole splitting (Fig. 2A and C). These data indicate that centrosome tethering by rootletin and C-NAP1, which links disengaged G1 phase centrioles and keeps duplicated centrosomes together,14 also opposes the splitting-off of individual centrioles that is facilitated by IR-induced centriole disengagement.
Figure 2. Centrosome cohesion blocks centriole splitting. (A) Immunoblot analysis of RNAi efficiency. Cells were treated with the indicated siRNAs for 24 h prior to 5 Gy IR, where indicated, so that irradiated and unirradiated cells were subjected to siRNA treatment for up to 72 h in both cases. Actin was used as a loading control. (B) Flow cytometry analysis of cell cycle in siRNA-treated hTERT-RPE1 cells. Cells were treated as in (A) and fixed at the indicated times. Plots show the DNA content of cells visualized using propidium iodide. (C) Quantitation of centriole splitting in siRNA-treated hTERT-RPE1 cells. Cells were treated as in (A) then stained for centrin2 and γ-tubulin at the timepoints shown. Data points are mean of ± s.e.m of three separate experiments, in which at least 100 cells were counted. ***p ≤ 0.01; **p ≤ 0.05; *p ≤ 0.055, in comparison with irradiated or unirradiated scrambled siRNA controls. (D) Immunoblot analysis of centrosome components after IR. hTERT-RPE1 cells were harvested at the indicated timepoints after 5 Gy IR and analyzed by immunoblotting using the indicated antibodies. Actin was used as a loading control. (E) Micrograph showing the impact of C-NAP1 depletion on the composition of the split centrioles. Scale bar, 10 μm.
We then examined what happens to levels of these proteins after IR. As shown in Figure 2D, levels of NEK2, CHK1 and kizuna declined, with C-NAP1 and rootletin remaining relatively constant. These data show that RPE1 cells retain cohesion proteins that oppose centriole splitting and lose positive regulators of such splitting after IR. Next, we tested whether centrosome cohesion proteins play a role in the composition of the isolated centrioles. We scored G2 phase cells in which we saw both a cluster of at least two centrioles and individual centriole(s). As shown in Table 1, centriolar and centrosomal markers were unaffected in IR-split centrioles after NEK2 or control knockdowns, except for a decline in the number of split centrioles positive for ninein in NEK2-depleted cells. Very few isolated centrioles were seen after kizuna depletion. However, in the C-NAP1 siRNA-treated samples, isolated centrioles were almost entirely devoid of CEP170 and ninein (Table 1 and Fig. 2E). A similar, marked reduction in rootletin was described in previous experiments, where C-NAP1 was depleted.14 A less pronounced phenotype was seen in rootletin-deficient cells, in which fewer split centrioles carried CEP170, but in which ninein was unaffected (Table 1). The isolated centrioles remained devoid of cenexin, and their associated PCM devoid of kizuna under all conditions (Table 1). Neither C-NAP1 nor rootletin depletion had any impact on the localization of kizuna, which was consistently absent from the disengaged centriole. These observations suggest that the disengaged centrioles carry an immature PCM and are thus likely to be daughter centrioles.29,41 As these centrioles contained all the other centrosomal markers, and as C-NAP1-depleted cells still showed normal CEP170 and ninein localization in the other associated centrioles, we also conclude that the loss of centrosomal cohesion, in particular, of C-NAP1, affects the proximal region of the daughter centriole(s) that will split.
