Abstract
Aspergillus fumigatus has two chitin synthases (CSMA and CSMB) with a myosin motor-like domain (MMD) arranged in a head-to-head configuration. To understand the function of these chitin synthases, single and double csm mutant strains were constructed and analyzed. Although there was a slight reduction in mycelial growth of the mutants, the total chitin synthase activity and the cell wall chitin content were similar in the mycelium of all of the mutants and the parental strain. In the conidia, chitin content in the ΔcsmA strain cell wall was less than half the amount found in the parental strain. In contrast, the ΔcsmB mutant strain and, unexpectedly, the ΔcsmA/ΔcsmB mutant strain did not show any modification of chitin content in their conidial cell walls. In contrast to the hydrophobic conidia of the parental strain, conidia of all of the csm mutants were hydrophilic due to the presence of an amorphous material covering the hydrophobic surface-rodlet layer. The deletion of CSM genes also resulted in an increased susceptibility of resting and germinating conidia to echinocandins. These results show that the deletion of the CSMA and CSMB genes induced a significant disorganization of the cell wall structure, even though they contribute only weakly to the overall cell wall chitin synthesis.
INTRODUCTION
Chitin, a microfibrillar homopolymer of β-(1,4)-linked N-acetylglucosamine (GlcNAc) residues, is one of the major components of the fungal cell wall, contributing to the shape and mechanical strength of the fungal cell. Since this polymer is essential for fungal growth and development, its synthesis has been studied for decades. Moreover, it has been considered an excellent target for the design of new antifungal agents (5). Chitin synthesis occurs at the plasma membrane with the extrusion of nascent chitin chains into the cell wall space. Chitin synthases (CHS), the proteins involved in this process, are highly variable, with a conserved catalytic domain bordered by transmembrane regions. Based on the sequences of their conserved regions, CHS proteins have been classified into two families that include seven classes (see Fig. S1 in the supplemental material) (38, 41). Family I includes classes I to III, while classes IV to VII belong to Family II (9, 55). The families are structurally different: in Family I, the catalytic site is bordered by transmembrane regions on each side, whereas in Family II, the central protein core is bound to the membrane through multiple helices at the C terminus (32, 41).
In fungi, the number of CHS genes varies from 1 (class III in Encephalitozoon cuniculi) to 3 (Saccharomyces cerevisiae, classes I, II, and IV) to >10 in filamentous fungi encompassing all of the CHS classes, with the number of CHS genes and families found in fungi being mostly correlated with the amount of cell wall chitin (25, 31, 50). In addition, classes III, V, VI, and VII have only been identified in filamentous fungi, suggesting that these enzymes play a role in hyphal growth. The function of the different CHS genes is only understood in S. cerevisiae, where it has been shown that each class of CHS proteins has a different function. Chs1p acts as a repair enzyme when mother and daughter cells separate (6). Chs2p synthesizes the chitin of the septum between mother and daughter cells (44). Chs3p is responsible for the synthesis of the chitin present in the lateral cell wall and at the chitin ring at the base of an emergent bud (43). In other fungi, the function of all individual CHS proteins and their specific involvement and interactions in the synthesis of the cell wall chitin remain poorly understood. It is especially puzzling to see that some chitin synthase mutants, such as chs1 of Neurospora crassa or chs3 of Exophiala dermatitidis, show a reduction in chitin synthase activity in vitro that can reach 90% of the total activity without any reduction in cell wall chitin content; in contrast, chs4 mutants of N. crassa and E. dermatitidis with an amount of chitin between 50 and 75% of the wild-type strain had normal chitin synthesis activity in vitro (15, 54).
Eight chitin synthase genes were identified in Aspergillus fumigatus (CHSA, CHSB, CHSC, CHSD, CHSG, CHSF, CSMA [earlier called CHSE], and CSMB). To date, six simple and three double mutants were constructed (G/C, A/C, and G/E) (32–35, 40). Inactivation of the Family I genes, CHSA (class I), CHSB (class II), and CHSC and CHSG (class III), individually did not lead to any growth phenotype, with the exception of CHSG. Characterization of Family II CHS remained incomplete in A. fumigatus; only CSMA (class V) and CHSD (class VI) were analyzed (32). Disruption of CSMA (class V) leads to an altered growth phenotype (poor conidiation, reduction in the colony radial growth rate, and decrease in chitin synthase activity), whereas the deletion of CHSD (class VI) did not result in any phenotype modification (2, 32). Although class V and class VII enzymes are required for correct morphogenesis in several filamentous fungi, there was no direct experimental evidence of the role of these gene products in the polysaccharide synthase activities as well as in the modification of the structural organization of the cell wall chitin (20, 21, 24, 27, 29, 55, 56). To date it has been shown only that these enzymes have a conserved myosin-17 myosin motor-like domain (MMD) that drives them to the tip of the hyphal cell (19, 42).
To understand better the function of A. fumigatus MMD-chitin synthases, single and double csm mutants were constructed and analyzed. Although CSMA and CSMB are arranged in a head-to-head configuration, their functions in cell wall construction were found to be different. Class V CsmA CHS protein was involved in the conidial chitin synthesis. In contrast, class VII CsmB CHS protein did not play a significant role in chitin synthesis in the conidial cell wall. Even though the deletion of CSMA and CSMB was not associated with a modification of the mycelial chitin level, the overall organization of the cell wall polysaccharides was perturbed, leading to an increased sensitivity to antifungal drugs.
MATERIALS AND METHODS
Strains and culture conditions.
Aspergillus fumigatus strains used in this work are listed in Table S1 in the supplemental material. They were grown at 37°C in either Aspergillus minimal medium (AMM) (11) containing 1% glucose and 5 mM ammonium tartrate, YG (1% glucose, 0.5% yeast extract, 1% trace elements), Sabouraud (2% glucose, 1% neopeptone) (Difco), PDA (potato dextrose agar) (Difco), or 2% malt agar (Cristomalt). When necessary, 6% KCl or 1 M sucrose was added to the medium to enhance conidiation. Conidia were collected from agar medium plates after 10 days of growth at 37°C using water containing 0.05% Tween 80.
Deletion of CSM genes.
Two different strategies were used to create single and double deletion of the CSMA and CSMB genes. For strategy 1, plasmids pCJ-E4, pCJ-Eb4, and pCJ-D2 were used for the construction of the ΔcsmA, ΔcsmB, and ΔcsmA/ΔcsmB mutants, respectively. These plasmids carry a DNA fragment designed to delete most of the open reading frames (ORFs) of the CSMA and CSMB genes. The strategy outlined in Fig. S2 in the supplemental material was used to produce strains with nonfunctional, disrupted A. fumigatus CSM (AfCSM) genes. The knockout Δcsm plasmids (pCJ-E4, pCJ-Eb4, and pCJ-D2) were constructed in the pGEMT vector (Promega), which contained two fragments of AfCSM genes separated by the 3.88-kb hisG-pyrG-hisG cassette obtained from the pPYRG2 plasmid (51). The two AfCSM fragments were obtained by PCR with A. fumigatus genomic DNA as a template and two sets of primers (AfV-A1/AfV-A2 and AfV-A5/AfV-A6) flanked by restriction enzymes for cloning (see Table S1 in the supplemental material). For the disruption of CSMA, protoplasts of a CEA17 strain of A. fumigatus were transformed using a 7.4-kb linear fragment released from pCJ-E4 by NotI digestion. The construction of the ΔcsmB strain was carried out by transforming CEA17 protoplasts with a 7.1-kb linear DNA fragment (obtained by PCR using the primers AfVB-B1/AfVB-B2 and AfVB-B5/AfVB-B6) released from pCJ-Eb4 by NotI digestion. For the construction of the double mutant ΔcsmA/ΔcsmB strain, a simultaneous deletion of both genes was carried out using a 7.2-kb linear DNA fragment (obtained using the primers AfVB-B1/AfVB-B2 and AfV-A5/AfV-A6) released from pCJ-D2 by NotI digestion. The transformation of A. fumigatus strain CEA17 using pCJ-E4, pCJ-Eb4, or pCJ-D2 was accomplished using the protoplast procedure described previously (51). After 3 days at 37°C, the pyrG+ transformants obtained were isolated from the AMM plates and were analyzed by PCR with different sets of primers, with one oligonucleotide primer inside the hisG-pyrG-hisG deletion cassette and another one upstream from the ORF of the gene of interest using genomic DNA obtained from spores as described previously (51). Integration of the cassette at the correct locus was confirmed by Southern blotting with genomic DNA digested by XmnI for the ΔcsmA strain, NcoI for the ΔcsmB strain, and BglII for the double mutant strain (see Fig. S2).
