Abstract
Resveratrol is a polyphenol contained in red wine that has been amply investigated for its beneficial effects on organismal metabolism, in particular in the context of the so-called “French paradox,” i.e., the relatively low incidence of coronary heart disease exhibited by a population with a high dietary intake of cholesterol and saturated fats. At least part of the beneficial effect of resveratrol on human health stems from its capacity to promote autophagy by activating the NAD-dependent deacetylase sirtuin 1. However, the concentration of resveratrol found in red wine is excessively low to account alone for the French paradox. Here, we investigated the possibility that other mono- and polyphenols contained in red wine might induce autophagy while affecting the acetylation levels of cellular proteins. Phenolic compounds found in red wine, including anthocyanins (oenin), stilbenoids (piceatannol), monophenols (caffeic acid, gallic acid) glucosides (delphinidin, kuronamin, peonidin) and flavonoids (catechin, epicatechin, quercetin, myricetin), were all capable of stimulating autophagy, although with dissimilar potencies. Importantly, a robust negative correlation could be established between autophagy induction and the acetylation levels of cytoplasmic proteins, as determined by a novel immunofluorescence staining protocol that allows for the exclusion of nuclear components from the analysis. Inhibition of sirtuin 1 by both pharmacological and genetic means abolished protein deacetylation and autophagy as stimulated by resveratrol, but not by piceatannol, indicating that these compounds act through distinct molecular pathways. In support of this notion, resveratrol and piceatannol synergized in inducing autophagy as well as in promoting cytoplasmic protein deacetylation. Our results highlight a cause-effect relationship between the deacetylation of cytoplasmic proteins and autophagy induction by red wine components.
Keywords: Beclin 1, LC3, longevity, p62/SQSTM1, trichostatin A, U2OS
Introduction
Autophagy is a catabolic pathway leading to the lysosomal degradation of intracellular material, including organelles and portions of the cytoplasm. Baseline levels of autophagy play a prominent homeostatic role and contribute to organismal development.1 Moreover, the autophagic flow is significantly upregulated as an adaptive response to distinct adverse conditions such as metabolic stress (nutrient deprivation and hypoxia) and infection by intracellular pathogens.2,3 In line with this notion, autophagic defects underlie a panel of clinically relevant pathologies encompassing heart failure, hereditary myopathies, neurodegenerative diseases, chronic inflammatory states, steatosis/steatohepatitis and cancer.2,3 Conversely, the pharmacological or genetic stimulation of autophagy increases the capacity of cells to withstand metabolic stress, as it facilitates the maintenance of high ATP levels,4,5 sustains genomic stability6 and reduces the abundance of potentially toxic cellular components, such as permeabilized mitochondria as well as protein aggregates that may promote neurodegeneration.7,8
Rapamycin, the best characterized pharmacological inducer of autophagy, acts by inhibiting the mammalian target of rapamycin complex 1 (mTORC1)9 and has been shown to prolong the lifespan of all organisms studied so far, including mice.10-14 In Saccharomyces cerevisiae, Caenorhabditis elegans and Drosophila melanogaster, the lifespan-extending activity of rapamycin is lost in conditions in which autophagy cannot be induced.10,15,16 Of note, rapamycin and the so-called rapalogs are the most effective inducers of autophagy currently used in the clinics, yet have severe immunosuppressive effects.17 Thus, the pharmacological profile of alternative, non-toxic pro-autophagic compounds (such as rilmenidine and carbamazepine) is being characterized in suitable preclinical models of chronic degenerative diseases.18,19
Non-toxic compounds such as resveratrol and spermidine are also being evaluated for their potential to induce autophagy in vivo and to improve healthy aging.20-23 Resveratrol is a natural polyphenol found in grapes, red wine, berries, knotweed, peanuts and other plants. This molecule first attracted interest as it was believed to explain the so-called “French paradox,” that is, the relatively low incidence of coronary heart disease manifested by a population with a high dietary intake of cholesterol and saturated fats.24,25 However, the concentrations of resveratrol found in red wine are excessively low to entirely account for this phenomenon.24 Besides operating as an antioxidant, resveratrol is a potent inducer of autophagy,26 an effect that stems from the activation of the NAD+-dependent deacetylase sirtuin 1 (SIRT1).21,27,28 Resveratrol was initially proposed to directly activate SIRT1,24,25 although less direct mechanisms may actually be preponderant.29,30 SIRT1 was originally characterized for its activity of histone deacetylase.31 However, at least two lines of evidence indicate that SIRT1 operates on cytoplasmic, rather than nuclear, substrates to mediate resveratrol-induced autophagy. First, the pro-autophagic activity of resveratrol is fully preserved in enucleated cells (i.e., cytoplasts). Second, a SIRT1 variant that cannot be imported into the nucleus (owing to the deletion of its nuclear localization signal) is as proficient at inducing autophagy as its wild-type counterpart.21
Recent proteomic studies revealed that 5–10% of mammalian and bacterial proteins undergo reversible lysine acetylation, a post-translational modification consisting of the addition of an acetyl group to the ε amino group of lysine residues.32,33 (De)acetylation reactions de facto rival (de)phosphorylation for importance as post-translational mechanisms for the regulation of protein function.33 Indeed, the (de)acetylation of multiple distinct proteins affects transcriptional programs (in particular when histones and transcription factors are involved) as well as multiple transcription-independent processes, including chromosome separation during mitosis [following the (de)acetylation of cohesin],34 cytoskeletal dynamics,35 DNA repair,36 RNA processing,37 receptor signaling,38 metabolic circuitries39 and cell death.40,41
In mammals, (de)acetylation reactions are catalyzed by at least 30 acetyltransferases and approximately 18 deacetylases,42,43 several of which have been involved in the control of autophagy: SIRT1,44 SIRT2,45 histone deacetylase 6 (HDAC6)46 and p300.47 However, attempts to identify one or a few critical substrates whose (de)acetylation would entirely account for the regulation of autophagy by these enzymes have failed. For instance, p300 can acetylate several autophagy-relevant proteins, including ATG5, ATG7, ATG12 and LC3B,47 while SIRT1 can catalyze the deacetylation of ATG5, ATG7, LC3B,48 as well as of the transcription factor forkhead box O3 (FOXO3), which, in turn, controls the expression of several pro-autophagic proteins.49 Moreover, resveratrol as well as spermidine (a natural polyamine found in citrus fruits and soybeans) provoke the (de)acetylation of several dozens of autophagy-relevant proteins,21,50 suggesting that (de)acetylation reactions regulate autophagy by a multipronged effect.