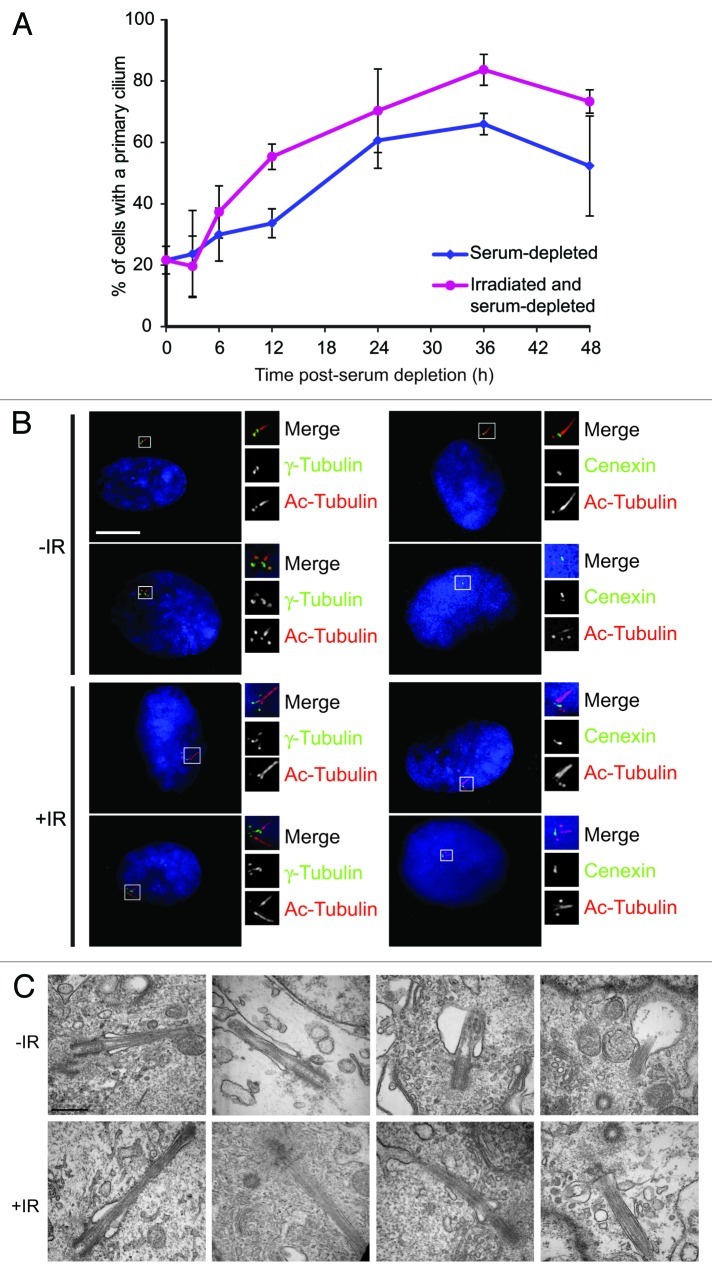
Given the role of the mother centriole in establishing the basal body, we then asked whether IR might affect ciliogenesis. As shown in Figure 3A and B, we saw slightly increased levels of primary cilium formation in RPE1 cells after irradiation. These cilia were apparently normal in structure, as determined by light and electron microscopy (Fig. 3B and C), with clearly visible ciliary pockets at the base of a microtubule structure that emerged from the mother centriole. We occasionally observed multiple cilia after irradiation, often emerging side by side, possibly from a single ciliary pocket, as has been observed recently, where multiple cilia were formed after Plk4 overexpression induced supernumerary centrioles.42 These multiple cilia only ever grew from mature centrioles, as confirmed by cenexin staining (Fig. 3B).
Figure 3. Apparently normal cilia form after irradiation. (A) Levels of ciliogenesis following serum depletion in unirradiated or 5 Gy IR-treated hTERT-RPE1 cells. Cells irradiated immediately before serum depletion. They were then fixed and stained with acetylated tubulin and γ-tubulin and scored for primary cilia. Data show the mean of ± s.e.m of three separate experiments, in which at least 100 cells were counted. (B) RPE1 cells stained for acetylated tubulin (“Ac-Tubulin”) and γ-tubulin or cenexin to show G1 and G2 phase centrosomes with a primary cilium, or multiple cilia after IR. Where indicated, cells were treated with 5 Gy IR, 48 h prior to being serum-starved for 24 h, then fixed and analyzed. Scale bar, 10 μm. (C) Electron micrographs of primary cilia in unirradiated and irradiated RPE1 cells treated as in (B). Scale bar, 500 nm.
