Abstract
In Pseudomonas aeruginosa, the quorum-sensing (QS) system is closely related to biofilm formation. We previously demonstrated that 14-alpha-lipoyl andrographolide (AL-1) has synergistic effects on antibiofilm and antivirulence factors (pyocyanin and exopolysaccharide) of P. aeruginosa when combined with conventional antibiotics, while it has little inhibitory effect on its growth. However, its molecular mechanism remains elusive. Here we investigated the effect of AL-1 on QS systems, especially the Las and Rhl systems. This investigation showed that AL-1 can inhibit LasR–3-oxo-C12-homoserine lactone (HSL) interactions and repress the transcriptional level of QS-regulated genes. Reverse transcription (RT)-PCR data showed that AL-1 significantly reduced the expression levels of lasR, lasI, rhlR, and rhlI in a dose-dependent manner. AL-1 not only decreased the expression level of Psl, which is positively regulated by the Las system, but also increased the level of secretion of ExoS, which is negatively regulated by the Rhl system, indicating that AL-1 has multiple effects on both the Las and Rhl systems. It is no wonder that AL-1 showed synergistic effects with other antimicrobial agents in the treatment of P. aeruginosa infections.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic human pathogen responsible for severe infections in immunocompromised and cystic fibrosis (CF) patients (15, 30). Due to its frequent occurrence in hospital water-supplying pipes and its capacity to persist on medical devices, P. aeruginosa is a leading cause of life-threatening infections (48). In addition, P. aeruginosa is notorious for the vigorous development of biofilm, which adds difficulties in antibiotic therapy and makes wounds unable to heal (12). Biofilm formation is believed to be one of the major causes of persistent infections.
Biofilm formation by P. aeruginosa is regulated by a complex network of signals that includes quorum sensing (QS), small RNAs, and nutritional cues (26). QS controls important functions, including biofilm formation and pathogenicity (53). P. aeruginosa has two acylated homoserine lactone (AHL)-based QS systems (Las and Rhl) and a Pseudomonas quinolone signal (PQS) (2-heptyl-3-hydroxy-4-quinolone)-based signaling pathway. The transcription factors LasR and RhlR interact with and are activated by 3-oxo-C12-HSL (N-3-oxo-dodecanoyl-homoserine lactone) and C4-HSL (N-butyryl-l-homoserine lactone), respectively. PqsR is a LasR-RhlR homolog, which responds to the PQS (54). It was reported previously that P. aeruginosa QS systems control up to 11% of its genome (47, 55, 56). Of these QS systems, the LasR–3-oxo-C12-HSL system is the dominant regulator, because it is a turning-on system of the P. aeruginosa QS cascade that triggers the successive activation of other QS systems, including the RhlR–C4-HSL and PqsR-PQS systems (41).
Exopolysaccharides (EPSs) are key matrix components of biofilms, as they contribute to the overall biofilm architecture and resistance (1, 31, 45). The Psl polysaccharide is an essential matrix component that is required for P. aeruginosa to initiate and maintain biofilms (13, 23, 32, 37). In P. aeruginosa, pslA to pslL are positively regulated by the Las system, according to work reported previously by Gilbert et al. (14).
P. aeruginosa has another important virulence component, called the type III secretion system (TS33), which is negatively regulated by QS. TS33 is a needlelike complex which secretes a number of cytotoxins, including ExoS, ExoT, ExoU, and ExoY (44). These products have been shown to have a cytotoxic effect in vitro. ExoS and ExoT are bifunctional proteins which have both N-terminal GTPase-activating protein (GAP) activity and C-terminal ADP ribosyltransferase (ADPRT) activity (16).
Multidrug resistance is now a worldwide problem. Novel small-molecule inhibitors for P. aeruginosa are urgently needed. Natural products are notable not only for their potent therapeutic activities but also for the fact that they frequently possess the desirable pharmacokinetic properties required for clinical development (62). Many natural products have been widely used in the clinic, a testimony to the remarkable ability of microorganisms to produce drug-like small molecules (4, 27, 61). We have developed a high-throughput synergy screening platform to realize the full potential of natural products (63).
Andrographolide (Andro) is extracted from an herb, Andrographis paniculata Nees. We reported previously that 14-alpha-lipoyl andrographolide (AL-1), a derivative of Andro, inhibited biofilm formation and sensitized the bacterium P. aeruginosa to a variety of antibiotics for distinct synergistic effects (59). However, how this QS inhibitor exerts its effects on biofilm formation is still elusive.
