Abstract
Growing RNA chains fold cotranscriptionally as they are synthesized by RNA polymerase (RNAP). Riboswitches, which regulate gene expression by adopting alternative RNA folds, are sensitive to cotranscriptional events. We developed an optical-trapping assay to follow the cotranscriptional folding of a nascent RNA, and used it to monitor individual transcripts of the pbuE adenine riboswitch, visualizing distinct folding transitions. We report a particular folding signature for the riboswitch aptamer whose presence directs the gene-regulatory transcription outcome, and we measured the termination frequency as a function of adenine level and tension applied to the RNA. Our results demonstrate that the outcome is kinetically controlled. These experiments furnish a means to observe conformational switching in real time, and enable the precise mapping of events during cotranscriptional folding.
Structured RNAs function during transcription or translation to influence a variety of processes. In vivo, nascent RNAs fold as they are transcribed, and this sequential process affects the folding efficiency (1-4) and the predominant conformation (5-9). The study of cotranscriptional folding has heretofore been largely indirect, with most approaches monitoring the final RNA product (1-10). Using single-molecule spectroscopy, we developed a system to visualize RNA folding in an individual, nascent transcript directly, and used it to record the functional switching of a riboswitch. The conformation of the riboswitch ligand-binding aptamer affects the structure of the downstream RNA through changes in the availability of nucleotides shared between the aptamer and an ‘expression platform’, leading to structure-dependent gene regulation (11, 12). The pbuE riboswitch from Bacillus subtilis regulates adenine levels, controlling transcription of downstream genes by forming an aptamer, which binds adenine and acts as an anti-terminator, or an expression platform, consisting of a terminator hairpin that halts transcription (13) (Fig. 1A). Previous work has considered the post-transcriptional folding of this aptamer (13-16). Here, we consider the functional consequences of the aptamer and terminator domains acting in concert in real time. Following transcription of the full riboswitch, only the folded terminator has previously been observed (15-18), as expected given the far greater energetic stability of this domain (19). Any switching behavior must therefore be studied in the context of active transcription.
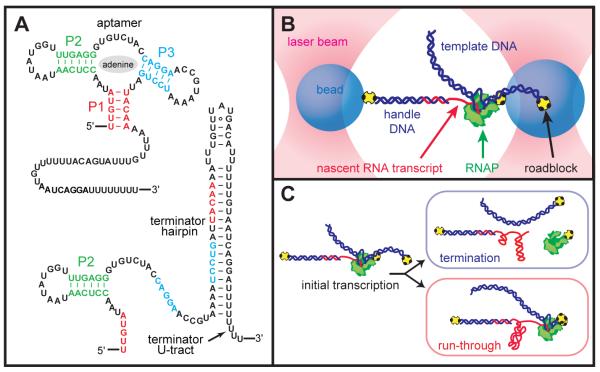
Fig. 1.
(A) Secondary-structure of competing aptamer and terminator conformations of the pbuE riboswitch. (B) Schematic of the “dumbbell” assay showing the experimental geometry, with the nascent RNA transcript transcribed in situ (not to scale). (C) Elongation produces the riboswitch, causing RNAP to dissociate at the terminator element or to run through to the roadblock at the template end.
In our assay (14, 20), a transcriptionally stalled molecule of Escherichia coli RNAP carrying a short initial transcript was tethered in a dual-beam optical tweezers apparatus in a “dumbbell” configuration (19), with RNAP linked to one bead and the transcript linked to the other via hybridization to a complementary, cohesive end of a dsDNA “handle” (14, 21) (Fig. 1B). Transcription was restarted by the addition of nucleoside triphosphates (1 mM NTPs) in the presence or absence of saturating levels of adenine base (300 μM), and the separation between beads was measured. Transcription proceeded through the riboswitch, either finishing at the position of the terminator—where the dumbbell assembly dissociated as RNAP released the RNA— or continuing to the end of the template, where RNAP was arrested by a streptavidin roadblock—at which point the dumbbell tether remained intact (Fig. 1C).