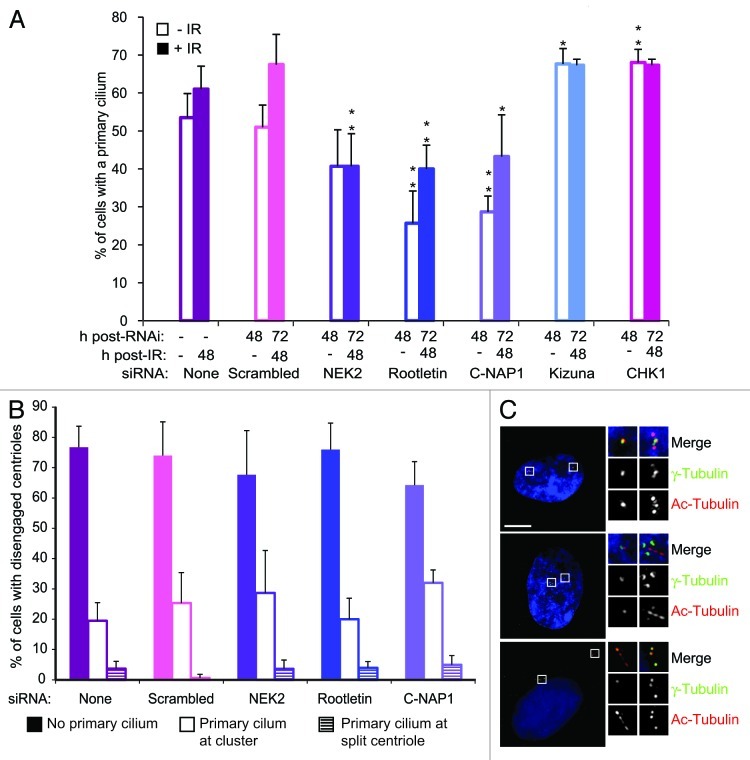
Next, we asked how IR-induced centriole splitting impacts on ciliogenesis. As shown in Figure 4A, knockdown of rootletin or C-NAP1 significantly reduced the number of cells that formed a primary cilium, prior to or after irradiation, while NEK2 depletion reduced the number of ciliated cells after IR treatment. Conversely, knockdown of either kizuna or CHK1 led to increased levels of ciliated cells. Overall, these observations suggest that increased centriole splitting reduces ciliogenesis capacity. Previous experiments depleting C-NAP1 and rootletin reported no effect on ciliogenesis.28 To explore why we obtained different results, we repeated our experiments under the other authors’ conditions, which had involved different culture conditions and siRNAs. Using the previously published siRNAs, we found no significant effect of C-NAP1 or rootletin depletion under either our conditions or those of the previous study (Fig. S2). Using the siRNAs featured in this study under the previously published conditions, we saw a reduction of ciliogenesis only with rootletin depletion (Fig. S2). Importantly, we found that Cep164 depletion reduced ciliogenesis to 17% in a single control experiment (data not shown), so that our data are entirely consistent with those that have been previously published, under those conditions.28 As we see efficient knockdown of our targets with the previously published siRNAs (Fig. S2), we conclude that our serum starvation conditions, where we include 0.2% newborn calf serum to alleviate the impact of irradiation and siRNA knockdown, allow detection of the effects of C-NAP1 or rootletin depletion that were not seen in the previous work. Similar technical differences have been discussed, where a decline in ciliogenesis was seen with NEK2 depletion in one study28 but not another.43
Figure 4. Centrosome cohesion proteins promote ciliogenesis. (A) RPE1 cells were siRNA treated for 24 h before being serum starved for 24 h, then fixed and stained for acetylated tubulin and γ-tubulin. Irradiated cells were treated with 5 Gy IR, 24 h prior to serum starvation. Data show the mean ± s.e.m of three separate experiments, where at least 100 cells were counted. **p ≤ 0.01; *p ≤ 0.05 compared with irradiated or unirradiated scrambled siRNA control, as appropriate. (B) Levels and location of primary cilia following centriole disengagement. hTERT-RPE1 cells were treated with siRNA, fixed and stained as in (A). Data show the mean ± s.e.m of three separate experiments in which at least 50 cells were counted. (C) Immunofluorescence micrographs of split centrioles in hTERT-RPE1 cells with and without primary cilia. Cells were treated and stained as in (A). Scale bar, 10 μm.