Based on these premises, we wondered whether chemical compounds contained in red wine other than resveratrol might induce autophagy and stimulate protein deacetylation. Driven by the published literature,51-53 we focused our attention on a series of polyphenols and monophenols found in red wine that are suspected to mediate beneficial effects on health. Here, we report that several of these compounds indeed operate as autophagy inducers, correlating with their capacity to reduce the acetylation of cytoplasmic proteins.
Results and Discussion
Autophagy induction by mono- and polyphenols
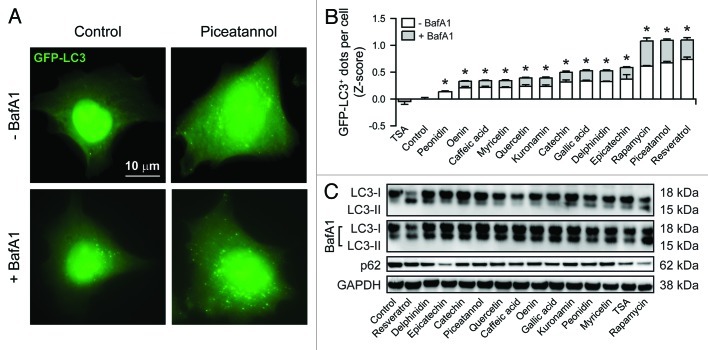
For the purpose of this study, we chose a series of natural phenols contained in red wine or other plant products that have been suggested to mediate beneficial effects on human health. These compounds include one anthocyanin (oenin), one stilbenoid (piceatannol), two monophenols (caffeic acid, gallic acid), three glucosides (delphinidin, kuronamin, peonidin) and four flavonoids (catechin, epicatechin, quercetin, myricetin). As a positive control for autophagy induction, we selected rapamycin, a natural macrolide produced by Streptomyces hygroscopicus that potently inhibits mTORC1.9 Thus, human osteosarcoma U2OS cells stably expressing a GFP-LC3 chimera were exposed for a short time (6 h) to the abovementioned phenols (final concentration = 30 µM) or to 1 µM rapamycin, and then the number of cytoplasmic GFP-LC3+ puncta per cell was assessed by quantitative fluorescence microscopy (Fig. 1A and B).54,55 As an alternative readout, we employed immunoblotting to monitor the lipidation of LC3 (resulting in the generation of a variant with increased electrophorectic mobility, i.e., LC3-II) as well as the degradation of the autophagic substrate p62 (Fig. 1C). As recommended by widely accepted guidelines,54,56 these analyses were performed in the absence as well as in the presence of bafilomycin A1, a selective inhibitor of the vacuolar ATPase that prevents the fusion between autophagosomes and lysosomes, hence blocking the turnover of GFP-LC3+ vesicles (and the degradation of autophagic substrates).57 Beyond resveratrol, which is a well-established inducer of autophagy,24 all tested components of red wine provoked the accumulation of cytoplasmic GFP-LC3+ puncta, although to a variable extent (Fig. 1A and B). Along similar lines, all phenolic compounds mentioned above were capable of increasing the abundance of LC3-II relative to that of its slowly migrating counterpart LC3-I and of stimulating the degradation of p62, which is indicative of a functional autophagic flux (Fig. 1C). In support of this notion, both the accumulation of GFP-LC3+ dots and LC3 lipidation following the administration of phenolic compounds were exacerbated by bafilomycin A1 (Fig. 1B and C). Of note, piceatannol, which is structurally very similar to resveratrol (it just contains one additional hydroxyl group on the second benzyl ring), turned out to be as efficient as resveratrol in triggering autophagy. Altogether, these results demonstrate that phenolic compounds contained in red wine other than resveratrol induce bona fide autophagy.