Adenine-dependent termination was measured both in bulk (no force) and for single molecules (Fig. 2A, B). Termination was scored as the termination efficiency (TE), defined as the fraction of complexes dissociating at the terminator position. Bulk assays gave high levels of termination in the absence of adenine (89%) that decreased in its presence (49%, saturating adenine). For single molecules, the TE depends upon force, because folded conformations of the aptamer and terminator are destabilized by increasing loads. At low forces, where these structures can form (<7 pN), TEs were similar to those in bulk. Without adenine (Fig. 2B, blue), most transcripts terminated at low force, but TEs decreased under loads approaching the unfolding force of the terminator hairpin (~13 pN). This behavior is identical to that reported for intrinsic terminators alone, and fit by a quantitative model (19, 21). The upstream aptamer sequence in the absence of adenine only slightly lowered the TE (at F = 0) relative to that of the isolated terminator. Under saturating adenine levels (Fig. 2B, red), the TE remained near 50% throughout the low-force regime, where the aptamer folds readily, but defaulted to the identical load dependence as data acquired without adenine for forces beyond ~7 pN, the equilibrium value for folding of the aptamer region alone. These results imply that, absent an adenine-reinforced aptamer caused by either (a) a lack of adenine or (b) a load-dependent destabilization of the aptamer, the riboswitch behaves as an unhindered terminator.
Fig. 2.
Termination efficiency (mean ± SEM). (A) TE from a gel-based assay with termination (T) and run-through (R) bands, and (B) as a function of tension applied to the RNA. Data under load were from single-molecule records (filled symbols); zero-force points were from gel assays (open symbols). Blocking RNA upstream of the terminator with a DNA oligomer suppressed aptamer formation and created an isolated terminator, raising TE slightly (gray open square). Data without adenine were fit by a model of intrinsic termination (blue curve). The equilibrium unfolding forces of the adenine-bound aptamer and terminator hairpin are indicated (orange, gray shading and dashed lines, respectively; mean ± SD). (C) Cotranscriptional folding of a hairpin (left; 72 bp, 12 pN load). Stages during transcription are illustrated (right; not to scale): (1) Extension remains constant before transcription re-start, then (2) increases (outward arrows) during elongation until (3) the duplex nucleates, causing extension to decrease (inward arrows) as (4) the hairpin forms. Once the hairpin is completed, (5) elongation resumes until RNAP reaches the roadblock (6).
We observed transcription directly, including cotranscriptional folding events, by monitoring transcript extension change over time. By shortening RNA tethers into the range of hundreds of bases, which increases stiffness and reduces noise, and lowering the tension below levels sufficient to open short hairpins, we were able to visualize structure formation as it took place. We also revised our protocols to measure transcription without significant delay from the re-start of transcription (19). Two antagonistic effects control end-to-end transcript length: the extension tends to increase overall as RNA gets synthesized, but may decrease whenever the nascent chain folds. Fig. 2C illustrates such lengthening as the first part of the stem and loop of a simple hairpin are synthesized, followed by shortening as the complementary portion of the stem is generated, until the folding completes, whereupon lengthening resumes until the roadblock is reached.
In transcription records of the adenine riboswitch obtained at comparatively high loads (F = 8.1 pN; Fig. 3A), extension increased monotonically during the synthesis of the early, unstructured sequence, followed by a decrease at the first fold, corresponding to the folding of the P2 stem-loop. Transcription continued until the terminator was reached, at which point its hairpin folded and transcription was terminated (releasing the RNA) or RNAP ran through to the end of the template. At this high force, the aptamer rarely forms, so the TEs and shapes of records were similar in the presence or absence of adenine (Fig. 3A and Fig. S3).
Fig. 3.