This study aims to investigate how AL-1 inhibits P. aeruginosa PAO1 biofilm formation. Since LasR play a critical role in biofilm development, we tested the effects of AL-1 on LasR using an AHL-deficient strain. We then investigated the effects of AL-1 on the expression levels of seven QS-related genes (lasI, lasR, rhlI, rhlR, pqsA, pqsH, and pqsR) of P. aeruginosa using luminescent reporters. The anti-QS activity of AL-1 was validated further by reverse transcription (RT)-PCR. Psl provides a hydrated scaffolding to stabilize the structure of the biofilm. This led to the hypothesis that AL-1 may decrease the amount of biofilm matrix Psl. To elucidate the effect of AL-1 on Psl, we used Psl immunoblots. A β-galactosidase assay further suggested that AL-1 has effects on Psl at both transcriptional and translational levels. A complete understanding of the effect of AL-1 on the P. aeruginosa biofilm matrix may help us in the development of novel therapeutics.
MATERIALS AND METHODS
Reagents.
All reagents were obtained from Sigma Chemical Co. (St. Louis, MO). AL-1 was chemically synthesized (see Fig. S1 in the supplemental material), as we previously reported (25).
Bacterial strains and growth conditions.
Pseudomonas putida F117(pKRC12) (3) was kindly provided by Jo Handelsman, from the University of Wisconsin—Madison. P. aeruginosa CIM45/46 (22) was provided by Luyan Ma, from the Institute of Microbiology, Chinese Academy of Sciences, China.
P. putida F117(pKRC12) cells were grown at 28°C in Luria-Bertani (LB) medium. All P. aeruginosa strains were grown at 37°C in LB medium without NaCl (LBNS) or in Jensen's medium (24) and are listed in Table 1. Plasmid pMS402 carrying a promoterless luxCDABE reporter gene cluster was used to construct promoter-luxCDABE reporter fusions of seven genes (lasI, lasR, rhlI, rhlR, pqsA, pqsH, and pqsR), as reported previously (11, 29). Antibiotics were added, as required, at final concentrations of trimethoprim (TMP) of 300 μg/ml and gentamicin (GEN) of 25 μg/ml.
Table 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa PAO1 | Nonmucoid P. aeruginosa prototroph | Laboratory stock |
| Pseudomonas putida F117(pKRC12) | AHL-deficient derivative of P. putida IsoF; ΔppuI; pKR-C12/pBBR1MCS-5 carrying PlasB-gfp (ASV)-Plac-lasR; based on components of the P. aeruginosa las quorum-sensing system; Genr | 3 |
| P. aeruginosa CIM45 | PAO1 with E88 (lacZ::pPslA TRO) at the attB1 site | 22 |
| P. aeruginosa CIM46 | PAO1 with E89 (lacZ EB::pPslA TRO) at the attB1 site | 22 |
| Escherichia coli DH5α | recA1 and endA1 cloning strain | Invitrogen |
| Plasmids | ||
| pMS402 | Reporter vector carrying promoterless luxCDABE; Kanr Tmpr | 19 |
| pKD-lasR | pMS402 containing the lasR promoter region | 11 |
| pKD-lasI | pMS402 containing the lasI promoter region | 11 |
| pKD-rhlR | pMS402 containing the rhlR promoter region | 11 |
| pKD-rhlI | pMS402 containing the rhlI promoter region | 11 |
| pKD-pqsA | pMS402 containing the pqsA promoter region | 29 |
| pKD-pqsR | pMS402 containing the pqsR promoter region | 29 |
| pKD-pqsH | pMS402 containing the pqsH promoter region | 29 |
gfp(ASV) encodes mutant Gfpmut3* proteins (unstable Gfp) with the C-terminal extension sequence RPAANDENYAASV.
Inhibitory activity of AL-1 in reporter strains with LasR.
We used P. putida strain F117(pKRC12), an AHL-deficient strain that has been engineered to produce green fluorescent protein (GFP) upon the activation of LasR by 3-oxo-C12-HSL (3). P. putida F117(pKRC12) cells were grown overnight and diluted with LB medium to achieve an optical density at 595 nm (OD595) of 0.05, and 100-μl aliquots of cells were added to 96-well plates with dimethyl sulfoxide (DMSO) or AL-1 preincubated with 3-oxo-C12-HSL at a final concentration of 50 nM or 1,000 nM for 30 min. Fluorescence was measured at regular intervals after 4 h by using the EnVision plate reader (PerkinElmer Life and Analytical Sciences, Wellesley, MA).
Luciferase activity-based bioassay for QS inhibitors.
A chemiluminometric assay was developed to study the effects of AL-1 on expression levels of genes. Using lux-based reporters which indicate luciferase activity, gene expression in liquid cultures was measured as light production (in counts per second) with a Victor3 multilabel plate reader (PerkinElmer Life and Analytical Sciences, Wellesley, MA). Cultures of the reporter strains grown overnight were diluted to an optical density at 620 nm (OD620) of 0.2 and cultivated for an additional 2 h before use. The cultures were inoculated into parallel wells in a 96-well black plate with a transparent bottom. A fresh culture (5 μl) was inoculated into the wells containing a total of 95 μl medium plus other components (the OD620 in the wells was ∼0.07). Filter-sterilized mineral oil (60 μl) was added to prevent evaporation during the assay. Promoter activity was measured every 30 min for 24 h. Bacterial growth was monitored at the same time by measuring the OD595 with the Victor3 multilabel plate reader.
qRT-PCR.