Figure 1. Autophagy induction by mono- and polyphenols. GFP-LC3-expressing human osteosarcoma U2OS cells (A and B) or their wild-type counterparts (C) were either left untreated or treated with 1 µM rapamycin, 10 µM trichostatin A (TSA) or 30 µM of the indicated phenolic compounds, alone or in combination with 50 nM bafilomycin A1 (BafA1) for 6 h. Thereafter, cells were either processed for the immunofluorescence microscopy-assisted quantification of cytoplasmic GFP-LC3+ dots (A and B) or for the immunoblotting-based detection of LC3 lipidation and p62 degradation (C). Representative images are reported in (A) (scale bar = 10 µm) and (C) (GAPDH levels were monitored to ensure equal loading of lanes). In (B), quantitative data are reported. White and light gray columns represent the Z-score of the number of GFP-LC3+ dots detected per cell in the absence and in the presence of BafA1, respectively. Results from one representative experiments are presented as means ± SEM (n = 500 cells/condition). *p < 0.05 (unpaired, two-tailed Student’s t-test), as compared with untreated cells.
Deacetylation of cytoplasmic proteins in response to mono- and polyphenols
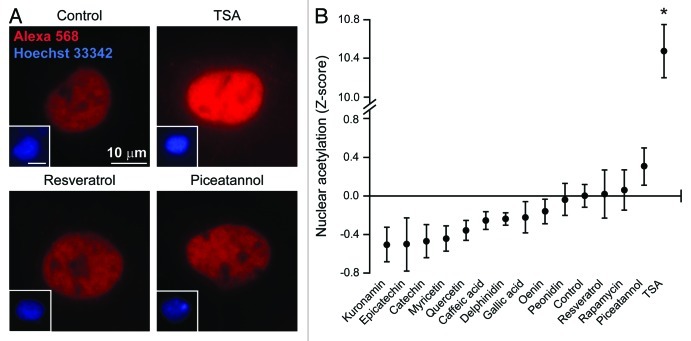
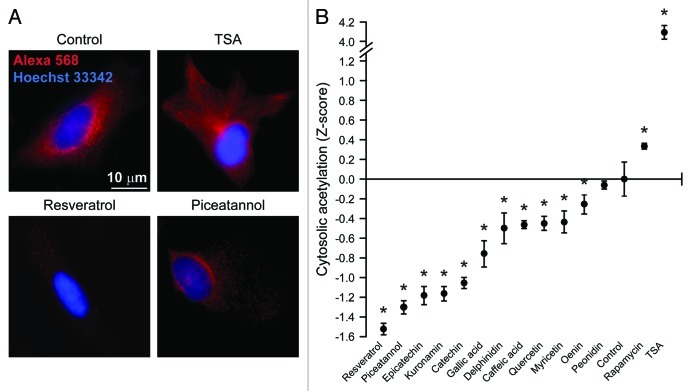
Since the regulation of autophagy by natural compounds including resveratrol and spermidine has been related to the modulation of acetyltransferase and deacetylases,50 we explored whether the aforementioned phenolic compounds would be able to alter the acetylation of cellular proteins, focusing on the nuclear vs. the cytoplasmic cell compartment. Under standard permeabilization conditions, the immunofluorescence microscopy-based quantification of acetylated lysines virtually generates results for the nuclear compartment only due to the sky-scraping abundance of (normally hyperacetylated) histones, which de facto covers any cytoplasmic signal. In this setting, trichostatin A, an inhibitor of class I and II histone deacetylases that we employed as positive control, robustly increased the fluorescent signal obtained with a monoclonal antibody specific for acetylated lysines. Conversely, we failed to detect any significant effect of red wine components and rapamycin on the acetylation of nuclear proteins (Fig. 2A and B). To further investigate the impact of pro-autophagic phenols on protein acetylation, we developed a protocol in which plasma membranes but not nuclear envelopes get permeabilized, allowing for the selective staining of cytoplasmic components. By this method, we observed that both trichostatin A and rapamycin increase the levels of protein acetylation in the cytoplasm. Conversely, cytoplasmic protein acetylation was reduced by a large array of mono- and polyphenols (Fig. 3A and B).
Figure 2. Effect of phenolic compounds on the acetylation of nuclear proteins. Wild-type human osteosarcoma U2OS cells were either left untreated or treated with 1 µM rapamycin, 10 µM trichostatin A (TSA) or 30 µM of the indicated phenolic compounds for 6h, and then processed according to standard procedures for the immunofluorescence-assisted detection of acetylated lysine residues (red signal from the Alexa 568 fluorochrome). This protocol mostly yields a nuclear staining, as confirmed by the counterstaining of chromatin with Hoechst 33342 (blue signal). Representative images are reported in (A) (scale bars = 10 µm) and quantitative data in (B). Results from one representative experiments are presented as Z-score means ± SEM (n = 500 cells/condition). *p < 0.05 (unpaired, two-tailed Student’s t-test), as compared with untreated cells.
Figure 3. Effect of phenolic compounds on the acetylation of cytoplasmic proteins. Wild-type human osteosarcoma U2OS cells were either left untreated or treated with 1 µM rapamycin, 10 µM trichostatin A (TSA) or 30 µM of the indicated phenolic compounds for 6 h. Thereafter, cells were fixed with paraformaldehyde, but not permeabilized, and immunostained with an antibody that specifically recognizes acetylated lysine residues (red signal from the Alexa 568 fluorochrome). This protocol yields a near-to-completely cytoplasmic staining, as confirmed by the counterstaining of chromatin with Hoechst 33342 (blue signal). Representative images are reported in (A) (scale bars = 10 µm) and quantitative data in (B). Results from one representative experiments are presented as Z-score means ± SEM (n = 500 cells/condition). *p < 0.05 (unpaired, two-tailed Student’s t-test), as compared with untreated cells.
