Summary
This article develops a variety of influence measures for carrying out perturbation (or sensitivity) analysis to joint models of longitudinal and survival data (JMLS) in Bayesian analysis. A perturbation model is introduced to characterize individual and global perturbations to the three components of a Bayesian model, including the data points, the prior distribution, and the sampling distribution. Local influence measures are proposed to quantify the degree of these perturbations to the JMLS. The proposed methods allow the detection of outliers or influential observations and the assessment of the sensitivity of inferences to various unverifiable assumptions on the Bayesian analysis of JMLS. Simulation studies and a real data set are used to highlight the broad spectrum of applications for our Bayesian influence methods.
Keywords: Bayesian influence measure, Longitudinal, Perturbation model, Sensitivity analysis, Survival
1. Introduction
There has been extensive research literature on joint modeling of longitudinal and survival data (JMLS) by using either frequentist or Bayesian methods. For instance, the early development of joint models for longitudinal and survival data was primarily motivated by characterizing the relationship between features of CD4 or viral load profiles and time-to-event in HIV/AIDS clinical trials. JMLS has been further developed in other types of biomedical applications, such as cancer vaccine (immunotherepy) trials and quality of life studies. References include Pawitan and Self (1993); De Gruttola and Tu (1994); Tsiatis, DeGruttola, and Wulfsohn (1995); Faucett and Thomas (1996); Wulfsohn and Tsiatis (1997); Henderson, Diggle, and Dobson (2000); Wang and Taylor (2001); Xu and Zeger (2001); Law, Taylor, and Sandler (2002); Song, Davidian, and Tsiatis (2002); Chen, Ibrahim, and Sinha (2002, 2004a); Brown and Ibrahim (2003a; Brown and Ibrahim (2003b); Brown, Ibrahim, and DeGruttola (2005) and Chi and Ibrahim (2006, 2007) among many others. Nice overviews of JMLS are given in Tsiatis and Davidian (2004) and Yu et al. (2004) from a frequentist perspective, and in Ibrahim, Chen, and Sinha (2001) as well as Hanson, Branscum, and Johnson (2011) from a Bayesian perspective.
Recent advances in computation and prior elicitation have made Bayesian analysis of these “complex” JMLS feasible. For instance, some Bayesian semiparametric approaches for longitudinal profiles include Gaussian processes, and functional Dirichlet processes among others. Nonparametric prior processes for the baseline cumulative hazard function, include the gamma process prior, the correlated gamma process, and the Dirichlet process prior among others. However, very little has been done on developing a general Bayesian influence approach to detect influential points and to assess various unverifiable assumptions underlying the JMLS, which is the focus of this article.
Bayesian local and global influence (or robustness) approaches have been widely used to perturb the data, the prior and the sampling distribution and assess the influence of these perturbations on the posterior distribution and the associated posterior quantities. However, these Bayesian local and global influence approaches are not directly applicable to complex JMLS. Although there are some frequentist diagnostic tools (Dobson and Henderson, 2003; Rizopoulos, Verbeke, and Molenberghs, 2008; Rizopoulos and Ghosh, 2011) developed specifically for some JMLS, they are not sufficient for carrying out sensitivity analysis (e.g., prior) of complex Bayesian analysis of JMLS.
The development of the proposed methodology was primarily motivated by a clinical trial conducted by the International Breast Cancer Study Group (IBCSG) (Chi and Ibrahim, 2006). A subset of the IBCSG data set contains the longitudinal measurements for quality of life (QOL) measured at baseline and at months 3 and 18 after randomization. In addition, we have bivariate failure times, including disease-free survival (DFS) and overall survival (OS), which were collected from n = 832 patients from Switzerland, Sweden, and New Zealand/Australia. The covariates are different therapeutic procedures, age, estrogen receptor (ER) status (negative/positive), and the number of positive nodes of the tumor. Although a Bayesian analysis of joint longitudinal and bivariate survival models have been used to fit this data set (Chi and Ibrahim, 2006), a general diagnostic framework for assessing such a model fit to the IBCSG data is completely lacking. There is a great need to develop various diagnostic measures for the detection of outliers and/or influential observations, and the assessment of the sensitivity of inferences to the prior distributions and other unverifiable assumptions on the JMLS.
The article is organized as follows. In Section 2, we introduce a general model for jointly modeling multivariate longitudinal and survival data. In Section 3, we discuss various perturbation models and then calculate their associated Bayesian influence measures to quantify the effects of perturbing the data, the prior, and the sampling distribution on possible posterior quantities of interest. We present a detailed analysis of the IBCSG data in Section 4.
2. Joint Models of Longitudinal and Survival Data
Consider data from n independent subjects. For each subject, we observe a K × 1 vector of multiple longitudinal responses and an M × 1 vector of multivariate time-to-event outcomes. For the ith subject, let yik (tijk) be an assessment of the kth longitudinal response measured at time tijk and let Yik = (yik (ti1k), …, yik (tinikk))T denote the observed longitudinal process for the kth response for i = 1, …, n, j = 1, …, nik, and k = 1, …, K. Moreover, for the ith subject, we observe the event time and the event indicator
for the mth time-to-event outcome for m = 1, …, M, where 1(A) is an indicator function of an event A and and Cim denote the true event time and the censoring time, respectively.
We consider a general shared parameter model for jointly modeling the longitudinal and survival data as follows. Let bi = (bi1, …, biK) be time-independent random effects underlying both the longitudinal and survival processes for the ith subject. Conditional on bi, all components of the longitudinal outcomes and the time-to-event outcomes are independent. Let Yi = (Yi1, …, YiK), Ti = (Ti1, …, TiM)T, and δi = (δi1, …, δiM)T. The shared parameter model is defined by
| (1) |
where p(… | …) and p(…) denote the appropriate conditional density and density functions, respectively, and is the vector containing all unknown parameters corresponding to each of the submodels. Moreover, corresponding to the partition of can be further decomposed as a finite-dimensional parameter vector and a vector of infinite-dimensional parameters , such as the baseline hazard function or cumulative baseline hazard function, for each of the submodels. This class of shared parameter models in equation (1) includes most JMLS in the existing literature as a special case (Ibrahim et al., 2001; Tsiatis and Davidian, 2004; Hanson et al., 2011).