Due to unexpected results with the ΔcsmA/ΔcsmB double mutant, we decided to undertake another replacement strategy in a different genetic background, termed strategy 2. The ΔcsmA/ΔcsmB double deletion mutant was constructed in the CEA17_ΔakuBKU80 background (13) using the β-rec/six site-specific recombination system (18). The self-excising β-rec/six blaster cassette containing the hygromycin resistance marker was released from the plasmid pSK529 via FspI restriction enzyme. Using the GeneArt Seamless Cloning and Assembly system (Life Technologies, Carlsbad, CA), the csmA and csmB replacement cassette containing the marker module flanked by 5′ and 3′ homologous regions was generated and cloned in the pUC19 vector. The corresponding replacement cassette of 6,796 bp for csmA and 6,806 bp for cmsB were released from the resulting vector via EcoRV and FspI, respectively. The CEA17ΔakuBKU80 parental strain was transformed with the csmA replacement cassette by electroporation. Transformants obtained were analyzed by diagnostic PCR with oligonucleotides Forw csmA and Sv630 (see Table S1 in the supplemental material). The ΔcsmA deletion mutant obtained was cultivated in the presence of 2% xylose-containing minimal medium that allows the excision of the selection marker by recombination of the six recognition regions. The proper integration of the csmA replacement cassette and the excision of the selection marker in the ΔcsmAx strain were then verified by Southern blot analysis (see Fig. S3). To obtain the ΔcsmA/ΔcsmB double deletion mutant, the csmB replacement cassette was transformed in the ΔcsmAx recipient strain. Transformants obtained were analyzed by diagnostic PCR with oligonucleotides Forw csmB and Sv630 and Southern blot analysis (see Table S1).
Construction of revertant strains.
Complementation of the Δcsm mutants was obtained by a transformation strategy using a wild-type (WT) copy of the genes in the recombinant plasmid pPYRGQ3, which contains the pyrG resistance marker. Spontaneous pyrG− fungal strains from a pyrG+ independent clone were selected on AMM containing uracil (0.05%), uridine (0.12%), and 5-fluoroorotic acid (1 mg/ml). These pyrG− fungal strains were then used to generate the complemented strains using primers shown in Table S1 in the supplemental material. ORFs of the genes CSMA and CSMB were amplified from genomic DNA of the CEA17 pyrG1+ (AF14) strain and inserted into pPYRGQ3 that had been digested with XbaI and XmaI to yield pPYRQ3_CSMA and pPYRQ3_CSMB. These plasmids were linearized with NotI and transformed into the corresponding pyrG− strains of A. fumigatus. The presence of the WT copy of the genes was confirmed by PCR and Southern blotting using the probes amplified with the primers sE_F/sE_R for CSMA complementation and sEb_F/sEb_R for CSMB complementation (see Fig. S4).
DNA isolation and hybridization.
DNA was isolated from A. fumigatus using the extraction procedure described by Calera and coworkers (7). For Southern blot analyses, 10 μg of DNA per lane was loaded onto a 0.8% agarose gel and transferred by capillarity to positively charged nylon membranes by following standard protocols. Probes for AfCSMA and AfCSMB were obtained by restriction enzyme digestion of the appropriate clones. The Rediprime Random Prime labeling system kit (Amersham) was used to label DNA probes according to the manufacturer's instructions.
Real-time reverse transcription-PCR (RT-PCR).
Vegetative mycelium was obtained after 16 h of culture in a glucose (3%)-yeast extract (YE; 1%) liquid medium. Mycelial growth (for 24 h) on malt agar covered with a cellophane membrane (DryEase cellophane; Invitrogen) corresponded to the onset of conidiating morphotype. Fungal material was disrupted with 0.5-mm-diameter glass beads in 500 μl, and then RNA was isolated as described earlier (36) or by using the Qiagen RNeasy minikit. Quantitative PCR assays were performed as described in Mouyna et al. (36). The expression ratios were normalized to TEF1 expression and calculated according to the 2ΔCT method (28). To verify the absence of genomic DNA contamination, negative controls in which reverse transcriptase was omitted were used for each gene set. Three independent biological replicates were performed. The expression of the eight chitin synthase genes (CHSA to CHSG) has been analyzed. Primers used for these genes and the gene accession numbers are given in Table S2 in the supplemental material.
Antifungal assays.
Ten-fold dilutions, starting at 2 × 106 spores as the highest concentration, were spotted onto YG plates containing different concentrations of echinocandins (caspofungin, anidulafungin, and micafungin), calcofluor white, itraconazole, voriconazole, or amphotericin B. Plates were incubated for 48 h at 37°C in a humid atmosphere. Antifungal activity was also assessed in YG-RPMI-morpholinepropanesulfonic acid (MOPS) liquid medium using the resazurin method (10). The effect of nikkomycin Z was tested by the CLSI M38-A2 protocol and the resazurin method (10).
GS and CHS activity assays.
β-(1,3)-Glucan synthase (GS) activity was measured according to a previously described procedure using an excess of GTPγS (4). CHS activity measurements were carried out with crude membrane extracts obtained as follows: flasks with 200 ml of Sabouraud medium were inoculated with 5 × 105 conidia per ml of the different strains and incubated for 17 h at 37°C in an orbital incubator. Mycelia were collected by filtration under vacuum, washed with water, and disrupted in an MSK (Braun) cell homogenizer in 50 mM Tris-HCl buffer (pH 7.5) containing 50 mM EDTA and 1 mM phenylmethylsulfonyl fluoride (PMSF) for 1 min at 4°C in the presence of glass beads (0.45-mm diameter). The disrupted mycelial suspension was centrifuged (8,000 × g, 10 min), and the supernatant was centrifuged for 35 min at 100,000 × g. Membranes were then washed in 50 mM Tris-HCl (pH 7.5) containing 5 mM magnesium acetate, centrifuged again for 35 min at 100,000 × g, resuspended in the same buffer containing 30% glycerol, and stored at −80°C. CHS activity was measured by the incorporation of UDP-N-acetylglucosamine (UDP-GlcNAc) into chitin with trypsin treatment using a modification of the protocol described by Choi and Cabib (8). The assay system for CHS activity consisted of 50 μg of membranes, 32 mM GlcNAc, 1.1 mM UDP-[14C]GlcNAc (293 mCi mmol−1; Amersham Life Science), and 4.3 mM magnesium acetate in 30 mM morpholineethanesulfonic acid (MES; pH 6.5) in a total volume of 50 μl. For the proteolytic activation step, 2 μl of trypsin (1 to 3 mg/ml) was added to the reaction medium, and proteolysis activation was stopped after 15 min of incubation by the addition of soybean trypsin inhibitor solution. Mixtures were incubated at 30°C for 90 min. Newly synthesized chitin was determined by measuring the radioactivity incorporated into the insoluble material after the addition of 10% trichloroacetic acid and filtration through GF/C glass fiber filters (Whatman). The radioactivity was counted in a Wallac 1409 scintillation apparatus. Specific activity was expressed as nmol of GlcNAc incorporated per h per mg of protein.
Carbohydrate analysis of the cell wall fractions.
Parental (WT) and chitin synthase mutant conidia were either used directly or were grown in medium containing 3% glucose and 1% YE for 24 or 36 h for the cell wall polysaccharide analyses. The monosaccharide composition in the cell wall fractions was determined as described earlier (39).
AFM.
Atomic force microscopy (AFM) images were obtained in contact mode at room temperature (20°C) in phosphate-buffered saline (PBS) using a Nanoscope V Multimode atomic force microscope with oxide-sharpened microfabricated Si3N4 cantilevers with 0.01 N/m spring constants (Bruker, Santa Barbara, CA). For live cell imaging, conidia were immobilized by mechanical trapping into polycarbonate porous membranes (3-μm diameter; It4ip). After filtering a cell suspension (200 μl; 2.5 × 106 cells/ml), the filter was carefully rinsed in PBS, cut (1 by 1 cm), and attached to a steel sample puck using a small piece of adhesive tape, and then the mounted sample was transferred into the AFM liquid cell. The imaging force was kept as low as possible (250 pN) to minimize sample damage (12).
Light microscopic analysis of the fungal morphotypes.