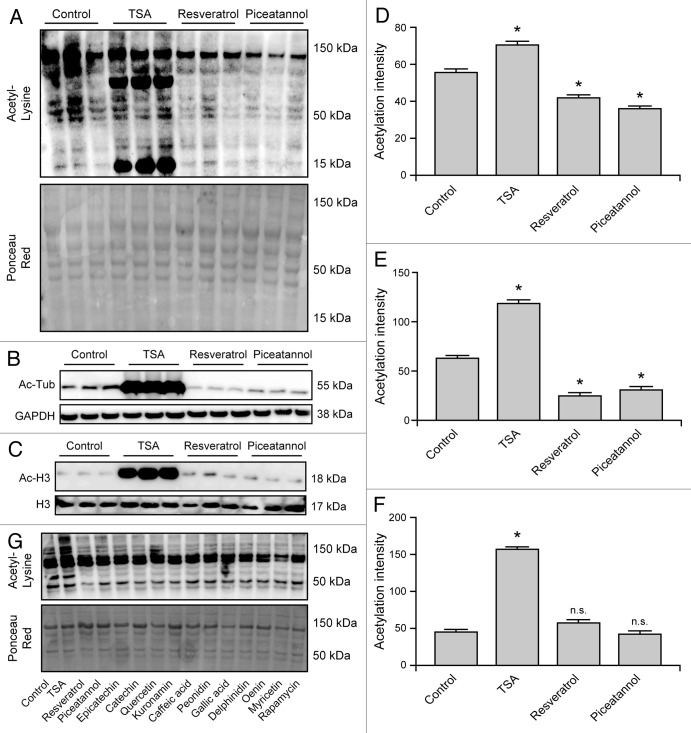
To validate these results by means of alternative techniques, U2OS whole-cell extracts were subjected to immunoblotting procedures based on antibodies specific for acetylated lysine residues (Fig. 4A), α tubulin acetylated on lysine 40, as a representative of the cytoplasmic compartment (Fig. 4B), and histone 3 (H3) acetylated on lysine 9, as a representative of the nuclear compartment (Fig. 4C). Confirming the results obtained by immunofluorescence microscopy, TSA increased the acetylation of all proteins, including α tubulin and H3. Conversely, resveratrol and piceatannol reduced the global levels of protein acetylation, yet affected cytoplasmic (α tubulin), but not nuclear (H3) proteins (Fig. 4A–F). Of note, all phenols included in this study were able to reduce the global levels of protein acetylation (Fig. 4G).
Figure 4. Immunoblotting-assisted detection of protein acetylation in cells responding to mono- and polyphenols. Wild-type human osteosarcoma U2OS cells were either left untreated or treated with 1 µM rapamycin, 10 µM trichostatin A (TSA) or 30 µM of the indicated phenolic compounds, encompassing resveratrol and piceatannol, for 6 h (A–G). Thereafter, cells were subjected to the immunoblotting-assisted quantification of global protein acetylation (A, D and G), α tubulin (Tub) acetylation (as a representative of cytoplasmic proteins) (B and E) and histone 3 (H3) acetylation (as a representative of nuclear proteins) (C and F). Images from one representative experiment are depicted in (A–C and G) (Ponceau Red staining or the levels of GAPDH and H3 were used to monitor equal loading of lanes), while quantitative results are reported in (D–F). Results from n = 3 independent experiments are reported as means ± SEM *p < 0.05, n.s. = non-significant (unpaired, two-tailed Student’s t-test), as compared with untreated cells.
Correlation between autophagy induction and deacetylation
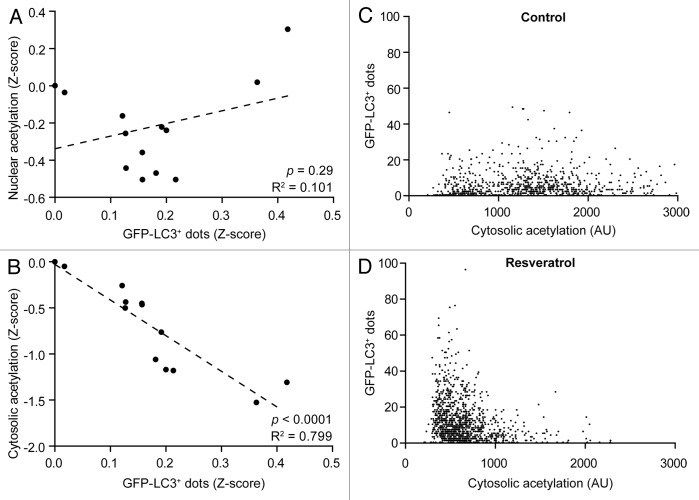
Based on the observations that mono- and polyphenols contained in red wine induce autophagy and reduce the acetylation of cellular proteins, we determined to which extent these two phenomena would correlate. While there was no statistically significant relationship between the number of GFP-LC3+ puncta and the levels of nuclear protein acetylation resulting from the administration to U2OS cells of the phenolic compounds used in this study (Fig. 5A), a highly significant negative correlation between the number of GFP-LC3+ puncta and the levels of cytosolic protein acetylation could be detected (Fig. 5B). Thus, on a cell population level, we could measure a decrease in cytoplasmic acetylation in response to resveratrol that was proportional to the accumulation of GFP-LC3+ dots (Fig. 5B). Conversely, there was no correlation between the levels of cytoplasmic protein acetylation and autophagy induction at the single-cell level, neither in untreated cells nor in cells receiving resveratrol (Fig. 5C and D).24,25
Figure 5. Negative correlation between the acetylation of cytoplasmic proteins and autophagy induction. GFP-LC3-expressing human osteosarcoma U2OS cells were left untreated or treated with 30 µM of the phenolic compounds used in this study, including resveratrol, for 6 h. Thereafter, cells were processed for the immunofluorescence microscopy-assisted quantification of GFP-LC3+ dots and nuclear (A) or cytoplasmic (B–D) protein acetylation levels. In (A and B), population-based results (each dot representing the mean of n = 500 cells) are reported, together with linear regression curves and the corresponding statistical indicators (p values and determination coefficients, R2). In (C and D), results from one representative experiment are reported (each dot representing a single cell). In this latter scenario, p values and determination coefficients were invariably were > 0.2 and < 0.1, respectively.