We specify each of the submodels in equation (1) as follows. First, we consider a multivariate generalized linear mixed model for the longitudinal process. Specifically, all components of Yik conditional on bik are independent and the conditional distribution of each yik (tijk) given bik is a member of the exponential family with a nonlinear link function given by
| (2) |
where gk (·) is a known monotonic link function and ηik (t, bik) is a parametric or nonparametric function of the random effects and t. Also, ηik (t, bik) may depend on other covariates of interest, such as gender. Furthermore, we consider a general form of ηik (t, bik) given by
| (3) |
where Rik (t) and Wik (t) are, respectively, the fixed effects and random effects design matrices, and βk and bik are vectors of the corresponding fixed and random effects parameters.
For the specification in equation (3), we may consider a random varying-coefficient model as follows:
| (4) |
where xi = (xi1, …, xip)T is a vector of covariates and fs (t) are known basis functions, such as B-splines. If θikl,s = βkl,s + bikl,s with E(bikl,s) = 0, then βk includes all coefficients βkl,s and bik includes all bikl,s for all k, l, and s.
Second, we consider a general multivariate survival model for the survival process as follows. Let S(t1, …, tM |zi, Hi (t, bi), bi) be the joint survivor function of (Ti1, …, TiM) given (zi, Hi (t, bi), bi), where Hi (t, bi) = {ηik (t̃, bik) : t̃ ∈ [0, t), k = 1, …, K} and zi is a vector of time-independent covariates. It is assumed that S(t1, …, tM |zi, Hi (t, bi), bi) takes the form
| (5) |
where F (…) is a known function, φ is a vector of unknown parameters for characterizing the dependence or association structure, and Sm (t|zi, Hi (t, bi), bi) for m = 1, …, M are the marginal survival functions given (zi, Hi (t, bi), bi). For bivariate time-to-event outcomes, Chi and Ibrahim (2006) have proposed a joint survivor function that is a special case of (5).
For the mth time-to-event outcome, we assume that the marginal hazard function of the ith subject is given by
| (6) |
where λm0(t) is an unknown baseline hazard function and θm,T contains all unknown parameters in λm (t|zi, Hi (t, bi), bi) except λm0(t). Moreover, g̃mk (·; ·) for k = 0, …, K are prespecified functions that characterize the effect of the kth longitudinal profile on the mth time-to-event outcome. Then, we calculate . In the literature, it is common to assume that
| (7) |
where γm is a vector of unknown parameters for the time-independent covariates and αmk are unknown parameters.
Third, we consider a multivariate model for the random effects bi as follows. Specifically, let p0(bi ; θb) be a prespecified density function, such as multivariate Gaussian. The density of bi, denoted by p(bi ; θb), is assumed to take the form p0(bi ; θb)ψ(bi ; θb), where ψ(·; ·) is a known and nonnegative function such that ∫ p(bi ; θb)dbi = 1. For instance, ψ(bi ; θb) can be the square of a polynomial function of individual components of θb and the density of a copula function. We may further consider nonparametric alternatives to the parametric model p(bi ; θb), such as a Dirichlet process.
A formal Bayesian analysis of (θ, b) also involves the specification of a prior distribution p(θ), where b = (b1, …, bn). A typical joint prior specification is to assume p(θ) = p(θF)p(θI), where p(θF) and p(θI), respectively, denote parametric prior distributions for the components of θF and non-parametric prior distributions for the components of θI. Let Do = {(Yi, Ti, δi): i = 1, …, n}. Then, we use Markov chain Monte Carlo (MCMC) methods to obtain samples from the joint posterior distribution of (θ, b), which is given by
| (8) |
We focus on nonparametric priors for the nonparametric components in ηik (t, bik) and the baseline hazard or cumulative baseline hazard function. We can take different prior distributions, including a Gaussian prior, zero-inflated priors, and stick-breaking priors among others, for the coefficients {θikl,s} in model (4). For instance, a Dirichlet process for {θikl,s }, denoted by DP(αP0), usually clusters all the longitudinal profiles into one of k ≤ n clusters, where P0 is the base probability measure and α is the confidence parameter. See a nice review of Bayesian nonparametric methods for functional data in Dunson (2009).
Different prior distributions for the baseline hazard λm0(·) or cumulative baseline hazard Λm0(·) include a piecewise constant hazards model, Gamma process model, Beta process model, or a Dirichlet process model. As an illustration, we construct the piecewise constant hazards model for λm0(·). We start with a finite partition of the time axis, 0 < cm,1 < · · · < cm,L, with cm,L > Tim for all i and then set λm0(t) = hmℓ for t ∈ Imℓ = (cm, ℓ−1, cm, ℓ]. Furthermore, a first-order autoregressive prior or an independent gamma prior can be taken for hm = (hm1, …, hmL)T (Sinha, 1993; Arjas and Gasbarra, 1994; Ibrahim et al., 2001). An important alternative is the gamma process prior for Λm0(·), that is, , where c0 is a fixed constant and is a known increasing function with .
Our aim is to carry out Bayesian inference about parameters of interest, which requires a reasonably “robust” prior p(θ) and the correct specification of p(Yi, Ti, δi; θ). A nonrobust prior p(θ), the presence of outliers, and the misspecification of the JMLS may introduce serious bias in the estimation and inference on θ. Thus, it is crucial to assess the sensitivity of statistical inference to the prior, the sampling distribution, and outliers. We note that existing frequentist diagnostic tools are not sufficient for this endeavor (Dobson and Henderson, 2003; Rizopoulos et al., 2008; Rizopoulos and Ghosh, 2011).
Example 1
For the purposes of illustration, we consider an example with two longitudinal markers and bivariate survival times. In this case, K = M = 2. Specifically, each longitudinal response is given by
| (9) |
for i = 1, …, 100, k = 1, 2, and j = 1, …, ni, where the ri s represent a baseline covariate in the longitudinal model. Moreover, it is assumed that tij1 = tij2 for all i and j, εij = (εij1, εij2)T are independently and identically distributed as N2(0, Σ), and the random effects are distributed as N4(0, Φ), where bik = (bik0, bik1)T for k = 1, 2. Here Σ and Φ for k = 1, 2 are covariance matrices. Conditional on bi, the two event and censoring times are assumed to be independent and their marginal hazard functions are given by
| (10) |
for m = 1, 2, where zi = (zi1, zi2)T is a vector of time-independent covariates. Let Yi (t) = (yi1(t), yi2(t))T and ηi (t, bi) = (ηi1(t, bi1), ηi2(t, bi2))T. The density of (Yi, Ti, δi, bi) given θ for the ith subject, denoted by p(Yi, Ti, δi, bi ; θ), is given by
where C0 is a constant independent of θ.