Fungi grown for 3 h up to several days on a 2% malt agar were observed by light microscopy. Measurement of the size of conidia and intercalary hyphal cells, as well as the width of the mycelium (the number of hyphae measured for width was 30), was performed after staining of the mycelium with calcofluor white (aqueous solution at a final concentration of 5 μg/ml). Resting and germinating conidia were stained with Trypan blue and calcofluor white (0.4% and 5 μg/ml, respectively, for 5 min). The percentage of stained fungal cells is an index of cell wall permeability, since none of these dyes was able to penetrate a wild-type conidium.
Extraction of conidial surface hydrophobin (RodAp).
Freeze-dried conidia were incubated with 48% hydrofluoric (HF) acid (72 h, 4°C). The extract obtained was dried under N2 and reconstituted in H2O (1), and the protein was quantified using Bio-Rad protein assay reagent according to the manufacturer's instructions. An aliquot was subjected to SDS-PAGE (15%), and the proteins were visualized by silver staining.
Conidiation and germination.
Conidial suspensions (100 μl, 104/ml) were inoculated into three tubes of malt agar (2%) or malt agar containing 6% KCl (10 ml/tube). After 10 days at 25°C, conidia were recovered with 2 ml water containing 0.01% Tween 20 and counted using a hemocytometer. The percentage of germination was quantified after spotting 5 × 103 conidia on Sabouraud agar medium or PDA spread over the glass slides, incubating at 37°C, and counting the germ tubes microscopically every 1 h between 7 and 12 h.
Statistical analysis.
At least three biological replicates were performed per experiment; the statistical significance of the results was evaluated by one-way variance analysis using JMP1 software (SAS Institute, Cary, NC).
RESULTS
Sequence analysis of the two CHS genes with an MMD in Aspergillus fumigatus and construction of single and double mutants.
BLAST analysis with the class V gene of A. nidulans showed that two orthologous CHS genes with a myosin motor-like domain (MMD) were located nearby in chromosome 2 of A. fumigatus. For better homology to the A. nidulans nomenclature, the genes AFUA_2G13440 (CHSE) and AFUA_2G13430 were named AfCSMA and AfCSMB, respectively (viz., chitin synthase with myosin motor-like domain A and B, respectively). AfCSMA and AfCSMB were arranged in a head-to-head configuration within the A. fumigatus genome at a distance of 3,221 bp between their translational start points (Fig. 1). The analysis of the DNA sequence of the CSMB gene revealed an ORF of 5,446 bp encoding a 1,756-amino-acid polypeptide organized in four exons interrupted by three introns. Originally described as CHSE (2), the full length of the CSMA gene was 5,740 bp and encoded a protein with 1,848 amino acids. The proteins encoded by CSMA and CSMB genes showed an overall similarity of only 25% but presented Family II chitin synthase domains (CSD) at their C-terminal ends that displayed 60% similarity between them. In their N-terminal ends, the CsmA and CsmB proteins had an MMD which displayed only 11% similarity. The ATP-binding motifs, such as the P-loop [GXXGXGK(T/S)], Switch I [TASKAG], and Switch II [DFPGF] in the CsmAp MMD, thought to be essential for ATPase and motor activities (45, 48), were not conserved in the MMD of CsmBp. This head-to-head configuration as well as the absence of ATP binding motifs has previously been described for CSM orthologs in other ascomycetes (30, 31, 45).
Fig 1.
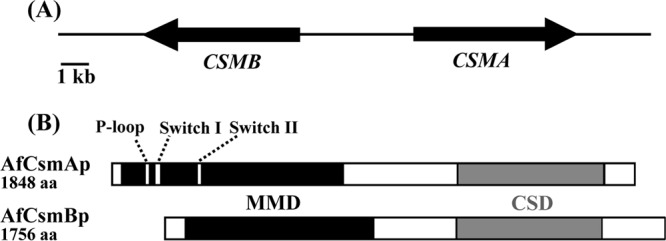
(A) Organization of the CSMA and CSMB genes in a head-to-head configuration in A. fumigatus chromosome 2. The orientation of transcription is shown by arrowheads. The distance between both translational start points is 3,221 bp. (B) Structures of CsmA and CsmB proteins. The myosin motor-like domains (MMD) and the chitin synthase domains (CSD) are indicated by solid black and gray boxes, respectively. The P-loop, Switch I, and Switch II motifs are also shown.
Deletion of the CSMA and CSMB genes was performed using two different strategies (see Fig. S2 and S3 in the supplemental material). The integration of the disrupting cassette at the right locus for the mutant as well as the ectopic integration of a copy of the CSMA and CSMB genes in the complemented strains was verified by Southern blotting. All of the mutants and the revertants constructed are shown in Table 1. To confirm the null mutation and the complementation of the different mutants, the expression of the corresponding genes in the respective mutants was assessed by RT-PCR and Southern blotting (see Fig. S4). Clearly, single mutants did not express the deleted gene and the double mutant failed to express both genes, whereas the revertant strains showed the expression of the complemented genes (data not shown).
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| CEA17 | CBS 144.89 pyrG− (auxotrophic pyrG1) | 14 |
| AF14 | CEA17 pyrG1+ (prototrophic wild type; isogenic CEA17) | 51 |
| CBS 144.89 | WT | 46 |
| CEA17_ΔakuBKU80 | CEA17 ΔakuBKU80 | 13 |
| A30 | CEA17 ΔcsmA::hisG-pyrG-hisG | This study |
| B1 | CEA17 ΔcsmB::hisG-pyrG-hisG | This study |
| D78 | CEA17 ΔcsmA ΔcsmB::hisG-pyrG-hisG | This study |
| A30 pyrG− | CEA17 ΔcsmA::hisG | This study |
| B1 pyrG− | CEA17 ΔcsmB::hisG | This study |
| D78 pyrG− | CEA17 ΔcsmA ΔcsmB::hisG | This study |
| R1 | CEA17 ΔcsmA::hisG csmA::pyrG | This study |
| S5 | CEA17 ΔcsmB::hisG csmB::pyrG | This study |
| pA1 | CEA17 ΔcsmA ΔcsmB::hisG csmA::pyrG | This study |
| pB1 | CEA17 ΔcsmA ΔcsmB::hisG csmB::pyrG | This study |
| ΔcsmA | CEA17_ΔakuBKU80 ΔcsmA::six-β-rec-hygroR-six | This study |
| ΔcsmAx | CEA17_ΔakuBKU80 ΔcsmA::six | This study |
| ΔcsmA ΔcsmB | CEA17_ΔakuBKU80 ΔcsmA::six ΔcsmB::six-β-rec-hygroR-six | This study |
Phenotype of the Δcsm mutants.
All stages of the biological cycle of single and double mutants of A. fumigatus were analyzed to try to understand the role of MMD-CHS in chitin synthesis.
(i) Chitin synthase activity and cell wall chitin content of the mycelium.
Previous studies have shown that the major chitin synthase activity in A. fumigatus was seen after proteolytic treatment of the mycelial membrane fractions (34). Under these conditions, the overall chitin synthase activity quantified in vitro in mycelial extracts was not affected by the deletion of the CSMA and/or CSMB genes (Fig. 2). Moreover, the chitin content of the mycelial cell wall (at both 24 and 36 h) was not statistically different in the single and double mutant strains compared to the parental strain (Fig. 2). The apex and septa of the mycelium were labeled with calcofluor white at the same intensity in the parental and mutant strains. In addition, deletion of the CSMA and/or CSMB genes was not compensated for by an increase in the expression of other CHS genes, as shown by the quantitative RT-PCR (qRT-PCR) data (see Fig. S5 in the supplemental material).
Fig 2.
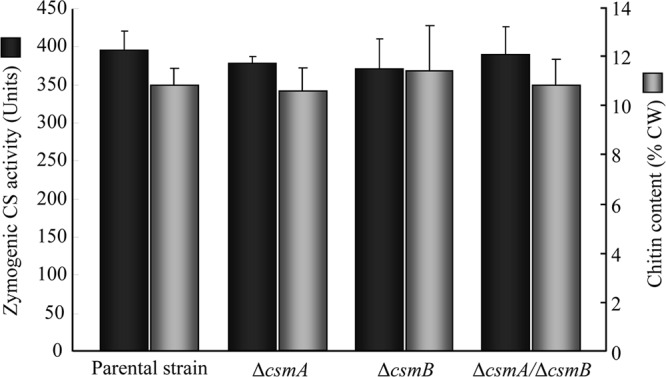
Mycelial chitin content (percent in the cell wall [CW] after 24 h of growth; data are means ± standard deviations [SD] from four individual experiments) and zymogenic chitin synthase (CS; means ± SD from three different experiments) activity of the parental strain and the single and double Δcsm mutant strains.