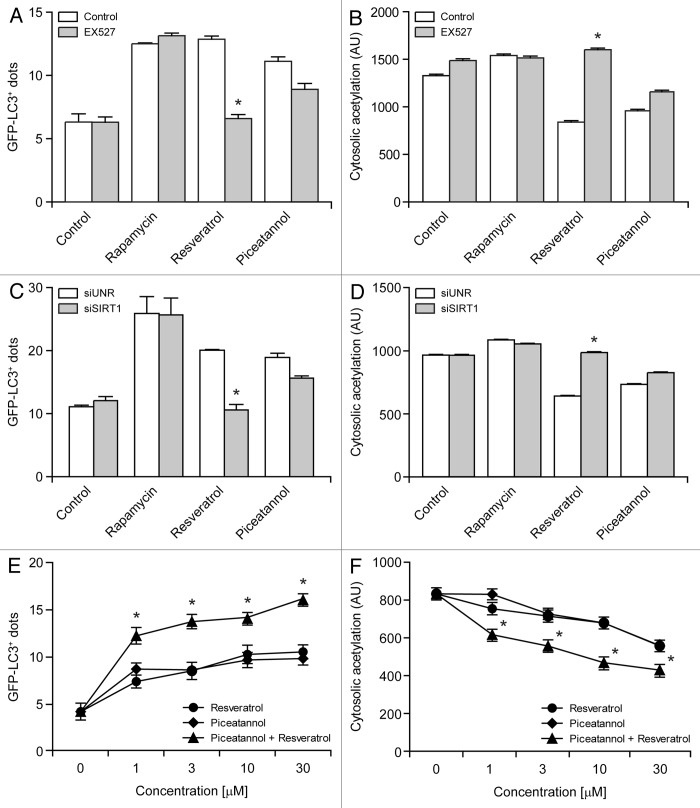
Resveratrol is known to induce autophagy by activating sirtuin 1,24,25 which can be inhibited by the metabolically stable indolic compound EX527.58,59 Importantly, both the accumulation of GFP-LC3+ puncta and the deacetylation of cytoplasmic proteins as stimulated in U2OS cells by resveratrol could be inhibited by EX527. Conversely, EX527 failed to affect autophagy induction as well as cytoplasmic deacetylation reactions as stimulated by piceatannol (Fig. 6A and B). Results obtained upon the depletion of SIRT1 by means of a previously validated siRNA60 confirm that, contrary to piceatannol, resveratrol stimulates autophagy and the deacetylation of cytoplasmic proteins by a SIRT1-dependent mechanism (Fig. 6C and D). This notion is further strenghtened by the observations that resveratrol and piceatannol combined in growing concentrations were more efficient at inducing the accumulation of GFP-LC3+ puncta as well as the deacetylation of cytoplasmic proteins in U2OS cells than either compound alone (Fig. 6E and F). Altogether, our findings suggest a cause-effect relationship between autophagy induction and the deacetylation of cytoplasmic proteins.
Figure 6. Resveratrol and piceatannol operate via distinct mechanisms and synergize in the induction of autophagy. (A–D). Differential effects of sirtuin 1 (SIRT1) inhibition on resveratrol- and piceatannol-induced autophagy. GFP-LC3-expressing human osteosarcoma U2OS cells were left untreated or treated with 1 µM rapamycin, 30 µM resveratrol and 30 µM piceatannol, alone or in combination with 10 µM EX527 for 6 h (A and B). Alternatively, GFP-LC3-expressing U2OS cells were transfected with a control siRNA (siUNR) or with a SIRT1-targeting siRNA (siSIRT1) for 24 h, and then left untreated or treated with 30 µM resveratrol or 30 µM piceatannol for additional 6 h (C and D). Finally, cells were processed for the immunofluorescence microscopy-assisted quantification of GFP-LC3+ dots (A and C) or cytoplasmic protein acetylation (B and D). Results from n = 3 independent experiments are reported as means ± SEM *p < 0.05 (unpaired, two-tailed Student’s t-test), as compared with cells treated with the same phenolic compound alone in control conditions (no EX527 or siUNR transfection). (E and F). Synergistic effects of resveratrol and piceatannol on the induction of autophagy and on the deacetylation of cytoplasmic proteins. U2OS cells were incubated with resveratrol and piceatannol, alone or in combination (the total concentration of polyphenols is indicated), for 6 h, followed by the immunofluorescence microscopy-assisted quantification of GFP-LC3+ dots (E) or cytoplasmic protein acetylation (F). Results from n = 3 independent experiments are reported as means ± SEM *p < 0.05 (unpaired, two-tailed Student’s t-test), as compared with cells receiving the same concentration of either compound alone.