P. aeruginosa PAO1 cells were grown in LBNS medium with shaking at 37°C overnight and diluted with LBNS medium to achieve an OD595 of 0.05. A total of 0.5 mM AL-1 or DMSO was added. After 5 h, total RNA was extracted by using a total RNA miniprep kit (Axygen). Residual DNA was removed by DNase I treatment (Fermentas), as recommended by the manufacturer. cDNA synthesis was performed by using SuperScript III first-strand synthesis (Invitrogen), according to the manufacturer's protocol, using random hexamers. Quantitative reverse transcription-PCR (qRT-PCR) was performed with SYBR green qPCR master mix (Fermentas). To calculate the relative expression levels of target genes, the expression level of the 16S rRNA gene was used as an internal control. Primers are listed in Table S1 in the supplemental material. The data presented below are the results obtained from three independent experiments.
Immunoblotting of Psl polysaccharide extracts.
Psl immunoblots were performed as described previously (6), with the following changes. P. aeruginosa PAO1 cells were grown in LBNS medium with shaking at 37°C overnight and treated with 0.5 mM AL-1 or DMSO. Crude polysaccharide extracts were obtained by spinning down the culture to 10 OD units, resuspending the extracts in 100 μl of 0.5 M EDTA, and boiling the extracts for 5 min at 100°C. The supernatant fraction was treated with proteinase K (final concentration, 0.5 mg/ml) for 60 min at 60°C, followed by proteinase K inactivation for 30 min at 80°C. Five microliters of the sample was spotted onto a nitrocellulose membrane. Blocking was done with 10% nonfat milk in TBST (20 mM Tris, 137 mM NaCl, 0.1% Tween 20 [pH 7.6]) for 1 h at room temperature. Psl was detected by using anti-Psl antibodies (1:25,000 dilution) and 1:10,000-diluted goat anti-rabbit IgG-conjugated secondary antibody (Thermo-Scientific). Nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) were added for detection.
β-Galactosidase assay.
β-Galactosidase activity was measured as described previously by Miller (38) and is expressed in Miller units (MU). Cell lysates were assayed for both β-galactosidase activities as well as protein content by a bicinchoninic acid (BCA) protein assay (Thermo-Scientific). The data presented below are the results obtained from three independent experiments. The variance is indicated by error bars in the figures.
Western blotting.
P. aeruginosa PAO1 cells grown in LBNS medium overnight at 37°C were diluted 1,000-fold in fresh LB medium supplemented with 200 mM NaCl containing DMSO, AL-1, or nitrilotriacetic acid (NTA) for 6 h at 37°C. The culture supernatant was collected by centrifugation, and the secretion proteins were concentrated by ultrafiltration. Proteins were separated by 12% SDS-PAGE and then blotted onto a polyvinylidene difluoride (PVDF) membrane by using a Trans-Blot SD semidry transfer cell (Bio-Rad Laboratories, Hercules, CA) and subjected to immunodetection. After being blocked with 5% nonfat milk in phosphate-buffered saline (PBS) overnight, the membrane was incubated in PBS with an anti-ExoS polyclonal antibody (Accurate Chemical & Scientific Corp., Westbury, NY) for 1 h. After being washed three times with PBS containing 0.3% (vol/vol) Triton X-100, the membrane was incubated in PBS with an anti-chicken IgG(H+L) conjugated with alkaline phosphatase (AP) (Southern Biotech, Birmingham, AL) for another 1 h. After being washed three times, the membrane was incubated with AP reaction buffer (100 mM Tris base [pH 9.5], 100 mM NaCl, and 50 mM MgCl2) for 5 min, and ExoS was detected by the chromogenic method.
Statistical data analysis.
The scientific statistical software Statistical Package for the Social Sciences (SPSS), version 17.0, was used to evaluate the significance of differences between groups. Each experimental value is expressed as the mean ± standard deviation (SD). A P value of <0.01 or a P value of <0.05 was taken to indicate a statistically distinct significance or significance.
RESULTS
Inhibitory activity of AL-1 in reporter strains with LasR.