When we then examined whether cilia form from the disengaged centriole, we found that the majority of the cilia arose at one of the clustered centrioles and not at the disengaged one, irrespective of the status of centrosome cohesion apparatus (Fig. 4B and C). These data show that centrosome cohesion proteins, which prevent centriole splitting, also affect the level of primary ciliogenesis.
Discussion
During the normal cell cycle, there exist two molecular linkages between centrioles. One connects a mother centriole to its daughter and is resolved through the action of anaphase-activated separase on cohesin and pericentrin B/ kendrin.8,11,44 This has been described as “centriole engagement”45 and as an “S-M linker” in a recent review.4 This review referred to the second linkage as a “G1-G2 tether,” as it connects the proximal ends of the parental centrioles from when they begin to serve as mothers until they move apart to establish the mitotic spindle. Tethering involves C-NAP1 and rootletin and is resolved through the localized activation of NEK2 kinase.14,16,46 Despite the proteinaceous nature of tethering, it is not a highly constrained linkage, as live-cell imaging has demonstrated a good deal of inter-centriolar movement within the cytoplasm during interphase.2
Our analysis sought to test which centriolar linkages were being affected by irradiation. The release of the daughter centriole in the majority of cases suggests that IR leads to centriole disengagement within G2 centrosomes. This is consistent with centrosome amplification being facilitated during G2 phase, as we have previously suggested.47 This is also consistent with DNA damage-induced activation of separase,48 which would then allow disengagement. Notably, the mother centrioles remain tethered after IR, with C-NAP1 and rootletin levels being unaffected. DNA damage has previously been shown to block G2 phase centrosome separation by inhibiting NEK2, thus blocking the phosphorylation of C-NAP1 and rootletin.21,49 While NEK2 depletion has a relatively minor impact on centriole splitting, C-NAP1 or rootletin knockdown greatly increase it. Together, these findings suggest that C-NAP1 and rootletin repress centriole splitting, and that their phosphorylation by NEK2 is not required for it. However, we do not see any increase in the number of mother centrioles that become isolated after C-NAP1 or rootletin depletion. If the daughter centrioles can only split when allowed to do so by centriole disengagement, we propose that the tethering/ cohesion components either regulate disengagement, per se, or the extent to which disengaged centrioles can move apart.
Current models for primary cilium formation identify the mother centriole as the origin of the basal body because of the requirement for fully functional appendages in efficient ciliogenesis.28,50 Key early electron microscopy analyses established the migration of centrioles/ basal bodies to the cell surface as a key step in cilium formation.51,52 We found that very few cilia arose from the individualized, split centrioles, consistent with these being daughter centrioles. The absence of cenexin from the split centriole would further impede ciliogenesis, as it has been proposed as a rate-limiting determinant of cilium formation.53 Another possibility is that the ability to act as a basal body requires a mature PCM.54 However, despite the increased level of centriole splitting caused by irradiation, irradiated cells were fully capable of ciliogenesis, suggesting that the isolation of daughter centrioles does not impact on ciliogenesis.
However, C-NAP1 or rootletin depletion caused elevated centriole splitting, which was consistently accompanied by reduced levels of ciliogenesis. NEK2 knockdown also impaired ciliogenesis. To resolve the apparent discrepancy between the lack of effect of centriole splitting on ciliogenesis after irradiation and the reduced ciliogenesis seen when splitting is increased by depletion of centrosome tethering proteins, it is worth noting that the composition of the centriole that disengages and splits after IR depends on C-NAP1 and, to a lesser extent, rootletin, in terms of Cep170 and ninein localization. NEK2 is no longer recruited to the proximal end of centrioles upon C-NAP1 depletion,43 demonstrating that the loss of C-NAP1 has a marked effect on centriole composition. NEK2 loss also impacts on the ciliation capacity of the mother centriole,43 although its loss after irradiation did not diminish ciliation capacity in our experiments. Interestingly, the ALMS1 protein, whose depletion leads to a reduction of C-NAP1 levels at the proximal end of the centriole, is required for the formation and maintenance of the primary cilium.28,55 A change in centriole composition may therefore be how loss of the centrosome cohesion proteins affects ciliogenesis. In conclusion, our data suggest that the proteins that tether G2 centrosomes also control disengaged centriole behavior and ciliogenesis after irradiation. The effects of DNA damage on centriole/ basal body activities are thus focused on the proximal ends of the centrioles.