Riboswitch transcription showing cotranscriptional folding. Transcript extension generally increased during transcription, with local decreases induced by folding events. Terminating traces led to RNA release and a sharp upward displacement, corresponding to tether breakage; run-through traces remained at a fixed extension once RNAP encountered the terminal roadblock. Records were collected in 0 μM or 300 μM adenine (saturating). (A) At high forces, where the aptamer rarely folds (8.1 pN), folding is adenine-independent (Fig. S3); there is no outcome-correlated difference in folding patterns (purple vs. green traces). Records are offset for display. (B) At lower forces (5.8 pN), run-through traces displayed the characteristic signature of bound aptamer formation. A run-through record is superimposed against a simple folding model (grey; see Supplementary Materials), with the events indicated. (C) Records displayed distinct motifs correlated with the transcription outcome. Records overlayed: absent adenine (top panel), run-through events were rare and records did not display an aptamer folding signature following P2 formation, appearing indistinguishable from terminating records. With adenine (bottom panel), aptamer folding led to transcriptional run-through.
At lower loads, adenine-dependent folding occurred in a way that decided transcriptional fate. The signature of aptamer folding appeared as a relatively large decrease in extension, after P2 folded and before the terminator hairpin was generated: this resulted in a stable fold, during which other folding events were not observed. A simplified model of cotranscriptional folding serves as a useful guide to track the folding events (Fig. 3B). P2 folded first, followed closely by P3 and then the aptamer. With the aptamer formed, RNAP is able to transcribe past the terminator hairpin without it folding. However, even in such cases, the adenine-bound aptamer was eventually observed to unfold and become replaced by an equilibrium structure with the terminator hairpin folded. However, because RNAP had already moved beyond the terminator position, the RNA was not released.
Comparisons at lower force (5.8 pN) showed that in the presence of adenine, the vast majority of records destined to run through displayed a distinct folding signature, attributable to aptamer folding. Aptamer folding signatures were not observed in records that ultimately terminated, nor in rare run-through records obtained without adenine (Fig. 3C and Fig. S4). With adenine present, the chance of run-through in a record carrying the aptamer-folding signature was at least 95%. Conversely, the probability of transcriptional termination when the aptamer-fold signature was not scored but adenine was present was ~85%. The probability of run-through under such conditions (~15%) agrees closely with the probability of run-through in the absence of adenine.
Based upon the short time window for folding/binding in the pbuE aptamer prior to the termination decision (~2 s), and upon the relatively long lifetime of the adenine-bound aptamer (~10 s) (16, 19), as well as the dependence of TE on transcription rate (18), it has been conjectured that kinetic (as opposed to thermodynamic) effects would play a dominant role in controlling transcriptional fate. We find unambiguous evidence that the pbuE riboswitch is kinetically controlled, because aptamers were observed to fold and bind adenine once, or not at all, precluding any possible thermodynamic equilibration between conformations during the binding window, as transcription proceeded rapidly (~20 nt/s). Once formed, the adenine-bound aptamer rarely unfolded before the termination decision (Fig. S5A), and it was observed to fold multiple times only in one unusually slow record (Fig. S5B). The probability of forming an unproductive, aptamer-like fold (i.e., where the aptamer folds and unfolds at least once before influencing the outcome) is therefore low: 5% or fewer transcripts generated an unproductive, adenine-associated fold.
The FMN riboswitch sequence contains two pause sites thought to be important for allowing FMN additional time to bind (7), and possibly increasing the time window for folding. In the pbuE riboswitch, two short series of U residues, situated before the U-tract of the terminator, have similarly been proposed to act as pause sites (16), with pausing reported in one of these regions, but only observed under limiting NTP conditions (18). In our experiments using physiological levels of NTPs, long pauses were not observed at either of these sites; pauses, if any, were too brief to identify (<1 s). This discrepancy might be attributed to the differences in NTP concentrations, to transcription by the cognate B. subtilis RNAP (although this is unlikely to increase pausing), or to the involvement of other factors (7, 18, 22), as discussed (19).