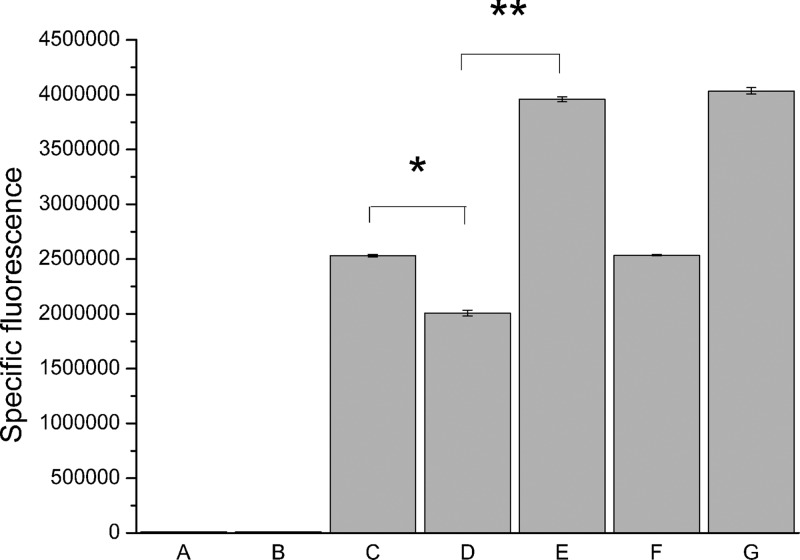
Our previous report showed that AL-1 can inhibit the biofilm formation and virulence factors of P. aeruginosa. As both of them are controlled by the QS system, we hypothesized that AL-1 interfered with the Las system of P. aeruginosa. To understand whether AL-1 interacts with the Las system, a 3-oxo-C12-HSL sensor strain engineered with a LasR transcriptional activator was used. In our experiment, AL-1 inhibited biosensor activity by nearly 20% at a concentration of 0.5 mM against 3-oxo-C12-HSL (P < 0.05) (Fig. 1). This result demonstrated that AL-1 interferes with the Las system via inhibiting the LasR–3-oxo-C12-HSL interaction. To further evaluate the activity of AL-1, 3-oxo-C12-HSL was added at a final concentration of 1 μM, and inhibition was not detected, suggesting that the competitive interaction between AL-1 and 3-oxo-C12-HSL for LasR binding did exist (Fig. 1).
Fig 1.
Specific fluorescence activity of P. putida F117(pKRC12) after 4 h when challenged with 0.5 mM AL-1 and induced with 50 nM 3-oxo-C12-HSL or 1,000 nM 3-oxo-C12-HSL. (A) DMSO only; (B) AL-1 only; (C) DMSO and induction with 50 nM 3-oxo-C12-HSL; (D) AL-1 and induction with 50 nM 3-oxo-C12-HSL; (E) AL-1 and induction with 1,000 nM 3-oxo-C12-HSL; (F) 50 nM 3-oxo-C12-HSL only; (G) 1,000 nM 3-oxo-C12-HSL only. Values reported are the means of values from three replicates with the deduction of LB fluorescence. Error bars indicate standard deviations. *, statistical significance compared with controls (P < 0.05); **, distinct significance (P < 0.01) compared with controls.
AL-1 depressed the expression levels of the lasI, lasR, rhlI, and rhlR genes.
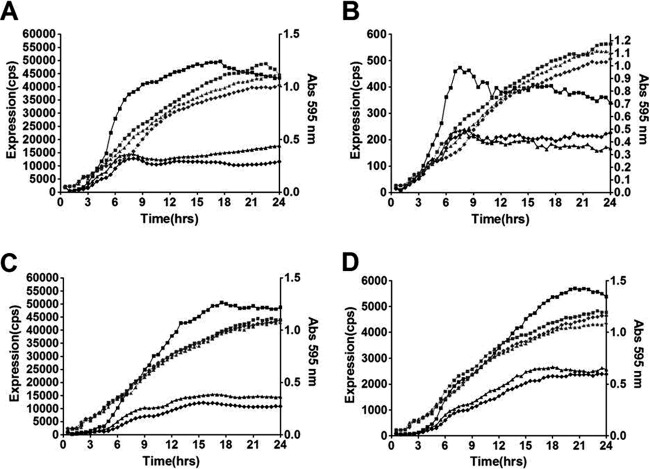
Since AL-1 inhibits not only the production of protease and pyocyanin but also the development of biofilms (59), LasR–3-oxo-C12-HSL is at the top of the hierarchical regulatory pathway in QS. We expected that AL-1 would affect QS-related genes such as las, rhl, and pqs. The results showed that AL-1 decreased the expression levels of lasI, lasR, rhlI, and rhlR in a dose-dependent manner (Fig. 2). The expression levels of the lasI and rhlI genes were reduced more than 3-fold, and the expression levels of the lasR and rhlR genes were decreased nearly 2-fold, when measured as the ratio of maximal levels of expression in the presence of 1 mM AL-1. The inhibitory effects of AL-1 on lasI, lasR, rhlI, and rhlR could be reversed by exogenous 3-oxo-C12-HSL at a final concentration of 1 μM (see Fig. S2 in the supplemental material). These results further indicate that AL-1 can inhibit LasR–3-oxo-C12-HSL interactions and repress the transcriptional levels of the las and rhl genes. However, the other tested genes (pqsA, pqsH, and pqsR) were not significantly influenced by AL-1, even at 10 mM (data not shown). In all experiments, no significant effects were observed for the growth of P. aeruginosa cells when treated with AL-1. The transcript levels of QS-related genes were also measured by using qRT-PCR. Consistent with the data obtained from luciferase reporters, the lasR, lasI, rhlR, and rhlI transcript levels in AL-1-treated strains decreased about 2.5-fold, 2-fold, 2-fold, and 3-fold, respectively, compared with the control, while pqsA, pqsB, pqsC, pqsD, and pqsE were not influenced by AL-1 (see Fig. S3 in the supplemental material).