Materials and Methods
Cells and cell culture
Human hTERT-RPE1, a non-transformed, telomerase-immortalized retinal epithelial cell line, was obtained from ATCC. Cells were grown in DMEM-F12 medium with 10% fetal bovine serum. Irradiations were performed using a 137Cs source at 23.5 Gy/ minute (Mainance Engineering). For flow cytometry, cells were fixed in 70% ethanol, then resuspended in PBS containing 100 μg/ ml RNase A and 40 μg/ml propidium iodide and incubated for 20 min. Cells were passed through a 27 gauge needle immediately before analysis. Cell cycle analysis was performed on a FACScalibur (BD Biosciences). To deplete cells of serum, cells were washed three times in warm PBS before adding DMEM-F12 medium with 0.2% newborn calf serum (NCS), unless otherwise indicated.
Immunoblotting
Primary monoclonal mouse antibodies against NEK2 (BD Transduction laboratories), C-NAP-1 (Clone 42, BD Transduction laboratories and sc-74347, Santa Cruz) and CHK1 (DCS310, Sigma-Aldrich) antibodies were used at 1:250, 1:500, 1:250 and 1:1,000, respectively, in immunoblot analyses. A polyclonal goat antibody against rootletin, (sc-67824, Santa Cruz) was used at 1:500. Polyclonal rabbit antibodies against Kizuna29; and pericentrin (Abcam) were used at 1:1,000 and anti-actin (Sigma-Aldrich) at 1:5,000.
RNA-mediated interference
hTERT-RPE1 cells were transfected with ON-TARGETplus SMART pools of RNA duplexes inhibitory to CHK1, C-NAP1 (CEP250), Rootletin (CROCC), NEK2 and KIZUNA (PLK1S1) and an ON-TARGETplus non-targeting short interfering RNA pool (Table S1; Dharmacon), or with custom siRNA targeting C-NAP1 (CEP250), Rootletin (CROCC) and CEP164 from Qiagen (Table S2) using oligofectamine (Invitrogen). Fifty nmol of siRNA were complexed with oligofectamine in serum-free Optimem (Invitrogen) and added to cells at 20–30% confluency. Serum was added 4 h after transfection and fresh media 24 h after transfection. Cells were analyzed 24 h, 48 h and 72 h after transfection. Where indicated, cells were irradiated 24 h after transfection.
Microscopy
hTERT-RPE1 cells were grown on glass coverslips and fixed in methanol/5 mM EGTA at -20°C for 10 min and stained as described.56 Prior to fixation and staining with acetylated tubulin, cells were incubated on ice for 20 min to depolymerize the microtubules unless otherwise indicated. Monoclonal antibodies were used as follows: Centrin3, 3E6 (Abnova, 1:1000), NEDD134 (a gift from A. Merdes, 1:500), γ-tubulin GTU88 (Sigma-Aldrich, 1:500), glutamylated-tubulin (a gift from C. Janke, 1:500), acetylated tubulin, T6793 (Sigma-Aldrich, 1:2000) and C-NAP1 and NEK2 (both BD Transduction Laboratories, 1:250). Polyclonal antibodies were used against CEP17038 (a gift from G. Guarguaglini, 1:1000), CEP164 (NBP1–77006, Novus Biologicals), Centrin-2 (N-17; Santa Cruz, 1:500), γ-tubulin (T3559; Sigma, 1:1000), Kizuna (29; a gift from M. Ohsugi, 1:500), Pericentrin (ab4448, Abcam, 1:2000), Centrobin (ab70448, Abcam, 1:500), Cenexin (ab43840, Abcam, 1:200), Acetylated tubulin (T7693, Calbiochem, 1:2000), Rootletin (sc-67824, Santa Cruz, 1:100), Ninein (a gift from A. Merdes, 1:200). Imaging was performed with an Olympus BX51 microscope, 100× objective, NA 1.35, using Openlab software (Improvision). Deconvolved images (Nearest Neighbor DCI) were saved as Adobe Photoshop CS files (version 8.0).