To carry out a Bayesian analysis, we take a joint prior distribution for θ as follows:
| (11) |
for k, m = 1, 2, where , R0, ρ0, , and ρ0 are prespecified hyperparameters. For the baseline hazard λm0(·), we take a piecewise constant hazards model with 250 subintervals with equal lengths such that , where the cm, ls are prespecified constants. We take hm l ~ Γ(τ0l, τ1l) for l = 1, …, 250 and m = 1, 2. We use MCMC methods to conduct our Bayesian influence analysis on θ and b.
3. Bayesian Influence Analysis
We address three issues related to Bayesian influence analysis of JMLS: perturbation models for perturbing the JMLS, appropriate perturbations, and Bayesian influence measures.
3.1. Perturbation Models and Appropriate Perturbations
We introduce three classes of perturbation models to formally perturb JMLS. Let ω be a perturbation vector in a set Ω ⊂ RW, which represents a Euclidean space of dimension W, where W is an integer. The perturbed model
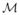 = {p(Do, b; θ, ω): ω ∈ Ω} characterizes various perturbations to the assumed density of p(Do, b; θ) such that ∫ p(Do, b; θ, ω)dDo db = 1 and p(Do, b; θ, ω0) = p(Do, b; θ) for a unique ω0, which represents no perturbation. The first class of perturbations is to individually perturb a subject’s longitudinal profile, repeated measures within each subject, covariates, survival time, and censoring indicator. For instance, we introduce a perturbation vector ωy,i into p(Yi |bi; θy) to perturb Yi such that the perturbed density is given by
= {p(Do, b; θ, ω): ω ∈ Ω} characterizes various perturbations to the assumed density of p(Do, b; θ) such that ∫ p(Do, b; θ, ω)dDo db = 1 and p(Do, b; θ, ω0) = p(Do, b; θ) for a unique ω0, which represents no perturbation. The first class of perturbations is to individually perturb a subject’s longitudinal profile, repeated measures within each subject, covariates, survival time, and censoring indicator. For instance, we introduce a perturbation vector ωy,i into p(Yi |bi; θy) to perturb Yi such that the perturbed density is given by
| (12) |
in which ω = {ωy,1, …, ωy,n }. This single-case perturbation equation (12) to the individual’s longitudinal profile is primarily designed to detect one or a few influential subjects, whose longitudinal profiles differ significantly from the other subjects in the evolution process. Furthermore, we may introduce a perturbation vector ωi into p(Ti, δi |bi; θT) to perturb (Ti, δi) such that the perturbed density is given by
| (13) |
in which . This single-case perturbation equation (13) to the survival data is designed to reveal influential values of survival times relative to other survival times.
The second class is to perturb the shared random effects that underly both the longitudinal measurement and survival processes. For instance, we introduce a perturbation vector ωb,i to simultaneously perturb (Yi, Ti, δi) with the presence of bi such that
| (14) |
In equation (14), we use the same ωb,i to simultaneously perturb p(Yi |bi ; θy) and p(Ti, δi |bi ; θT) to delineate large discrepancies between the longitudinal profile and the corresponding survival time, each of which may not be influential by themselves. This single-case perturbation equation (14) is primarily designed to detect influential survival times, whose occurrence is low relative to their subject-specific longitudinal profiles. An alternative perturbation is to introduce a perturbation ωb,i into p(bi ; θb) such that
| (15) |
Perturbation equation (15) is designed to detect “influential” random effects bi, whereas their corresponding Ti and Yi may be not influential. Furthermore, by setting ωb,i = ωb, we can formally assess the parametric distributional assumptions of the random effects bi and the amount of such perturbations to statistical inferences, such as their impact on parameter estimation.
The third class includes perturbations to the prior p(θ) and the simultaneous perturbations to all three components of the Bayesian model. A fundamental issue associated with any Bayesian analysis is how much posterior quantities, such as the Bayes factor, parameter estimates, and credible (or highest posterior density) intervals, are sensitive to changes in the prior distribution. Thus, it is important to assess both Bayesian semiparametric assumptions for the longitudinal profiles and perturbations regarding the nonparametric prior processes for the cumulative baseline hazard function. For instance, we consider a prior perturbation to Λm0(t) ~
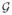 P (c0Λ*(t), c0) by assuming Λm0(t) ~
P (c0Λ*(t), c0) by assuming Λm0(t) ~
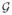 P (c0Λ*(t, ωP), c0) such that
and Λ*(t, 0) = Λ*(t). Combining the first two classes with the third class, we can obtain various simultaneous perturbations to the prior, the sampling distribution, and the data, which allows us to assess the simultaneous sensitivity of all components of a Bayesian analysis.
P (c0Λ*(t, ωP), c0) such that
and Λ*(t, 0) = Λ*(t). Combining the first two classes with the third class, we can obtain various simultaneous perturbations to the prior, the sampling distribution, and the data, which allows us to assess the simultaneous sensitivity of all components of a Bayesian analysis.
Example 1 (continued)
We consider some simultaneous perturbations as follows:
| (16) |
Thus, the perturbed (unnormalized) log-posterior is given by
| (17) |
In this case, ω contains (ωy,1, …, ωy,n, ωb,1, …, ωb,n, ωα1, ωα2, ωγ1, ωγ2)T and ω0 = (0, …, 0, 1, …, 1, 1, 0, 1, 0)T represents no perturbation.
After perturbing the JMLS, we need to quantify the amount of perturbation introduced by each perturbation, the extent to which each component of a perturbation vector contributes to, and the degree of orthogonality for the components of a perturbation vector (Amari, 1990; Zhu, Ibrahim, and Tang, 2011). This is very critical for us to properly pinpoint the cause (e.g., prior) of a large effect. Specifically, we regard the perturbed model p(Do, b, θ; ω) as the probability density of (Do, b, θ) for ω and then calculate its score function ∂ωhℓ(ω), where ∂ωh = ∂/∂ωh, ℓ(ω) = log p(Do, b, θ; ω), and ωh is the hth component of ω. Thus, the Fisher information matrix with respect to ω, denoted by G(ω) = (gh s (ω)), is a W × W matrix with its (h, s) element given by , for h, s = 1, …, W, where Eω denotes the expectation taken with respect to p(Do, b, θ; ω).
We call p(Do, b, θ; ω) an appropriate perturbation model if G(ω0) equals cIW, where c is any positive scalar and IW is a W × W identity matrix (Zhu et al., 2007). Specifically, ghh (ω) can be regarded as the amount of perturbation introduced by ωh, whereas the correlation indicates an association between ωh and ωs. The diagonal structure of G(ω) implies that all components of ω are orthogonal to each other. Orthogonal subcomponents of ω allow for easy detection of the cause of a large effect. If G(ω0) is not diagonal, then we choose a new perturbation vector ω̃, defined by
| (18) |
Based on ω̃, we can obtain a new perturbation model p(Do, b, θ; ω̃) such that G(ω̃) evaluated at ω0 equals IW.