Fig 2.
Inhibition of QS genes in P. aeruginosa PAO1 by AL-1. Expression profiles and corresponding growth curves are shown for lasI (A), lasR (B), rhlI (C), and rhlR (D). The black lines represent the expression levels of the promoters. The blue lines represent the growth of the strain. The data for the control treatment (without drug) (■), treatment with 0.5 mM AL-1 (▲), and treatment with 1 mM AL-1 (□) are shown. The assays were independently repeated at least three times, and the data shown are representative of comparable results (cps, counts per second).
AL-1 reduced the production of Psl polysaccharide.
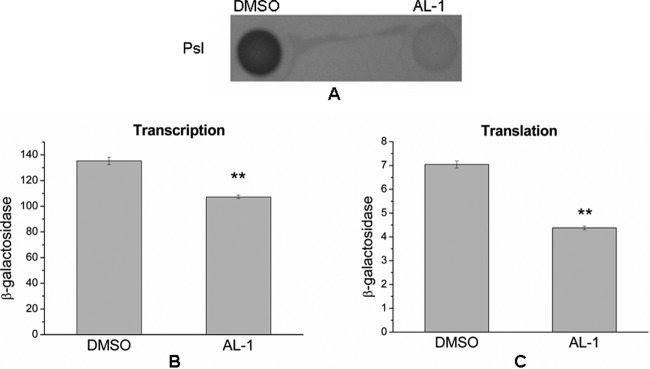
As AL-1 significantly reduces the production of EPS in P. aeruginosa (59), and the Psl polysaccharide is the primary matrix structural polysaccharide, AL-1 may also inhibit biofilm formation by decreasing the level of production of Psl, which is the key biofilm matrix polysaccharide in P. aeruginosa. By using Psl antiserum, it is easy to determine that AL-1 did reduce the level of Psl production (Fig. 3A). To further investigate the effect of AL-1 at the psl gene transcriptional or translational level, we utilized pslA chromosomal transcriptional and translational lacZ fusion reporter strains. The result showed that AL-1 decreased psl expression levels at both transcriptional (Fig. 3B) and translational (Fig. 3C) levels (P < 0.01).
Fig 3.
(A) Effects of AL-1 on production of Psl. The concentrations of AL-1 were 0.5 mM. (B and C) Transcriptional (B) and translational (C) lacZ fusion constructs assayed for β-galactosidase activities showed a deregulation of pslA mediated by AL-1 compared with DMSO. Data represent the means of data from duplicate β-galactosidase activity assays from three separate experiments, and activity is expressed as Miller units. **, a P value of <0.01 was taken to indicate a statistically distinct significance.
AL-1 increased the secretion of T3SS proteins.
Since the TS33 is negatively regulated by QS, it might also influence ExoS secretion. Western blotting showed that AL-1 led to a severe increase in the level of ExoS at 1 mM (Fig. 4). qRT-PCR was also performed: AL-1 treatment increased the levels of exoS, exoY, and exoT by 2.5-fold, 1.6-fold, and 2-fold, respectively (see Fig. S3 in the supplemental material). Similar to data from previous reports, ExoT was detected in the supernatant (Fig. 4). The anti-ExoS antibody cross-reacts with ExoT, which may be responsible for this phenomenon, as previously described (8, 58).
Fig 4.
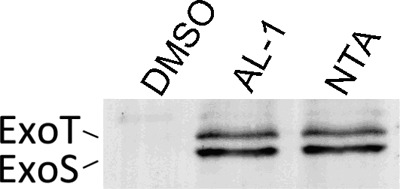
Effects of AL-1 on the T3SS effector ExoS. P. aeruginosa PAO1 cells were grown in the presence of 1 mM AL-1 and 10 mM NTA. The same volume of DMSO was added to the culture as a negative control.
DISCUSSION
In many pathogenic bacteria, QS systems regulate a variety of physiological processes, such as antibiotic biosynthesis, biofilm formation, and the production of virulence factors. In P. aeruginosa, the QS regulators LasR and RhlR control the expressions of hundreds of genes (47), many of which encode central metabolic functions. Controlling the virulence of P. aeruginosa is one of the most important issues in medicine. QS systems have been used as effective antimicrobial drug targets by altering the tolerance of biofilms to antibiotics. The development of QS system-targeted antivirulence compounds is urgently needed.