Electron microscopy
hTERT-RPE1 cells were grown on 10 cm dishes and serum starved or irradiated as described. Before fixation, they were washed in PBS and then scraped into Eppendorf tubes before being pelleted at 250 g for 5 min. They were then processed for transmission EM (TEM) as described.57 Cells were fixed in 2% glutaraldehyde/2% PFA in 0.1 M cacodylate buffer, pH 7.2, then post-fixed in a solution of 2% osmium tetroxide/0.1 M cacodylate buffer. Cell pellets were then dehydrated through a graded series of ethanol (30, 60, 90 and 100%) before propylene oxide was added to the pellet. Next, cell pellets were embedded in Agar Low Viscosity Resin and sections cut using a Reichert-Jung Ultracut E microtome (Leica). Cells were stained with uranyl acetate and lead citrate and then viewed on an electron microscope (H-7000; Hitachi). Images were taken with an ORCA-HRL camera (Hamamatsu Photonics) and processed using AMT version 6 (AMT Imaging).
Statistical analysis
Statistical analyses were performed with Prism v5.0 (GraphPad).
Supplementary Material
Acknowledgments
We thank Giulia Guarguaglini, Carsten Janke, Laurence Haren and Andreas Merdes, Miho Ohsugi and Tadashi Yamamoto, William Tsang and Brian Dynlacht for antibodies and Suzanna Prosser for critical reading of the manuscript. We are indebted to a PRTLI4 grant to the National Biophotonics and Imaging Platform Ireland (www.nbipireland.ie) for the TEM. This work was supported by Science Foundation Ireland Principal Investigator awards 08/IN.1/B1029 and 10/IN.1/B2972, a Government of Ireland PhD studentship and an award from the Thomas Crawford Hayes Memorial Fund (to C.S.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental material may be found here: www.landesbioscience.com/journals/cc/article/21986
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21986
References
- 1.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–63. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 2.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–30. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kochanski RS, Borisy GG. Mode of centriole duplication and distribution. J Cell Biol. 1990;110:1599–605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–60. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardin BR, Schiebel E. Breaking the ties that bind: new advances in centrosome biology. J Cell Biol. 2012;197:11–8. doi: 10.1083/jcb.201108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderhub SJ, Krämer A, Maier B. Centrosome amplification in tumorigenesis. Cancer Lett. 2012;322:8–17. doi: 10.1016/j.canlet.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, et al. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–9. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–54. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hut HM, Lemstra W, Blaauw EH, Van Cappellen GW, Kampinga HH, Sibon OC. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol Biol Cell. 2003;14:1993–2004. doi: 10.1091/mbc.E02-08-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saladino C, Bourke E, Conroy PC, Morrison CG. Centriole separation in DNA damage-induced centrosome amplification. Environ Mol Mutagen. 2009;50:725–32. doi: 10.1002/em.20477. [DOI] [PubMed] [Google Scholar]
- 11.Schöckel L, Möckel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol. 2011;13:966–72. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- 12.Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108:107–28. doi: 10.1016/1047-8477(92)90011-X. [DOI] [PubMed] [Google Scholar]
- 13.Vorobjev IA, Chentsov YuS Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93:938–49. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141:1563–74. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–46. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14:2876–89. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17:470–81. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH, Jr., O’Toole ET, Winey M, et al. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J Cell Sci. 2001;114:3749–57. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- 21.Mi J, Guo C, Brautigan DL, Larner JM. Protein phosphatase-1alpha regulates centrosome splitting through Nek2. Cancer Res. 2007;67:1082–9. doi: 10.1158/0008-5472.CAN-06-3071. [DOI] [PubMed] [Google Scholar]
- 22.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 23.Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–34. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 24.Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH, Jr., et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–61. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Chen MH, et al. Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–6. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 26.Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/S0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 28.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–30. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol. 2006;8:1095–101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- 30.Prosser SL, Straatman KR, Fry AM. Molecular dissection of the centrosome overduplication pathway in S-phase-arrested cells. Mol Cell Biol. 2009;29:1760–73. doi: 10.1128/MCB.