Example 1 (continued)
Let for i = 1, …, n. It follows from the definition of G(ω0) that . The diagonal structure of G(ω0) indicates that all components of ω are orthogonal to each other. However, because G(ω0) does not take the form of cIW, this indicates that different components of ω introduce different amounts of perturbations. For instance, a large gy,ii indicates that ωy,i introduces a large perturbation for a subject with more repeated measures (ni). In practice, we can always choose the appropriate perturbation scheme in equation (18).
3.2. Local Influence Measures
Let f(p(Do, b, θ; ω)) = f (ω) be a real objective function (e.g., Bayes factor) of the perturbed model, which defines the aspect of inference of interest for sensitivity analysis. We use f(ω) to measure the effect of a small perturbation ω to a JMLS around ω0. Specifically, we consider a smooth curve of the perturbed model p(Do, b, θ; ω(t)) such that p(Do, b, θ; ω(0)) = p(Do, b, θ). Then, the score function of ℓ(ω (t)) with respect to t is equal to ∂t ℓ(ω(t)) = hT∂ωℓ(ω(t)), where ∂t = ∂/∂t, ∂ω = ∂/∂ω, and ∂t ω(t)|t =0 = h ∈ RW. Then, we quantify the effects of introducing ω(t) to perturb the JMLS by using {f (ω(t)) − f (ω(0))}2 relative to the Kullback–Leibler divergence between p(Do, b, θ; ω(0)) and p(Do, b, θ; ω(t)), denoted by S(ω(0), ω(t)).
For small t, it follows from a Taylor’s series expansion that S(ω(0), ω(t)) ≈ 0.5t2hT G(ω0)h and f (ω(t)) − f (ω(0)) = ḟh(0)t + O(t2), where , in which ∇f = ∂ωf(ω0). If ∇f ≠ 0, we use a quantity FIf,h called the first-order influence measure (FI) in the direction h ∈ RW for the objective function f (ω), which is given by
| (19) |
where G = G(ω0). For the appropriate perturbation ω̃(ω) given in equation (18), FIf,h reduces to with the constraint hT h = 1.
We use the maximum value of FIf,h and its associated eigenvector, denoted by hmax, as influence measures to quantify the largest degree and influential direction of local influence of ω̃ to the JMLS. It can be easily shown that FIf,hmax equals and . In particular, hmax can be used to detect robustness of priors or identify influential observations and incorrect sampling distributional assumptions for single-case and global perturbations (Cook, 1986). Following Zhu and Lee (2001) and Zhu et al. (2007), we also suggest inspecting FIf, ei to identify the most significant components of ω̃, where ei is a W × 1 vector with a 1 for the ith element and 0 otherwise. For instance, we consider the Bayes factor given by f(ω) = log p(Do; ω) − log p(Do; ω0), where p(Do; ω) = ∫ p(Do, b, θ; ω)dbdθ. Thus, under some smoothness conditions, it can be shown that ∇f = Eω<sup>0</sup>[∂ωlog p(Do, b, θ; ω0)|Do ] and . To calculate the local influence measures associated with f(ω), we just need to compute ∇f and G. In practice, we can use MCMC methods to draw samples {(θ(s), b(s)): s = 1, …, S0} from p(θ, b; Do, ω0) to approximate ∇f via .
We can also carry out Bayesian local influence when ∇f = 0. Because f (ω(t)) = f (ω(0)) + 0.5hT Hf ht2 + O(t3), where , we introduce a second-order influence measure (SI) in the direction h ∈ RW, given by
| (20) |
For ω̃ (ω) in equation (18), SIf,h reduces to hTG−1/2HfG−1/2h, where hT h = 1. Moreover, we also use SIf,ei and the eigenvalue–eigenvector pairs of G−1/2Hf G−1/2 as our influence measures. We use the eigenvector of the largest eigenvalue of G−1/2Hf G−1/2, denoted by hmax, as influence measures to quantify the influential direction of local influence of ω̃ to the JMLS.
Finally, we examine the influence measures associated with three common objective functions, these being the φ–divergence, the posterior mean distance, and the Bayes factor, and include the detailed formulas in the supplementary document. Although all three objective functions can assess the local influence of a perturbation vector ω to the JMLS, there is a conceptual difference among these measures. The φ–divergence and the Bayes factor quantify the effects of introducing ω on the overall posterior distribution, whereas the posterior mean distance quantifies the effect of ω on the posterior mean of θ. Because the perturbation vector ω may influence various characteristics of the posterior distribution, such as the shape, mode, and mean, the φ–divergence, and the Bayes factor can be more sensitive to some perturbations of the posterior distribution compared to the posterior mean for certain perturbation schemes ω. In contrast, the posterior mean distance may be more sensitive to perturbations which have a dramatic effect on the posterior mean.
4. Application to the IBCSG Data
We applied our proposed methodology to both simulated data and the IBCSG data discussed in the Introduction Section. For the sake of space, we only present some influence analysis results for the IBCSG data here. We refer the reader to the supplementary document for further details.
For the IBCSG data, we considered a JMLS for jointly investigating the relationship between the multidimensional QOL and the bivariate failure time variables DFS and OS. To satisfy the normality assumption for the four considered longitudinal QOL indicators (appetite, denoted as y1; perceived coping, denoted as y2; mood, denoted as y3; and physical well-being, denoted as y4), we transformed their corresponding observed values of QOL to (Chi and Ibrahim, 2006). The transformed QOLs decreased over time and were scaled between 0 and 10 with smaller values reflecting better QOL. There were 832 patients from Switzerland, Sweden, and New Zealand/Australia with a total of 2154 QOL observations being included in this analysis.
Let yi1(tij1), …, yi4(tij4) be the observed values of the transformed QOLs for the ith patient at the jth time point, respectively. We considered the following JMLS:
| (21) |
for i = 1, …, 832, j = 1, 2, 3, k = 1, …, 4, and m = 1, 2, where xi = (xi1, …, xi6)T includes the number of initial cycles, reintroduction, interaction of the number of initial cycles and reintroduction, age, residency for Switzerland, and residency for Sweden. Moreover, zi includes the number of initial cycles, reintroduction, interaction of the number of initial cycles and reintroduction, age, number of positive nodes, and ER status. We assumed that εij = (εij1, …, εij4)T are independently and identically distributed as N4(0, Σ), and the random effects bik = (bik0, bik1)T are independently and identically distributed as N2(0, φk) for i = 1, …, 832, j = 1, 2, 3, and k = 1, …, 4. For the baseline hazard λm0(·), we take the piecewise constant hazards model with 250 subintervals with equal lengths such that with L = 250, where the cm,l s are prespecified constants.