Previously, several natural compounds were reported to decrease the virulence and antibiotic-resistant biofilm formation of P. aeruginosa without affecting its growth. For example, furanones prevent AHLs from binding to the luxR homologs and eventually cause a rapid turnover of these proteins (33, 34). Baicalein significantly inhibits the biofilm formation of P. aeruginosa at 20 μM without affecting its growth. Its mode of action is to promote the proteolysis of the signal receptor TraR protein at 4 to 40 mM (60), whereas PD12 and V-06-018 inhibit LasR-dependent gene expression (39). However, the applications of these compounds have been hindered by either their low solubility or their high toxicity (60). In the present study, we reported the efficient effects of AL-1 on QS-related genes and biofilm development, which is a low-toxic compound in animal experiments (the 50% lethal dose [LD50] of AL-1 was 1,243 mg/kg of body weight/day) (7).
The present study demonstrated that AL-1 affected the Las and Rhl systems. Recent research revealed that the Las and Rhl systems are key areas for base infection treatments (20, 51). The Las system controls biofilm formation (10, 43), and the Rhl system is responsible for the production of rhamnolipids, pyocyanin, and elastase. Rhamnolipids play multiple roles in the establishment and maintenance of P. aeruginosa biofilms, while pyocyanin and elastase are related to the pathogenesis of P. aeruginosa. LasR is a hierarchical regulator coregulated with the RhlR. This “dense-overlapping regulon” makes exceptional adaptability of the QS response to different environmental conditions (46). Considering that AL-1 can influence the Las and Rhl systems, it could become an efficient compound for the treatment of P. aeruginosa-related infections.
EPS is an important constituent of the P. aeruginosa biofilm and is required for bacterial cells to adhere to a substratum and maintain biofilm structure (35). P. aeruginosa EPS was tested by using a phenol solution-sulfuric acid method, as previously described (9, 36). After being treated with AL-1, the amount of P. aeruginosa EPS was significantly reduced (59). The psl cluster plays a role in biofilm development, so an immunoblotting assay was used to investigate the effect of AL-1 on psl. The result showed that AL-1 can decrease the level of Psl production. β-Galactosidase activity also suggested that the levels of psl transcription and translation are reduced by AL-1. It was suggested previously that psl may be transcriptionally regulated by LasR. RsmA, a small RNA-binding protein, is known to negatively regulate pathogenicity determinants such as motility, AHLs, and secondary metabolite production (5, 18, 42). Previous reports concluded that RsmA was acting as a translational repressor of psl (22). These results strictly corroborate our data obtained by qRT-PCR: the level of rsmA was increased about 3-fold with respect to the level of the control (see Fig. S3 in the supplemental material). It is possible to postulate that the decreased psl translational level may be due to the increased level of rsmA mediated by AL-1. RsmA is controlled by a complex regulatory system, including sensor kinases, response regulators, and the small RNAs rsmZ and rsmY (52). LadS and RetS control biofilm and virulence phenotypes through the two-component regulatory system GacS/A, LadS promotes the phosphorylation of GacA, the phosphorylated GacA then activates the transcriptions of rsmZ and rsmY, and the small RNAs bind to rsmA, which eventually affects biofilm formation and the T3SS (17, 52, 56). RetS exerts opposite effects in this system (52). Future studies should be done to find out the effects of AL-1 on the global regulatory networks.
The decrease in the Psl expression level mediated by AL-1 in the immunoblotting assay appears to be far greater than psl transcription and translation in β-galactosidase assays. It was speculated that AL-1 may have posttranslational effects on Psl.
Meanwhile, AL-1 can increase the level of secretion of ExoS. This could be due to the effect of AL-1 on the QS system. Interestingly, a previous study showed that the treatment of P. aeruginosa with azithromycin (AZM) can inhibit the QS system but increase the expression levels of T3SS genes (49). The secretion of ExoS in an rhlI mutant showed that exoS was submitted to negative RhlR–C4-HSL-dependent control (2). Hogardt et al. also reported that exoS is negatively regulated by the Rhl system (21). Mutations in T3SS genes result in enhanced biofilm formation in strain PAO1 (28). These data provided evidence that AL-1 downregulates the rhl gene and possibly upregulates the type III effectors during biofilm inhibition. RsmA exerted a negative effect on the synthesis of both 3-oxo-C12-HSL and C4-HSL (42). Mulcahy et al. reported previously that RsmA is required for ExoS secretion (40). The increased level of ExoS secretion may be due to the elevated level of rsmA transcripts mediated by AL-1. Overall, the T3SS and the QS system were connected through both the Rhl system and RsmA. As mentioned above, we think that the potential benefits outweigh the risk. The increased ExoS level may be due to the exchanged life-style of P. aeruginosa. AL-1 inhibits biofilm formation and makes P. aeruginosa planktonic, and the bacteria may express virulence factors, such as T3SS effectors, for self-protection.