Our data show that the folded aptamer can persist briefly after transcription of the terminator hairpin, temporarily trapping the riboswitch far from equilibrium. Taking U8 of the terminator U-tract as the position where the terminator is largely completed, we find that in the presence of adenine, run-through records unfolded beyond this point with a dwell time averaging 2.6 s (Fig. S7). In the functional riboswitch, the aptamer persisted for a shorter time than it would have without any competing structure (~10 s for the adenine-bound aptamer; Fig. S7B), implying that some mechanism, for example branch migration, may facilitate conversion between states (5, 6). The unfolding aptamer gave way to the terminator conformation instantaneously on the timescale of our apparatus (~50 ms), and the RNA remained thereafter in this equilibrium state (Fig. S6).
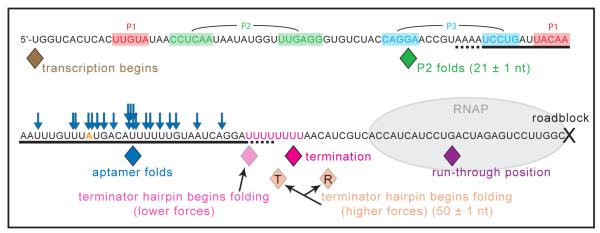
Using data collected over a range of forces in the presence and absence of adenine, we can construct a map of cotranscriptional folding (Fig. 4; Table S2). P2 is the first significant structure to fold, sequestering 21 ± 1 nt (as expected, based on sequence) once RNAP reached a position 12 ± 1 nt after the last nucleotide of P2 was transcribed, which agrees closely with the ~12 nt RNA footprint estimated for RNAP (23, 24). The aptamer region is transcribed by nt 74, and it folded, on average, when RNAP reached nt 90 ± 1, soon after the RNAP footprint cleared the aptamer. At low forces, if the aptamer failed to fold, the terminator hairpin formed later in transcription, once a segment of its complementary RNA hairpin sequence became available. Termination subsequently occurred at nt 112 ± 1, around U7 of the U-tract, which is typical for intrinsic terminators (25). Notably, at higher forces for records that terminated, the terminator hairpin folded at nt 110 ± 1, whereas for records that ran through, the terminator folded later, at nt 117 ± 1 (after transcription of the downstream U-tract), supporting the notion that the energy of hairpin formation assists in releasing the RNA. For loads close to the unfolding force of the terminator, records showed no evidence of cotranscriptional hairpin folding, and the vast majority ran through. The few records that terminated appeared to do so at the ‘slippery’ U-tract (21). In all records that ran through, RNAP arrested at nt 134 ± 1, a position ~15 nt upstream of the roadblock, consistent with the downstream footprint of RNAP (26, 27).
Fig. 4.
Map of cotranscriptional folding events. The average computed position of the last nucleotide transcribed at the specified event is indicated (solid diamonds; SEM ±~1 nt, approximately equal to the symbol width), beginning with the resumption of transcription at the 5′-U, ending with termination or run-through and roadblock arrest. The numbers of nt associated with folding elements are indicated. Adenine-bound aptamer folding (blue diamond, average position; blue arrows, sampled individual observations) occurred between P1 and complete terminator transcription. For forces below 9 pN (5.8 pN; 8.1 pN), the terminator hairpin (underlined) was observed to begin folding (light pink diamond) before the termination position (dark pink diamond; transcript release). At higher forces (10.4 pN), the average folding position differed between traces that terminated (T, orange diamond) or ran through (R, orange diamond), occurring too late in the latter to produce successful termination. See Table S2 for additional details.