To conduct a Bayesian analysis, we specified the following prior distributions:
| (22) |
for m = 1 and 2, and k = 1, …, 4, where h = {hml: m = 1, 2, l = 1, …, L}, , R0, ρ0, , τ0l, τ1l, , and are prespecified hyperparameters. Moreover, , R0, and were set to their Bayesian posterior means obtained from MCMC methods based on noninformative prior distributions for αm, γm, Σ−1, β, and φk. We used MCMC methods, whose key steps are described in the supplementary document, to carry out the Bayesian analysis.
To illustrate our influence analysis, we considered five different perturbations to the JMLS and carried out the associated influence analysis. Specifically, for each perturbation ω, we calculated the metric tensor G and then took the new appropriate perturbation ω̃ (ω) given in equation (18). Detailed derivations of influence quantities, such as G(ω0), can be found in the supplementary document. We chose the Bayes factor as the objective function and then calculated the local influence measure hmax of the Bayes factor for each perturbation scheme by using the MCMC output. Specifically, a total of 5,000 iterations after 5,000 burn-in samples were used to compute all local influence measures.
The first perturbation is a single-case perturbation, which is obtained by perturbing each subject’s longitudinal profile as follows:
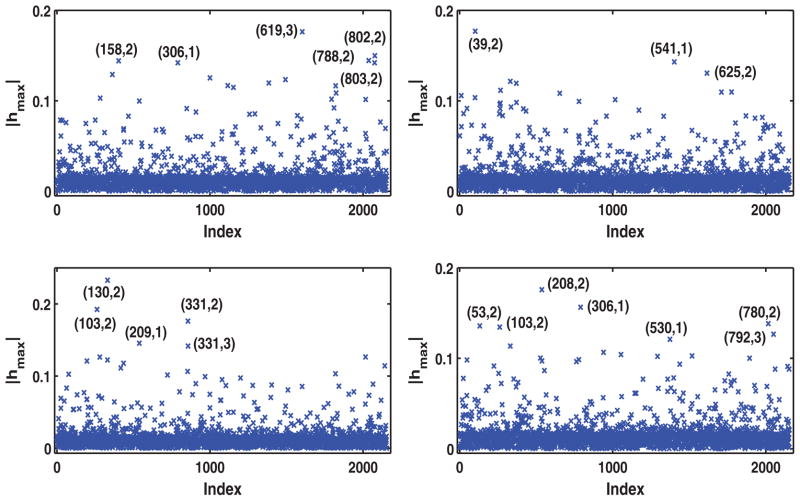
In this case, , in which ωij = (ωij1, …, ωij4)T for i = 1, …, n = 832, and j = 1, …, 3, and ω0 = 0 presents no perturbation. This single-case perturbation is designed to detect influential transformed QOLs of the longitudinal profiles in the evolution process. Figure 1 presents the subcomponents of hmax corresponding to all cases (i, j) for k = 1, 2, 3, and 4, where (i, j) represents the ith patient at the jth time point. Inspecting Figure 1 reveals the most influential cases as (158,2), (306,1), (619,3), (788,2), (802,2), and (803,2) for k = 1; (39,2), (541,1), and (625,2) for k = 2; (103,2), (130,2), (209,1), (331,2), and (331,3) for k = 3; and (53,2), (103,2), (208,2), (306,1), (530,1), (780,2), and (792,3) for k = 4.
Figure 1.
Results of single-case perturbation to longitudinal profiles for the IBCSG data set: Index plots of the local influence measure |hmax| corresponding to k = 1 (left upper panel), k = 2 (right upper panel), k = 3 (left lower panel), and k = 4 (right lower panel).
The second perturbation is also a single-case perturbation, which perturbs each marginal hazard function as follows:
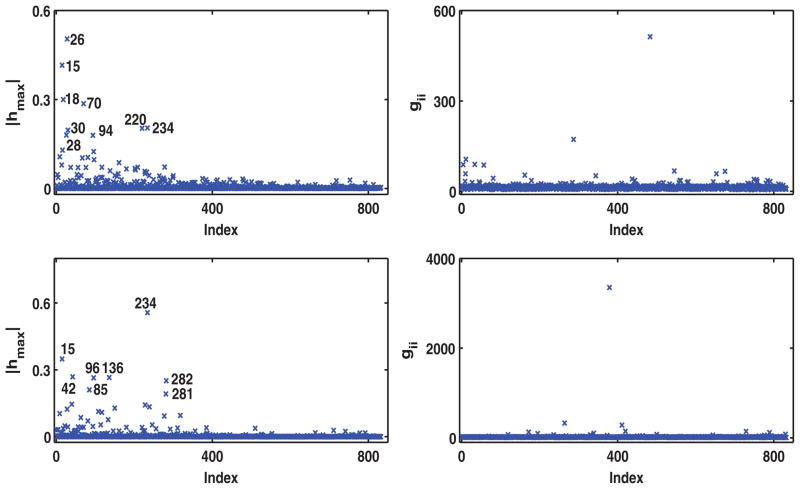
for i = 1, …, n and m = 1, 2. In this case, ω = (ω11, ω21, …, ωn1, ωn2) with ω0 = 1 representing no perturbation. This single-case perturbation is used to detect influential disease-free survival and overall survival times in the survival process. Inspecting Figure 2 reveals patients 15, 18, and 28 as most influential for m = 1, and patients 15, 42, 85, 96, 136, 234, 281, and 282 for m = 2 by our local influence measure hmax for the Bayes factor.
Figure 2.
Results of single-case perturbation to marginal hazard functions for the IBCSG data set: Index plots of the local influence measure hmax for ωins with m = 1 (left upper panel), and for ωins with m = 2 (left lower panel), and gii with m = 1 (right upper panel), and m = 2 (right lower panel).