Researchers investigating the antibiotic resistance of bacteria in biofilms thought that bacterial biofilms may cause a slow or incomplete penetration of antibiotics (50). However, if the antibiotic can permeate the biofilm, some of the bacteria may differentiate into a protected phenotype, and the altered chemical microenvironment within the biofilm also makes the antibiotic less effective (50). AL-1 has a synergistic effect with traditional antibiotics; the underlying mechanism may be mediated by the markedly reduced biofilm formation.
In summary, AL-1 inhibits P. aeruginosa PAO1 biofilm formation by repressing the QS system (Fig. 5). Clearly, AL-1 is an interesting compound due to its mode of action and synergistic effects with antibiotics and may address the potential use of narrow-spectrum antibiotics for the treatment of chronic P. aeruginosa infections.
Fig 5.
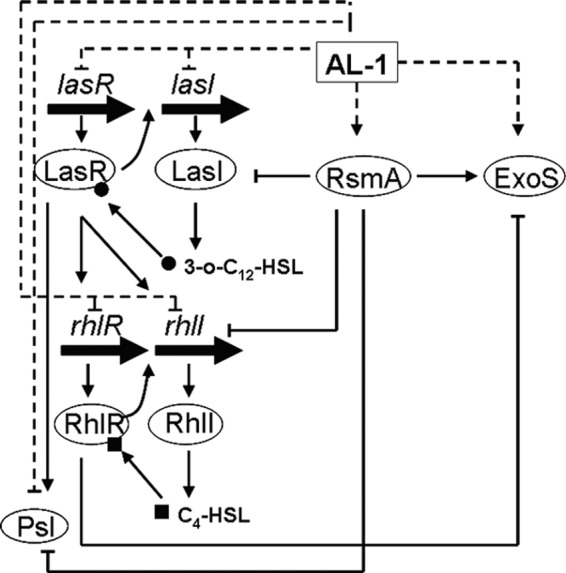
Proposed model of the effect of AL-1 on QS-related genes and exoS. Dashed lines indicate the effects of AL-1, and solid lines indicate the QS network. Arrowheads, activation; flat arrowheads, repression.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Natural Science Foundation of China (grants 31100075, 81102362, 31170095, and 31000004 to L.Z. and U1032007 to Y.W.), the CAS Pillar Program (grant XDA04074000), the Ministry of Science and Technology of China (grants 2011ZX11102-011-11 and 2007DFB31620), the National Science Foundation for Young Scientists of China (grant 31000049), the Research Growth Initiative of the University of Wisconsin—Milwaukee (UWM), and a catalyst grant of the UWM Research Foundation. L.Z. is an awardee of the National Distinguished Young Scholar Program in China.
Footnotes
Published ahead of print 16 July 2012
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1. Allesen-Holm M, et al. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128 [DOI] [PubMed] [Google Scholar]
- 2. Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. 2005. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J. Bacteriol. 187:3898–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borlee BR, Geske GD, Blackwell HE, Handelsman J. 2010. Identification of synthetic inducers and inhibitors of the quorum-sensing regulator LasR in Pseudomonas aeruginosa by high-throughput screening. Appl. Environ. Microbiol. 76:8255–8258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bull AT, Stach JE. 2007. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 15:491–499 [DOI] [PubMed] [Google Scholar]
- 5. Burrowes E, Abbas A, O'Neill A, Adams C, O'Gara F. 2005. Characterisation of the regulatory RNA RsmB from Pseudomonas aeruginosa PAO1. Res. Microbiol. 156:7–16 [DOI] [PubMed] [Google Scholar]
- 6. Byrd MS, et al. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73:622–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen JX, et al. 2009. Activity of andrographolide and its derivatives against influenza virus in vivo and in vitro. Biol. Pharm. Bull. 32:1385–1391 [DOI] [PubMed] [Google Scholar]
- 8. Cowell BA, Twining SS, Hobden JA, Kwong MS, Fleiszig SM. 2003. Mutation of lasA and lasB reduces Pseudomonas aeruginosa invasion of epithelial cells. Microbiology 149:2291–2299 [DOI] [PubMed] [Google Scholar]
- 9. Cuesta G, Suarez N, Bessio MI, Ferreira F, Massaldi H. 2003. Quantitative determination of pneumococcal capsular polysaccharide serotype using a modification of phenol-sulfuric acid method. J. Microbiol. Methods 52:69–73 [DOI] [PubMed] [Google Scholar]
- 10. Davies DG, et al. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295–298 [DOI] [PubMed] [Google Scholar]
- 11. Duan K, Surette MG. 2007. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J. Bacteriol. 189:4827–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fogerty MD, et al. 2008. Risk factors for pressure ulcers in acute care hospitals. Wound Repair Regen. 16:11–18 [DOI] [PubMed] [Google Scholar]
- 13. Friedman L, Kolter R. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. 2009. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 73:1072–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7:654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heeb S, Blumer C, Haas D. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heurlier K, et al. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:2936–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72 [DOI] [PubMed] [Google Scholar]
- 20. Hoang TT, Schweizer HP. 1999. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181:5489–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hogardt M, Roeder M, Schreff AM, Eberl L, Heesemann J. 2004. Expression of Pseudomonas aeruginosa exoS is controlled by quorum sensing and RpoS. Microbiology 150:843–851 [DOI] [PubMed] [Google Scholar]
- 22. Irie Y, et al. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78:158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jensen SE, Fecycz IT, Campbell JN. 1980. Nutritional factors controlling exocellular protease production by Pseudomonas aeruginosa. J. Bacteriol. 144:844–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang X, et al. 2009. Synthesis and evaluation of antibacterial activities of andrographolide analogues. Eur. J. Med. Chem. 44:2936–2943 [DOI] [PubMed] [Google Scholar]