(ii) Mycelial growth.
On agar media, there was a slight but significant reduction of the colony diameter among the mutant and parental strains when the medium was inoculated with 10-day-old conidia (Fig. 3A). In contrast, in liquid YG medium under shaking, no difference in mycelial dry weight was seen among the mutant and parental strains (data not shown). After 24 h of growth, the mycelial morphology was similar in the parental and mutant strains, except that the mycelia were significantly wider in the mutant strains (in μm, 1.2 ± 0.2, 2.0 ± 0.3, 1.6 ± 0.2, and 1.9 ± 0.3 for the parental and ΔcsmA, ΔcsmB, and ΔcsmA/ΔcsmB mutant strains, respectively). However, in the later stages (32 to 48 h) of growth, the differences in the mycelial morphology of the mutant and the parental strains was accentuated (in μm, 2.0 ± 0.1, 3.3 ± 0.2, 2.5 ± 0.1, and 2.9 ± 0.1 for the parental and ΔcsmA, ΔcsmB, and ΔcsmA/ΔcsmB mutant strains, respectively), and there was an intrahyphal growth in the csm mutants; the frequency of such intrahyphal growth was 70 to 80% in the ΔcsmA and ΔcsmA/ΔcsmB mutants and 10 to 20% in the ΔcsmB mutant (Fig. 3B).
Fig 3.
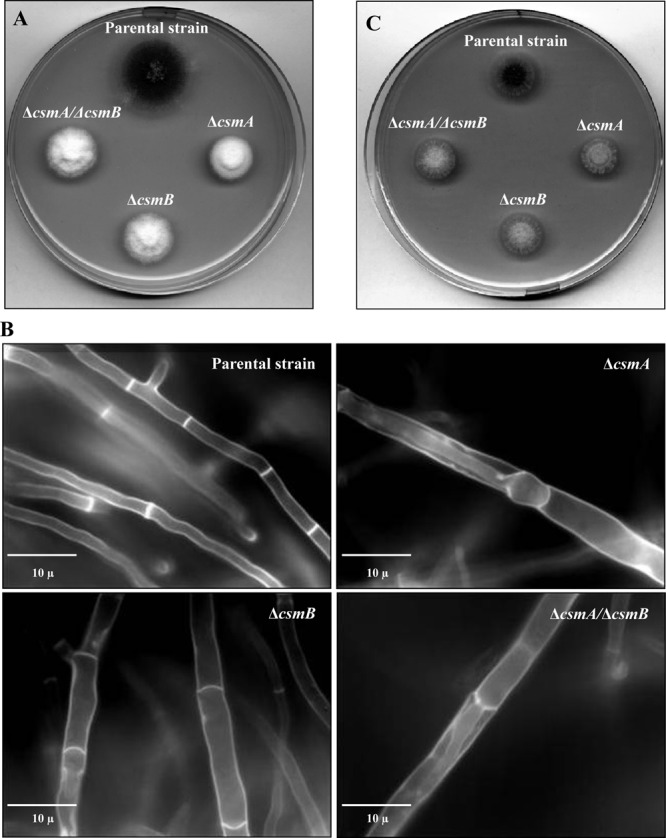
Colony growth of the parental and Δcsm mutant strains in rich media without (A) and with (C) an osmotic stabilizer (KCl) after 48 h at 37°C. (B) Calcofluor white staining of the parental and the Δcsm mutant strains' mycelia after 32 h of growth in liquid Sabouraud culture medium at 37°C.
(iii) Conidiation.
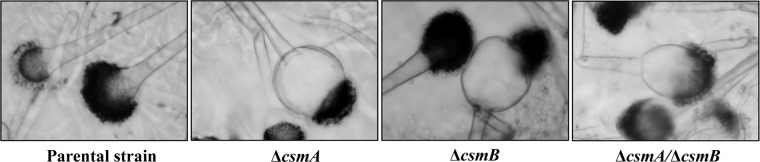
The colonies of ΔcsmA, ΔcsmB, and ΔcsmA/ΔcsmB mutant strains were always white in the different agar media tested, in clear contrast to the greenish color of the parental strain, indicating an altered conidiation (Fig. 3A). The reduction in the amount of conidia produced is shown in Table 2. The Δcsm mutants sporulated very poorly, with no more than 2% of the conidiation level of the parental strain. This conidiation defect resulted from the reduction in total numbers of conidiophores and also from the formation of morphologically altered heterogeneous conidiophores: ∼10 to 15% of all of the mutant conidiophores showed enlarged vesicles with fewer phialides and a reduced number of conidia (Fig. 4). Incubation in media stabilized osmotically with KCl (or sucrose) only partially restored the conidiation capacity of the mutants (Table 2 and Fig. 3C).
Table 2.
Conidiation of the ΔcsmA, ΔcsmB, ΔcsmA/ΔcsmB mutants and the parental strain in the presence or absence of KCl
| Strain | Growth ina: |
|
|---|---|---|
| Malt agar (2%) | Malt agar + KCl (6%) | |
| Parental | (7.8 × 108) ± 0.72 | (4.4 × 108) ± 0.53 |
| ΔcsmA | (3.53 × 105) ± 0.31 | (1.2 × 107) ± 0.2 |
| ΔcsmB | (1.33 × 107) ± 0.31 | (3.2 × 107) ± 0.2 |
| ΔcsmA ΔcsmB | (1.53 × 107) ± 0.31 | (4.73 × 107) ± 0.61 |
Data are means ± SD.
Fig 4.
Morphology of the abnormal conidiophores of the Δcsm mutant strains after 4 days of growth on malt (2%) plus KCl (6%) agar medium at 37°C. Note that in the ΔcsmB strain, of the two conidiophores, one is similar to that of the parental strain.
(iv) Conidial germination.
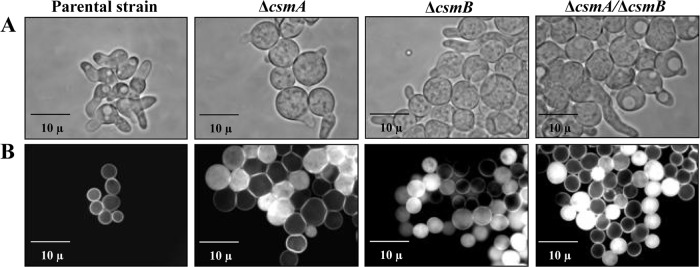
Although they had a size similar to that of the parental strain in the resting stage, the volume of the conidia increased significantly above the parental strain during the course of germination. After 6 h of growth, their diameter was approximately 2 to 2.5 times larger than that of the parental strain (Fig. 5A). However, these morphological changes did not affect the percentage of conidial germination. This increase in volume occurred in all of the media tested, and in AMM it was accompanied by a slight delay in the germination time of the Δcsm mutants. In addition, the use of different dyes indicated that the cell walls of the mutant conidia (10 days old) were more permeable than those of the parental strain. Intracellular labeling of 20 to 25% of the swollen conidia was seen after incubation of the conidia for 5 min in the presence of calcofluor white (Fig. 5B) or Trypan blue (0.4%), whereas there was no labeling of the parental strain morphotypes.
Fig 5.
(A) Conidial germination of the parental and the Δcsm mutant strains showing that the diameters of the mutant conidia are larger (grown in liquid Sabouraud culture medium at 37°C for 7.5 h). (B) Calcofluor white staining of swollen conidia of the parental and the Δcsm mutant strains (grown in liquid Sabouraud culture medium at 37°C for 4.5 h) showing the permeability of swollen conidia to the dye only in the Δcsm mutants.
(v) Conidial phenotype.