The third perturbation is to simultaneously perturb the shared random effects bi in both the longitudinal profiles and the marginal hazard functions:
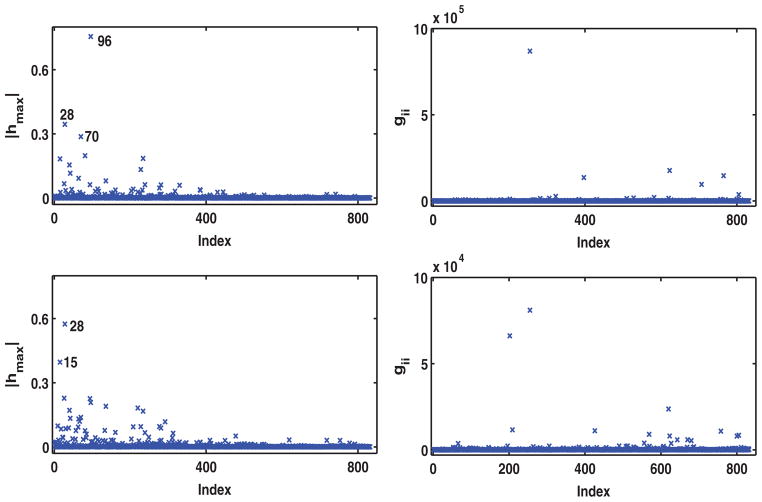
where ωi = (ωi1, …, ωi4). In this case, ω = {ω11, …, ω14, …, ωn1, …, ωn4) and ω0 = 1 represents no perturbation. This single-case perturbation is used to detect these influential subjects, whose survival times have a low chance of occurrence given their subject-specific longitudinal profiles. Inspecting Figures 3 and 4 reveal the following influential subjects. Specifically, in Figure 3, patients 15, 28, 40, 70, 81, 96, 117, 136, 228, and 282 were detected to be the most influential for k = 1 and patients 15, 28, 30, 40, 70, 94, 136, 220, and 234 were detected to be the most influential for k = 2. Figure 4 shows that patients 15, 28, 70, 81, 136, and 150 were detected to be the most influential for k = 3 and patients 9, 15, 18, 28, 70, 94, 96, and 136 were detected to be the most influential for k = 4.
Figure 3.
Results of perturbing the shared random effects for the IBCSG data set: Index plots of the local influence measure |hmax| for perturbing bik with k = 1 (left upper panel), and k = 2 (left lower panel), gii with k = 1 (right upper panel) and k = 2 (right lower panel).
Figure 4.
Results of perturbing the shared random effects for the IBCSG data set: Index plots of the local influence measure |hmax| for perturbing bik with k = 3 (left upper panel), and k = 4 (left lower panel), gii with k = 3 (right upper panel) and k = 4 (right lower panel).
The fourth perturbation involves perturbing the prior distributions as follows:
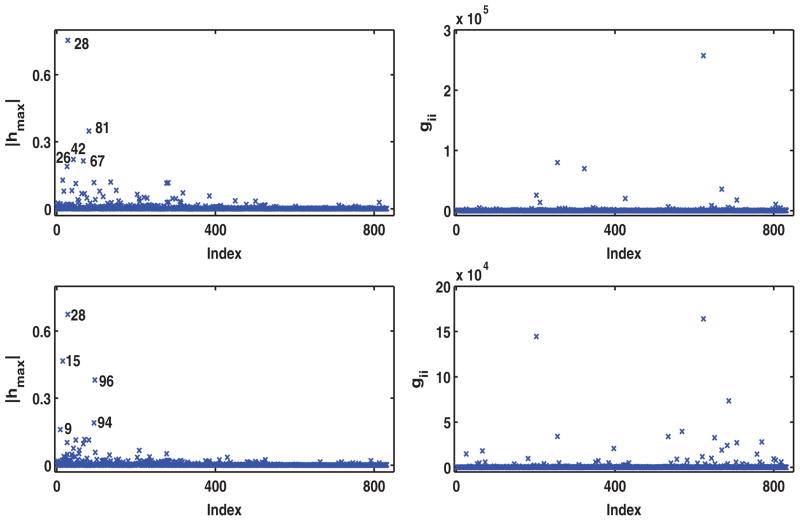
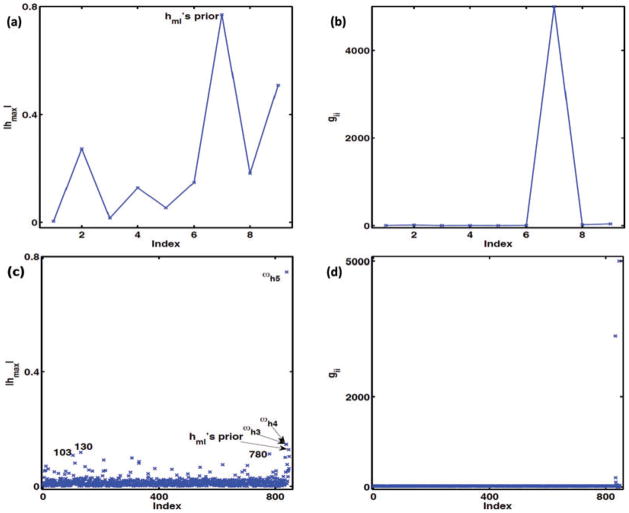
In this case, ω = {ωβ0, ωβ1, ωα0, ωα1, ωγ0, ωγ1, ωλ, ωΣ, ωφ}, and ω0 = {0, 1, 0, 1, 0, 1, 1, 1, 1} represents no perturbation. This perturbation assesses the sensitivity of the Bayes factor to minor changes in the prior distributions. Based on the local influence measures |hmax| for the Bayes factor and the diagonal elements of G(ω0), Figures 5(a) and (b) shows that perturbing the prior of hml has a large impact on the Bayesian analysis.
Figure 5.
Results of the IBCSG data set. Perturbing the prior distributions: (a) index plot of the local influence measure |hmax| (the left panel); and (b) index plot of gii (the right panel) for perturbing priors of the parameters βk, αm, γm, hm l, Σ and Φ1, …, Φ4. The simultaneous perturbation: (c) index plot of the local influence measure |hmax| (the left panel); and (d) index plot of gii (the right panel).
The last perturbation is a simultaneous perturbation. Specifically, we consider the following perturbation scheme:
and a subset of the s and the priors of βk, αm, γm, hml, Σ−1 and are pertarbed. In this case, we have
and ω0 = {1, …, 1, 1, 0, 1, 1, …, 1, 0, 1, 0, 1, 0, 1, 1, 1, 1} represents no perturbation. We are interested in examining the sensitivity of all components of the Bayesian analysis to such simultaneous perturbations. Based on all the subcomponents of |hmax| and gii, Figure 5(c) identifies influential perturbations , hml s and the prior distribution of the hml s as well as the three most influential patients 103, 130, and 780. Finally, we deleted the three influential subjects 103, 130, and 780 and recalculated the posterior estimates of the parameters for the IBCSG data (Table 1). Inspecting Table 1 indicates that many subcomponents of βk and αm are very sensitive to the deletion of these three subjects.