- 26. Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73:310–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knight V, et al. 2003. Diversifying microbial natural products for drug discovery. Appl. Microbiol. Biotechnol. 62:446–458 [DOI] [PubMed] [Google Scholar]
- 28. Kuchma SL, Connolly JP, O'Toole GA. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1441–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang H, Li L, Dong Z, Surette MG, Duan K. 2008. The YebC family protein PA0964 negatively regulates the Pseudomonas aeruginosa quinolone signal system and pyocyanin production. J. Bacteriol. 190:6217–6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lyczak JB, Cannon C, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 31. Ma L, et al. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354 doi:10.1371/journal.ppat.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. 2006. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188:8213–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manefield M, et al. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145:283–291 [DOI] [PubMed] [Google Scholar]
- 34. Manefield M, et al. 2002. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148:1119–1127 [DOI] [PubMed] [Google Scholar]
- 35. Marcotte L, Kegelaer G, Sandt C, Barbeau J, Lafleur M. 2007. An alternative infrared spectroscopy assay for the quantification of polysaccharides in bacterial samples. Anal. Biochem. 361:7–14 [DOI] [PubMed] [Google Scholar]
- 36. Masuko T, et al. 2005. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal. Biochem. 339:69–72 [DOI] [PubMed] [Google Scholar]
- 37. Matsukawa M, Greenberg EP. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Müh U, et al. 2006. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 50:3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mulcahy H, O'Callaghan J, O'Grady EP, Adams C, O'Gara F. 2006. The posttranscriptional regulator RsmA plays a role in the interaction between Pseudomonas aeruginosa and human airway epithelial cells by positively regulating the type III secretion system. Infect. Immun. 74:3012–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pesci EC, Pearson JP, Seed PC, Iglewski BH. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pessi G, et al. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Richards JJ, Ballard TE, Huigens RW, III, Melander C. 2008. Synthesis and screening of an oroidin library against Pseudomonas aeruginosa biofilms. Chembiochem 9:1267–1279 [DOI] [PubMed] [Google Scholar]
- 44. Roy-Burman A, et al. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767–1774 [DOI] [PubMed] [Google Scholar]
- 45. Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10:644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schuster M, Greenberg EP. 2007. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics 8:287 doi:10.1186/1471-2164-8-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh R, Paul D, Jain RK. 2006. Biofilms: implications in bioremediation. Trends Microbiol. 14:389–397 [DOI] [PubMed] [Google Scholar]
- 49. Skindersoe ME, et al. 2008. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52:3648–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138 [DOI] [PubMed] [Google Scholar]
- 51. Vannini A, et al. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ventre I, et al. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 103:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Venturi V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30:274–291 [DOI] [PubMed] [Google Scholar]
- 54. Wade DS, et al. 2005. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 187:4372–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Whiteley M, Lee KM, Greenberg EP. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reference deleted.
- 58. Yamazaki A, et al. 2012. Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS-GacA two-component signal transduction system. Antimicrob. Agents Chemother. 56:36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zeng X, et al. 2011. Synergistic effect of 14-alpha-lipoyl andrographolide and various antibiotics on the formation of biofilms and production of exopolysaccharide and pyocyanin by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55:3015–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zeng Z, et al. 2008. Virtual screening for novel quorum sensing inhibitors to eradicate biofilm formation of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 79:119–126 [DOI] [PubMed] [Google Scholar]
- 61. Zhang L, et al. 2005. Exploring novel bioactive compounds from marine microbes. Curr. Opin. Microbiol. 8:276–281 [DOI] [PubMed] [Google Scholar]
- 62. Zhang L, Arnold LD. 2005. Integrated approaches for discovering novel drugs from microbial natural products, p 33–56 In Zhang LX, Arnold LD. (ed), Natural products: drug discovery and therapeutics medicines. Humana Press Inc, Totowa, NJ [Google Scholar]
- 63. Zhang L, et al. 2007. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc. Natl. Acad. Sci. U. S. A. 104:4606–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.