In contrast to the mycelium, the ΔcsmA mutant showed a high reduction (>65%) in the amount of chitin in the conidial cell wall. The ΔcsmB strain and, unexpectedly, the ΔcsmA/ΔcsmB double mutant had levels of chitin similar to those of the parental strain. Moreover, the CSMA revertant in the ΔcsmA/ΔcsmB double mutant had chitin levels similar to those of the parental strain and single ΔcsmB mutant strain (Table 3), confirming that the reduction of chitin in the ΔcsmA mutant was specifically due to the CSMA deletion. Most importantly, the overall cell wall polysaccharide composition of all mutant strains was modified (Table 3). The decrease of the AI/AS ratio in the cell wall [due both to a reduction in the β-(1,3)-glucan contents and an increase in the α-(1,3)-glucan contents] indicated an alteration of the overall cell wall structural organization. However, transmission electron microscopic (TEM) analysis of the mutant conidia did not show a difference in the cell wall structure and width compared to that of the parental strain (data not shown). As was the case for the mycelium, qRT-PCR data did not show a difference in the expression of other CHS genes in the conidiating mycelia of the CSMA and CSMA/CSMB deletion mutants (see Fig. S6 in the supplemental material). This result suggests that CSMA and CSMB controlled two different interconnected pathways associated with chitin synthesis, at least during conidial development.
Table 3.
Monosaccharide composition of the parental, mutant, and revertant strain conidial cell wallsa
| Strain | Alkali-insoluble (AI) fraction |
Alkali-soluble (AS) fraction |
AI/AS | |||||
|---|---|---|---|---|---|---|---|---|
| Man | Glu | Gal | GlcN | Man | Glu | Gal | ||
| WT | 8.6 ± 0.6 | 53.6 ± 0.9 | 2.9 ± 0.4 | 4.1 ± 0.2 | 4.5 ± 0.5 | 24.4 ± 1.1 | 1.9 ± 0.2 | 2.25 ± 0.1 |
| ΔcsmA | 3.8 ± 0.2 | 42.6 ± 1.7 | 1.4 ± 0.4 | 1.3 ± 0.1 | 6.0 ± 0.8 | 43.0 ± 1.0 | 2.0 ± 0.7 | 0.96 ± 0.1 |
| ΔcsmB | 4.6 ± 1.0 | 43.6 ± 0.9 | 2.5 ± 0.8 | 3.5 ± 0.2 | 4.7 ± 0.9 | 39.6 ± 0.9 | 1.5 ± 0.5 | 1.18 ± 0.1 |
| ΔcsmA ΔcsmB | 4.3 ± 0.4 | 39.4 ± 0.9 | 2.9 ± 0.4 | 4.4 ± 0.2 | 5.6 ± 0.8 | 41.7 ± 1.4 | 1.8 ± 0.3 | 1.04 ± 0.0 |
| ΔcsmA::CSMA | 7.5 ± 2.3 | 53.4 ± 1.7 | 2.8 ± 0.5 | 5.3 ± 0.5 | 5.2 ± 0.2 | 24.1 ± 1.6 | 1.7 ± 0.1 | 2.22 ± 0.2 |
| ΔcsmB::CSMB | 7.8 ± 0.4 | 50.9 ± 0.4 | 3.6 ± 1.0 | 4.4 ± 0.2 | 4.6 ± 0.4 | 26.3 ± 0.1 | 2.5 ± 0.8 | 2.00 ± 0.1 |
| ΔcsmA ΔcsmB::CSMA | 3.0 ± 0.3 | 43.4 ± 0.2 | 1.9 ± 0.1 | 3.2 ± 0.2 | 4.0 ± 0.7 | 42.8 ± 0.8 | 1.6 ± 0.0 | 1.06 ± 0.0 |
| ΔcsmA ΔcsmB::CSMB | 3.6 ± 0.9 | 44.4 ± 2.7 | 2.6 ± 0.6 | 1.4 ± 0.2 | 5.4 ± 1.0 | 39.6 ± 0.8 | 3.0 ± 0.5 | 1.08 ± 0.1 |
Data are means ± SD. Man, mannose; Glu, glucose; Gal, galactose; GlcN, glucosamine.
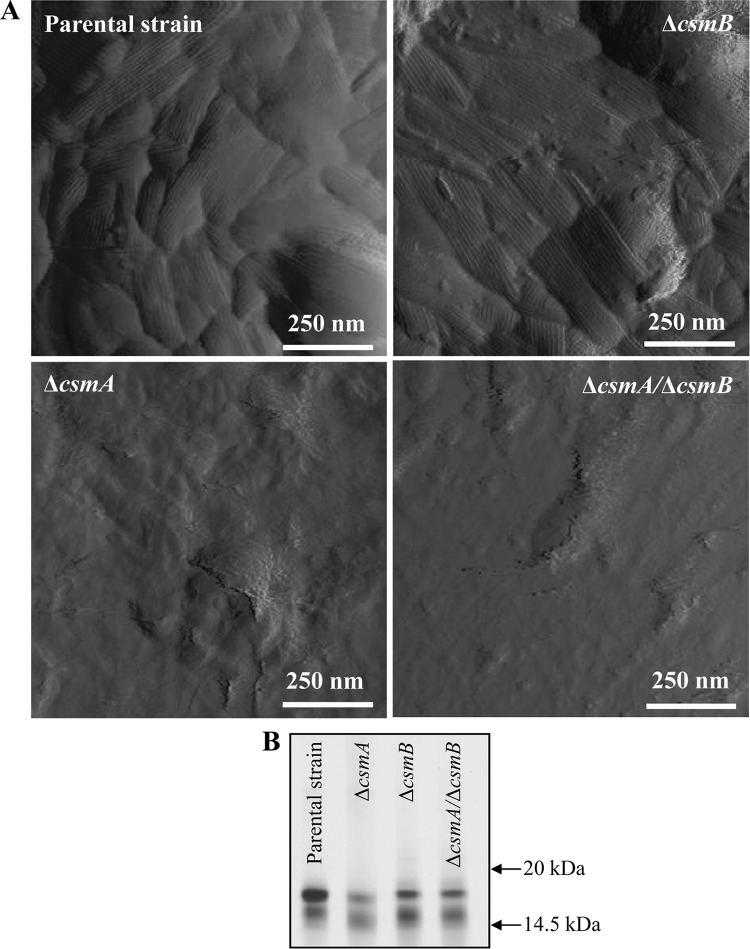
High-resolution atomic force microscopic (AFM) imaging was used to gain insight into the surface ultrastructure of resting conidia (16, 37). As can be seen in Fig. 6, high-resolution images revealed the presence of homogeneous layers of rodlets on the surface of the parental strain and ΔcsmB mutant. However, ΔcsmB mutant conidial surfaces showed amorphous structures in places. In contrast, the surfaces of the ΔcsmA and ΔcsmA/ΔcsmB mutant strains were different from those of the parental strain, i.e., rodlet structures were either completely lacking or heterogeneous depending on the single cell that was analyzed. The surface of most of these mutant conidia consisted essentially of smooth and/or granular amorphous structures (Fig. 6A). These images are in apparent contrast to biochemical analyses of HF extraction of the conidia showing the presence of RodAp (the hydrophobic protein forming the rodlet layer) in all of the strains, although the amount of RodAp extracted from the ΔcsmA strain was less than that of the parental strain (Fig. 6B). The presence of a rodlet layer underneath the amorphous layer was confirmed by scratching the csm mutant conidial surface with the AFM probe (data not shown). Thus, the combination of the AFM and biochemical data provided direct evidence that rodlets were not missing from the ΔcsmA and ΔcsmA/ΔcsmB mutants but rather were overlaid by an amorphous layer.
Fig 6.
(A) AFM deflection images in buffer of A. fumigatus conidia of the parental strain and ΔcsmB mutant showing the presence of a rodlet layer on its surface; the ΔcsmB mutant shows an amorphous layer in places. AFM deflection images of A. fumigatus conidia of ΔcsmA mutant and ΔcsmA/ΔcsmB double mutant strains revealing amorphous layers devoid of rodlets. For every strain, the images shown are representative of at least 10 conidia. (B) HF extracts from the ΔcsmA, ΔcsmB, and ΔcsmA ΔcsmB mutant and parental strain conidia showing the presence of RodAp (conidial surface protein, hydrophobin, responsible for the rodlet structure) in all of the strains.
Another proof of the cell wall disorganization was the loss of viability of the conidia over time. Beyond 1 month of storage of conidia in aerial conditions at room temperature, conidia started to be inviable (see Fig. S7 in the supplemental material). This loss of viability was exacerbated in the ΔcsmA/ΔcsmB double mutant. In addition, 90% of the conidia of the ΔcsmA and ΔcsmA/ΔcsmB mutant strains failed to germinate after storage at 4°C overnight in water (data not shown). The reduced survival of the conidia over time can lead to artificial differences seen in the size of the colony when the petri plates are inoculated with old conidia or conidia stored improperly (see Fig. S7).
The A. fumigatus Δcsm mutants displayed an exquisite sensitivity to echinocandins.