Concluding remarks
The results presented in this work point to a strong negative correlation between the capacity of different mono- and polyphenols to trigger autophagy and their potential to cause the deacetylation of cytoplasmic proteins. Spermidine-induced autophagy correlates with its inhibitory effects on histone acetylases and resveratrol-induced autophagy has been linked to its capacity to activate the protein deacetylase sirtuin 1.25,50 In accord with previous reports,50 the pharmacological or genetic inhibition of sirtuin 1 reduced both autophagy induction and cytoplasmic protein deacetylation as triggered by resveratrol, hence establishing a bona fide cause-effect relationship between these two phenomena. Surprisingly, a large panel of mono- and polyphenols contained in red wine and many other food preparations that are suggested to have beneficial effects on human health was able to coordinately promote the deacetylation of cytoplasmic proteins and the autophagic flux. These observations are in line with the speculation that chemical compounds that extend the lifespan (or at least the healthy lifespan), including natural polyphenols,61,62 do so (at least in part) by inducing autophagy.63 Interestingly, the phenolic compounds found in red wine may induce autophagy through heterogeneous, yet-to-be elucidated mechanisms. Of note, piceatannol, the metabolic product resulting from the oxidation of resveratrol by human cytochrome P450 enzyme CYP1B1,64 induced autophagy (while promoting the deacetylation of cytoplasmic proteins) through a yet elusive mechanism that appears to be virtually independent from SIRT1. Accordingly, piceatannol (as quercetin) has been reported to bind and activate SIRT1 with a KM much higher than that of resveratrol.62 Intriguingly, we found that resveratrol and piceatannol combined in growing concentrations induce autophagy and stimulate the deacetylation of cytoplasmic proteins more efficiently than either compound separately, suggesting that distinct polyphenols may synergize in the activation of autophagy. Whether such a synergistic effect would account for the French paradox remains an open conundrum.65,66
In this work, we developed a simple assay to measure the acetylation of cytoplasmic (as opposed to nuclear) proteins. This protocol is based on a relatively mild fixation/permeabilization step (which allows antibodies to access the cytoplasmic, but not the nuclear, compartment) in combination with immunofluorescence microscopy based on an antibody that specifically recognizes proteins containing acetylated lysine residues. When this method is coupled to quantitative imaging (to define cytoplasmic regions of interest and to exclude false-positive nuclei, as resulting, for instance, from mitosis or apoptosis-associated nuclear breakdown),67-69 it accurately reflects the level of cytoplasmic protein acetylation as measured with other, more laborious techniques. We surmise that this method can be adapted to distinct cell types, including primary leukocytes and epithelial cells, thus offering a cost-effective alternative to measure the metabolic (and pro-autophagic) status of cells. Irrespective of these speculative considerations, our data establish a strong correlation and cause-effect relationship between cytoplasmic deacetylation reactions and autophagy induction.
Materials and Methods
Chemical, cell lines and culture conditions
Resveratrol, piceatannol, epicatechin, caffeic acid, gallic acid, quercetin and bafilomycin A1 were purchased from Sigma-Aldrich; oenin chloride, kuronamin chloride, peonidin-3-O-glucoside chloride and delphinidin-3-O-glucoside chloride from Extrasynthese, rapamycin and EX527 from Tocris Bioscience and myricetin from Indofine Chemical. Culture media and supplements for cell culture were obtained from Gibco-Invitrogen and conventional plasticware from Corning. 96-well black/clear imaging plates were purchased from BD Falcon. Wild-type human osteosarcoma U2OS cells and their GFP-LC3-expressing derivatives were cultured in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 mg/L sodium pyruvate, 10 mM HEPES buffer, 100 units/mL penicillin G sodium and 100 µg/mL streptomycin sulfate (37°C, 5% CO2). Cells were seeded in 6- or 96-well plates and let adapt to the culture substrate for 12–24 h prior to experimental assessments.
RNA interference in human cell cultures
U2OS cells stably expressing GFP-LC3 were grown in 96-well imaging plates until ~50% confluence and then subjected to siRNA transfection by means of the Oligofectamine™ transfection reagent (Invitrogen), following the manufacturer's recommendations. The following siRNA duplexes were employed, both purchased from Sigma-Aldrich: the SIRT1 targeting siRNA siSIRT1 (sense 5′-ACUUUGCUGUAACCCUGUA-3′)60 and the irrelevant, non-targeting siRNA siUNR (sense 5′-GCCGGUAUGCCGGUUAAGU-3′),70,71 employed as a negative control. Cells were subjected to experimental procedures no earlier than 24 h after transfection.
Immunofluorescence microscopy
In all cases but for the specific detection of acetylated lysines in the cytoplasm, cells were fixed with 4% paraformaldeyde (PFA, w:v in PBS) for 15 min at room temperature and then permeabilized with 0.1% TritonX-100 (v:v in PBS) for 10 min, as previously reported.72 Conversely, for the cytoplasm-restricted detection of acetylated lysines, fixation in PFA was not followed by any type of permeabilization. In both experimental settings, non-specific binding sites were then blocked with 5% FBS (v:v in PBS), followed by overnight incubation at 4°C with a primary antibody specifically recognizing lysine residues that have been post-translationally modified by acetylation at the ε amino group (Cell Signaling Technology). Revelation was performed with appropriate AlexaFluor™ conjugates (Molecular Probes-Invitrogen). Nuclear counterstaining was obtained with 10 μ Hoechst 33342 (Molecular Probes-Invitrogen). Non-automated fluorescence microscopy assessments were performed on an IRE2 microscope (Leica Microsystems) equipped with a DC300F camera.