Table 1.
The IBCSG data set: Posterior means (Mean) of the parameters and their posterior standard deviations (SD) with and without influential patients 103, 130, and 780.
| Par. | With
|
Without
|
Par. | With
|
Without
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | ||
| α11 | 0.263 | 0.055 | 0.725 | 0.141 | β10 | 3.481 | 0.147 | 4.285 | 0.052 |
| α12 | −0.838 | 0.042 | 0.090 | 0.142 | β11 | −0.051 | 0.154 | −0.265 | 0.082 |
| α13 | −1.081 | 0.061 | 0.077 | 0.129 | β12 | −0.090 | 0.162 | −0.106 | 0.097 |
| α14 | 1.245 | 0.076 | −1.655 | 0.215 | β13 | −0.050 | 0.222 | 0.349 | 0.086 |
| α21 | 0.642 | 0.096 | 1.132 | 0.146 | β14 | 0.585 | 0.127 | −0.150 | 0.052 |
| α22 | −1.363 | 0.067 | 0.062 | 0.179 | β15 | 0.140 | 0.182 | −0.320 | 0.059 |
| α23 | −1.525 | 0.056 | 0.069 | 0.156 | β16 | 0.174 | 0.141 | −0.154 | 0.091 |
| α24 | 1.650 | 0.121 | −2.276 | 0.334 | β17 | −0.735 | 0.056 | −0.903 | 0.019 |
| γ11 | 0.047 | 0.240 | 0.062 | 0.214 | β20 | 5.177 | 0.604 | 5.806 | 0.140 |
| γ12 | −0.437 | 0.235 | −0.398 | 0.201 | β21 | 0.268 | 0.124 | −0.195 | 0.154 |
| γ13 | −0.149 | 0.346 | 0.084 | 0.399 | β22 | 0.152 | 0.200 | −0.197 | 0.153 |
| γ14 | −1.185 | 0.243 | −0.250 | 0.164 | β23 | −0.329 | 0.219 | 0.440 | 0.205 |
| γ15 | 1.304 | 0.166 | 1.058 | 0.159 | β24 | 0.344 | 0.246 | 0.140 | 0.158 |
| γ16 | −0.790 | 0.212 | −0.322 | 0.154 | β25 | 0.298 | 0.135 | 0.204 | 0.130 |
| γ21 | −1.280 | 0.480 | −1.044 | 0.367 | β26 | 0.377 | 0.254 | 0.012 | 0.122 |
| γ22 | −1.851 | 0.449 | −1.685 | 0.377 | β27 | −0.863 | 0.112 | −1.014 | 0.048 |
| γ23 | 1.525 | 0.682 | 1.695 | 0.723 | β30 | 4.289 | 0.162 | 5.048 | 0.120 |
| γ24 | −2.911 | 0.542 | −1.317 | 0.300 | β31 | 0.347 | 0.094 | 0.059 | 0.118 |
| γ25 | 2.129 | 0.245 | 1.589 | 0.334 | β32 | 0.213 | 0.136 | 0.158 | 0.125 |
| γ26 | −2.929 | 0.430 | −1.726 | 0.239 | β33 | −0.428 | 0.205 | −0.089 | 0.180 |
| σ11 | 3.941 | 0.165 | 4.259 | 0.205 | β34 | 0.594 | 0.120 | 0.148 | 0.109 |
| σ12 | 1.588 | 0.115 | 1.642 | 0.120 | β35 | 0.268 | 0.161 | −0.061 | 0.119 |
| σ13 | 2.444 | 0.125 | 2.524 | 0.131 | β36 | 0.820 | 0.104 | 0.159 | 0.098 |
| σ14 | 2.537 | 0.122 | 2.533 | 0.130 | β37 | −0.823 | 0.037 | −0.855 | 0.057 |
| σ22 | 4.163 | 0.173 | 3.660 | 0.164 | β40 | 3.974 | 0.623 | 4.560 | 0.071 |
| σ23 | 2.716 | 0.128 | 2.386 | 0.129 | β41 | 0.047 | 0.192 | 0.017 | 0.058 |
| σ24 | 2.112 | 0.131 | 2.322 | 0.129 | β42 | −0.182 | 0.241 | −0.029 | 0.065 |
| σ33 | 4.672 | 0.171 | 4.449 | 0.170 | β43 | −0.019 | 0.293 | 0.130 | 0.076 |
| σ34 | 3.129 | 0.137 | 3.395 | 0.142 | β44 | 0.853 | 0.290 | 0.215 | 0.036 |
| σ44 | 4.695 | 0.173 | 5.243 | 0.177 | β45 | 0.151 | 0.102 | −0.170 | 0.055 |
| φ1,11 | 1.516 | 0.179 | 1.073 | 0.207 | β46 | 0.724 | 0.140 | 0.158 | 0.028 |
| φ1,12 | −0.567 | 0.110 | −0.356 | 0.098 | β47 | −0.655 | 0.115 | −0.661 | 0.011 |
| φ1,22 | 0.479 | 0.078 | 0.380 | 0.074 | φ3,11 | 0.685 | 0.081 | 1.067 | 0.117 |
| φ2,11 | 1.163 | 0.133 | 1.539 | 0.156 | φ3,12 | −0.267 | 0.056 | −0.376 | 0.076 |
| φ2,12 | −0.241 | 0.079 | −0.289 | 0.121 | φ3,22 | 0.626 | 0.079 | 0.417 | 0.068 |
| φ2,22 | 0.654 | 0.084 | 0.644 | 0.139 | |||||
| φ4,11 | 0.537 | 0.088 | 0.328 | 0.072 | |||||
| φ4,12 | −0.144 | 0.056 | −0.090 | 0.053 | |||||
| φ4,22 | 0.403 | 0.057 | 0.404 | 0.057 | |||||
Supplementary Material
Footnotes
The web-based supplementary document referenced in Sections 3 and 4 is available with this article at the Biometrics website on the Wiley Online Library.
