Abstract
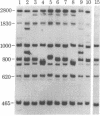
Mitochondrial DNA samples from each of 21 humans of diverse racial and geographic origin were digested with each of 18 restriction endonucleases. The sizes of the resulting DNA fragments were compared after gel electrophoresis. No differences among the samples were detected in digest with 7 of the enzymes. Analysis of digests with the remaining enzymes showed one or more differences. Each of the 21 samples could be characterized individually on the basis of these digests. All between-sample differences could be explained by single base substitutions. No evidence for sequence rearrangements (inversions, transpositions) was obtained. Fourteen of the site alterations were shared by two or more samples; six of these were shared between races. The data indicate that individuals differ from a postulated ancestral mtDNA sequence at 0.18% of their base pairs. On the basis of an estimated rate for base substitution of 1% per 10(6) years [Brown, W. M., George, M., Jr. & Wilson, A. C. (1979) Proc. Natl. Acad. Sci. USA 76, 1967-1971], Homo sapiens could have speciated or passed through a severe population constriction as recently as 180,000 years ago. The data suggest that group-specific patterns of cleavage exist. The high resolution and precision afforded by this method of analysis makes possible the investigation of many questions concerning human population genetics, evolution, and recent history.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avise J. C., Giblin-Davidson C., Laerm J., Patton J. C., Lansman R. A. Mitochondrial DNA clones and matriarchal phylogeny within and among geographic populations of the pocket gopher, Geomys pinetis. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6694–6698. doi: 10.1073/pnas.76.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise J. C., Lansman R. A., Shade R. O. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics. 1979 May;92(1):279–295. doi: 10.1093/genetics/92.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrell B. G., Bankier A. T., Drouin J. A different genetic code in human mitochondria. Nature. 1979 Nov 8;282(5735):189–194. doi: 10.1038/282189a0. [DOI] [PubMed] [Google Scholar]
- Battey J., Clayton D. A. The transcription map of mouse mitochondrial DNA. Cell. 1978 May;14(1):143–156. doi: 10.1016/0092-8674(78)90309-4. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. Polynucleotide kinase exchange reaction: quantitave assay for restriction endonuclease-generated 5'-phosphoroyl termini in DNA. J Biol Chem. 1977 May 25;252(10):3176–3184. [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bird A. P. Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J Mol Biol. 1978 Jan 5;118(1):49–60. doi: 10.1016/0022-2836(78)90243-7. [DOI] [PubMed] [Google Scholar]
- Brack C., Eberle H., Bickle T. A., Yuan R. A map of the sites on bacteriophage PM2 DNA for the restriction endonucleases HindIII and HpaII. J Mol Biol. 1976 Jun 14;104(1):305–309. doi: 10.1016/0022-2836(76)90016-4. [DOI] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Shine J., Goodman H. M. Human mitochondrial DNA: analysis of 7S DNA from the origin of replication. Proc Natl Acad Sci U S A. 1978 Feb;75(2):735–739. doi: 10.1073/pnas.75.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Vinograd J. Restriction endonuclease cleavage maps of animal mitochondrial DNAs. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4617–4621. doi: 10.1073/pnas.71.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. M., Watson R. M., Vinograd J., Tait K. M., Boyer H. W., Goodman H. M. The structures and fidelity of replication of mouse mitochondrial DNA-pSC 101 EcoRI recombinant plasmids grown in E. coli K12. Cell. 1976 Apr;7(4):517–530. doi: 10.1016/0092-8674(76)90202-6. [DOI] [PubMed] [Google Scholar]
- Brown W. M., Wright J. W. Mitochondrial DNA analyses and the origin and relative age of parthenogenetic lizards (genus Cnemidophorus). Science. 1979 Mar 23;203(4386):1247–1249. doi: 10.1126/science.424751. [DOI] [PubMed] [Google Scholar]
- Dawid I. B. Evolution of mitochondrial DNA sequences in Xenopus. Dev Biol. 1972 Oct;29(2):139–151. doi: 10.1016/0012-1606(72)90051-6. [DOI] [PubMed] [Google Scholar]
- Jacobsen H., Klenow H., Overgaard-Hansen K. The N-terminal amino-acid sequences of DNA polymerase I from Escherichia coli and of the large and the small fragments obtained by a limited proteolysis. Eur J Biochem. 1974 Jun 15;45(2):623–627. doi: 10.1111/j.1432-1033.1974.tb03588.x. [DOI] [PubMed] [Google Scholar]
- Jakovcic S., Casey J., Rabinowitz M. Sequence homology of the mitochondrial leucyl-tRNA cistron in different organisms. Biochemistry. 1975 May 20;14(10):2037–2042. doi: 10.1021/bi00681a001. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Nei M., Li W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D., Attardi G. Precise localization of the origin of replication in a physical map of HeLa cell mitochondrial DNA and isolation of a small fragment that contains it. J Mol Biol. 1978 Jul 5;122(3):301–319. doi: 10.1016/0022-2836(78)90192-4. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S. S., Newbold J. E., Hutchison C. A., 3rd, Edgell M. H. Specific cleavage analysis of mammalian mitochondrial DNA. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4496–4500. doi: 10.1073/pnas.72.11.4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J. L., Dawid I. B. Mapping of mitochondrial DNA in Xenopus laevis and X. borealis: the positions of ribosomal genes and D-loops. J Mol Biol. 1978 Feb 15;119(1):133–146. doi: 10.1016/0022-2836(78)90273-5. [DOI] [PubMed] [Google Scholar]
- Richardson C. C. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc Natl Acad Sci U S A. 1965 Jul;54(1):158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1980 Jan 11;8(1):r63–r80. doi: 10.1093/nar/8.1.197-d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., Shank P. R. X-Ray Intensifying Screens Greatly Enhance the Detection by Autoradiography of the Radioactive Isotopes 32P and 125I. Anal Biochem. 1978 May;86(1):184–192. doi: 10.1016/0003-2697(78)90333-0. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Upholt W. B. Estimation of DNA sequence divergence from comparison of restriction endonuclease digests. Nucleic Acids Res. 1977;4(5):1257–1265. doi: 10.1093/nar/4.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]