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moudjou M, Paintrand M, Vigues B, Bornens M. A human centrosomal protein is immunologically related to basal body-associated proteins from lower eucaryotes and is involved in the nucleation of microtubules. J Cell Biol. 1991;115:129–40. doi: 10.1083/jcb.115.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, et al. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol. 2005;171:437–45. doi: 10.1083/jcb.200506185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bobinnec Y, Moudjou M, Fouquet JP, Desbruyères E, Eddé B, Bornens M. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton. 1998;39:223–32. doi: 10.1002/(SICI)1097-0169(1998)39:3<223::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 34.Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol. 2006;172:505–15. doi: 10.1083/jcb.200510028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lüders J, Patel UK, Stearns T. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol. 2006;8:137–47. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- 36.Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–50. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 37.Fletcher L, Cerniglia GJ, Nigg EA, Yend TJ, Muschel RJ. Inhibition of centrosome separation after DNA damage: a role for Nek2. Radiat Res. 2004;162:128–35. doi: 10.1667/RR3211. [DOI] [PubMed] [Google Scholar]
- 38.Guarguaglini G, Duncan PI, Stierhof YD, Holmström T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16:1095–107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113:3013–23. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell. 2001;12:1687–97. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oshimori N, Li X, Ohsugi M, Yamamoto T. Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J. 2009;28:2066–76. doi: 10.1038/emboj.2009.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahjoub MR, Stearns T. Supernumerary Centrosomes Nucleate Extra Cilia and Compromise Primary Cilium Signaling. Curr Biol. 2012 doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spalluto C, Wilson DI, Hearn T. Nek2 localises to the distal portion of the mother centriole/basal body and is required for timely cilium disassembly at the G2/M transition. Eur J Cell Biol. 2012;91:675–86. doi: 10.1016/j.ejcb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 44.Matsuo K, Ohsumi K, Iwabuchi M, Kawamata T, Ono Y, Takahashi M. Kendrin is a novel substrate for separase involved in the licensing of centriole duplication. Curr Biol. 2012;22:915–21. doi: 10.1016/j.cub.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 45.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–51. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 46.Yang J, Adamian M, Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol Biol Cell. 2006;17:1033–40. doi: 10.1091/mbc.E05-10-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inanç B, Dodson H, Morrison CG. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol Biol Cell. 2010;21:3866–77. doi: 10.1091/mbc.E10-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prosser SL, Samant MD, Baxter JE, Morrison CG, Fry AM. Oscillation of APC/C activity during cell cycle arrest promotes centrosome amplification. J Cell Sci. 2012 doi: 10.1242/jcs.106096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Fletcher L, Muschel RJ. The role of Polo-like kinase 1 in the inhibition of centrosome separation after ionizing radiation. J Biol Chem. 2005;280:42994–9. doi: 10.1074/jbc.M505450200. [DOI] [PubMed] [Google Scholar]
- 50.Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–24. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 51.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–30. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 52.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–77. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson CT, Stearns T. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr Biol. 2009;19:1498–502. doi: 10.1016/j.cub.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moser JJ, Fritzler MJ, Ou Y, Rattner JB. The PCM-basal body/primary cilium coalition. Semin Cell Dev Biol. 2010;21:148–55. doi: 10.1016/j.semcdb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 55.Knorz VJ, Spalluto C, Lessard M, Purvis TL, Adigun FF, Collin GB, et al. Centriolar association of ALMS1 and likely centrosomal functions of the ALMS motif-containing proteins C10orf90 and KIAA1731. Mol Biol Cell. 2010;21:3617–29. doi: 10.1091/mbc.E10-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, et al. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–73. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liptrot C, Gull K. Detection of viruses in recombinant cells by electron microscopy. In: Spiers RE, Griffiths JB, MacDonald C, eds. Animal Cell Technology: Development, Processes and Production. Oxford: Butterworth-Heinemann, 1992:653–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.