The sensitivity of the mutants to different well-known cell wall-disturbing compounds was tested on agar plates. Among the eight CHS mutants, only the csm mutants showed hypersensitivity to caspofungin (Fig. 7A and B). The Δcsm (both single and double) mutants showed an exquisite hypersensitivity to all of the echinocandins (caspofungin, micafungin, and anidulafungin), which are specific inhibitors of the β-(1,3)-glucan synthesis in fungi (see Fig. S8 in the supplemental material). After 24 h of incubation of the conidia with the drug, conidia of the Δcsm mutants were abnormally enlarged and collapsed (Fig. 7C). MICs calculated for all Δcsm mutants in the YG medium were 12 ng/ml, and no paradoxical effect was seen with these mutants that were truly killed in vitro by echinocandins (Fig. 7C). The lack of germ tube formation in the Δcsm mutants indicated that their resting conidia were susceptible to the echinocandins. In addition, preincubation of the mutant resting conidia and germinating conidia, grown either in liquid Sabouraud medium (8 h at 37°C) or in RPMI-MOPS medium (12 h at 37°C) for 1 h in the caspofungin solution (0.25 μg/ml), resulted in the death of both morphotypes (as determined by the resazurin method [10]), confirming that the cell wall structures of both conidia and hyphae were altered by the CSM deletion. In contrast, the Δcsm mutants were not more susceptible to nikkomycin Z than the parental strain. Susceptibility to drugs was indeed found to be specific to echinocandins, since the Δcsm mutants were only slightly more sensitive to azoles and not sensitive to calcofluor white and amphotericin B (see Fig. S8 in the supplemental material).
Fig 7.
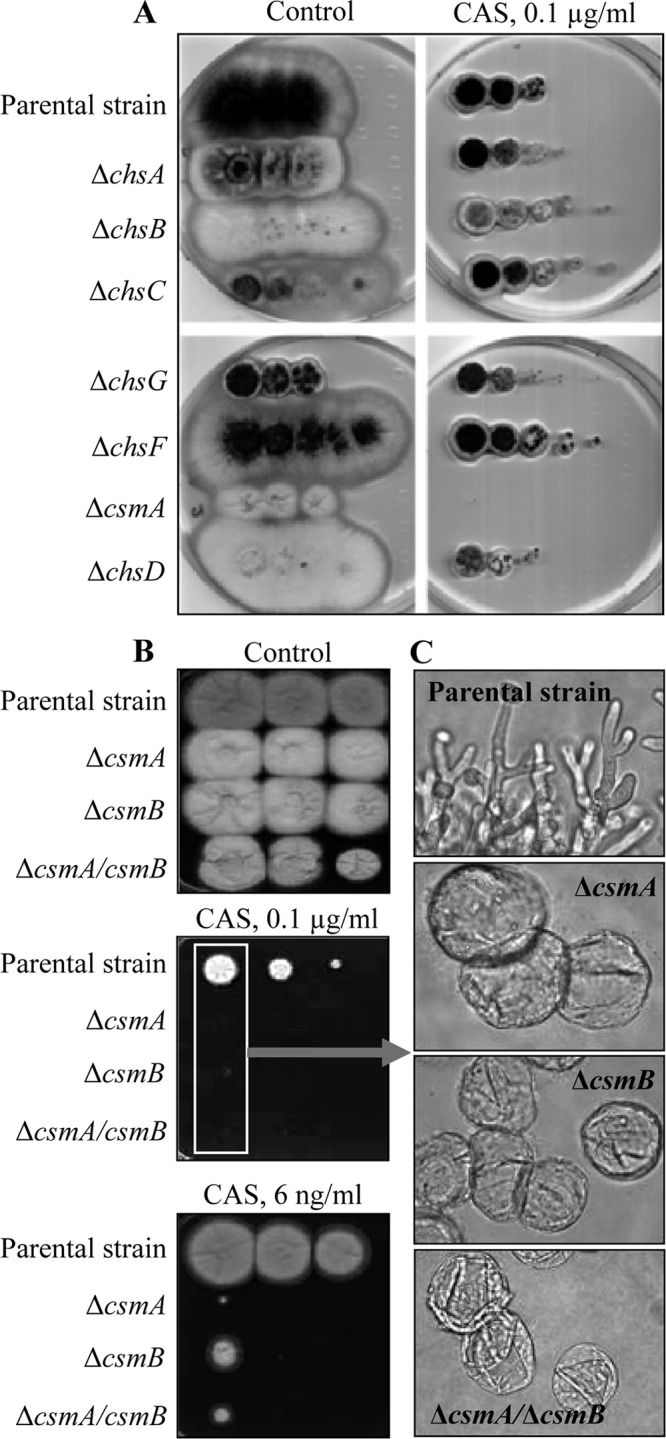
(A) YG plates containing 0.1 μg/ml of caspofungin inoculated with serial 10-fold dilutions of conidia (2 × 106) from all single Δchs mutants of A. fumigatus (but not the ΔcsmB mutant). (B) Ten-fold conidial dilutions (2 × 106) of ΔcsmA, ΔcsmB, and ΔcsmA ΔcsmB mutant strains and the parental strain were spotted on YG plates with 0.1 μg (6 ng/ml) caspofungin. Plates were incubated for 3 days at 28°C. (C) Microscopic analyses of the conidia abnormally swollen on the plates containing 0.1 μg caspofungin at 37°C for 24 h.
To investigate if the increased susceptibility of the mutants to echinocandins was due to an alteration in β-(1, 3)-glucan synthase activity, this enzymatic activity was measured in vitro using membrane preparations. No difference was found in the β-(1,3)-glucan synthase activity in the mutant strains compared to the parental strain (data not shown). In addition, there was no difference in the mycelial β-(1,3)-glucan content of the ΔcsmB and ΔcsmA/ΔcsmB mutants compared to that of the parental strain, with only a very slight decrease in the ΔcsmA mutant (data not shown). These results suggest that the sensitivity of the Δcsm mutants to echinocandins was not associated with a modification in the β-(1,3)-glucan synthase activity or the β-(1,3)-glucan content of the cell wall.
DISCUSSION
Complexity of the chitin synthase family in filamentous fungi.
Chitin synthases belong to a very complex protein family in filamentous fungi, and the precise biological functions of the individual family members remain poorly understood due to their insufficient biochemical characterization. To date, none of these transmembrane enzymes has been purified, and enzymatic analysis still relies on the use of crude membrane preparation producing chitin from radiolabeled UDP-GlcNAc. Moreover, no direct correlation exists between the CHS activity measured in vitro using membrane preparations and the end product of the activity, the chitin content of the cell wall. One of the difficulties for understanding the role of CHS proteins is the fact that phenotypes of the mutants resulting from the deletion of orthologous genes in different fungal species are often very different (see Fig. S9 in the supplemental material). For example, in S. cerevisiae, Cryptococcus neoformans (3), and Ustilago maydis (55), chitin synthase of class IV is responsible for the synthesis of most of the cell wall chitin. In contrast, studies in filamentous Ascomycetes, like A. fumigatus or Neurospora crassa, have not implicated the class IV chitin synthases but have implicated those of class III (CHSB of A. nidulans or CHSG of A. fumigatus) (15, 34). These comparative analyses have also shown that high levels of sequence similarities between two chitin synthases in the same species or taxonomically close species are not synonymous with similar biological functions. For example, in A. fumigatus, two class III genes have been identified (CHSC and CHSG), and the ΔchsC ΔchsG double mutant is not more affected in growth than the ΔchsG single mutant (34).
Role of MMD-CS in chitin synthesis.
The data obtained here, showing the lack of difference in chitin content of the mycelium of the parental and Δcsm mutant strains, was in agreement with previous data from Mellado and coworkers (34), but somehow it contradicted another previous report (2); however, the small decrease in chitin synthesis reported for the ΔchsE (csmA) mutant in the study of Aufauvre-Brown and coworkers (2) is technically questionable based on the methodology used by them (i.e., the enzymatic method, the efficiency of which is influenced by the structural organization of the cell wall, in contrast to the chemical hydrolysis method used in the present study). These differences also could be due to the genetic background, since the periodic hyphal swelling reported by Aufauvre-Brown and coworkers (2) using strain AF273 as the parental strain was not observed with the CBS144-89-ku80 background strain used in our study. Clearly, CSM proteins are not responsible for the bulk of chitin synthesis in the fungal systems studied. However, specific analytical methods to quantify the chitin defects in situ at the cellular level are still lacking, and the current chemical analyses are unable to reveal a difference in a specific location of chitin in the cell wall or in the modification of the types of chitin microfibrils associated with separate CHS genes, as seen in C. albicans (26). Such analysis could be related to the effect of the CSM deletion on the reduction of the nonzymogenic activity (which accounted for less than 10% of the total chitin synthase activity). This activity, quantified without trypsin at pH 8 and in the presence of Co+2 and Ni+2 (22), was reduced in the single and double Δcsm mutants (data not shown), suggested that the differential effect of cations seen on chitin synthase activities in yeast (49) was also found in filamentous fungi.