References
- Amari SI. Lecture Notes in Statistics. 2. Vol. 28. Berlin: Springer-Verlag; 1990. Differential-Geometrical Methods in Statistics. [Google Scholar]
- Arjas E, Gasbarra D. Nonparametric bayesian inference from right censored survival data, using the gibbs sampler. Statistica Sinica. 1994;4:505–524. [Google Scholar]
- Brown ER, lbrahim JG. A bayesian semiparametric joint hierarchical model for longitudinal and survival data. Biometrics. 2003a;59:221–228. doi: 10.1111/1541-0420.00028. [DOI] [PubMed] [Google Scholar]
- Brown ER, lbrahim JG. Bayesian approaches to joint cure rate and longitudinal models with applications to cancer vaccine trials. Biometrics. 2003b;59:686–693. doi: 10.1111/1541-0420.00079. [DOI] [PubMed] [Google Scholar]
- Brown ER, Ibrahim JG, DeGruttola V. A flexible B-spline model for multiple longitudinal biomarkers and survival. Biometrics. 2005;61:64–73. doi: 10.1111/j.0006-341X.2005.030929.x. [DOI] [PubMed] [Google Scholar]
- Chen MH, Ibrahim JG, Sinha D. Bayesian inference for multivariate survival data with a surviving fraction. Journal of Multivariate Analysis. 2002;80:101–126. [Google Scholar]
- Chen MH, Ibrahim JG, Sinha D. A new joint model for longitudinal and survival data with a cure fraction. Journal of Multivariate Analysis. 2004;91:18–34. [Google Scholar]
- Chi YY, Ibrahim JG. Joint models for multivariate longitudinal and multivariate survival data. Biometrics. 2006;62:432–445. doi: 10.1111/j.1541-0420.2005.00448.x. [DOI] [PubMed] [Google Scholar]
- Chi Y, Ibrahim JG. A New class of joint models for longitudinal and survival data accomodating zero and non-zero cure fractions: A case study of an international breast cancer study group trial. Statistica Sinica. 2007;17:445–462. [Google Scholar]
- Cook RD. Assessment of local influence (with Discussion) Journal of the Royal Statistical Society, Series B: Methodological. 1986;48:133–169. [Google Scholar]
- De Gruttola V, Tu XM. Modelling progression of cd-4 lymphocyte count and its relationship to survival time. Biometrics. 1994;50:1003–1014. [PubMed] [Google Scholar]
- Dobson A, Henderson R. Diagnostics for joint longitudinal and dropout time modeling. Biometrics. 2003;59:741–751. doi: 10.1111/j.0006-341x.2003.00087.x. [DOI] [PubMed] [Google Scholar]
- Dunson D. Nonparametric Bayes applications to biostatistics. In: Hjort N, Holmes C, Muller P, Walker S, editors. Bayesian Nonparametrics in Practice. Cambridge, UK: Cambridge University Press; 2009. pp. 223–273. [Google Scholar]
- Faucett C, Thomas D. Simultaneously modeling censored survival data and repeatedly measured covariates: A gibbs sampling approach. Statistics in Medicine. 1996;15:1663–1685. doi: 10.1002/(SICI)1097-0258(19960815)15:15<1663::AID-SIM294>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Hanson T, Branscum A, Johnson W. Predictive comparison of joint longitudinal-survival modeling: A case study illustrating competing approaches (with discussion) Lifetime Data Analysis. 2011;17:3–28. doi: 10.1007/s10985-010-9162-0. [DOI] [PubMed] [Google Scholar]
- Henderson R, Diggle P, Dobson A. Joint modeling of longitudinal measurements and event time data. Biostatistics. 2000;4:465–480. doi: 10.1093/biostatistics/1.4.465. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG, Chen MH, Sinha D. Bayesian Survival Analysis. Springer Series in Statistics. New York: Springer-Verlag; 2001. [Google Scholar]
- Ibrahim JG, Chen MH, Sinha D. Bayesian methods for joint modeling of longitudinal and survival data with applications to cancer vaccine studies. Statistica Sinica. 2004;14:863–883. [Google Scholar]
- Law NJ, Taylor JMG, Sandler HM. The joint modeling of a longitudinal disease progression marker and the failure time process in the presence of cure. Biostatistics. 2002;3:547–563. doi: 10.1093/biostatistics/3.4.547. [DOI] [PubMed] [Google Scholar]
- Pawitan Y, Self S. Modeling disease marker processes in aids. Journal of the American Statistical Association. 1993;83:719–726. [Google Scholar]
- Rizopoulos D, Ghosh P. A Bayesian semiparametric multivariate joint model for multiple longitudinal outcomes and a time-to-event. Statistics in Medicine. 2011;30:1366–1380. doi: 10.1002/sim.4205. [DOI] [PubMed] [Google Scholar]
- Rizopoulos D, Verbeke G, Molenberghs G. Shared parameter models under random effects misspecification. Biometrika. 2008;95:63–74. [Google Scholar]
- Sinha D. Semiparametric Bayesian analysis of multiple event time data. Journal of the American Statistical Association. 1993;88:979–983. [Google Scholar]
- Song X, Davidian M, Tsiatis AA. A semiparametric likelihood approach to joint modeling of longitudinal and time-to-event data. Biometrics. 2002;58:742–753. doi: 10.1111/j.0006-341x.2002.00742.x. [DOI] [PubMed] [Google Scholar]
- Tsiatis A, Davidian M. An overview of joint modeling of longitudinal and time-to-event data. Statistica Sinica. 2004;14:793–818. [Google Scholar]
- Tsiatis AA, DeGruttola V, Wulfsohn MS. Modeling the relationship of survival to longitudinal data measured with error. Applications to survival and cd4 counts in patients with aids. Journal of the American Statistical Association. 1995;90:27–37. [Google Scholar]
- Wang Y, Taylor JMG. Jointly modeling longitudinal and event time data with application to acquired immunodeficiency syndrome. Journal of the American Statistical Association. 2001;96:895–905. [Google Scholar]
- Wulfsohn MS, Tsiatis AA. A joint model for survival and longitudinal data measured with error. Biometrics. 1997;53:330–339. [PubMed] [Google Scholar]
- Xu J, Zeger SL. The evaluation of multiple surrogate endpoints. Biometrics. 2001;57:81–87. doi: 10.1111/j.0006-341x.2001.00081.x. [DOI] [PubMed] [Google Scholar]
- Yu M, Law NJ, Taylor JMG, Sandler HM. Joint longitudinal-survival-cure models and their application to prostate cancer. Statistica Sinica. 2004;14:835–862. [Google Scholar]
- Zhu H, Ibrahim J, Lee SY, Zhang H. Perturbation selection and influence measures in local influence analysis. Annals of Statistics. 2007;35:2565–2588. [Google Scholar]
- Zhu H, Ibrahim JG, Tang NS. Bayesian local influence analysis: a geometric approach. Biometrika. 2011;98:307–323. doi: 10.1093/biomet/asr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Lee SY. Local influence for incomplete-data models. Journal of the Royal Statistical Society, Series B: Statistical Methodology. 2001;63:111–126. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.