Automated immunofluorescence microscopy
Images from plates processed as described above were acquired using a BD pathway 855 automated microscope (BD Imaging Systems) equipped with a 40X objective (Olympus) and coupled to a robotized Twister II plate handler (Caliper Life Sciences). Images were then analyzed either for the presence of GFP-LC3+ (green) puncta in the cytoplasm or for the intensity of protein acetylation (red) in the cytoplasm and the nucleus by means of the BD Attovision software (BD Imaging Systems). To this aim, the surface of each cell was segmented and subdivided into a cytoplasmic and a nuclear region according to manufacturer’s standard proceedings. The RB 2x2 and Marr-Hildreth algorithms were used to recognize cytoplasmic GFP-LC3+ dots.
Immunoblotting
Approximately 5 × 105 cells were washed with cold PBS and lysed following standard procedures.73,74 Thereafter, 25 μg of proteins were separated according to molecular weight on NuPAGE Novex Bis-Tris 4–12% pre-cast gels (Invitrogen) and electrotransferred to Immobilon™ PVDF membranes (Bio-Rad). Non-specific binding sites were blocked with 5% non-fat powdered milk (w:v) plus 0.05% Tween 20 (v:v) in TBS for 1 h, followed by overnight incubation at 4°C with the following primary antibodies: anti-Acetylated-Lysine (Cell Signaling Technology), anti-Acetylated-Histone 3 (Lys9, Cell Signaling Technology), anti-Acetylated-Tubulin (Lys40, Cell Signaling Technology), anti-LC3B (Cell Signaling Technology) and anti-p62 (Abnova). Revelation was performed with appropriate horseradish peroxidase (HRP)-labeled secondary antibodies (Southern Biotech) coupled with the SuperSignal West Pico chemoluminiscent substrate (Thermo Scientific-Pierce). Ponceau Red staining or antibodies recognizing glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Chemicon International) or histone 3 (Cell Signaling Technology) were used to monitor equal loading of lanes. Densitometry was performed by means of the open-source software ImageJ (freely available at http://rsbweb.nih.gov/ij/download.html). To this aim, the intensity of the band of interest was normalized to that of loading controls.
Statistical analysis
Unless otherwise specified, experiments invariably entailed triplicate parallel assessments and were repeated at least twice. Data were analyzed using the Prism 5 software (GraphPad Software) and statistical significance was assessed by means of unpaired, two-tailed Student’s t-tests. The correlation between data sets was evaluated by means of determination coefficients (R2). p values < 0.05 were considered statistically significant.
Acknowledgments
G.K. is supported by the Ligue Nationale contre le Cancer (LNC, Equipe labelisée), Agence Nationale pour la Recherche (ANR), European Commission (Active p53, Apo-Sys, ChemoRes, ApopTrain), Fondation pour la Recherche Médicale (FRM), Institut National du Cancer (INCa), Cancéropôle Ile-de-France, Fondation Bettencourt-Schueller and the LabEx Immuno-Oncology. G.M. and M.N.S. are supported by an EMBO fellowship and a postdoctoral contract from the Junta de Extremadura, respectively. S.A.K. is the recipient of a grant from the Higher Education Commission (HEC) of Pakistan. L.G. is financed by the LabEx Immuno-Oncology.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22027
References
- 1.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–12. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madeo F, Eisenberg T, Kroemer G. Autophagy for the avoidance of neurodegeneration. Genes Dev. 2009;23:2253–9. doi: 10.1101/gad.1858009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38:768–74. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blagosklonny MV. Rapamycin and quasi-programmed aging: four years later. Cell Cycle. 2010;9:1859–62. doi: 10.4161/cc.9.10.11872. [DOI] [PubMed] [Google Scholar]
- 12.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–90. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, et al. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 2011;10:4230–6. doi: 10.4161/cc.10.24.18486. [DOI] [PubMed] [Google Scholar]
- 15.Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA, Jr., Aris JP. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5:847–9. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–9. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 17.Hartford CM, Ratain MJ. Rapamycin: something old, something new, sometimes borrowed and now renewed. Clin Pharmacol Ther. 2007;82:381–8. doi: 10.1038/sj.clpt.6100317. [DOI] [PubMed] [Google Scholar]
- 18.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–32. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 19.Rose C, Menzies FM, Renna M, Acevedo-Arozena A, Corrochano S, Sadiq O, et al. Rilmenidine attenuates toxicity of polyglutamine expansions in a mouse model of Huntington’s disease. Hum Mol Genet. 2010;19:2144–53. doi: 10.1093/hmg/ddq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 21.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knab B, Osiewacz HD. Methylation of polyphenols with vicinal hydroxyl groups: A protection pathway increasing organismal lifespan. Cell Cycle. 2010;9:3387–8. doi: 10.4161/cc.9.17.13016. [DOI] [PubMed] [Google Scholar]
- 23.Lu JY, Lin YY, Zhu H, Chuang LM, Boeke JD. Protein acetylation and aging. Aging (Albany NY) 2011;3:911–2. doi: 10.18632/aging.100398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–29. doi: 10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 27.Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, Tavernarakis N, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009;1:961–70. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein S, Matter CM. Protective roles of SIRT1 in atherosclerosis. Cell Cycle. 2011;10:640–7. doi: 10.4161/cc.10.4.14863. [DOI] [PubMed] [Google Scholar]