As in F. oxysporum, Fusarium verticillioides, or Gibberella zeae, the csm double mutant of A. fumigatus is fully viable, whereas the CSMA and CSMB double gene deletion was synthetically lethal in A. nidulans (45), questioning whether it is an experimental artifact or a biological adaptation of this species. The growth phenotype of the Δcsm mutant in the different species analyzed was also variable and goes from severely reduced growth in G. zeae or F. oxysporum mutants to growth almost identical to that of the parental strain in A. fumigatus. The ΔcsmA mutant showed a decrease in the conidial cell wall chitin content. Unexpectedly, the deletion of CSMB in the csmA mutant restored a normal amount of chitin in the conidial cell wall. We were very cautious to verify that this result was well founded by constructing single and double Δcsm mutant strains, for which we followed two different deletion strategies using different parental strains. It was also verified that the two genes AFUA_2G13430 and AFUA_2G13450 at the 3′ and 5′ end of the CSMA and CSMB operons were expressed, showing that the deletion did not affect the neighboring genes. Similarly and unexpectedly, the deletion of class V and VII chitin synthases of F. oxysporum led to a 40% increase in the cell wall chitin content compared to that of the parental strain. We cannot explain presently the presence of a wild-type level of chitin in the double csm mutants of A. fumigatus, since this double deletion does not seem to be associated with compensatory expression of CHS from other families (see Fig. S5 and S6 in the supplemental material).
The present results show that CSMA and CSMB do not have overlapping functions and do not compensate for each other, as already suggested by previous studies of G. zeae or F. verticilloides. Although the direct role of CSM proteins in chitin synthesis has not been elucidated, the function of the MMD in cellular trafficking of the chitin synthase has been extremely well dissected recently (42, 47). Using U. maydis as a model, Weber and coworkers showed that CHS with a myosin-17 motor domain travels along both central microtubules and peripheral filamentous actin (55). This transport is mediated by kinesin-1 and myosin-5; only a small percentage of the vesicles get exocystosed, whereas the majority is returned to the central core by the motor dynein. As shown earlier by the group of Horiuchi (19), successful exocytosis at the hyphal tip requires the MMD.
Chitin synthesis in vegetative hyphae and aerial structures uses two different pathways.
In A. fumigatus, the major morphological perturbations resulting from the MMD-CHS deletion was associated with conidiogenesis. Single and double Δcsm mutant strains of A. fumigatus sporulated poorly due to the formation of abnormal conidiophores that contained very few conidia, whereas conidiation was normal in all of the other chitin synthase mutants, suggesting that chitin is essential for the maintenance of the conidiophores in an aerial position and for the production of conidia. The role of CSM proteins in the production of aerial hyphae, appressorium formation, conidiogenesis, and sexual reproduction has been documented already in other filamentous species (23, 24, 32, 46). In spite of this phenotype, the amount of chitin in the conidium and conidiating structures or appressorium has not been investigated previously in other fungal species. These data suggest that CSM proteins are more important for hyphal specialization than for development. However, the specific role of individual chitin synthases in the construction of the conidial cell wall remains unknown.
The deletion of CSM-CHS affects A. fumigatus susceptibility to antifungal drugs.
Even though CSM deletions did not modify the amount of chitin in the mycelial cell wall, they altered the cell wall structural organization. In the case of the Δcsm mutant, modifications in the cell wall polysaccharides that were associated with a loss of viability and permeability changes could be responsible for an abnormal swelling of conidia during germination and an increased drug uptake. It would be very peculiar for these modifications to promote the susceptibility of the Δcsm mutants to echinocandins and not to other drugs. The chemical nature of the structural modifications as well as the mechanisms altering the cell wall permeability should be investigated, because they could lead to the discovery of new antifungal targets.
The increased sensitivity of the Δcsm mutants to caspofungin was in agreement with the synergistic antifungal effect of a combination of a chitin synthesis inhibitor such as nikkomycin and a β-(1,3)-glucan synthase inhibitor such as caspofungin. However, this increase in the sensitivity to echinocandins is only seen with the class V and VII mutants, whereas other single CHS deletion mutant strains have sensitivity to echinocandins similar to that of the parental strain. It is well known that modifications of the cell wall chitin are associated with a modification of the susceptibility to echinocandins (52). In A. fumigatus, compared to the parental strain, calcineurin mutants contain smaller amounts of β-glucan and chitin in the presence of caspofungin and are more sensitive to this drug (17). In contrast, although the Δras mutant contains a smaller amount of β-glucans, it is more resistant to caspofungin due to an increase in the cell wall chitin content (17). Similarly, in C. albicans, an increase of cell wall chitin consecutive with growth in the presence of calcium and calcofluor white is also associated with a reduction in the sensitivity to caspofungin (53). However, Δpmt2 mutant strains are also extremely sensitive to echinocandins without any modifications of the chitin levels (36). These data suggested that echinocandin influx is controlled by several mechanisms which have yet to be investigated.
Looking at the chitin synthase family in A. fumigatus is like working on a giant puzzle where many pieces are still missing. Results obtained to date show that the biochemical and cellular functions of each CHS gene as well as the interactions and compensatory effects among all CHS proteins are extremely complex and remain poorly understood. Therefore, the acquisition of new bits of information is absolutely required to assemble the full scenario for chitin synthesis in filamentous ascomycete biology.
Supplementary Material
ACKNOWLEDGMENTS
Work in the Aspergillus unit was partly supported by European grants Fungwall, Antifun, and ESF Fuminomics. Work at the Université Catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université Catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté Française de Belgique (Concerted Research Action). Y.F.D. and D.A. are Senior Research Associates and Postdoctoral Research Fellows, respectively, of the FRS-FNRS. Research at the laboratory of C.R. was supported by CICYT grants BIO2007-60779 and BFU2010-18632.