These experiments provide a real-time window into transcript formation and functional switching in a riboswitch. Cotranscriptional folding is fundamental to the action of the pbuE adenine riboswitch, which exerts its modulatory effects far from equilibrium. Once the aptamer sequence is formed, the adenine-bound aptamer is able to briefly lock down the riboswitch conformation until RNAP has successfully passed the termination signal. Conversely, if the ligand-bound aptamer fails to become stabilized by the time RNAP reaches the decision point, the terminator hairpin forms instead, leading to prompt transcript release at the terminator U-tract. Cotranscriptional folding events follow a sequential progression, with associated structures forming within seconds (or less) of the times that their sequences clear the RNAP footprint. Nascent transcript structures that get trapped out of equilibrium are later found to switch conformations nearly instantly. The sensitivity of this system to kinetic details therefore poses a serious challenge in the quest for synthetic mimics of riboswitches in a bioengineering context. The experimental approaches presented here provide the opportunity to explore the dynamics of cotranscriptional folding directly in a variety of RNA structures. We anticipate that force spectroscopy will become a useful tool in the study of cotranscriptional events, and possibly cotranslational events as well.
Supplementary Material
One Sentence Summary.
Folding events were observed cotranscriptionally as individual RNA molecules of the pbuE riboswitch were transcribed, and different folding patterns were shown to switch the termination outcome in a kinetically-controlled fashion.
Acknowledgements
We thank R. Landick for providing purified RNAP, and C. García-García, V. Schweikhard, W. Greenleaf, P. Anthony, and other members of the Block lab for useful discussions. The data described in this manuscript are tabulated in the main paper and in the supplementary materials. This research was supported by a grant from the NIH/NIGMS (S.M.B.), and an NSF Graduate Research Fellowship and Stanford Graduate Fellowship (K.L.F.).
References and Notes
- 1.Heilman-Miller SL, Woodson SA. RNA. 2003;9:722. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan T, Fang X, Sosnick T. Journal of Molecular Biology. 1999;286:721. doi: 10.1006/jmbi.1998.2516. [DOI] [PubMed] [Google Scholar]
- 3.Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Proc Natl Acad Sci USA. 1999;96:9545. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong TN, Sosnick TR, Pan T. Proc Natl Acad Sci USA. 2007;104:17995. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xayaphoummine A, Viasnoff V, Harlepp S, Isambert H. Nucleic Acids Research. 2007;35:614. doi: 10.1093/nar/gkl1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. PLoS Biology. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. Molecular Cell. 2005;18:49. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. Genes Dev. 2009 Nov 15;23:2650. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perdrizet GA, 2nd, Artsimovitch I, Furman R, Sosnick TR, Pan T. Proc Natl Acad Sci USA. 2012 Feb 28;109:3323. doi: 10.1073/pnas.1113086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan T, Sosnick T. Annu. Rev. Biophys. Biomol. Struct. 2006;35:161. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 11.Henkin TM. Genes Dev. 2008;22:3383. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth A, Breaker RR. Annu Rev Biochem. 2009;78:305. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandal M, Breaker RR. Nature Structural & Molecular Biology. 2004;11:29. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 14.Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Science. 2008;319:630. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemay JF, Penedo JC, Tremblay R, Lilley DMJ, Lafontaine DA. Chemistry & Biology. 2006;13:857. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. Biochemistry. 2005;44:13404. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 17.Rieder R, Lang K, Graber D, Micura R. ChemBioChem. 2007;8:896. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 18.Lemay JF, et al. PLoS Genetics. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supplementary materials on Science Online.
- 20.Dalal RV, et al. Molecular Cell. 2006;23:231. doi: 10.1016/j.molcel.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson MH, Greenleaf WJ, Landick R, Block SM. Cell. 2008;132:971. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R. Journal of Bacteriology. 2000;182:6027. doi: 10.1128/jb.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monforte JA, Kahn JD, Hearst JE. Biochemistry. 1990;29:7882. doi: 10.1021/bi00486a015. [DOI] [PubMed] [Google Scholar]
- 24.Komissarova N, Kashlev M. Proc Natl Acad Sci USA. 1998;95:14699. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.d’Aubenton Carafa Y, Brody E, Thermes C. Journal of Molecular Biology. 1990;216:835. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 26.Darst SA. Current Opinion in Structural Biology. 2001;11:155. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 27.Greive SJ, von Hippel PH. Nature Reviews Molecular Cell Biology. 2005;6:221. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.