- 29.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–51. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vetterli L, Maechler P. Resveratrol-activated SIRT1 in liver and pancreatic β-cells: a Janus head looking to the same direction of metabolic homeostasis. Aging (Albany NY) 2011;3:444–9. doi: 10.18632/aging.100304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–38. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim GW, Yang XJ. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem Sci. 2011;36:211–20. doi: 10.1016/j.tibs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–6. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 35.Perdiz D, Mackeh R, Poüs C, Baillet A. The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell Signal. 2011;23:763–71. doi: 10.1016/j.cellsig.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–53. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Muth V, Nadaud S, Grummt I, Voit R. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20:1353–62. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, et al. Acetylation-dependent signal transduction for type I interferon receptor. Cell. 2007;131:93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–7. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–20. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011;36:108–16. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirschey MD, Shimazu T, Capra JA, Pollard KS, Verdin E. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging (Albany NY) 2011;3:635–42. doi: 10.18632/aging.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salminen A, Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell Signal. 2009;21:1356–60. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 46.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–80. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–8. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–55. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–29. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aires DJ, Rockwell G, Wang T, Frontera J, Wick J, Wang W, et al. Potentiation of dietary restriction-induced lifespan extension by polyphenols. Biochim Biophys Acta. 2012;1822:522–6. doi: 10.1016/j.bbadis.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burke MF, Khera AV, Rader DJ. Polyphenols and cholesterol efflux: is coffee the next red wine? Circ Res. 2010;106:627–9. doi: 10.1161/CIRCRESAHA.109.215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pallauf K, Rimbach G. Autophagy, polyphenols and healthy ageing. Ageing Res Rev. 2012 doi: 10.1016/j.arr.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tasdemir E, Galluzzi L, Maiuri MC, Criollo A, Vitale I, Hangen E, et al. Methods for assessing autophagy and autophagic cell death. Methods Mol Biol. 2008;445:29–76. doi: 10.1007/978-1-59745-157-4_3. [DOI] [PubMed] [Google Scholar]
- 56.Kepp O, Galluzzi L, Lipinski M, Yuan J, Kroemer G. Cell death assays for drug discovery. Nat Rev Drug Discov. 2011;10:221–37. doi: 10.1038/nrd3373. [DOI] [PubMed] [Google Scholar]
- 57.Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006;281:36303–16. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- 58.Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010;9:844–55. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 59.Schlicker C, Boanca G, Lakshminarasimhan M, Steegborn C. Structure-based development of novel sirtuin inhibitors. Aging (Albany NY) 2011;3:852–72. doi: 10.18632/aging.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–63. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- 61.Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov. 2012;11:443–61. doi: 10.1038/nrd3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 63.Nair S, Ren J. Autophagy and cardiovascular aging: lesson learned from rapamycin. Cell Cycle. 2012;11:2092–9. doi: 10.4161/cc.20317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Potter GA, Patterson LH, Wanogho E, Perry PJ, Butler PC, Ijaz T, et al. The cancer preventative agent resveratrol is converted to the anticancer agent piceatannol by the cytochrome P450 enzyme CYP1B1. Br J Cancer. 2002;86:774–8. doi: 10.1038/sj.bjc.6600197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galluzzi L, Kepp O, Kroemer G. TP53 and MTOR crosstalk to regulate cellular senescence. Aging (Albany NY) 2010;2:535–7. doi: 10.18632/aging.100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martins I, Galluzzi L, Kroemer G. Hormesis, cell death and aging. Aging (Albany NY) 2011;3:821–8. doi: 10.18632/aging.100380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12:385–92. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- 68.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 69.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Nomenclature Committee on Cell Death 2009 Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de La Motte Rouge T, Galluzzi L, Olaussen KA, Zermati Y, Tasdemir E, Robert T, et al. A novel epidermal growth factor receptor inhibitor promotes apoptosis in non-small cell lung cancer cells resistant to erlotinib. Cancer Res. 2007;67:6253–62. doi: 10.1158/0008-5472.CAN-07-0538. [DOI] [PubMed] [Google Scholar]
- 71.Galluzzi L, Vitale I, Senovilla L, Olaussen KA, Pinna G, Eisenberg T, et al. Prognostic impact of vitamin B6 metabolism in lung cancer. Cell Rep. 2012 doi: 10.1016/j.celrep.2012.06.017. In press. [DOI] [PubMed] [Google Scholar]
- 72.Morselli E, Tasdemir E, Maiuri MC, Galluzzi L, Kepp O, Criollo A, et al. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle. 2008;7:3056–61. doi: 10.4161/cc.7.19.6751. [DOI] [PubMed] [Google Scholar]
- 73.Criollo A, Galluzzi L, Maiuri MC, Tasdemir E, Lavandero S, Kroemer G. Mitochondrial control of cell death induced by hyperosmotic stress. Apoptosis. 2007;12:3–18. doi: 10.1007/s10495-006-0328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, et al. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]