Footnotes
Published ahead of print 10 September 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Aimanianda V, et al. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117–1121 [DOI] [PubMed] [Google Scholar]
- 2. Aufauvre-Brown A, Mellado E, Gow NA, Holden DW. 1997. Aspergillus fumigatus chsE: a gene related to CHS3 of Saccharomyces cerevisiae and important for hyphal growth and conidiophore development but not pathogenicity. Fungal Genet. Biol. 21:141–152 [DOI] [PubMed] [Google Scholar]
- 3. Banks IR, et al. 2005. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4:1902–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beauvais A, Drake R, Ng K, Diaquin M, Latge JP. 1993. Characterization of the 1,3-beta-glucan synthase of Aspergillus fumigatus. J. Gen. Microbiol. 139:3071–3078 [DOI] [PubMed] [Google Scholar]
- 5. Bowman JC, et al. 2006. Efficacy of caspofungin against Aspergillus flavus, Aspergillus terreus, and Aspergillus nidulans. Antimicrob. Agents Chemother. 50:4202–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cabib E, Silverman SJ, Shaw JA. 1992. Chitinase and chitin synthase 1: counterbalancing activities in cell separation of Saccharomyces cerevisiae. J. Gen. Microbiol. 138:97–102 [DOI] [PubMed] [Google Scholar]
- 7. Calera JA, et al. 1997. Characterization of the Aspergillus nidulans aspnd1 gene demonstrates that the ASPND1 antigen, which it encodes, and several Aspergillus fumigatus immunodominant antigens belongs to the same family. Infect. Immun. 65:1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi WJ, Cabib E. 1994. The use of divalent cations and pH for the determination of specific yeast chitin synthetases. Anal. Biochem. 219:368–372 [DOI] [PubMed] [Google Scholar]
- 9. Choquer M, Boccara M, Goncalves IR, Soulie MC, Vidal-Cros A. 2004. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 271:2153–2164 [DOI] [PubMed] [Google Scholar]
- 10. Clavaud C, Beauvais A, Barbin L, Munier-Lehmann H, Latge JP. 2012. The composition of the culture medium influences the beta-1,3-glucan metabolism of Aspergillus fumigatus and the antifungal activity of inhibitors of beta-1,3-glucan synthesis. Antimicrob. Agents Chemother. 56:3428–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cove DJ. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:51–56 [DOI] [PubMed] [Google Scholar]
- 12. Dague E, et al. 2007. Chemical force microscopy of single live cells. Nano Lett. 7:3026–3030 [DOI] [PubMed] [Google Scholar]
- 13. da Silva Ferreira ME, et al. 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5:207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. d'Enfert C. 1996. Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr. Genet. 30:76–82 [DOI] [PubMed] [Google Scholar]
- 15. Din AB, Specht CA, Robbins PW, Yarden O. 1996. chs-4, a class IV chitin synthase gene from Neurospora crassa. Mol. Gen. Genet. 250:214–222 [DOI] [PubMed] [Google Scholar]
- 16. Dupres V, Alsteens D, Andre G, Dufrene YF. 2010. Microbial nanoscopy: a closer look at microbial cell surfaces. Trends Microbiol. 18:397–405 [DOI] [PubMed] [Google Scholar]
- 17. Fortwendel JR, et al. 2009. Differential effects of inhibiting chitin and 1,3-β-d-glucan synthesis in ras and calcineurin mutants of Aspergillus fumigatus. Antimicrob. Agents Chemother. 53:476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hartmann T, et al. 2010. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system. Appl. Environ. Microbiol. 76:6313–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Horiuchi H. 2009. Functional diversity of chitin synthases of Aspergillus nidulans in hyphal growth, conidiophore development and septum formation. Med. Mycol. 47(Suppl. 1):S47–S52 [DOI] [PubMed] [Google Scholar]
- 20. Horiuchi H, Fujiwara M, Yamashita S, Ohta A, Takagi M. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 181:3721–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horiuchi H, Takagi M. 1999. Chitin synthase genes of Aspergillus species. Contrib. Microbiol. 2:193–204 [DOI] [PubMed] [Google Scholar]
- 22. Jimenez C, Sacristan C, Roncero MI, Roncero C. 2010. Amino acid divergence between the CHS domain contributes to the different intracellular behaviour of family II fungal chitin synthases in Saccharomyces cerevisiae. Fungal Genet. Biol. 47:1034–1043 [DOI] [PubMed] [Google Scholar]
- 23. Kim JE, et al. 2009. Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr. Genet. 55:449–459 [DOI] [PubMed] [Google Scholar]
- 24. Larson TM, Kendra DF, Busman M, Brown DW. 2011. Fusarium verticillioides chitin synthases CHS5 and CHS7 are required for normal growth and pathogenicity. Curr. Genet. 57:177–189 [DOI] [PubMed] [Google Scholar]
- 25. Latgé JP, Calderone R. 2005. The fungal cell wall, p 73–104 In Kües U, et al. (ed), The mycota. I. Growth, differentiation and sexuality. Springer-Verlag, Berlin, Germany [Google Scholar]
- 26. Lenardon MD, Whitton RK, Munro CA, Marshall D, Gow NA. 2007. Individual chitin synthase enzymes synthesize microfibrils of differing structure at specific locations in the Candida albicans cell wall. Mol. Microbiol. 66:1164–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu H, Kauffman S, Becker JM, Szaniszlo PJ. 2004. Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot. Cell 3:40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta CT) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 29. Madrid MP, Di Pietro A, Roncero MI. 2003. Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47:257–266 [DOI] [PubMed] [Google Scholar]
- 30. Mandel MA, Galgiani JN, Kroken S, Orbach MJ. 2006. Coccidioides posadasii contains single chitin synthase genes corresponding to classes I to VII. Fungal Genet. Biol. 43:775–788 [DOI] [PubMed] [Google Scholar]
- 31. Martin-Urdiroz M, Roncero MI, Gonzalez-Reyes JA, Ruiz-Roldan C. 2008. ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum. Eukaryot. Cell 7:112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mellado E, Aufauvre-Brown A, Gow NA, Holden DW. 1996. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol. Microbiol. 20:667–679 [DOI] [PubMed] [Google Scholar]
- 33. Mellado E, Aufauvre-Brown A, Specht CA, Robbins PW, Holden DW. 1995. A multigene family related to chitin synthase genes of yeast in the opportunistic pathogen Aspergillus fumigatus. Mol. Gen. Genet. 246:353–359 [DOI] [PubMed] [Google Scholar]
- 34. Mellado E, et al. 2003. Cell wall biogenesis in a double chitin synthase mutant (chsG-/chsE-) of Aspergillus fumigatus. Fungal Genet. Biol. 38:98–109 [DOI] [PubMed] [Google Scholar]
- 35. Mellado E, Specht CA, Robbins PW, Holden DW. 1996. Cloning and characterization of chsD, a chitin synthase-like gene of Aspergillus fumigatus. FEMS Microbiol. Lett. 143:69–76 [DOI] [PubMed] [Google Scholar]
- 36. Mouyna I, et al. 2010. Members of protein O-mannosyltransferase family in Aspergillus fumigatus differentially affect growth, morphogenesis and viability. Mol. Microbiol. 76:1205–1221 [DOI] [PubMed] [Google Scholar]
- 37. Muller DJ, Dufrene YF. 2011. Atomic force microscopy: a nanoscopic window on the cell surface. Trends Cell Biol. 21:461–469 [DOI] [PubMed] [Google Scholar]
- 38. Nino-Vega GA, Carrero L, San-Blas G. 2004. Isolation of the CHS4 gene of Paracoccidioides brasiliensis and its accommodation in a new class of chitin synthases. Med. Mycol. 42:51–57 [DOI] [PubMed] [Google Scholar]
- 39. Richie DL, et al. 2009. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 5:e1000258 doi:10.1371/journal.ppat.1000258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rogg LE, Fortwendel JR, Juvvadi PR, Lilley A, Steinbach WJ. 2011. The chitin synthase genes chsA and chsC are not required for cell wall stress responses in the human pathogen Aspergillus fumigatus. Biochem. Biophys. Res. Commun. 411:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roncero C. 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41:367–378 [DOI] [PubMed] [Google Scholar]
- 42. Schuster M, et al. 2012. Myosin-5, kinesin-1 and myosin-17 cooperate in secretion of fungal chitin synthase. EMBO J. 31:214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shaw JA, et al. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114:111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Silverman SJ, Sburlati A, Slater ML, Cabib E. 1988. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 85:4735–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takeshita N, Yamashita S, Ohta A, Horiuchi H. 2006. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 59:1380–1394 [DOI] [PubMed] [Google Scholar]
- 46. Thau N, et al. 1994. Rodletless mutants of Aspergillus fumigatus. Infect. Immun. 62:4380–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Treitschke S, Doehlemann G, Schuster M, Steinberg G. 2010. The myosin motor domain of fungal chitin synthase V is dispensable for vesicle motility but required for virulence of the maize pathogen Ustilago maydis. Plant Cell 22:2476–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsuizaki M, Takeshita N, Ohta A, Horiuchi H. 2009. Myosin motor-like domain of the class VI chitin synthase CsmB is essential to its functions in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 73:1163–1167 [DOI] [PubMed] [Google Scholar]
- 49. Valdivieso MH, Duran A, Roncero C. 1999. Chitin synthases in yeast and fungi. EXS 87:55–69 [DOI] [PubMed] [Google Scholar]
- 50. Valdivieso MH, Ferrario L, Vai M, Duran A, Popolo L. 2000. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 182:4752–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vicentefranqueira R, Moreno MA, Leal F, Calera JA. 2005. The zrfA and zrfB genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, zinc-depleted environment. Eukaryot. Cell 4:837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Walker LA, Gow NA, Munro CA. 2010. Fungal echinocandin resistance. Fungal Genet. Biol. 47:117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Walker LA, et al. 2008. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4:e1000040 doi:10.1371/journal.ppat.1000040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Z, Zheng L, Hauser M, Becker JM, Szaniszlo PJ. 1999. WdChs4p, a homolog of chitin synthase 3 in Saccharomyces cerevisiae, alone cannot support growth of Wangiella (Exophiala) dermatitidis at the temperature of infection. Infect. Immun. 67:6619–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weber I, Assmann D, Thines E, Steinberg G. 2006. Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18:225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Werner S, Sugui JA, Steinberg G, Deising HB. 2007. A chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant Microbe Interact. 20:1555–1567 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.